Abstract
Objectives
Hypermutable bacteria are causing a drastic problem via their enhanced ability to become resistant. Our objectives were to compare bacterial killing and resistance emergence between differently shaped tobramycin concentration–time profiles at a given fAUC/MIC, and determine the tobramycin exposure durations that prevent resistance.
Methods
Static concentration time–kill studies over 24 h used Pseudomonas aeruginosa WT strains (ATCC 27853 and PAO1) and hypermutable PAOΔmutS. fAUC/MIC values of 36, 72 and 168 were assessed at initial inocula of 106 and 104 cfu/mL (all strains) and 101.2 cfu/mL (PAOΔmutS only) in duplicate. Tobramycin was added at 0 h and removed at 1, 4, 10 or 24 h. Proportions of resistant bacteria and MICs were determined at 24 h. Mechanism-based modelling was conducted.
Results
For all strains, high tobramycin concentrations over 1 and 4 h resulted in more rapid and extensive initial killing compared with 10 and 24 h exposures at a given fAUC/MIC. No resistance emerged for 1 and 4 h durations of exposure, although extensive regrowth of susceptible bacteria occurred. The 24 h duration of exposure revealed less regrowth, but tobramycin-resistant populations had completely replaced susceptible bacteria by 24 h for the 106 cfu/mL inoculum. The hypermutable PAOΔmutS showed the highest numbers of resistant bacteria. Total and resistant bacterial counts were described well by novel mechanism-based modelling.
Conclusions
Extensive resistance emerged for 10 and 24 h durations of exposure, but not for shorter durations. The tobramycin concentration–time profile shape is vital for resistance prevention and should aid the introduction of optimized combination regimens.
Introduction
A global healthcare crisis is arising from Gram-negative bacteria, such as Pseudomonas aeruginosa, where effective antibiotics are increasingly scarce leading to difficult-to-treat life-threatening infections.1–4 The occurrence of hypermutator phenotypes in P. aeruginosa clinical isolates is exacerbating the problem because of their enhanced ability to become resistant.5,6 The fast-acting aminoglycoside tobramycin causes significant bacterial killing although extensive resistance may occur following monotherapy.7 Tobramycin is a protein synthesis inhibitor8 that has also been found to disrupt the bacterial outer membrane.9,10 The most important resistance mechanisms of P. aeruginosa against aminoglycosides are the up-regulation of the MexXY-OprM efflux pump,11–13 reduced permeability of the outer membrane, and enzymes that inactivate aminoglycosides intracellularly by phosphorylation, acetylation or adenylation.14
Hypermutable strains of P. aeruginosa are of major concern, particularly in chronic respiratory infections such as those in patients with cystic fibrosis (CF).15–17 Hypermutation is caused by defects in DNA or error repair systems18 and is most commonly due to mutations in the mutS gene, encoding a component of the methyl-directed mismatch repair system.5,19,20 It provides bacteria with an advantage through the ability to adapt quickly to stressful and fluctuating environments via rapidly gaining or enhancing resistance mechanisms.15,21,22
Two pharmacokinetic/pharmacodynamic indices are most commonly used as predictors for bacterial killing by aminoglycosides, being the fAUC/MIC and the fCmax/MIC.23–25 The fCmax/MIC relies on the concentration at a single timepoint within a dosage interval. The fAUC/MIC only considers the total (i.e. time-averaged) exposure across a 24 h period and suggests the same extent of bacterial killing regardless of the shape of the concentration–time profile. Once- and thrice-daily intravenous aminoglycoside therapy at the same daily dose were found to be equally effective clinically against pulmonary exacerbations of CF, but the emergence of resistance was not studied.26,27 However, in a small clinical study in 33 patients with CF, tobramycin Cmax/MIC was found as the best predictor of clinical outcome as measured by lung function;28 the investigators suggested that resistance was greater after once-daily dosing, but an alternative analysis may have led to a different conclusion. Recently we have shown that high ciprofloxacin concentrations for a short duration of exposure (at a specific fAUC/MIC) resulted in resistance prevention.29
The main objective of the current investigation was to evaluate the bacterial killing and emergence of resistance resulting from differently shaped tobramycin concentration–time profiles at a given fAUC/MIC. We aimed to determine whether short-duration, high concentrations were more efficient in bacterial killing and prevention of the emergence of resistance compared with low concentrations over longer durations. Furthermore, we sought to evaluate resistance prevention in the worst-case scenario of a hypermutable strain. To address these objectives we used in vitro time–kill studies to assess bacterial killing and resistance emergence for different concentration–time profiles, at given fAUC/MIC in hypermutable and non-hypermutable P. aeruginosa.
Materials and methods
Bacterial strains and media
The P. aeruginosa ATCC 27853 and PAO1 WT reference strains were used in this study. We also used the isogenic hypermutable P. aeruginosa PAOΔmutS strain that was constructed from the PAO1 WT reference strain by Mena et al.30 via deletion of the mutS gene. All susceptibility and time–kill studies were performed in CAMHB (containing 20–25 mg/L Ca2+ and 10–12.5 mg/L Mg2+; BD, Sparks, MD, USA). Viable counting was performed on cation-adjusted Mueller–Hinton agar (containing 25 mg/L Ca2+ and 12.5 mg/L Mg2+; Medium Preparation Unit, The University of Melbourne, Parkville, Victoria, Australia). Drug-containing agar plates were prepared using cation-adjusted Mueller–Hinton agar (BD) supplemented with the appropriate amount of tobramycin (AK Scientific, Union City, MD, USA). The antibiotic stock solution was prepared in Milli-Q water and subsequently filter-sterilized using a 0.22 μm PVDF syringe filter (Merck Millipore, Cork, Ireland).
Time–kill experiments
To assess bacterial killing and emergence of resistance, time–kill experiments were performed in duplicate as previously described29 for the different tobramycin exposure profiles. We studied three overall (i.e. time-averaged) tobramycin exposures, corresponding to fAUC/MIC of 36, 72 and 168; two exposures were above and one below the recommended fAUC/MIC exposure of 42 for the bactericidal effect.24 For MICs ≤1 mg/L (83% of P. aeruginosa isolates reported by EUCAST),31 the two lower exposures are achievable at common clinical doses, whereas fAUC/MIC of 168 requires a high clinical dose in ICU patients.32 The tobramycin agar dilution MIC using the CLSI method33 was 0.5 mg/L for PAO1 and ATCC 27853; while for PAOΔmutS it was 1 mg/L due to the resistant bacterial subpopulations.34
Overall exposures were achieved by exposing the bacteria to appropriate tobramycin concentrations for the durations of 1, 4, 10 and 24 h, as reported in Table 1. Tobramycin was dosed at 0 h and rapidly removed at the respective timepoint via two or three sequential centrifugation and resuspension steps, as we previously described.29 This method assured that the tobramycin concentrations were negligible (<0.16× MIC) after drug removal. These different exposures were studied at initial inocula of 106 and 104 cfu/mL (all three strains), as well as 101.2 cfu/mL (PAOΔmutS only). The probability of at least one pre-existing bacterial cell that was resistant to 2.5 mg/L tobramycin was ≤7.1% at 104 cfu/mL for ATCC 27853 and PAO1, and 2.3% at 101.2 cfu/mL for PAOΔmutS. In contrast, for all strains at 106 cfu/mL and PAOΔmutS at 104 cfu/mL, this probability was >96.7%. All studies included a growth control. Viability counts of bacteria as described previously29 were determined within 5 min prior to dosing and at 0.5, 2, 6, 10 (or 12) and 24 h after dosing as well as 5 min before and 10 min after drug removal to confirm minimal loss of bacteria via the drug removal procedure.
