ABSTRACT
H19 is a long noncoding RNA differentially expressed in many tumors and participates in tumorigenesis. This study aimed to investigate the expression and function of H19 in pancreatic ductal adenocarcinoma (PDAC). Pure malignant cells were isolated from frozen sections of 25 PDAC cases by laser captured microdessection, and H19 expression level was detected by qRT-PCR. Knockdown and overexpression were employed to manipulate H19 levels in pancreatic cancer cells, then cell viability, proliferation, apoptosis and cell cycle, and the growth of xenografts were evaluated. E2F-1 levels in PDAC tissues were detected by Western blot and immunohistochemical analysis. We found that H19 was overexpressed in PDAC tissues and correlated to histological grade of PDAC. Knockdown of H19 in T3M4 and PANC-1 cells with high H19 endogenous level suppressed cell viability, proliferation and tumor growth, while H19 overexpression in COLO357 and CAPAN-1 with low H19 endogenous level enhanced cell viability, proliferation and tumor growth. Knockdown of H19 led to G0/G1 arrest, accompanied by decreased levels of E2F-1 and its downstream targets. E2F-1 was overexpressed in PDAC tissues with possible correlation with H19 expression level. In conclusion, H19 is overexpressed and plays oncogenic role in PDAC through promoting cancer cell proliferation via the upregulation of E2F-1.
KEYWORDS: E2F-1, H19, laser captured microdissection, pancreatic dutal adenocarcinoma
Introduction
Pancreatic cancer ranks as the fourth leading cause of cancer death. The incidence of pancreatic cancer is dramatically increased after people reach their 40s.1 With recent advances in the diagnosis and treatment, the 5-year survival of pancreatic cancer patients has increased to 7%, but the mortality rate is still high.2 Pancreatic ductal adenocarcinoma (PDAC) accounts for almost 90% of pancreatic malignancies, and is quite difficult to be diagnosed at the early stage. Once PDAC is apparent in the clinical, the primary tumor has already invaded the adjacent tissues and even metastasized to distant organs to become unresectable.3 Furthermore, most PDAC have developed chemo-resistance which makes the prognosis even worse. Multiple genetic changes haven been found in PDAC, such as the mutations of BRCA1, BRCA2, PALB2, KRAS2, p16/CDKN2A, TP53, DPC4/SMAD4 and aberrant activation of Hedgehog signaling.4 Further understanding of molecular mechanisms of PDAC may help identify potential tumor biomarkers and develop novel therapeutic targets.
Long non-coding RNAs (lncRNAs), with length >200 nucleotides, are once considered as junk DNA and transcriptional noise.5 However, emerging evidences demonstrate that lncRNAs are important regulators of gene expression by participating in epigenetic modification, transcriptional control, RNA processing control, translational control and post-translational modification. An increasing number of lncRNAs have been identified to be differentially expressed in many human cancers and implicated in tumor progression and metastasis. Most lncRNAs are hard to be characterized because of their high turnover, poor conservation and different abundance. Only the stable, highly abundant and conserved lncRNAs are likely to become potential biomarkers and therapeutic targets of cancers and other diseases.6
H19 gene is located on human chromosome 11p15.5 with close proximity to insulin-like growth factor 2 (IGF2).7 H19 was abundantly expressed in almost all fetal tissues on maternal allele and imprinted on paternal allele. During this stage, H19 plays critical role in the differentiation of several vital organs. After birth there is a drastic reduction of H19 level, and it is only expressed in few specific tissues such as mammary gland and skeletal muscle.8,9 Nevertheless, differential expression of H19 is observed in several tumors. In cancer progression, H19 has a dual role as oncogene or tumor suppressor depending on cancer type and developmental stage. Due to the lack of H19 expression in Wilm's tumor, embryonic rhabdomyosarcoma and Beckwith-Wiedmann syndrome, H19 may function as a tumor suppressor in these tumors.10-13 In contrast, overexpression of H19 has been documented in breast cancer, bladder cancer, gastric cancer, colon cancer, esophageal cancer, lung cancer, osteosarcoma, epithelial ovarian cancer and cervical cancer.8,14-21 Aberrant expression of H19 in these tumors is related to increased cell proliferation and invasion and reduced apoptosis.
It is speculated that the loss of imprinting (LOI), loss of heterozygosity (LOH) of H19/IGF2 cluster, or epigenetic deregulation of IGF2/H19 imprinted domain could drive tumorigenesis. Biallelic H19 and IGF2 expression was observed in choriocarcinoma and ovarian cancer.22,23 Moreover, Berteaux et al. reported that H19 could promote breast cancer cell proliferation by recruiting E2F1 to H19 promoterv.8 Yang et al. found that H19 was related to p53-mediated apoptosis and growth inhibition, and it partially suppressed p53 activation in gastric cancer.24 Furthermore, H19 could encode the precursor for miR-675,25 and the targets of miR-675 such as RB and tumor suppressor Runt Domain Transcription Factor (RUNX1) are involved in colorectal cancer and gastric cancer.16,26 Although H19 has been proved to function as an oncogene in many malignances, the relationship between H19 and pancreatic cancer has not been fully elucidated. Ma et al. demonstrated that H19 promoted PDAC cell invasion and migration partially by increasing HMGA2 mediated epithelial-mesenchymal transition (EMT) through antagonizing let-7.27 We speculate that other targets of H19 may mediate oncogenic role of H19 in PDAC. In this study, we employed laser captured microdissection (LCM) technique to accurately detect the expression level of H19 in PDAC excluding the interference of adjacent non-tumor tissues, and used pancreatic cancer cell lines and xenograft nude mouse model to explore possible regulatory mechanism of H19 in the tumorigenesis of PDAC. We demonstrated that H19 was overexpressed in both PDAC microdessected tissues and pancreatic cancer cell lines. Knockdown of H19 in T3M4 and PANC-1 cells inhibited cell proliferation by inducing G0/G1 cell cycle arrest, while enforced expression of H19 in COLO357 and CAPAN-1 cells led to opposite effects. Similar results were observed in xenograft nude mice. Furthermore, we identified E2F transcription factor 1 (E2F-1) as a potential effector of H19. These data suggest that H19 plays an oncogenic role in PDAC through promoting cancer cell proliferation via the upregulation of E2F-1.
