ABSTRACT
The primary physiological function of blood platelets is to seal vascular lesions after injury and form hemostatic thrombi in order to prevent blood loss. This task relies on the formation of strong cellular-extracellular matrix interactions in the subendothelial lesions. The cytoskeleton of a platelet is key to all of its functions: its ability to spread, adhere and contract. Despite the medical significance of platelets, there is still no high-resolution structural information of their cytoskeleton. Here, we discuss and present 3-dimensional (3D) structural analysis of intact platelets by using cryo-electron tomography (cryo-ET) and atomic force microscopy (AFM). Cryo-ET provides in situ structural analysis and AFM gives stiffness maps of the platelets. In the future, combining high-resolution structural and mechanical techniques will bring new understanding of how structural changes modulate platelet stiffness during activation and adhesion.
Keywords: actin, atomic force microscopy, cryo-electron tomography, integrins, platelets
Introduction
Platelets are discoid cell fragments released into the blood circulation by bone marrow megakaryocytes. They are involved in processes such as hemostasis, thrombosis, wound healing, atherosclerosis and inflammation; however, their primary function is to prevent blood loss by sealing vascular injuries.1 Under normal conditions platelets patrol the vascular system in a non-adherent “resting” state but become rapidly activated at sites of vascular injury. Platelet activation is triggered by exposed extracellular matrix (ECM) molecules or by soluble platelet agonists, which induce intracellular signaling events via their respective receptors. Consequently, these signals initiate re-organization of the cytoskeleton, the release of prothrombotic granula content and the activation of platelet integrins, which mediate strong adhesion to the proteins of the ECM. Distinct morphological changes can be observed during platelet activation, starting from a discoid transforming to a spherical shape (rounding) and finally to a fried-egg shape which adheres to the surface (Fig. 1). A microtubule ring underlies the platelet membrane at its larger circumference and sustains the discoid shape.2 Upon platelet activation, the microtubule ring constricts and actin filaments are severed. This stage is followed by extensive actin polymerization forming multiple filopodia.3 Finally, the platelets spread, form lamellipodia and platelet adhesion progresses. Strong platelet adhesion requires a connection between the cytoskeletal filaments and the ECM. Thus, reorganization of the platelet cytoskeleton is simultaneously coordinated with the formation of adhesion plaques, which link the actin network to the ECM.4
Figure 1.
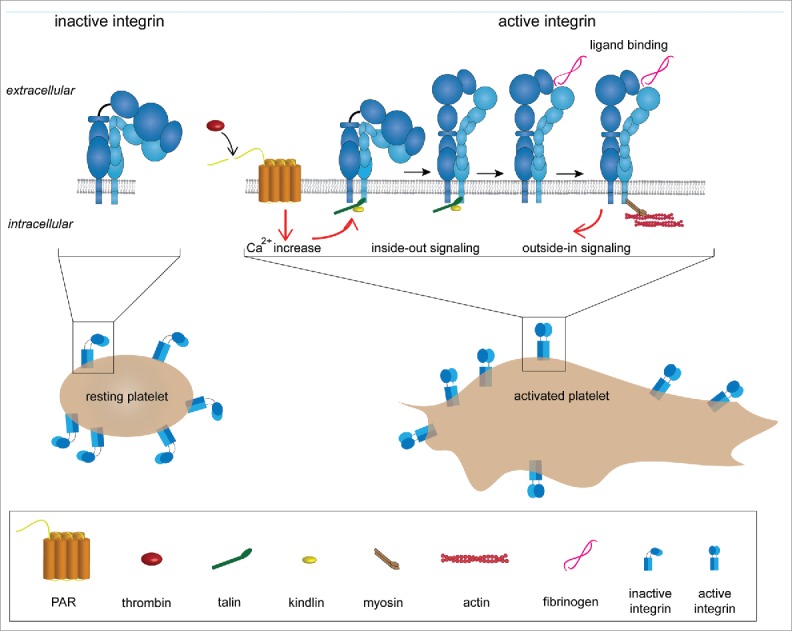
Adhesion system of platelets (platelet spreading and integrin activation). In resting platelets, αIIbβ3 integrins are in a bent and inactive conformation. Integrin activation can be triggered in 2 ways: (1) inside-out signaling or (2) outside-in signaling. In the first case integrin activation is initiated by platelet agonists such as thrombin. Thrombin activates the protease-activated GPCRs (G protein-coupled receptors), which leads to an increase in concentration of the cytosolic Ca2+. This triggers an intracellular signaling cascade that promotes talin binding to the cytoplasmic tail of the β2 chain. This event elicits integrin extension and activation. In the outside-in signaling, αIIbβ3 is activated by ligand binding (e.g., fibrinogen).