Table 1.
Tobramycin concentrations (mg/L) and durations of exposure for each studied fAUC/MIC against three strains, ATCC 27853 (MIC 0.5 mg/L), PAO1 (MIC 0.5 mg/L) and PAOΔmutS (MIC 1 mg/L)
| Drug exposure duration (h) | fAUC/MIC: 36 | fAUC/MIC: 72 | fAUC/MIC: 168 |
|---|---|---|---|
| ATCC 27853 and PAO1 control | 0 | 0 | 0 |
| 1 | 18 | 36 | 84 |
| 4 | 4.5 | 9 | 21 |
| 10 | 1.8 | 3.6 | 8.4 |
| 24 | 0.75 | 1.5 | 3.5 |
| PAOΔmutS control | 0 | 0 | 0 |
| 1 | 36 | 72 | 168 |
| 4 | 9 | 18 | 42 |
| 10 | 3.6 | 7.2 | 16.8 |
| 24 | 1.5 | 3 | 7 |
Emergence of resistance
The proportion of resistant bacteria (PRB) and MICs were determined at 0 (i.e. before treatment) and 24 h. Tobramycin was removed before evaluating emergence of resistance. Agar dilution MICs were determined once the bacterial suspensions were spectrophotometrically adjusted (i.e. dilution in fresh, pre-warmed, sterile CAMHB) to an inoculum of 106 cfu/mL, unless the suspension was already below this inoculum. Agar plates containing 1.25 mg/L (PAO1), 1.5 mg/L (ATCC 27853), 2.5 mg/L (all three strains) and 5 mg/L (PAOΔmutS) tobramycin were used for determining the PRB. Antibiotic-containing agar plates were incubated for 3 days and the log10 PRB was calculated as the difference, on log10 scale, between the viability of resistant bacteria on antibiotic-containing agar plates and total population viability on drug-free plates.
Some of the viable counts at 24 h were too low to quantify colonies on antibiotic-containing agar plates. These bacterial suspensions still provided information on the upper limit of the log10 PRB (e.g. log10 PRB ≤−6). To include these data we used the following reporting rules. If the PRB was not quantifiable, but the upper limit was within 1 log10 of the PRB for the growth control, we assumed the PRB was unchanged and used the value of the growth control. If the PRB was not quantifiable and the upper limit was >1 log10 higher than the PRB for the growth control, the PRB of this treatment was reported as missing.
Mechanism-based modelling of bacterial killing and resistance
Mechanism-based pharmacokinetic/pharmacodynamic models were developed to characterize the time-course of bacterial killing and emergence of tobramycin resistance.
Life cycle growth model
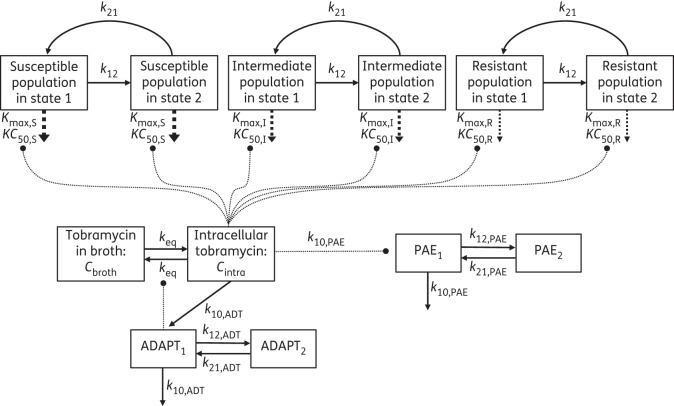
The growth and replication of P. aeruginosa was described by a life cycle growth model that accounts for the underlying biological processes.35–37 A diagram of the model structure is shown in Figure 1. Inclusion of three pre-existing bacterial populations (the bacterial populations present prior to treatment)—susceptible, intermediate and resistant—best described the observed data. For each of these populations, the model included two bacterial states: state 1 representing the bacteria preparing for replication and state 2 those immediately before replication;36–39 e.g. cfuS1 denotes the susceptible bacteria in state 1 and cfuS2 the susceptible bacteria in state 2.
Figure 1.
Model diagram of the life cycle growth model including three populations, susceptible, intermediate and resistant, with two states each to describe bacterial replication. The maximum killing rate constants (Kmax) and the antibiotic concentrations (KC50) causing 50% of Kmax are explained in Table 2. The concentration of tobramycin in broth (Cbroth), intracellular tobramycin (Cintra), ADAPT1, ADAPT2 and all corresponding rate constants are described in the Materials and methods section.
The total bacterial population (cfuall) was defined as the sum of bacteria in all subpopulations and bacterial states:
| (1) |
where the susceptible population in state 1 (cfuS1) was described by
| (2) |
and the susceptible population in state 2 (cfuS2) was described by
| (3) |
The intermediate (cfuI1, cfuI2) and resistant (cfuR1, cfuR2) populations were modelled similarly.
In Equations (2) and (3), the first-order growth rate constant k12 was defined as 60/MGT, with MGT representing the mean generation time for each of the bacterial populations. The first-order replication rate constant k21 was set to 50 h−1 as described previously.37 ALIVES was 1 while the estimated (cfuS1 + cfuS2) was ≥0.5 cells in the entire broth volume, otherwise ALIVES was 0. This part of the model allowed us to describe bacterial eradication. The replication factor REP defines the probability of successful replication, as described previously.37
| (4) |
REP approaches 2 when the total bacterial count (cfuall) is low, resulting in a 100% probability of successful replication. As cfuall approaches the maximum population size (cfumax), REP approaches 1, which reflects a 50% probability of successful replication and ensures that cfuall does not exceed cfumax. The term KILLSPAE1 describes bacterial killing by tobramycin including the post-antibiotic effect (PAE),40 as described in Equations (5) and (6) below.