Results
High H19 expression in PDAC tissues and pancreatic cancer cell lines
To explore the role of H19 in PDAC, we examined the expression of H19 in both PDAC tissues and pancreatic cancer cell lines. Tissue heterogeneity was observed in PDAC sections. Besides 20–30% malignant ducts, there were approximately 20–30% destroyed normal acinar and islet cells along with 30–40% fibroblast cells, lymphatic cells and neonatal blood vessels. Therefore, it was necessary to isolate cancer cells from non-tumor tissues by LCM technique (Fig. 1A). qPCR analysis showed that H19 was highly expressed in 68% PDAC tissues (18/25) compared with adjacent normal tissues (ANT) (Fig. 1B). We then analyzed the clinicopathological characteristics of 25 PDAC cases and found significant difference in H19 expression between the well-, moderate-differentiated tumor group and poorly-differentiated tumor group (P < 0.05, Table 1). In addition, compared to normal human pancreatic duct cells, the expression of H19 was remarkably upregulated in pancreatic cancer cell lines, especially in BxPC-3, T3M4 and PANC-1 cells (Fig. 1C).
Figure 1.
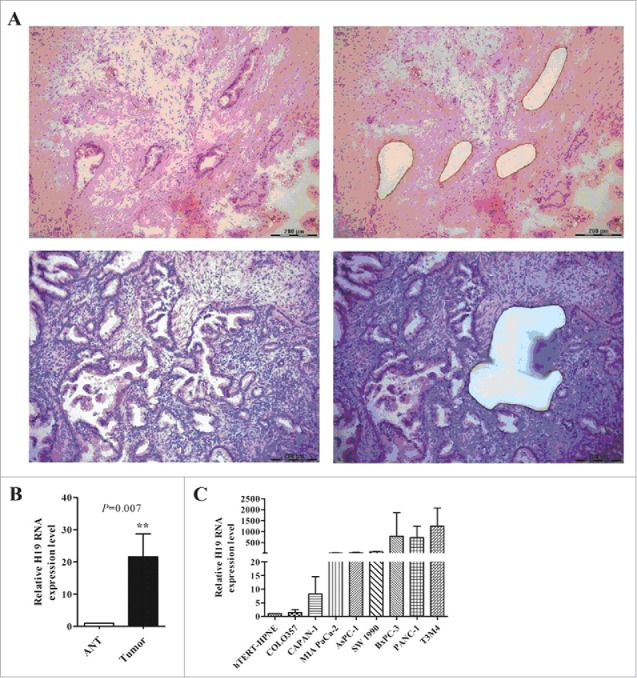
High expression of H19 in PDAC frozen sections and pancreatic cancer cells. (A) Laser captured microdissection of PDAC tissues. Left: before LCM, right: after LCM. (B) Expression of H19 in microdissected tissues was detected by qPCR. (C) Expression of H19 in pancreatic cancer cell lines was detected by qPCR. Results were expressed as mean ± s.d., n = 3, **P < 0.01.
Table 1.
Relationship between H19 and clinicopathological characteristics of PDAC.
| H19 expression |
||||
|---|---|---|---|---|
| Characteristics | Number of case (n = 25) | Low (n = 8) (%) | High (n = 17) (%) | P value |
| Age (years) | 0.999 | |||
| <60 | 12 | 4(50%) | 8(47.06%) | |
| ≥60 | 13 | 4(50%) | 9(52.94%) | |
| Gender | 0.999 | |||
| Male | 11 | 3(37.5%) | 8(47.06%) | |
| Female | 14 | 5(62.5%) | 9(52.94%) | |
| Tumor size | 0.394 | |||
| <4cm | 16 | 4(50%) | 12(70.59%) | |
| ≥4 cm | 9 | 4(50%) | 5(29.41%) | |
| Location | 0.359 | |||
| Head of pancreas | 17 | 4(50%) | 13(76.47%) | |
| Body and tail of pancreas | 8 | 4(50%) | 4(23.53%) | |
| Histological grade | 0.017 | |||
| Well and moderate | 7 | 5(62.5%) | 2(11.76%) | |
| Poor | 18 | 3(37.5%) | 15(88.23%) | |
| Primary tumor | 0.081 | |||
| T1, T2 | 4 | 3(37.5%) | 1(5.88%) | |
| T3, T4 | 21 | 5(62.5%) | 16(94.12%) | |
| Lymph node | 0.999 | |||
| N0 | 17 | 6(75%) | 11(64.71%) | |
| N1 | 8 | 2(25%) | 6(35.29%) | |
| Venous invasion | 0.667 | |||
| Absent | 16 | 6(75%) | 10(58.82%) | |
| Present | 9 | 2(25%) | 7(41.18%) | |
| Nervous invasion | 0.057 | |||
| Absent | 4 | 2(25%) | 2(11.76%) | |
| Present | 21 | 6(75%) | 15(88.24%) | |
| CA19-9 | 0.999 | |||
| <37 U/ml | 5 | 1(12.5%) | 4(23.53%) | |
| ≥37 U/ml | 20 | 7(87.5%) | 13(76.47%) | |
| CEA | 0.194 | |||
| <5 ng/ml | 15 | 3(37.5%) | 12(70.59%) | |
| ≥5 ng/ml | 10 | 5(62.5%) | 5(29.41%) | |
H19 regulates pancreatic cancer cell proliferation and apoptosis
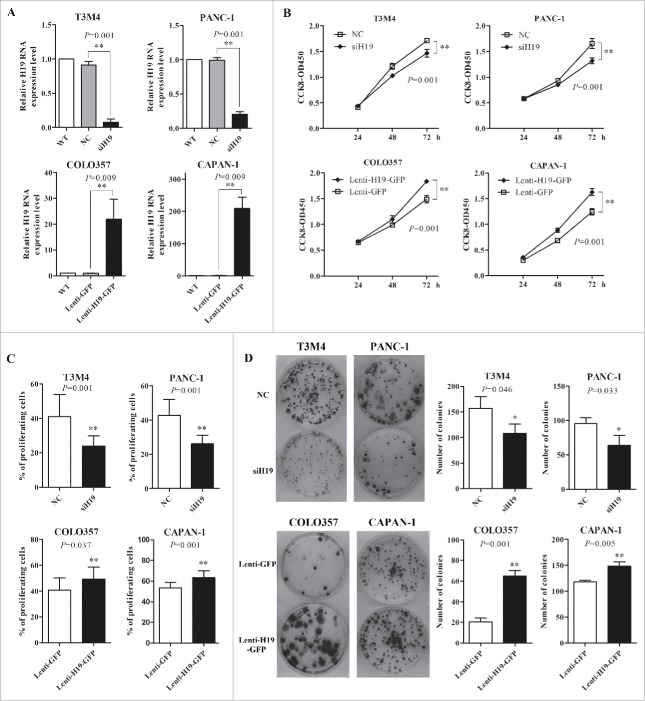
Next we investigated the function role of high expression of H19 in pancreatic cancer. We employed siH19 mixture of 3 different siRNAs (siH19-1, siH19-2 and siH19-3) to enhance efficiency of H19 knockdown in T3M4 and PANC-1 cells with high level of H19 expression, while used lentivirus to overexpress H19 in COLO357 and CAPAN-1 cells with low level of H19 expression (Fig. 2A). CCK-8 assay showed that knockdown of H19 significantly reduced the viability of T3M4 and PANC-1 cells while H19 overexpression significantly increased the viability of COLO357 and CAPAN-1 cells (Fig. 2B). EdU incorporation assay showed that knockdown of H19 significantly reduced the proliferation of T3M4 and PANC-1 cells while H19 overexpression significantly increased the proliferation of COLO357 and CAPAN-1 cells (Fig. 2C). Consistently, colony formation assay showed that knockdown of H19 significantly reduced the growth of T3M4 and PANC-1 cells while H19 overexpression significantly increased the growth of COLO357 and CAPAN-1 cells (Fig. 2D). Collectively, these data suggest that H19 enhances pancreatic cancer cell proliferation.