Integrins represent the major family of platelet adhesion receptors mediating firm adhesion to exposed matrix molecules of endothelial lesions.5 Integrins also facilitate platelet adhesion, aggregation and spreading via natural ligands, such as fibrinogen and fibronectin, and subsequent cytoskeleton reorganization. Since platelet integrins are constantly exposed to ligands, their activity has to be tightly controlled in order to circumvent deregulated thrombus formation. Integrins are kept in a resting state, i.e., low-affinity, on the surface of circulating platelets, and rapidly shift toward an active, high-affinity state upon activation (Fig. 1). This transition is regulated by intracellular events utilizing 2 cytoplasmic proteins, talin and kindlin, in the presence of platelet agonists.6 Both proteins directly bind the cytoplasmic tail of β-integrin subunits and thereby trigger integrin activation.
Cell attachment and spreading are intensively studied by fluorescence microscopy.7 However, due to the small size of resting platelets (~4 μm in diameter), studying their 3D cytoskeletal organization at high-resolution is still elusive. Here, we demonstrate the potential of structural analysis of intact platelets by cryo-electron tomography (cryo-ET). The current technology allows us to visualize the cytoskeletal architecture of spread platelets on ECM protein-coated (fibrinogen or collagen) surfaces, and analyze its most abundant integrin, the αIIbβ3. In addition, similar preparation is used to measure the stiffness of living platelets using the atomic force microscope (AFM) (Fig. 2). Combining the 2 approaches may eventually allow to directly correlate structure and function, e.g., how cytoskeleton architecture contributes to cellular stiffness.
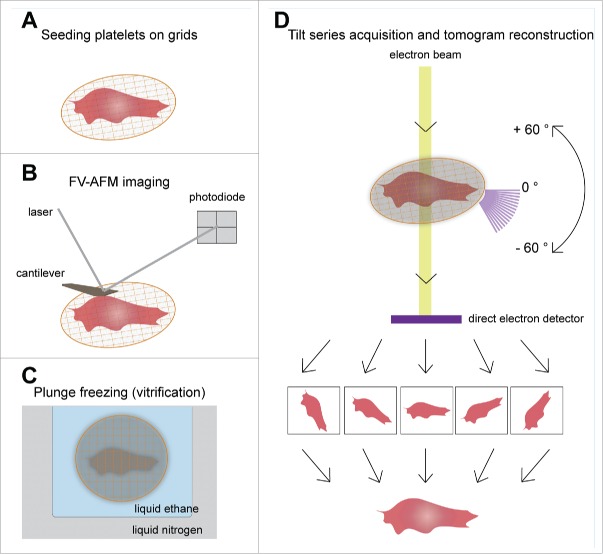
Figure 2.
Schematic overview of AFM-cryo ET correlative approach. (A) Purified platelets are seeded on ECM protein (fibrinogen or collagen type I) coated grids in the presence of 1 mM Ca2+ and Mg2+ to allow spreading. (B) Mechanical properties of the platelets are then measured by FV-AFM imaging. (C) After AFM, the sample could be vitrified by plunge freezing in liquid N2 cooled liquid ethane, and inserted into the cryo-electron microscope. (D) A series of images of the area of interest are acquired by tilting the stage from −60° to +60° with a 2° increment. The tilt series images are then back projected to reconstruct the 3D volume of the original platelet.
Visualizing cellular architecture by cryo-electron tomography
Technical advances in microscopic imaging techniques such as cryo-electron microscopy (cryo-EM) have led to unprecedented capabilities for examining cellular structures. In particular, cryo-ET has had a pivotal role in cellular biology since it can provide a 3D structural map of a specific cellular state in an unperturbed, vitrified sample, i.e., in a close-to-physiological state,8-10 whereas in conventional EM cells are fixed, dehydrated, stained with heavy metal and physically sliced.
Cryo-ET enables direct observation of macromolecular densities in situ due to the phase contrast between the biological material and the surrounding vitrified ice, bypassing the use of fixatives as well as commonly used contrasting agents, such as heavy metal salts.11 In practice, vitrified samples of suitable thickness (<1 μm) are rotated inside the transmission electron microscope (TEM) around a defined tilt axis and in discrete increments, covering a maximal range of 140° (between −70° and +70°).12 A series of 2D projections, i.e., a ‘tilt-series’, is collected under ‘low electron dose’ conditions (typically <60 e-/Å2) to prevent radiation damage to the sample.13,14 The tilt-series is subsequently aligned to a common frame in order to reconstruct the 3D volume of the specimen, namely a tomogram 12 (Fig. 2).