Bacterial killing by tobramycin including PAE
A prolonged PAE is a trait of tobramycin40 hence it is represented in the bacterial killing function KILLSPAE1 in Equation 5. The KILLSPAE2 (Equation 6) is in equilibrium with KILLSPAE1 and provides a delay such that bacterial killing occurs for some time after removal of tobramycin from the system.
| (5) |
| (6) |
The PAE rate constants , and represent the delayed killing that results from the PAE and were defined as k10,PAE = 1/MTT10,PAE, k12,PAE = 1/MTT12,PAE and k21,PAE = 1/MTT21,PAE, where MTT denotes mean turnover time. Modelling the PAO1 did not require this PAE.
The bacterial killing by tobramycin is represented by KILLS:
| (7) |
where is the (estimated) intracellular tobramycin concentration. Kmax,S is the maximum killing rate constant and KC50,S is the Cintra required to achieve 50% of Kmax,S.9 Bacterial killing and PAE of the I and R populations were described similarly. The Cintra is defined in Equation (8), as described previously.9
| (8) |
The rate constant keq is defined as ln(2)/t1/2eq, where t1/2eq is the equilibration half-life between tobramycin in broth and the intracellular space. The term ADAPT1 reflects adaptive resistance of P. aeruginosa against tobramycin. An increase in adaptive resistance (ADAPT1, defined below) decreases Cintra and thereby reduces the extent of bacterial killing by tobramycin.
Adaptive resistance to tobramycin
Incorporating adaptive resistance in the mechanism-based model was required to describe the observed bacterial count profiles in our experiments. Adaptive resistance against aminoglycosides is often caused by the overexpression of the MexXY-OprM efflux pump41 or inhibition of energy-dependent uptake in P. aeruginosa,42 both of which would decrease Cintra, as described above. It was described by a two-compartment model for ADAPT1 (Equation 9) and ADAPT2 (Equation 10)9:
| (9) |
| (10) |
where ADAPTmax is the maximum extent of adaptive resistance and EC50,ADT is the Cintra that induces the half-maximum extent of adaptive resistance. The peripheral adaptation compartment ADAPT2 allows for the delayed decline of adaptive resistance after removal of tobramycin. The adaptive resistance rate constants k10,ADT, k12,ADT and k21,ADT describe the time-course of adaptive resistance and are defined as k10,ADT = 1/MTT10,ADT, k12,ADT = 1/MTT12,ADT and k21,ADT = 1/MTT21,ADT.
Resistant bacterial populations on antibiotic-containing agar plates
The viable counts on tobramycin-containing agar plates were modelled simultaneously with the total viable counts on drug-free agar plates. The fractions of subpopulations (susceptible, intermediate, resistant) that were able to grow on tobramycin-containing agar plates at different concentrations were estimated as described previously.43
Initial conditions
The total initial inocula (log10cfu0,4, log10cfu0,6) and the PRB for the intermediate (log10PRBI) and resistant (log10PRBR) populations were estimated (Table 2). The initial condition for the susceptible population was calculated by subtracting the initial conditions of the intermediate and the resistant populations from the respective total inoculum. All bacteria were initialized in state 1 and the initial conditions for cfuS2, cfuI2 and cfuR2 were set to 0. ALIVES, ALIVEI and ALIVER were initialized at 1 and all other equations at 0.
Table 2.
Population parameter estimates for tobramycin against three strains of P. aeruginosa
| Parameter | Symbol (unit) | Population estimate (SE%) for the strain |
||
|---|---|---|---|---|
| ATCC 27853 | PAO1 | PAOΔmutS | ||
| Bacterial growth and subpopulations | ||||
| log10 initial inoculum | ||||
| 104 cfu/mL | log10cfu0,4 | 4.20 (4.32) | 4.23 (1.49) | 4.50 (1.97) |
| 106 cfu/mL | log10cfu0,6 | 5.94 (2.84) | 5.56 (1.78) | 6.05 (1.82) |
| MGT | ||||
| susceptible population | MGTS (min) | 87.3 (7.24) | 41.6 (1.86)a | 63.9 (8.67) |
| intermediate population | MGTI (min) | 49.3 (3.59) | — | 53.5 (3.55) |
| resistant population | MGTR (min) | 79.9 (8.89) | — | 82.2 (6.75) |
| log10 maximum population size | log10cfumax | 9.09 (2.25) | 8.95 (1.25) | 9.16 (1.66) |
| log10 PRB | ||||
| intermediate population | log10PRBI | −4.25 (3.47) | −4.58 (2.43) | −4.12 (3.79) |
| resistant population | log10PRBR | −7.17 (2.73) | −6.39 (1.11) | −5.60 (4.64) |
| Bacterial killing by tobramycin | ||||
| equilibrium half-life between tobramycin in broth and the intracellular space | t1/2eq (min) | 25.0, fixed | 40.0 (3.17) | 25.0, fixed |
| maximum killing rate constant | ||||
| susceptible population | Kmax,S (h−1) | 78.9 (13.6) | 30.3 (3.25) | 24.8 (9.91) |
| intermediate population | Kmax,I (h−1) | 2.23 (20.9) | 2.07 (4.64) | 2.02 (13.1) |
| resistant population | Kmax,R (h−1) | 14.2 (12.0) | 7.38 (5.10) | 7.93 (10.3) |
| intracellular tobramycin concentration causing 50% of Kmax | ||||
| susceptible population | KC50,S (mg/L) | 0.154 (15.4) | 0.0206 (15.7) | 0.0410 (19.6) |
| intermediate population | KC50,I (mg/L) | 0.0295 (23.9) | 0.0270 (31.6) | 0.115 (31.9) |
| resistant population | KC50,R (mg/L) | 2.89 (26.4) | 0.574 (8.03) | 2.79 (15.3) |
| Adaptive resistance | ||||
| maximum extent of stimulation of adaptive resistance | ADAPTmax | 13.5 (20.5) | 14.2 (4.49) | 11.2 (28.1) |
| intracellular tobramycin concentration causing 50% of ADAPTmax | EC50,ADT (mg/L) | 6.06 (51.9) | 11.1 (4.82) | 28.8 (9.33) |
| MTT | ||||
| for adaptive resistance | MTT10,ADT (h) | 6.81 (23.2) | 7.37 (6.81) | 13.5 (9.72) |
| for distribution from the central to the peripheral adaptive compartment | MTT12,ADT (h) | 0.917 (50.6) | 0.972 (10.3) | 0.439 (32.2) |
| for distribution from the peripheral to the central adaptive compartment | MTT21,ADT (h) | 6.0, fixed | 6.0, fixed | 6.0, fixed |
| PAE | ||||
| MTT | ||||
| for the PAE | MTT10,PAE (h) | 0.0166 (56.9) | — | 0.0158 (29.2) |
| between the central and peripheral PAE compartment | MTT12,PAE (h) | 0.519 (43.3) | — | 1.06 (22.0) |
| between the peripheral and central PAE compartment | MTT21,PAE (h) | 0.662 (16.2) | — | 0.504 (12.6) |
| Residual variability | ||||
| SD of additive residual error on log10 scale for the | ||||
| total population | SDcfu | 0.302 | 0.455 | 0.459 |
| population on 2.5 mg/L tobramycin plates | SDcfu3 | 0.137 | 1.51 | 0.195 |
| population on 5 mg/L tobramycin plates | SDcfu5 | 0.307 | 1.79 | 0.399 |
aModel only contains one MGT.