Figure 2.
H19 regulates pancreatic cancer cell viability, proliferation, growth and cell cycle progression in vitro. (A) T3M4 and PANC-1 cells were transfected with siH19 mixture or siNC. COLO357 and CAPAN-1 cells were infected with Lenti-H19-GFP or Lenti-GFP. H19 expression level was verified by qPCR. (B) Cell viability was determined by CCK-8 assay. The cell proliferation curves were based on the absorbance at 450 nm measured every 24 h. (C) EdU incorporation assay of cell proliferation. The cells that incorporated EdU were detected by fluorescent microscopy and counted. (D) 200 cells were seeded in 6-well plates and cultured for 14 d and then the number of macroscopic colonies was counted.
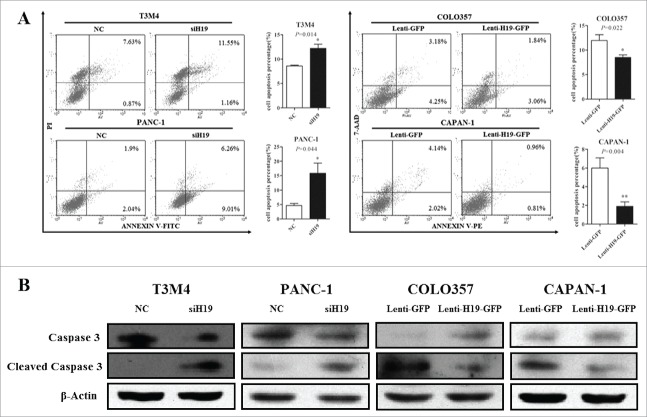
Furthermore, we examined the effects of H19 on the apoptosis of pancreatic cancer cells. Flow cytometry analysis showed that knockdown of H19 led to increased apoptosis of T3M4 and PANC-1 cells, while overexpression of H19 led to decreased apoptosis of COLO357 and CAPAN-1 cells (Fig. 3A). Consistently, knockdown of H19 led to increased cleavage of Caspase 3 in T3M4 and PANC-1 cells, while overexpression of H19 led to decreased cleavage of Caspase 3 in COLO357 and CAPAN-1 cells (Fig. 3B). Collectively, these data suggest that H19 inhibits pancreatic cancer cell apoptosis.
Figure 3.
H19 regulates pancreatic cancer cell apoptosis in vitro. (A) T3M4 and PANC-1 cells were transfected with siH19 mixture or siNC while COLO357 and CAPAN-1 cells were transfected with Lenti-H19-GFP or Lenti-GFP vector. The cells were stained with PI and Annexin and apoptosis was measured by flow cytometry. (B) The protein levels of caspase 3 and cleaved caspase 3 were determined by Western blot analysis. Results were expressed as mean ± s.d., n = 3, *P < 0.05, **P < 0.01.
H19 regulates tumorigenicity of pancreatic cancer cells in vivo
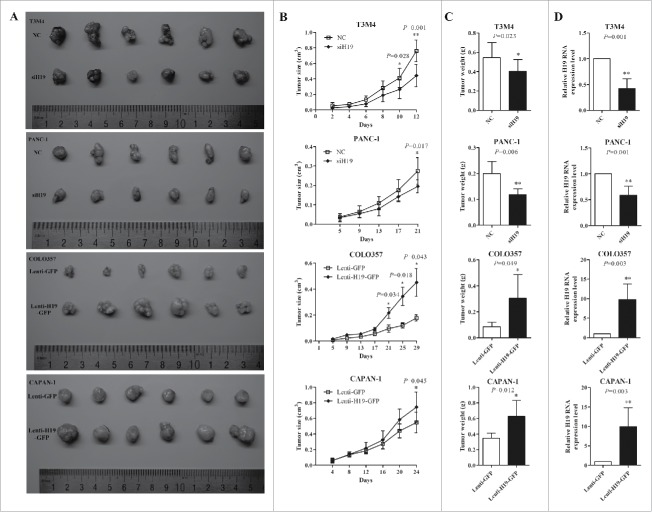
T3M4, PANC-1, COLO357 and CAPAN-1 cells transfected with siH19 mixture or infected with Lenti-H19-GFP were used to established nude mice xenograft model. After two to 3 weeks there were significant changes in both tumor volume and weight compared to negative controls (P < 0.05). The tumors in H19 knockdown group were smaller and grew more slowly than in NC group, while the tumors in H19 overexpression group grew bigger and faster, compared to NC group (Fig. 4A-C). H19 expression levels in xenograft tumor were verified by qPCR and were consistent with the in vitro experiments (Fig. 4D).
Figure 4.
H19 promotes the growth of xenograft pancreatic tumor in vivo. (A) Photograghs of tumors derived from siH19 group, Lenti-H19-GFP group and negative control group of nude mice. (B) The volume of xenograft tumors was measured every 2 d to draw tumor growth curves. (C) Tumor weight of xenograft tumors was calculated. (D) H19 expression in xenagraft tumor was determined by qPCR. Results were expressed as mean ± s.d., n = 6, *P < 0.05, **P < 0.01.