Technical advances over the last few years, such as improved sample preparation and electron detection methods, have been instrumental in obtaining data with unprecedented structural details.15,16 Direct electron detectors allow to retrieve high-frequency information at a high frame rate. As a consequence, multiple projections of the same area, a few milliseconds long, can be computationally compensated for sample motion thereby increasing the quality of the data. This presents an exciting opportunity to explore the molecular architecture of platelets and the cell adhesion machinery at nanometer to subnanometer resolution.
Zooming into intact platelets with cryo-electron tomography
Cryo-ET has been used for high resolution imaging of the cytoskeleton in eukaryotic cells. For example, structural characterization of focal adhesions in fibroblasts17,18 revealed that actin filaments are orientated parallel to the long axis of the focal adhesion, whereas the parallel orientation is not preserved at the periphery of the adhesion sites. Importantly, these studies indicated that individual actin filaments interact with integrins via protein assemblies.17,19
Adhering platelets spread to form thin cellular structures, typically <1µm in thickness, ideal for imaging with cryo-ET. Here, platelets were allowed to spread on fibrinogen-coated EM grids and were subsequently vitrified at liquid nitrogen temperature and imaged by cryo-ET (Fig. 3A and B).20
Figure 3.
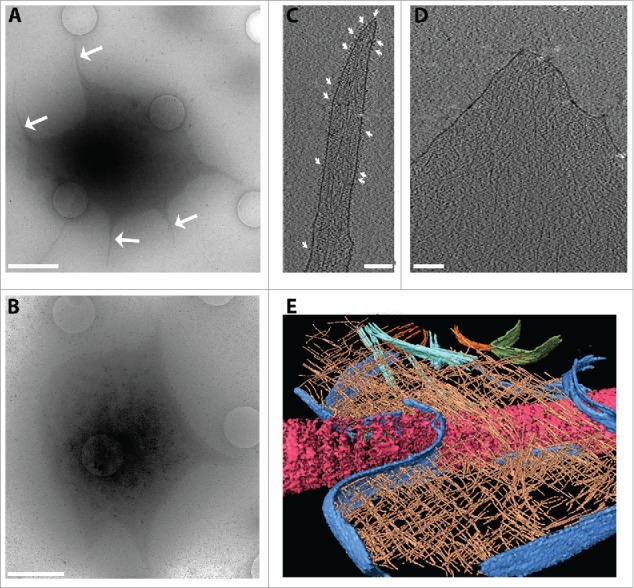
Analysis of platelet ultrastructure with cryo-electron tomography. Platelets were seeded on fibrinogen-coated (50 µg/ml) silicon grids in the presence of 1 mM Ca2+ and Mg2+ to allow spreading. (A, B) Overview images of platelets (4800x) at 30 µm defocus. In (A) the white arrows point to the filopodia emanating from the platelet during spreading. (B) An adhering platelet with a lamellopodium. Scale bars: 2 µm. (C) An x-y slice through a tomogram of a platelet filopodium. White arrows indicate densities assigned to integrins embedded in a continuous plasma membrane. Actin filaments are seen enclosed by the plasma membrane. (D) An x-y slice through a tomogram of a platelet lamellipodium. (E) A rendered view of a platelet spread on collagen fiber. Collagen (pink), plasma membrane (blue), actin (brown), microtubules (cyan) and mitochondria outer membrane (green) were segmented and rendered using Amira software (FEI). The cell thickness in these regions is typically 70–150 nm. Tomograms were acquired on an FEI Titan Krios, 300 kV, equipped with a Gatan Quantum Energy Filter and a K2 Summit direct electron detection camera, at 0.34 nm/pixel at the specimen level. The tilt series was acquired from −60o to +60o in 2° increments, at 8 µm defocus. The cumulative electron dose was ~40 e- Å-2.
Upon platelet activation, filopodia-like structures form prior to cell adhesion and spreading21 (Fig. 3A, arrows). We investigated one such region with cryo-ET. Platelet filopodia showed an architecture similar to that observed in filopodia formed by fibroblasts, which are usually composed of long filamentous actin bundles that push the distal membrane forward (Fig. 3C).22 The platelet filopodia consist of actin filaments that are shorter than an entire filopod and are in parallel or oblique to the filopod's axis. Furthermore, a large number of densities are detected at the plasma membrane (Fig. 3C, arrows). Those densities are presumed to be membrane bound receptors (see below).