Observation model
The log10 viability counts were fitted using an additive residual error model on log10 scale. A previously described residual error model was utilized to fit directly the number of colonies on a plate when there were less than two colonies per plate observed.44 Viable counts below the limit of counting and model predictions <0 log10 cfu/mL were plotted as 0.
Estimation
The model parameters were estimated simultaneously using the viable counts on drug-free and tobramycin-containing plates for each of the strains at the 104 and 106 cfu/mL inocula using the importance sampling algorithm (pmethod = 4) in parallelized S-ADAPT (version 1.57), facilitated by SADAPT-TRAN.45 A coefficient of variation of 15% during the end of estimation allowed the between-curve variability of the parameters to be fixed.44 The objective function, standard diagnostic plots, plausibility of the parameter estimates and visual predictive checks were utilized to assess competing models.
Results and discussion
In the present study, we demonstrated that different shapes of the concentration–time profile had an important impact on bacterial regrowth and emergence of resistance in P. aeruginosa ATCC 27853, PAO1 and PAOΔmutS. The log10 PRB for ATCC 27853 was −5.36 on 1.5 mg/L tobramycin plates and −6.43 on 2.5 mg/L tobramycin plates before treatment. PAO1 had a log10 PRB of −5.34 on 1.25 mg/L and −6.77 on 2.5 mg/L tobramycin plates before treatment. The hypermutable PAOΔmutS had a log10 PRB before treatment of −4.14 on 2.5 mg/L and −5.62 on 5 mg/L tobramycin plates. These results show a dramatic difference in PRB before treatment observed between the PAOΔmutS and the two WT strains (ATCC 27853 and PAO1) on 2.5 mg/L tobramycin plates, which was expected due to the hypermutable strain having an increased likelihood of mutating.
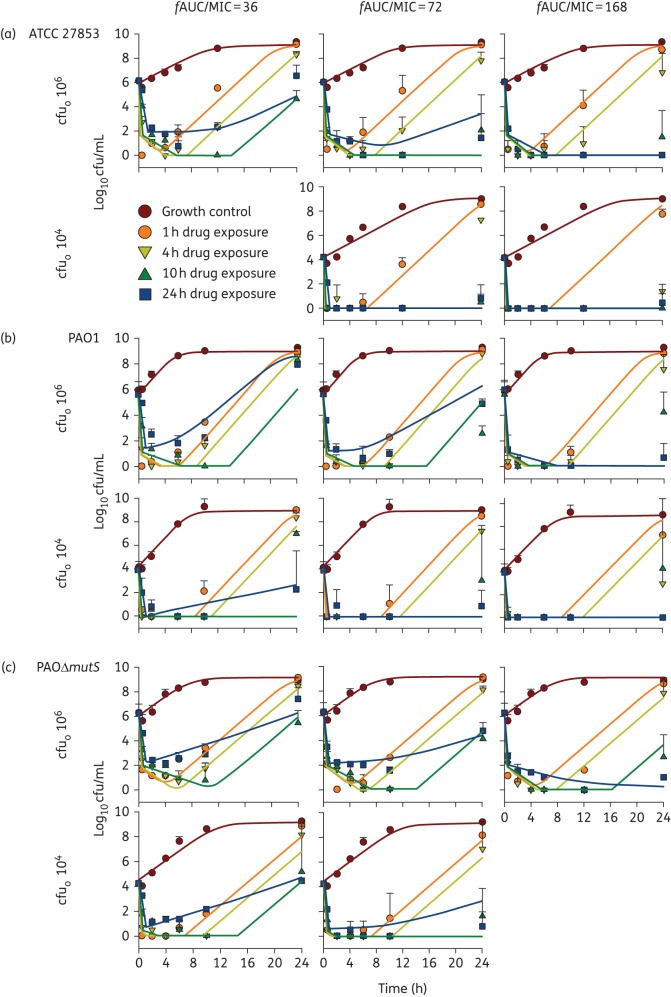
The extent of initial killing of P. aeruginosa increased with tobramycin exposure (fAUC/MIC) for all three strains, as expected for this fast-acting aminoglycoside. At a given fAUC/MIC, the extent of initial bacterial killing increased with concentration as the duration of exposure became shorter (Figure 2). In general, at a given fAUC/MIC, the high concentrations for short durations of exposure resulted in more extensive regrowth than exposure to lower concentrations over a longer time (Figure 2). It is of great concern that for all strains even the high fAUC/MIC of 168, i.e. four times the suggested fAUC/MIC breakpoint of 42 for bactericidal effect,24 did not inhibit the regrowth of bacteria at the initial inoculum of 106 cfu/mL. An fAUC/MIC of 168 would be expected to be achieved in patients for MICs up to ∼0.5 mg/L following a tobramycin dose of 5–6 mg/kg in critically and non-critically ill patients32,46 and 8–11 mg/kg in patients with CF (both based on a 70 kg patient).47,48 However, according to EUCAST, 46% of the 25 002 evaluated isolates had an MIC ≥1 mg/L.31
Figure 2.