E2F-1 mediates oncogenic effects of H19 in PDAC
Considering that E2F-1 is a well-known key transcription factor in G1-S transition, we wondered whether E2F-1 mediated the stimulatory effects of H19 on pancreatic cancer cell proliferation. Cell cycle analysis revealed that knockdown of H19 in T3M4 and PANC-1 cells caused a G0/G1 phase arrest, the percentage of cells in G0/G1 increased whereas the percentage of cells in G2/M decreased. In contrast, H19 overexpression in COLO357 and CAPAN-1 cells led to decreased cell number in G0/G1 phase and increased cell number in G2/M phase (Fig. 5A).
Figure 5.
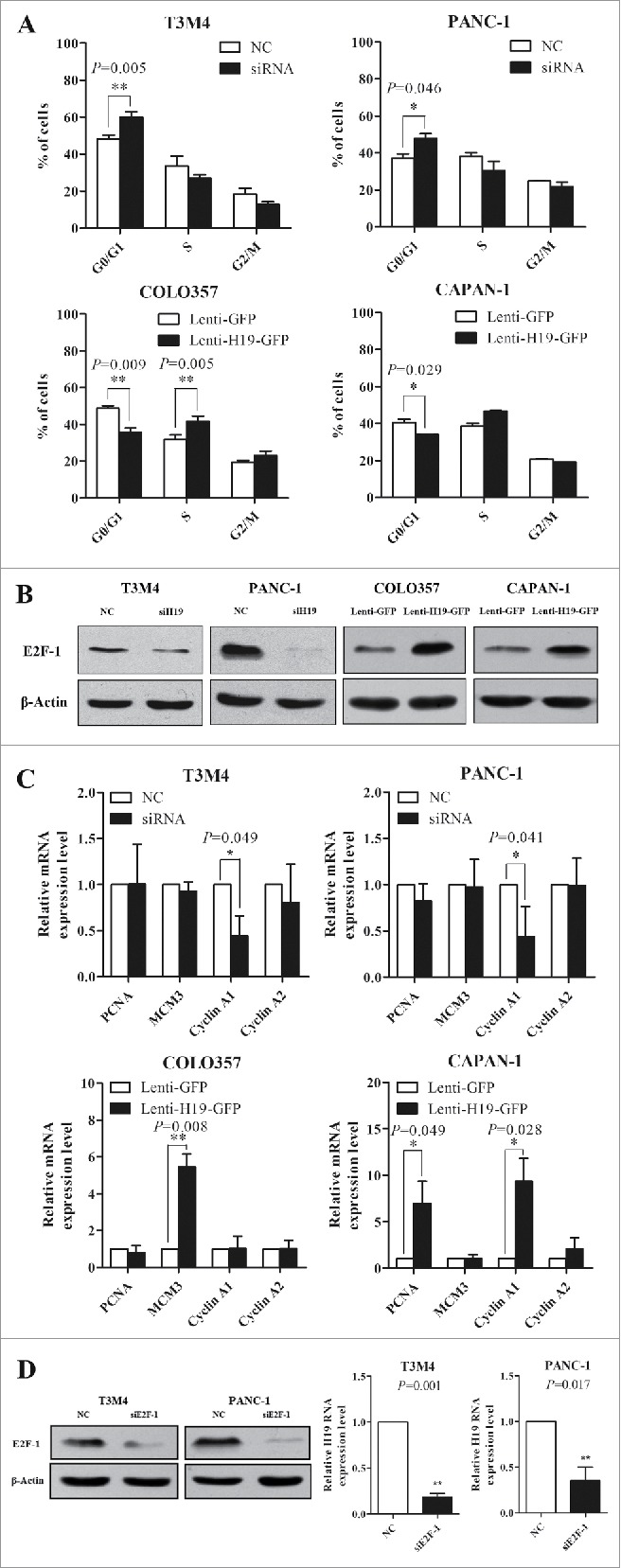
H19 regulates pancreatic cancer cell cycle progression via E2F-1 and the downstream targets. (A) Flow cytometry analysis of cell cycle progression. Results were expressed as mean ± s.d., n = 3, *P < 0.05, **P < 0.01. (B) Western blot analysis of E2F-1 protein level. (C) qRT-PCR analysis of mRNA levels of PCNA, MCM3, Cyclin A1 and Cyclin A2. Results were expressed as mean ± s.d., n = 3, *P < 0.05, **P < 0.01. (D) T3M4 and PANC-1 cells were transfected with siE2F-1 or siNC. The protein level of E2F-1 was determined by Western blot analysis. H19 expression was determined by qPCR. Results were expressed as mean ± s.d., n = 3.
Next, we examined protein level of E2F-1 and found that H19 knockdown in T3M4 and PANC-1 cells downregulated E2F-1 level, while overexpression of H19 in COLO357 and CAPAN-1 cells upregulated E2F-1 level (Fig. 5B). Furthermore, the downstream targets of E2F-1 implicated in cell proliferation such as PCNA, MCM3, Cyclin A1 and Cyclin A2 showed consistent changes in expression level with the upregulation or downregulation of E2F-1 (Fig. 5C). Interestingly, knockdown of E2F-1 in T3M4 and PANC-1 cells caused reduced level of H19 (Fig. 5D). These results indicate that E2F-1 and its downstream targets may mediate the stimulatory effects of H19 on pancreatic cell proliferation.