The reconstructed volumes of lamellae of platelets spread on a fibrinogen surface showed dense cytoskeletal networks (Fig. 3D). Images of a platelet attached to a collagen bundle also showed densely packed actin filaments arranged in a meshwork resembling the typical actin meshwork of eukaryotic lamellipodia (Fig. 3E).23
Toward in situ characterization and the structure of integrins
Besides cytoskeleton remodeling, integrin activation plays an important role in platelet adhesion. The αIIbβ3 integrin heterodimer constitutes the major population of surface proteins in platelets (~80% of ~100,000 receptors/cell).24 Therefore, we hypothesized that the globular densities on the extracellular side of platelets are αIIbβ3 integrins (Fig. 3C). In order to test this hypothesis, we labeled the platelets with a monoclonal antibody targeted to αIIbβ3 followed by a secondary antibody-gold cluster conjugate (Fig. 4A). The 6 nm gold colloids were detected exclusively in proximity to the platelet membrane, indicating specific binding. The gold nanoparticles were situated 15–18 nm away from the globular densities present in the platelet membrane, in agreement with the expected maximal distance of ~19 nm between a gold-labeled secondary antibody and its epitope.25 As a control, the secondary gold-labeled antibody was incubated with platelets alone but no gold nanoparticles were detected in the vicinity of the plasma membrane (data not shown).
Figure 4.
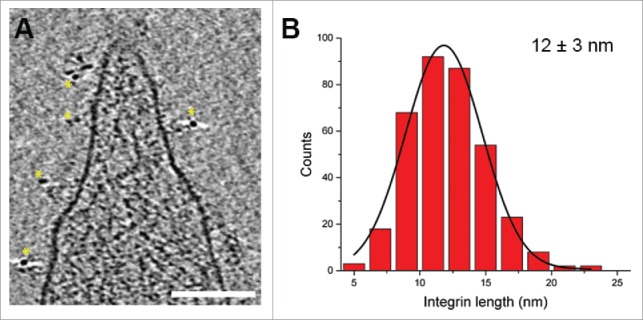
Analysis of platelet membrane receptors. (A) An x-y slice through a tomogram of a platelet, where integrin αIIbβ3 has been immunogold labeled.32 Yellow stars highlight the 6 nm gold nanoparticles, which were observed only in the proximity of the platelet membrane. Scale bar: 100 nm. (B) Histogram of integrin length distribution. The integrin lengths were measured in 5 tomograms (acquired as in Figs. 1C, D) for a total of 366 integrins.32 The black line is a Gaussian fit to the histogram.
The precise structural details of integrin in its different conformational states are debatable. It has been proposed that in the low-affinity non-active conformation, the 2 subunits are bent and closely interact with each other. Upon activation, integrins extend into an elongated conformation in which the 2 subunits separate.26 Growing evidence suggests that conformational changes in integrin require extracellular binding27 in addition to cytoplasmic binders. For example, talin and kindlin binding to the integrin cytoplasmic domain was associated with an extended integrin conformation.28-31
The observed integrin receptors on the platelet surface allowed us to address the question of integrin conformation. We measured the distance between the globular head domain of individual integrins and the cell membrane (Fig. 4B). The distribution of integrin extension suggests that integrins may exhibit a conformational heterogeneity ranging from a bent inactive conformation to a fully extended active conformation, i.e., a spectrum of intermediate state conformations. This was also recently shown for integrins embedded in the membranes of platelet-derived microparticles.32 Based on these results and our ongoing work, it is very likely that by applying cryo-ET the structural conformations of integrins may be resolved in situ.
Cellular structural mechanics
Nanotechnological tools, such as the atomic force microscope (AFM), are routinely employed to image reconstituted and cellular systems.33 Besides surface characterization, soft biological matter can be mechanically characterized, e.g., by measuring stiffness, Young's modulus, and viscoelastic properties of a cell. Quantitative measurements have shown that the material properties of tissues change with age and disease.34 Interestingly, normal, benign and malignant tissues were shown to exhibit different mechanical properties enabling a future possibility of applications in clinical diagnostics.35
A change in the material properties is bound to arise from a change in the macromolecular structures of a tissue or a cell. However, to understand the direct influence of macromolecular re-modeling of a cell in different states and its relation to the mechanical properties, a direct correlation between the mechanical properties of a cell and its structure is important. Key to achieving this is the assignment of the causality of changes in mechanical properties to specific intracellular structural elements. A complementary approach of cryo-ET and AFM is a logical step toward understanding how mechanical properties are influenced by dynamic structural changes in a cell.