Observed viable counts (mean ± SD) and population predicted profiles (continuous lines in corresponding colours) for P. aeruginosa ATCC 27853 (a), PAO1 (b) and PAOΔmutS (c) exposed to tobramycin at an fAUC/MIC of 36 (left column), 72 (middle column) and 168 (right column) delivered over 1, 4, 10 or 24 h durations of exposure, at initial inocula (cfuo) of 106 and 104 cfu/mL, excluding fAUC/MIC of 36 for ATCC 27853 and fAUC/MIC of 168 for PAOΔmutS for cfuo 104 cfu/mL. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Generally, with short durations of exposure (1 and 4 h) the ATCC 27853 strain displayed rapid initial killing of 4–6 log10 that was followed by extensive regrowth after drug removal, whilst the longer durations of exposure (10 and 24 h) had limited or no regrowth (Figure 2a). An exception was the low fAUC/MIC of 36 where the longer durations of exposure revealed 4 log10 regrowth. In addition, at the high fAUC/MIC of 168 the 4 h duration of exposure showed limited regrowth at the 104 cfu/mL initial inoculum that likely lacked pre-existing resistant bacteria (Figure 2a). The PAO1 strain showed similar results to the ATCC 27853 strain, with similar extents of initial killing, and mostly the shorter durations of exposure leading to extensive regrowth (Figure 2b). However, the PAO1 strain showed more extensive regrowth for the longer durations of exposure at an fAUC/MIC of 36 in comparison with ATCC 27853; extensive regrowth was observed for the 10 and 24 h durations of exposure for PAO1 at the 106 cfu/mL inoculum (Figure 2b). The initial killing and bacterial regrowth of PAOΔmutS was mostly comparable to its parental strain PAO1 and ATCC 27853 (Figure 2). At the very low initial inoculum of 101.2 cfu/mL complete eradication at 24 h was observed for all tobramycin exposures against PAOΔmutS, except for the 1 h duration of exposure at fAUC/MIC of 36 where there was ∼2 log10 regrowth (data not shown).
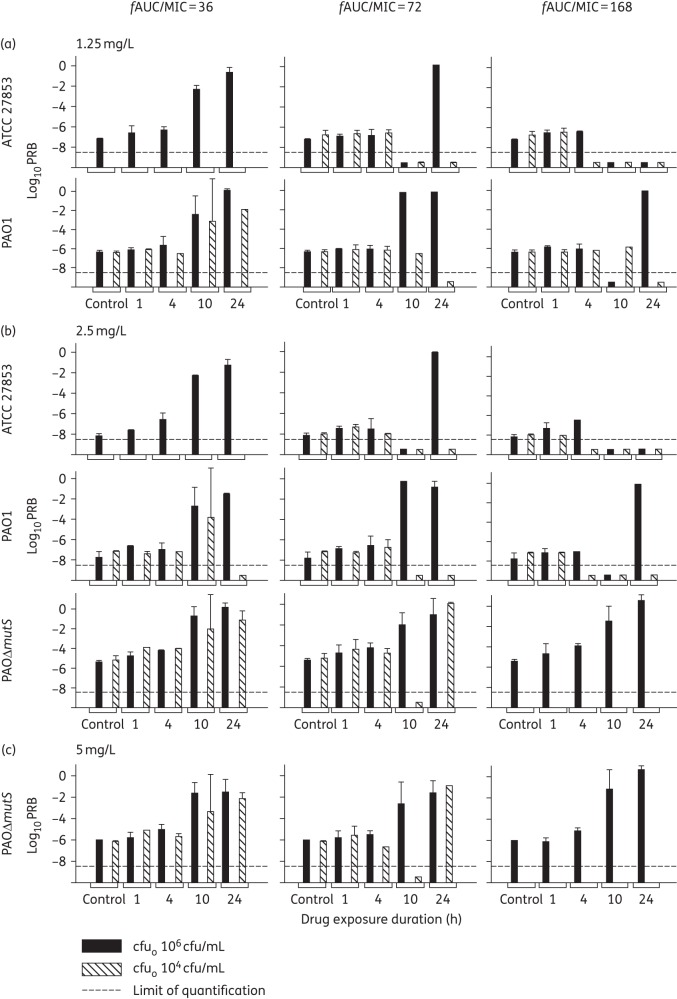
Bacterial regrowth was a common feature of the time–kill profiles for each strain (Figure 2). Notably, at the 106 cfu/mL inoculum, although the shorter durations of exposure (1 and 4 h) had more extensive regrowth, their PRB at 24 h were comparable to the growth control for all three strains, suggesting that this regrowth was a susceptible population (Figure 3). Consideration should be given to combination antibiotic therapy as an option for fighting this susceptible regrowth, i.e. using a second antibiotic to prevent the regrowth of the tobramycin-susceptible bacteria. In contrast, the longer durations of exposure (10 and 24 h) at the 106 cfu/mL inoculum frequently resulted in increased PRB at 24 h for all three strains at an fAUC/MIC of 36 and 72 (Figure 3). These increases in PRB were mostly supported by raised MIC at 24 h for the 10 and 24 h durations of exposure (Table 3). Overall, the hypermutable PAOΔmutS strain had considerably higher PRB values compared with the two WT strains (Figure 3).
Figure 3.
Log10 PRB (mean ± SD) at 24 h on agar plates containing 1.25 (1.5 for ATCC 27853; a), 2.5 (b) and 5 (c) mg/L tobramycin. This figure shows fAUC/MIC values of 36 (left column), 72 (middle column) and 168 (right column) for the P. aeruginosa ATCC 27853, PAO1 and PAOΔmutS delivered over 1, 4, 10 or 24 h durations of exposure, at initial inocula (cfuo) of 106 and 104 cfu/mL, excluding fAUC/MIC of 36 for ATCC 27853 and fAUC/MIC of 168 for PAOΔmutS for cfuo 104 cfu/mL. When a treatment yielded complete eradication or when there were no colonies on antibiotic-containing agar plates no PRB could be determined, and therefore is shown below the limit of quantification as −9.5 log10.
Table 3.
MIC (mg/L) at 24 h [geometric mean (range)] for tobramycin fAUC/MIC of 36, 72 and 168 delivered over various durations of exposure and initial inocula (cfuo) against P. aeruginosa ATCC 27853 (top), PAO1 (middle) and PAOΔmutS (bottom)
| Drug exposure duration (h) |
fAUC/MIC: 36 |
fAUC/MIC: 72 |
fAUC/MIC: 168 |
|||
|---|---|---|---|---|---|---|
| cfuo 1 × 106 | cfuo 1 × 104 | cfuo 1 × 106 | cfuo 1 × 104 | cfuo 1 × 106 | cfuo 1 × 104 | |
| ATCC 27853 control | 0.5 (0.5–0.5) | ND | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) |
| 1 | 0.5 (0.5–0.5) | ND | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) |
| 4 | 0.5 (0.5–0.5) | ND | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.25 |
| 10 | 1.0 (0.5–2.0) | ND | 0.5 | — | 0.25 | — |
| 24 | 4.0 (4.0–4.0) | ND | 4.0 | — | — | — |
| PAO1 control | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) |
| 1 | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.5 (0.5–0.5) | 0.7 (0.5–1.0) | 0.7 (0.5–1.0) | 0.5 (0.5–0.5) |
| 4 | 0.7 (0.5–1.0) | 0.5 (0.5–0.5) | 0.7 (0.5–1.0) | 0.7 (0.5–1.0) | 0.7 (0.5–1.0) | 0.5 |
| 10 | 2.0 (0.5–8.0) | 2.0 (0.5–8.0) | 0.25 | 1.0 | 0.5 (0.25–1.0) | — |
| 24 | 5.7 (4.0–8.0) | — | 4.0 (4–4) | — | — | — |
| PAOΔmutS control | 1.0 (1.0–1.0) | 1.0 (1.0–1.0) | 1.0 (1.0–1.0) | 1.0 (1.0–1.0) | 1.0 (1.0–1.0) | ND |
| 1 | 1.0 (1.0–1.0) | 2.8 (2.0–4.0) | 1.0 (1.0–1.0) | 2.0 (2.0–2.0) | 1.0 (1.0–1.0) | ND |
| 4 | 1.0 (1.0–1.0) | 2.0 (2.0–2.0) | 1.0 (1.0–1.0) | 1.0 (1.0–1.0) | 1.0 (1.0–1.0) | ND |
| 10 | 8.0 (8.0–8.0) | 1.0 | 0.7 (0.5–1.0) | — | — | ND |
| 24 | 11.3 (8.0–16.0) | 5.7 (4.0–8.0) | 5.7 (4.0–8.0) | — | — | ND |
ND indicates that this inoculum was not carried out for this strain in this study.