The expression levels of E2F-1 and H19 are correlated in PDAC
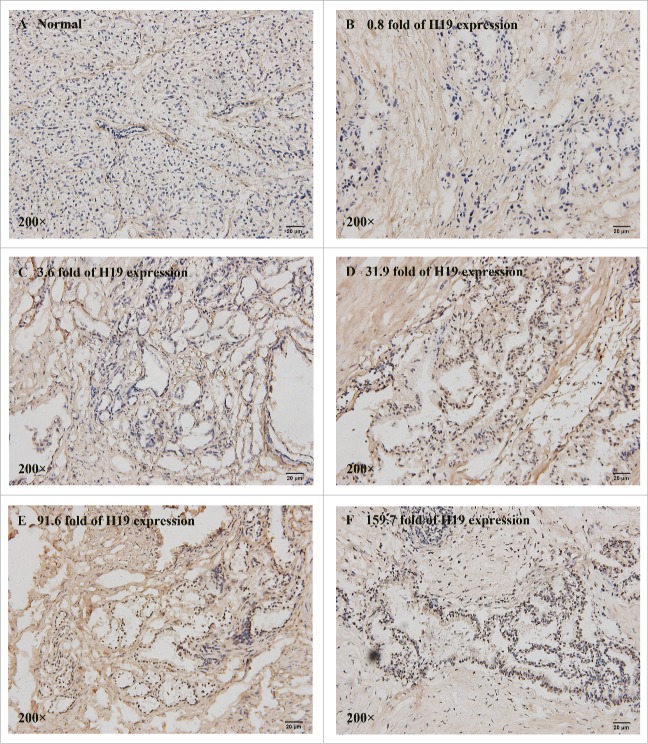
Next we explored whether there may be a correlation between E2F-1 and H19 expression levels in PDAC tissues. We examined 25 cases of PDAC tissues and 10 cases of adjacent normal tissues by immunohistochemical staining. All normal pancreatic tissues barely expressed E2F-1 (IHC score ≤1 ) (Fig. 6A). However, the nuclear staining intensity of E2F-1 was significantly increased in PDAC tissues, and strong E2F-1 staining was detected in 44% (11/25) PDAC tissues. There was a remarkable difference in E2F-1 expression between H19 high expression group and low expression group (P < 0.05, Table 2). 58.82% (10/17) of PDAC cases with high level of H19 exhibited E2F-1 overexpresssion, while only 12.5% (1/8) of PDAC cases with low level of H19 had strong staining of E2F-1.
Figure 6.
Correlation of E2F-1 and H19 expression levels in frozen sections of 25 PDAC tissues. The nuclear staining intensity and positively stained cells of E2F-1 were distinctly different in normal pancreatic tissue (A) (IHC score = 0) and PDAC tissues with various H19 level at 0.8-fold (B) (IHC score = 1), 3.6-fold (C) (IHC score = 2), 31.9-fold (D) (IHC score = 4), 91.6-fold (E) (IHC score = 5) and 159.7-fold (F) (IHC score = 6).
Table 2.
The correlation of E2F-1 and H19 expression levels in PDAC tissues.
| E2F-1 expression |
||||
|---|---|---|---|---|
| Groups | Number of case | Low (%) | High (%) | P value |
| Pathology | 0.015 | |||
| Normal pancreatic tissue | 10 | 10(41.67%) | 0(0%) | |
| PDAC | 25 | 14(58.3%) | 11(100%) | |
| H19 expression in PDAC | 0.042 | |||
| Low | 8 | 7(50%) | 1(9.09%) | |
| High | 17 | 7(50%) | 10(90.91%) | |
Spearman's rank correlation analysis showed no significant correlation between E2F-1 and H19 expression (r = 0.339, P = 0.098), but there was a trend that PDAC tissues have high level of E2F-1 and H19 expression. The IHC score of E2F-1 staining was 6 in PDAC specimen in which H19 expression increased by 159.7-fold, but was only 2 in the specimen in which H19 expression merely increased by 3.6-fold. The possible correlation between E2F-1 and H19 expression provides further evidence that E2F-1 mediates the effects of H19 on pancreatic tumorigenesis.
Discussion
In the present study, we explored the expression and function role of lncRNA H19 in pancreatic cancer. The constitution of pancreatic carcinoma is much more complicate than other solid tumors. Tissue heterogeneity may be the main biological feature of PDAC. Besides sporadic malignant cells, there are many destroyed normal ducal and acinar cells, pancreatic islets fibroblastic tissues and lymphatic cells. Moreover, most pancreatic tumors exhibit extensive fibroblastic stromal content, a pathological feature named desmoplasia.28 The malignant cells scattered in the non-tumor tissues and it is unreasonable to extract RNA or DNA from the whole pancreatic tumor tissue to analyze gene expression changes involved in tumorigenesis. Microdissection makes it possible to isolate homogenous, ultrapure samples from heterogeneous tissue sections without contamination from surrounding cells.29 LCM technique has helped identify molecular signatures such as biomarkers in many cancers.30
Using LCM we found that H19 was highly expressed in most PDAC and related to tumor differentiation. H19 expression was much higher in poorly-differentiated tumor which exhibited more severe malignant phenotypes than in well-differentiated tumor. Perhaps due to small sample size in this study, we found no significant correlations between H19 level and other clinicopahological features. Further studies that enroll more clinical samples are needed to analyze the clinical significance of H19 overexpression in PDAC. Compared with human pancreatic normal epithelial cell line, 7 pancreatic cancer cell lines exhibited high expression level of H19. In particular, pancreatic cancer cell lines with high proliferation rate such as T3M4 and PANC-1 had extremely high level of H19 expression. Taken together, these results strongly suggest that H19 plays an oncogenic role in PDAC tumorigenesis.
To understand the mechanism underlying the oncogenic role of H19 in PDAC, we employed loss and gain of function approaches to knockdown or overexpress H19 in pancreatic cancer cell lines. After knockdown of H19 in cells with high level of endogenous H19, the cell viability, proliferation and colony formation were significantly suppressed while apoptosis was significantly enhanced. In contrast, overexpression of H19 in cells with low level of endogenous H19 led to increased cell viability, proliferation and colony formation but decreased apoptosis. In xenograft nude mice we showed consistent results with in vitro cell experiments. Given that H19 is highly abundant in fetal tissues and essential for cell proliferation and regeneration,9 aberrant H19 overexpression after birth could promote tumor growth.