Mechanical characterization of platelets
Platelets from healthy donors were seeded on a fibrinogen-coated glass dish and allowed to adhere to the surface. A mechanical map of an entire platelet was then recorded by force-volume (FV) imaging. Instead of contouring the platelet as in the conventional contact mode, force-distance (F-D) curves were acquired by scrutinizing the platelet in the x-y plane. The advantage of this technique is that a soft material, such as a cell, is prevented from being damaged, which is sometimes difficult to avoid in the contact mode. Moreover, a single force-volume scan is capable of providing platelet topography and simultaneously map the mechanical (e.g., elasticity, deformation, Young's modulus, energy dissipation) and adhesive properties.
A mechanical map of a spread human platelet was created by collecting F-D curves (64 × 64 pixels); the curves were fitted to the Hertz model and Young's modulus determined (Fig. 5). The mechanical map clearly showed a softer central region (32 kPa) and an outer stiffer region (224 kPa) (Fig. 5C-E). A similar trend was observed previously, where the elastic moduli were measured in a range of 1–50 kPa.36 The resolution of the measurement also allowed us to observe mechanical gradients in the central and the outer regions. The soft nature of the central region is likely because of the presence of vesicles (also seen in tomograms; data not shown). The higher values of Young's moduli surrounding the central region can be attributed to the actin network and in some cases to the microtubule ring. An exact assignment of the stiffness to the cytoskeletal structure requires further precise correlation; the mechanical properties of platelets at different stages of activation are currently under investigation. Remodeling of cytoskeletal organization in diseased platelets (e.g., Glanzmann thrombasthenia, Bernard Soulier syndrome) and changes in their mechanical properties need to be studied in detail in the future.
Figure 5.
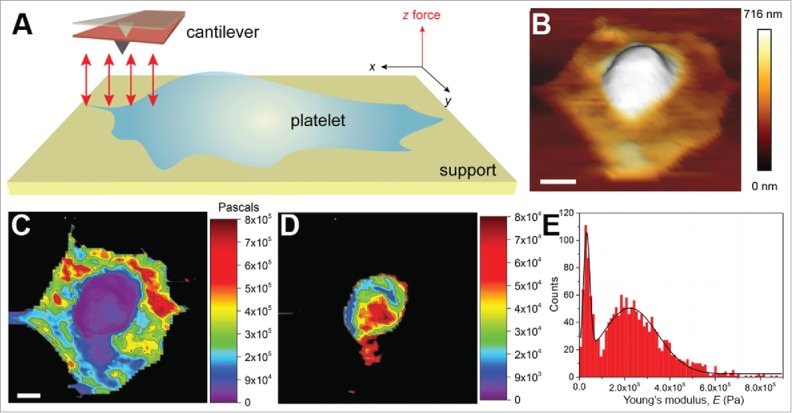
Mechanical investigation of platelets. (A) Schematic illustrating the principle of mechanical characterization of platelets. A platelet is allowed to spread on a fibrinogen-coated glass surface in a physiological buffer (e.g., Tyrode's buffer containing 1 mM each of Ca2+ and Mg2+ ions, ± thrombin). Unlike the conventional AFM contact mode imaging, here F-D curves are recorded (red arrows) in the x-y plane. The F-D curves are used to extract the desired material properties. (B) A 3D topograph of a platelet spread on a fibrinogen-coated surface. The platelet shows a typical fried-egg shape, a high central region and flat periphery, indicative of an adhering platelet. (C) F-D curves were acquired by indenting the platelet with a force of 500 pN at an approach-retract velocity of 5 µm/s. The extension curves were then fit to the Hertz model to obtain the Young's moduli, contour-mapped on the platelet surface. The central region was observed to be softer than the peripheral region. The stiff glass background is shown in black. (D) A resolution of ~137 nm (64 × 64 pixel map of 8.74 × 8.74 µm2) was reasonable to obtain a contour map specifically of the central region. (E) A histogram of the modulus values, 32 kPa for the central region and ~224 kPa for the peripheral region. Scale bars (B, C): 1 µm. C and D are the same scale.