MICs are in bold if they were at least 4-fold above baseline. No range is provided if only one replicate was available. PAOΔmutS at cfuo of 101.2 cfu/mL had MIC 1.0 mg/L with a range 1.0–1.0 mg/L for the growth control, whilst extensive bacterial killing did not allow determination of the MIC for any treated arms.
For ATCC 27853 and PAO1 the increase in PRB at longer durations could be prevented or considerably limited by the high fAUC/MIC of 168 at the 106 cfu/mL inoculum, except for the PAO1 at the 24 h duration of exposure. In contrast, for PAOΔmutS essentially the whole bacterial population was replaced by resistant bacteria even at the fAUC/MIC of 168. This difference was observed despite the same probability of >96.7% of at least one pre-existing resistant bacterial cell for all three strains. Thus our study demonstrates that resistance suppression is more challenging for the hypermutable strain compared with the two non-hypermutable strains, which is supported by literature reporting that hypermutation results in more difficult-to-eradicate infections.17,21
For the 104 cfu/mL inoculum, ATCC 27853 and PAO1 exposed to tobramycin for short durations (1 and 4 h) had limited to no increase in PRB and for ATCC 27853 the PRB was unquantifiable for long durations of exposure (Figure 3). These results were supported by the MIC at 24 h remaining unchanged for the 1 and 4 h durations of exposure (Table 3). However, for PAO1 the PRB slightly increased for the 10 and 24 h durations of exposure for the low fAUC/MIC of 36 (Figure 3); the 10 h duration of exposure was supported by a raised MIC (Table 3). Similarly, the PAOΔmutS at 104 cfu/mL inoculum revealed increases in PRB for the 10 h duration of exposure at an fAUC/MIC of 36 and the 24 h duration of exposure at an fAUC/MIC of both 36 and 72 (Figure 3). The low inoculum of 101.2 cfu/mL (∼300 bacteria in 20 mL) was required to minimize the probability of pre-existing resistant bacteria for the hypermutable PAOΔmutS strain. No PRB could be determined for this inoculum due to extensive bacterial killing and the very low numbers of bacteria present (data not shown). Although adaptive resistance mechanisms only require tobramycin exposure49 these may have been prevented by the extremely fast bacterial killing seen at the 101.2 cfu/mL inoculum.
The pre-existing resistant bacteria in the initial inoculum may have been playing a role in the emergence of resistance at the 106 cfu/mL inoculum for all strains and at 104 cfu/mL for PAOΔmutS. Treatment failure in P. aeruginosa infections often occurs from selection of resistant bacteria, e.g. due to overexpression of the MexXY-OprM efflux pump, aminoglycoside-modifying enzymes and decreased outer membrane permeability.11–14 For PAO1 it is likely that additional resistance pathways not involving amplification of pre-existing resistant bacteria may have played a role in resistance emergence at the 104 cfu/mL inoculum. The static concentrations with complete removal of tobramycin at the end of each exposure period would have prevented the development of resistance mechanisms that require sub-MIC concentrations to occur. Common mechanisms involved in the resistance observed in PAO1 could be adaptive resistance via the upregulation of the MexXY-OprM efflux pump,41,50 which is frequently caused by overexpression of the MexY component, or de novo mutations during treatment.41,51,52 Previous studies have found that a resistance mechanism unrelated to this efflux pump has played a role in aminoglycoside resistance for PAOΔmutS.6 Ultimately, molecular studies would identify these resistance mechanisms.
Our developed mechanism-based model successfully described simultaneously the time-course of viable counts for the total bacterial population and less susceptible bacterial populations growing on tobramycin-containing plates, for all three strains (Figure 2 and Figure S1, available as Supplementary data at JAC Online). The model included three bacterial subpopulations, i.e. susceptible, intermediate and resistant (Figure 1). Tobramycin causes bacterial killing via inhibiting protein synthesis,8 which is driven by the intracellular tobramycin concentration as described in the model.9 Aminoglycosides can also cause bacterial killing by disrupting the outer membrane;10,53,54 however, adding a second mechanism of killing was not required to describe our observed data. It has been suggested that the cellular recovery after tobramycin exposure causes a delay in bacterial regrowth, resulting in a PAE that was included in the model.40,55 Such a PAE may allow the suppression of regrowth of susceptible subpopulations in patients.56 The PAE duration has been found to be related to the antibacterial concentration,57,58 the duration of drug exposure and the inoculum size.59 A longer duration of exposure was found to result in a longer PAE.40 The ATCC 27853 and PAOΔmutS strains required a PAE to be present in the model, whilst PAO1 was represented by a simplified model that did not require a PAE.
The bacterial regrowth was also well captured by the model with only minor mispredictions of up to ∼2.5 log10. Only for 2 of 80 modelled profiles (ATCC 27853 at the fAUC/MIC of 72, 4 h duration of exposure and PAO1 at the fAUC/MIC of 36, 10 h duration of exposure, 104 cfu/mL inoculum) a larger misprediction occurred as the model predicted complete eradication before bacteria were able to regrow (Figure 2). Emergence of resistance as quantified via tobramycin-containing agar plates was adequately represented in the model (Figure S1). The inclusion of adaptive resistance in the model allowed us to consider the likely cause of emergence of aminoglycoside resistance at low initial inocula.42,60–63 Incorporating a function for adaptive resistance in addition to amplification of pre-existing resistant bacteria was required to describe best the observed data. Overall, the model accurately described the bacterial killing (Figure 2) and emergence of resistance (Figure S1) for all three strains. We recognize that both the in vitro experiments and mechanism-based model lack an immune system effect and therefore the results of this study would be most applicable to immunocompromised patients. In addition, the static nature of the concentration delivery for defined durations generated different concentration–time profiles to those that would be observed in patients.