Notably, we found that knockdown of H19 expression induced G0/G1 phase arrest in pancreatic cancer cells, indicating that H19 regulates the growth of PDAC through cell cycle modulation. As E2F family of transcription factors are crucial for the entry into S phase, we analyzed E2F-1 protein level and found that downregulation of H19 resulted in decreased E2F-1 expression, while upregulation of H19 led to increased E2F-1 expression. Furthermore, we observed consistent changes of mRNA levels of several genes known to be regulated by E2F-1, such as Cyclin A1 and MCM3. In addition, immunohistochemical staining of E2F-1 in PDAC specimens was significantly different between H19 high expression group and low expression group (P = 0.042), and was inclined to be positively correlated to H19 expression level (P = 0.098). There is a chance that the positive correlation between E2F-1 and H19 could be confirmed if we enlarge the sample size in our further studies. Berteaux et al. had revealed the possible link between H19 and E2F-1 in breast cancer. They found 2 putative E2F consensus sites in H19 minimal promoter, and showed that endogenous E2F-1 was recruited to H19 promoter to activate H19 expression. Silencing of E2F-1 could decrease endogenous H19 expression.8 In agreement with previous report, we found that knockdown of E2F-1 reduced the level of H19 in T3M4 and PANC-1 cells. Interestingly, Tsang et al. characterized H19/miR-675/RB pathway in colorectal cancer cells and found that RB was a potential target of H19-derived miR-675. H19 and miR-675 expression levels were upregulated in colorectal cancer tissues and cells, leading to decreased RB expression and increased cell growth and tumor formation.16 In this study, we provided clue that H19 may regulate E2F-1 expression as a positive feedback. Additional investigations are needed to elucidate how H19 upregulates E2F-1 expression in pancreatic cancer cells.
In conclusion, our present findings show that H19 is overexpressed in PDAC and plays oncogenic role in pancreatic tumorigensis. H19 regulates pancreatic cancer cell proliferation by promoting G1-S transition. In PDAC tissues and cells, E2F-1 protein level is positively correlated to H19 expression and could mediate oncogenic potential of H19.
Materials and methods
Human tissue specimens
Thirty PDAC cases were enrolled at Peking University First Hospital affiliated to Peking University Health Science Center. All the 30 patients underwent primary surgical resection of PDAC, and the medical records and follow-up data were complete. Both the tumor and adjacent normal tissue were immediately frozen in liquid nitrogen and stored at −80°C until further use. All patients provided written informed consent for the use of their tissues. This study was approved by the Ethics Committee of Peking University First Hospital affiliated to Peking University Health Science Center.
Cell culture
Human pancreatic cancer cell lines COLO357, CAPAN-1, MIA PaCa-2, AsPC-1, BxPC-3, PANC-1 and T3M4 were kindly provided by Dr. Marko Kornmann of University Hospital of Ulm (Ulm, Germany), SW 1990 and human pancreatic normal epithelial cell line hTERT-HPNE were purchased from American Type Culture Collection (Manassas, USA). COLO357, MIA PaCa-2 and PANC-1 cells were maintained in Dulbecco's modified Eagle's medium (Gibco, USA), CAPAN-1, BxPC-3, T3M4 and AsPC-1 were maintained in RPMI 1640 medium (Gibco, USA), SW 1990 was maintained in Leibovitz's L-15 Medium (KeyGEN BioTECH, China). These media were supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 µg/ml), and cells were cultured at 37°C with 5% CO2. hTERT-HPNE was maintained in 75% DMEM without glucose (Gibco, USA) supplemented with 10 ng/ml human recombinant EGF, 5.5 mM D-glucose (1g/L), 750 ng/ml puromycin, 5% fetal bovine serum and cultured at 37°C with 5% CO2.
Laser capture microdissection
PDAC tumor and adjacent normal tissues were flash-frozen after surgery followed by cryosectioning. Three to 4 frozen sections were cut at 15 μm thickness to ensure the good quality of sections and sufficient quantity of RNA, and placed on labeled, uncharged, pre-cleaned PEN glass microscope slides especially for LCM. Then sections were stained with hematoxylin and eosin (H&E). Leica LMD7000 (Germany) was used to isolate malignant pancreatic ducts from the frozen sections. LCM process consisted of 6 steps: 1) loading the slides and LCM caps, 2) locating the cells of interest, 3) LCM cap placement and laser location, 4) marking the cells of interest, 5) capturing the cells, and 6) unloading of the samples and caps containing the captured tissues.
Real-time quantitative PCR
Total RNA was extracted from microdissected tissues and pancreatic cancer cell lines using TRIzol (Invitrogen, USA). Reverse transcription (RT) was applied using ReverTra Ace® qPCR RT kit and real-time quantitative (qRCR) was conducted using SYBR Green PCR Master Mix (TOYOBO, Japan) on ABI7500 system (Applied Biosystems, USA). Eukaryotic 18S rRNA (18S) was used as endogenous control for human tissues and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for the cells and xenograft tumors. H19 primers were purchased from RiboBio (Guangzhou, China). Other primers were as follows: 5′-ACGTGACGTGTCAGGACCT-3′(sense) and 5′-GATCGGGCCTTGTTTGCTCTT-3′(antisense) for E2F-1, 5′-ACACTAAGGGCCGAAGATAACG-3′ (sense) and 5′-ACAGCATCTCCAATATGGCTGA-3′ (antisense) for PCNA, 5′-GGCCTCCATTGATGCTACCTA-3′ (sense) and 5′-ACTTTGGGACGAACTAGAGAACA-3′ (antisense) for MCM3, 5′-GCCTGGCAAACTATACTGTG-3′ (sense) and 5′-CTCCATGAGGGACACACACA-3′ (antisense) for CyclinA1, 5′-CAGTGTGAAGATGCCCTGGCTT-3′ (sense) and 5′-CAAGGATGGCCCGCATACTGTTA-3′ (antisense) for CyclinA2, 5′-GTAACCCGTTGAACCCCATT-3′(sense) and 5′-CCATCCAATCGGTAGTAGCG-3′ (antisense) for 18S, 5′-GTATTGGGCGCCTGGTCACC-3′(sense) and 5′-CGCTCCTGGAAGATGGTGATGG-3′ (antisense) for GAPDH. Each sample was analyzed in triplicate. All the data were analyzed by the comparative threshold cycle (CT) (2−ΔΔCT) method.
siRNA transfection and lentivirus infection.
T3M4 and PANC-1 cells with high expression of H19 were transfected with 3 interfering RNAs (siRNAs) targeting H19 or E2F-1, or negative control siRNA (NC) purchased from RiboBio (Guangzhou, China). The sequences of siRNAs were as follows: 5′ GGCCUUCCUGAACACCUUAdTdT 3′ (siH19-1), 5′ GACGUGACAAGCAGGACAUdTdT 3′ (siH19-2), 5′ CCUCUAGCUUGGAAAUGAA dTdT 3′ (siH19-3), 5′ TGGACCACCTGATGAATAT 3′ (siE2F1-1), 5′ GGCCCGATCGATGTTTTCC 3′ (siE2F1-2), 5′ CCTGATGAATATCTGTACT 3′ (siE2F1-3).
Full length H19 sequence was subcloned into lentivirus vector Lenti-GFP (GeneChem, Shanghai, China) to get Lenti-H19-GFP. COLO357 and CAPAN-1 cells with low expression of H19 were infected with Lenti-H19-GFP or Lenti-GFP according to the manufacturer's protocol.
Cell viability assay
Cell viability was determined using Cell Counting kit-8 (CCK-8) (Dojindo Laboratories, Tokyo, Japan). Pancreatic cells were seeded into 96-well plates at 3,500–5,000 cells/well depending on different cell types. Then at 24 h, 48 h and 72 h, WST-8 reagent (10 µl per well) was added into each well and the plates were incubated at 37˚C for 2 h. Absorbance was measured at 450 nm with a microplate reader (Bio-Rad Laboratories, USA). Three independent experiments were performed in quadruplicate.
Apoptosis detection assay
The transfected cells were seeded into 6-well plates at a density of approximately 1 × 105 in 60 mm dishes and grown to 80% confluence in the logarithmic growth phase. Then the cells were collected and stained by using Apoptosis Detection Kit according to the manufacturer's protocol (BD PharMingen, USA). Apoptosis was measured by flow cytometry (FACSCalibur, BD, USA), and the cells undergoing apoptosis were quantified by calculating the percentage of cells positive for Annexin V.
EdU incorporation assay
Cell proliferation was evaluated using EdU Apollo®567 In Vitro Imaging Kit (RiboBio, Guangzhou, China). Cells were seeded into 96-well plates at 3,000 cells/well. After 24 h, 5-ethynyl-20-deoxyuridine (EdU) was added to the media at 50 μM and the plates were incubated at 37˚C for 2 h. After labeling, the cells were fixed with 4% formaldehyde for 15 min and permeabilized with 0.5% Triton X-100 for 20 min at room temperature. After washing with PBS 3 times, the cells were incubated with 1 × Apollo® reaction cocktail and counterstained with DAPI for 30 min, then observed under fluorescence microscopy. Ten randomly chosen fields were observed and the proportion of EdU incorporated cells was calculated. Three independent experiments were performed in triplicate.
Colony formation assay
The cells were seeded into 6-well plates at 200 cells/well. Approximately 14 d later, the number of macroscopic colonies was counted, and images were obtained of the representative colonies.
Cell cycle analysis
Cells were seeded into 6-well plates and grown to 80% confluence in the logarithmic growth phase. The serum was deprived for 24 h to synchronize cells. The cells were washed 3 times with PBS and cultured in medium supplemented with 10% fetal bovine for another 6 h. The cells were harvested, washed with PBS and fixed with 70% pre-cooled ethanol at 4°C for at least 4 h. Then cells were incubated in PBS solution containing 50 μg/ml propidium iodide (PI) and 100 μg/ml RNase for 30 min at room temperature. Finally, cell cycle was analyzed by flow cytometry (FACSCalibur, BD, USA).
Western blot analysis
Cells were lysed in RIPA buffer and total protein concentration was measured using BCA Protein Assay Kit (Thermo Scientific, USA). Then proteins (25 μg/lane) were separated on 10% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked in 5% skim milk in TBST buffer at room temperature for 2 h and incubated with primary antibody against E2F-1 (1:2000, CST, USA), caspase 3 (1:2000, Abcam, USA), cleaved caspase 3 (1:1000, CST, USA) or β-actin (1:40000, MBL, Japan) at 4°C overnight. Then the membranes were incubated with HRP-conjugated secondary antibodies at 4°C for 2 h. The peroxidase activity was detected by using Immobilon Western Chemiluminescent HRP Substrate (Millipore, USA) and exposed to X-ray films.
Xenograft model
Two.5 × 106 T3M4 cells, 1 × 107 PANC-1 cells, 2 × 106 COLO357 cells or 2 × 106 CAPAN-1 cells were harvested and injected into the axillary fossa of male Balb/c (nu/nu) nude mice. Both the experimental group and control group had 6 mice. Tumor volumes were measured (length × width2 × 0.5) every 2 d. When the tumors grew to a certain size, the mice were executed and tumor weights were measured.
Immunohistochemical staining
The flash-frozen tissues were embedded and cut into 5-μm sections. The frozen sections were incubated with E2F-1 antibody (1:100, KeyGEN BioTECH, China) for 2 h at room temperature, followed by incubation with HRP conjugated secondary antibody. Protein staining was evaluated under a microscope at 200 × magnification by 2 independent experienced pathologists as 0 = no staining, 1 = weak staining (light yellow), 2 = moderate staining (yellow brown), and 3 = strong staining (brown). Tumor cells in 5 fields were randomly selected and scored based on the percentage of positively stained cells as 0 = no staining cells, 1 = less than 10% of positive cells, 2 = 10–49% of positive cells, and 3 = over 50% of positive cells. The final IHC score ranging from 0 to 6 was calculated by multiplying the intensity score with the percentage of positive cells. 0–3 was regarded as low expression and 4–6 was regarded as high expression.
Statistical analysis
All data were expressed as the mean values ± standard deviation (s.d.) from at least 3 independent experiments. The difference between 2 groups was analyzed by 2-tailed Student's t-test or paired t-test. The data from the clinicopathological characteristics analysis and immunohistochemistry were calculated by the Fisher exact probability test. The correlation of E2F-1 protein level with H19 expression was examined by Spearman's rank correlation. All analyses were performed using SPSS v.13.0 statistical software (SPSS, Inc.).The difference was considered significant for P value < 0.05.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from The National Natural Science Foundation of China (No. 81172184, 81372605, 81572339) and Beijing Natural Science Foundation (No. 7142162). We thank Prof. Zebin Mao at the Department of Biochemistry and Molecular Biology in Health Science Center, Peking University for the assistance and technical support.