In conclusion, the study allowed us to determine whether different tobramycin concentration–time profiles at a given overall exposure affect not only bacterial killing, but also resistance prevention. Our results for the 24 h duration of exposure demonstrated that, despite limited regrowth, there was complete replacement of susceptible bacteria with tobramycin-resistant populations when regrowth occurred. Emergence of resistance was suppressed for 1 and 4 h durations of exposure supporting once-daily dosing, although extensive regrowth of susceptible bacteria occurred. Therefore, investigation of dosage regimens involving short-duration, high tobramycin concentrations together with a second antibiotic to prevent tobramycin-susceptible regrowth is warranted. Our mechanism-based mathematical model would assist in the optimization of such antibiotic combinations. Combination dosage regimens may be particularly beneficial and are urgently required to combat hypermutable strains arising in patients. Studies in dynamic in vitro systems that simulate antibiotic concentration–time profiles as observed in patients are necessary to evaluate such innovative aminoglycoside combination dosage regimens and translate these regimens to benefit patients.
Funding
This work was supported by the Australian National Health and Medical Research Council (NHMRC; Career Development fellowship number 1084163 to J. B. B., Career Development fellowship number 1062509 to C. B. L., and Project grant numbers 1101553 and 1045105 to C. B. L., J. B. B., A. O. and R. L. N.) and the William Buckland Foundation (project reference CT 22943 to C. B. L.). B. T. T., R. L. N. and J. B. B. are supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award number R01AI111990).
Transparency declarations
None to declare.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary data
Figure S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
Part of this work was presented as a poster at the Fifty-fourth Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, USA, 2014 (A2-051), the Australasian Society of Clinical and Experimental Pharmacologists and Toxicologists—Molecular Pharmacology of GPCRs (ASCEPT-MPGPCR) Joint Scientific Meeting, Melbourne, Victoria, 2014 (Abstract 521) and the American Society for Microbiology (ASM)—MICROBE, Boston, MA, USA, 2016 (A-4065).
References
- 1.Hede K. Antibiotic resistance: an infectious arms race. Nature 2014; 509: S2–3. [DOI] [PubMed] [Google Scholar]
- 2.Chastre J. Evolving problems with resistant pathogens. Clin Microbiol Infect 2008; 14 Suppl 3: 3–14. [DOI] [PubMed] [Google Scholar]
- 3.Cully M. Public health: the politics of antibiotics. Nature 2014; 509: S16–7. [DOI] [PubMed] [Google Scholar]
- 4.Spellberg B, Blaser M, Guidos RJ et al. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis 2011; 52 Suppl 5: S397–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macia MD, Blanquer D, Togores B et al. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob Agents Chemother 2005; 49: 3382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plasencia V, Borrell N, Macia MD et al. Influence of high mutation rates on the mechanisms and dynamics of in vitro and in vivo resistance development to single or combined antipseudomonal agents. Antimicrob Agents Chemother 2007; 51: 2574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milatovic D, Braveny I. Development of resistance during antibiotic therapy. Eur J Clin Microbiol 1987; 6: 234–44. [DOI] [PubMed] [Google Scholar]
- 8.Lambert PA. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med 2002; 95: 22–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Bulitta JB, Ly NS, Landersdorfer CB et al. Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob Agents Chemother 2015; 59: 2315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loh B, Grant C, Hancock RE. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother 1984; 26: 546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morita Y, Tomida J, Kawamura Y. MexXY multidrug efflux system of Pseudomonas aeruginosa. Front Microbiol 2012; 3: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2011; 2: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guenard S, Muller C, Monlezun L et al. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2014; 58: 221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005; 49: 479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliver A. Mutators in cystic fibrosis chronic lung infection: prevalence, mechanisms, and consequences for antimicrobial therapy. Int J Med Microbiol 2010; 300: 563–72. [DOI] [PubMed] [Google Scholar]
- 16.Oliver A, Mena A. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect 2010; 16: 798–808. [DOI] [PubMed] [Google Scholar]
- 17.Oliver A, Canton R, Campo P et al. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2000; 288: 1251–4. [DOI] [PubMed] [Google Scholar]
- 18.Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol 1996; 50: 625–43. [DOI] [PubMed] [Google Scholar]
- 19.Oliver A, Baquero F, Blazquez J. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol Microbiol 2002; 43: 1641–50. [DOI] [PubMed] [Google Scholar]
- 20.Feliziani S, Lujan AM, Moyano AJ et al. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One 2010; 5: e12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferroni A, Guillemot D, Moumile K et al. Effect of mutator P. aeruginosa on antibiotic resistance acquisition and respiratory function in cystic fibrosis . Pediatr Pulmonol 2009; 44: 820–5. [DOI] [PubMed] [Google Scholar]
- 22.LeClerc JE, Li B, Payne WL et al. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 1996; 274: 1208–11. [DOI] [PubMed] [Google Scholar]
- 23.Craig WA. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis 1998; 26: 1–10; quiz 1-2. [DOI] [PubMed] [Google Scholar]
- 24.Ioannides-Demos LL, Liolios L, Wood P et al. Changes in MIC alter responses of Pseudomonas aeruginosa to tobramycin exposure. Antimicrob Agents Chemother 1998; 42: 1365–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambrose PG, Bhavnani SM, Rubino CM et al. Pharmacokinetics-pharmacodynamics of antimicrobial therapy: it's not just for mice anymore. Clin Infect Dis 2007; 44: 79–86. [DOI] [PubMed] [Google Scholar]
- 26.Smyth AR, Bhatt J. Once-daily versus multiple-daily dosing with intravenous aminoglycosides for cystic fibrosis. Cochrane Database Syst Rev 2014; issue 2: CD002009. [DOI] [PubMed] [Google Scholar]
- 27.Smyth A, Tan KH, Hyman-Taylor P et al. Once versus three-times daily regimens of tobramycin treatment for pulmonary exacerbations of cystic fibrosis—the TOPIC study: a randomised controlled trial. Lancet 2005; 365: 573–8. [DOI] [PubMed] [Google Scholar]
- 28.Burkhardt O, Lehmann C, Madabushi R et al. Once-daily tobramycin in cystic fibrosis: better for clinical outcome than thrice-daily tobramycin but more resistance development? J Antimicrob Chemother 2006; 58: 822–9. [DOI] [PubMed] [Google Scholar]
- 29.Rees VE, Bulitta JB, Nation RL et al. Shape does matter: short high-concentration exposure minimizes resistance emergence for fluoroquinolones in Pseudomonas aeruginosa. J Antimicrob Chemother 2015; 70: 818–26. [DOI] [PubMed] [Google Scholar]
- 30.Mena A, Smith EE, Burns JL et al. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J Bacteriol 2008; 190: 7910–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.EUCAST. Antimicrobial Wild Type Distributions of Microorganisms. http://www.eucast.org/mic_distributions_and_ecoffs/.