References
- 1.Lin QJ, Yang F, Jin C, Fu DL. Current status and progress of pancreatic cancer in China. World J Gastroenterol 2015. July 14; 21(26):7988-8003; PMID:26185370; http://dx.doi.org/0.3748/wjg.v21.i26.7988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65:5; PMID:25559415; http://dx.doi.org/ 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 3.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet 2011; 378:607; PMID:21620466; http://dx.doi.org/ 10.1016/S0140-6736(10)62307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin 2013; 63:318; PMID:23856911; http://dx.doi.org/ 10.3322/caac.21190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013; 193:651; PMID:23463798; http://dx.doi.org/ 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells Dev 2013; 22:2240; PMID:23528033; http://dx.doi.org/ 10.1089/scd.2013.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pachnis V, Belayew A, Tilghman SM. Locus unlinked to α-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci U S A 1984; 81:5523; PMID:6206499; http://dx.doi.org/ 10.1073/pnas.81.17.5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 2005; 280:29625; PMID:15985428; http://dx.doi.org/ 10.1074/jbc.M504033200 [DOI] [PubMed] [Google Scholar]
- 9.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 2014; 28:491; PMID:24532688; http://dx.doi.org/ 10.1101/gad.234419.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature 1993; 365:764; PMID:7692308; http://dx.doi.org/ 10.1038/365764a0 [DOI] [PubMed] [Google Scholar]
- 11.Chung WY, Yuan L, Feng L, Hensle T, Tycko B. Chromosome 11p15.5 regional imprinting: comparative analysis of KIP2 and H19 in human tissues and Wilms' tumors. Hum Mol Genet 1996; 5:1101; PMID:8842727; http://dx.doi.org/ 10.1093/hmg/5.8.1101 [DOI] [PubMed] [Google Scholar]
- 12.Lynch CA, Tycko B, Bestor TH, Walsh CP. Reactivation of a silenced H19 gene in human rhabdomyosarcoma by demethylation of DNA but not by histone hyperacetylation. Mol Cancer 2002; 1:2; PMID:12234381; http://dx.doi.org/ 10.1186/1476-4598-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H, et al.. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A 2008; 105:12417; PMID:18719115; http://dx.doi.org/ 10.1073/pnas.0801540105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ariel I, Sughayer M, Fellig Y, Pizov G, Ayesh S, Podeh D, Libdeh BA, Levy C, Birman T, Tykocinski ML, et al.. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol 2000; 53:320; PMID:11193051; http://dx.doi.org/ 10.1136/mp.53.6.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014; 5:2318; PMID:24810858; http://dx.doi.org/ 10.18632/oncotarget.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010; 31:350; PMID:19926638; http://dx.doi.org/ 10.1093/carcin/bgp181 [DOI] [PubMed] [Google Scholar]
- 17.Huang C, Cao L, Qiu L, Dai X, Ma L, Zhou Y, Li H, Gao M, Li W, Zhang Q, et al.. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett 2015; 10:291; PMID:26171017; PMID: 26171017; http://dx.doi.org/23711233 10.3892/ol.2015.3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu LL, Chang K, Lu LS, Zhao D, Han J, Zheng YR, Yan YH, Yi P, Guo JX, Zhou YG, et al.. Lentivirus-mediated RNA interference targeting the H19 gene inhibits cell proliferation and apoptosis in human choriocarcinoma cell line JAR. BMC Cell Biol 2013; 14:26; PMID:23711233; http://dx.doi.org/ 10.1186/1471-2121-14-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene 2014; 33:4857; PMID:24141783; http://dx.doi.org/ 10.1038/onc.2013.433 [DOI] [PubMed] [Google Scholar]
- 20.Zhu Z, Song L, He J, Sun Y, Liu X, Zou X. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol 2015; 8:10082; PMID:26617715 [PMC free article] [PubMed] [Google Scholar]
- 21.Douc-Rasy S, Barrois M, Fogel S, Ahomadegbe JC, Stehelin D, Coll J, Riou G. High incidence of loss of heterozygosity and abnormal imprinting of H19 and IGF2 genes in invasive cervical carcinomas. Uncoupling of H19 and IGF2 expression and biallelic hypomethylation of H19. Oncogene 1996; 12:423; PMID:8570220 [PubMed] [Google Scholar]
- 22.Lustig-Yariv O, Schulze E, Komitowski D, Erdmann V, Schneider T, de Groot N, Hochberg A. The expression of the imprinted genes H19 and IGF-2 in choriocarcinoma cell lines. Is H19 a tumor suppressor gene? Oncogene 1997; 15:169; PMID:9244352; http://dx.doi.org/ 10.1038/sj.onc.1201175 [DOI] [PubMed] [Google Scholar]
- 23.Kim HT, Choi BH, Niikawa N, Lee TS, Chang SI. Frequent loss of imprinting of the H19 and IGF-II genes in ovarian tumors. Am J Med Genet 1998; 80:391; PMID:9856569; http://dx.doi.org/ 10.1002/(SICI)1096-8628(19981204)80:4%3c391::AID-AJMG16%3e3.0.CO;2-H [DOI] [PubMed] [Google Scholar]
- 24.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J 2012; 279:3159; PMID:22776265; http://dx.doi.org/ 10.1111/j.1742-4658.2012.08694.x [DOI] [PubMed] [Google Scholar]
- 25.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007; 13:313; PMID:17237358; http://dx.doi.org/ 10.1261/rna.351707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun 2014; 448:315; PMID:24388988; http://dx.doi.org/ 10.1016/j.bbrc.2013.12.126 [DOI] [PubMed] [Google Scholar]
- 27.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7s suppression on its target HMGA2-mediated EMT. Tumour Biol 2014; 35:9163; PMID:24920070; http://dx.doi.org/ 10.1007/s13277-014-2185-5 [DOI] [PubMed] [Google Scholar]
- 28.Apte MV, Park S, Phillips PA, Santucci N, Goldstein D, Kumar RK, Ramm GA, Buchler M, Friess H, McCarroll JA, et al.. Desmoplastic reaction in pancreatic cancer: role of pancreatic stellate cells. Pancreas 2004; 29:179; PMID:15367883; http://dx.doi.org/ 10.1097/00006676-200410000-00002 [DOI] [PubMed] [Google Scholar]
- 29.Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Laser capture microdissection: molecular analysis of tissue. Science 1997; 278:1481-1483; PMID:9411767; http://dx.doi.org/ 10.1126/science.278.5342.1481 [DOI] [PubMed] [Google Scholar]
- 30.Cha S, Imielinski MB, Rejtar T, Richardson EA, Thakur D, Sgroi DC, Karger BL. In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection: annotation by protein set enrichment analysis and gene ontology. Mol Cell Proteomics 2010; 9:2529; PMID:20739354; http://dx.doi.org/ 10.1074/mcp.M110.000398 [DOI] [PMC free article] [PubMed] [Google Scholar]