- 32.Conil JM, Georges B, Ruiz S et al. Tobramycin disposition in ICU patients receiving a once daily regimen: population approach and dosage simulations. Br J Clin Pharmacol 2011; 71: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Ninth Edition: Approved Standard M07-A9. CLSI, Wayne, PA, USA, 2012. [Google Scholar]
- 34.Oliver A, Levin BR, Juan C et al. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob Agents Chemother 2004; 48: 4226–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maidhof H, Johannsen L, Labischinski H et al. Onset of penicillin-induced bacteriolysis in staphylococci is cell cycle dependent. J Bacteriol 1989; 171: 2252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landersdorfer CB, Ly NS, Xu H et al. Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob Agents Chemother 2013; 57: 2343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulitta JB, Ly NS, Yang JC et al. Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53: 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuji BT, Bulitta JB, Brown T et al. Pharmacodynamics of early, high-dose linezolid against vancomycin-resistant enterococci with elevated MICs and pre-existing genetic mutations. J Antimicrob Chemother 2012; 67: 2182–90. [DOI] [PubMed] [Google Scholar]
- 39.Lin HY, Landersdorfer CB, London D et al. Pharmacodynamic modeling of anti-cancer activity of tetraiodothyroacetic acid in a perfused cell culture system. PLoS Comput Biol 2011; 7: e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu ZY, Li RC. Impact of pharmacokinetics on the postantibiotic effect exhibited by Pseudomonas aeruginosa following tobramycin exposure: application of an in-vitro model. J Antimicrob Chemother 1998; 42: 61–5. [DOI] [PubMed] [Google Scholar]
- 41.Hocquet D, Vogne C, El Garch F et al. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 2003; 47: 1371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daikos GL, Jackson GG, Lolans VT et al. Adaptive resistance to aminoglycoside antibiotics from first-exposure down-regulation. J Infect Dis 1990; 162: 414–20. [DOI] [PubMed] [Google Scholar]
- 43.Ly NS, Bulitta JB, Rao GG et al. Colistin and doripenem combinations against Pseudomonas aeruginosa: profiling the time course of synergistic killing and prevention of resistance. J Antimicrob Chemother 2015; 70: 1434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulitta JB, Yang JC, Yohonn L et al. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother 2010; 54: 2051–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulitta JB, Bingolbali A, Shin BS et al. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J 2011; 13: 201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matthews I, Kirkpatrick C, Holford N. Quantitative justification for target concentration intervention-parameter variability and predictive performance using population pharmacokinetic models for aminoglycosides. Br J Clin Pharmacol 2004; 58: 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hennig S, Norris R, Kirkpatrick CM. Target concentration intervention is needed for tobramycin dosing in paediatric patients with cystic fibrosis—a population pharmacokinetic study. Br J Clin Pharmacol 2008; 65: 502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hennig S, Standing JF, Staatz CE et al. Population pharmacokinetics of tobramycin in patients with and without cystic fibrosis. Clin Pharmacokinet 2013; 52: 289–301. [DOI] [PubMed] [Google Scholar]
- 49.Pagkalis S, Mantadakis E, Mavros MN et al. Pharmacological considerations for the proper clinical use of aminoglycosides. Drugs 2011; 71: 2277–94. [DOI] [PubMed] [Google Scholar]
- 50.Vogne C, Aires JR, Bailly C et al. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother 2004; 48: 1676–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeannot K, Sobel ML, El Garch F et al. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J Bacteriol 2005; 187: 5341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hay T, Fraud S, Lau CH et al. Antibiotic inducibility of the mexXY multidrug efflux operon of Pseudomonas aeruginosa: involvement of the MexZ anti-repressor ArmZ. PLoS One 2013; 8: e56858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hancock RE, Raffle VJ, Nicas TI. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1981; 19: 777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadurugamuwa JL, Clarke AJ, Beveridge TJ. Surface action of gentamicin on Pseudomonas aeruginosa. J Bacteriol 1993; 175: 5798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li RC, Lee SW, Kong CH. Correlation between bactericidal activity and postantibiotic effect for five antibiotics with different mechanisms of action. J Antimicrob Chemother 1997; 40: 39–45. [DOI] [PubMed] [Google Scholar]
- 56.Craig WA. Post-antibiotic effects in experimental infection models: relationship to in-vitro phenomena and to treatment of infections in man. J Antimicrob Chemother 1993; 31 Suppl D: 149–58. [DOI] [PubMed] [Google Scholar]
- 57.Isaksson B, Nilsson L, Maller R et al. Postantibiotic effect of aminoglycosides on gram-negative bacteria evaluated by a new method. J Antimicrob Chemother 1988; 22: 23–33. [DOI] [PubMed] [Google Scholar]
- 58.Karlowsky JA, Zhanel GG, Davidson RJ et al. Postantibiotic effect in Pseudomonas aeruginosa following single and multiple aminoglycoside exposures in vitro. J Antimicrob Chemother 1994; 33: 937–47. [DOI] [PubMed] [Google Scholar]
- 59.Bundtzen RW, Gerber AU, Cohn DL et al. Postantibiotic suppression of bacterial growth. Rev Infect Dis 1981; 3: 28–37. [DOI] [PubMed] [Google Scholar]
- 60.Barclay ML, Begg EJ. Aminoglycoside adaptive resistance: importance for effective dosage regimens. Drugs 2001; 61: 713–21. [DOI] [PubMed] [Google Scholar]
- 61.Barclay ML, Begg EJ, Chambers ST. Adaptive resistance following single doses of gentamicin in a dynamic in vitro model. Antimicrob Agents Chemother 1992; 36: 1951–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barclay ML, Begg EJ, Chambers ST et al. The effect of aminoglycoside-induced adaptive resistance on the antibacterial activity of other antibiotics against Pseudomonas aeruginosa in vitro. J Antimicrob Chemother 1996; 38: 853–8. [DOI] [PubMed] [Google Scholar]
- 63.Daikos GL, Lolans VT, Jackson GG. First-exposure adaptive resistance to aminoglycoside antibiotics in vivo with meaning for optimal clinical use. Antimicrob Agents Chemother 1991; 35: 117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.