Future perspectives
Cryo-ET provides high-resolution snapshots of (intra)cellular structures arrested at a specific time. In situ structural identification of macromolecular complexes is therefore possible. Following the conformational change of integrin receptors, adhesion complexes and the cytoskeleton re-organization in platelets at sub-nanometer resolution will likely increase our understanding of platelet spreading and adhesion. Pertinent questions are: What are the structural changes in the cytoskeletal network and the integrins during spreading and adhesion? How do the mechanical properties of a platelet correlate to the structural changes?
The AFM cantilever can be used as a nano-indenter to determine the mechanical parameters of a native cell either dynamically or at a specific stage (e.g., upon chemical fixation).37 To understand the interplay of mechanical forces and adhesion in platelets, the mechanical measurements need to be quantitatively correlated with the structural data. The two techniques can be used in tandem—AFM for nano-mechanics and cryo-ET to look at exactly those regions for structural insight. To achieve an unambiguous correlation between the mechanical properties and associated structural elements of platelets, a correlative assay needs to be developed so that the same platelet sample used for FV-AFM imaging can be inspected by cryo-ET (Fig. 2). For example, a sample on an EM grid could be mechanically characterized with AFM, and then vitrified to perform cryo-ET analysis. Developing this approach will contribute to a comprehensive understanding of platelet adhesion mechanism, e.g., following from initial to final stages of the adhesion process. Characterizing how mechanical and adhesive properties go hand-in-hand with structural alterations will impart important insight to platelet and cell research from a new perspective.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Wolfgang Siess and Janina Jamasbi for the help in Fig. 3E, and Rossitza Irobalieva for comments. We thank the Center for Microscopy and Image Analysis at the University of Zurich.
Funding
This study was supported by a grant from an ERC Starting Grant (243047 INCEL), Novartis foundation (14B093) and Mäxi Foundation to O.M.
References
- [1].Savage B, Almus-Jacobs F, Ruggeri ZM. Specific synergy of multiple substrate-receptor interactions in platelet thrombus formation under flow. Cell 1998; 94:657-66; PMID:9741630; http://dx.doi.org/ 10.1016/S0092-8674(00)81607-4 [DOI] [PubMed] [Google Scholar]
- [2].Winokur R, Hartwig JH. Mechanism of shape change in chilled human platelets. Blood 1995; 85:1796-804; PMID:7703486. [PubMed] [Google Scholar]
- [3].Hartwig JH. The platelet: form and function. Semin Hematol 2006; 43:S94-100; PMID:16427392; http://dx.doi.org/ 10.1053/j.seminhematol.2005.11.004 [DOI] [PubMed] [Google Scholar]
- [4].Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol 2011; 3:pii: a005033; PMID:21441590; http://dx.doi.org/ 10.1101/cshperspect.a005033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ablooglu AJ, Kang J, Petrich BG, Ginsberg MH, Shattil SJ. Antithrombotic effects of targeting alpha IIb beta 3 signaling in platelets. Blood 2009; 113:3585-92; PMID:19005179; http://dx.doi.org/ 10.1182/blood-2008-09-180687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science 2009; 324:895-9; PMID:19443776; http://dx.doi.org/ 10.1126/science.1163865 [DOI] [PubMed] [Google Scholar]
- [7].Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science 2009; 326:1208-12; PMID:19965462; http://dx.doi.org/ 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Irobalieva RN, Martins B, Medalia O. Cellular structural biology as revealed by cryo-electron tomography. J Cell Sci 2016; PMID:26787742. [DOI] [PubMed] [Google Scholar]
- [9].Harapin J, Eibauer M, Medalia O. Structural analysis of supramolecular assemblies by cryo-electron tomography. Structure 2013; 21:1522-30; PMID:24010711; http://dx.doi.org/ 10.1016/j.str.2013.08.003 [DOI] [PubMed] [Google Scholar]
- [10].Fridman K, Mader A, Zwerger M, Elia N, Medalia O. Advances in tomography: probing the molecular architecture of cells. Nat Rev Mol Cell Biol 2012; 13(11):736-42; PMID:23047735. [DOI] [PubMed] [Google Scholar]
- [11].Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 2002; 298:1209-13; PMID:12424373; http://dx.doi.org/ 10.1126/science.1076184 [DOI] [PubMed] [Google Scholar]
- [12].Frank J. Introduction:Principles of Electron Tomography. : Frank J, ed. ELECTRON TOMOGRAPHY. New York: Plenum Press, 1992:1-13. [Google Scholar]
- [13].Dierksen K, Typke D, Hegerl R, Koster AJ, Baumeister W. Towards Automatic Electron Tomography. Ultramicroscopy 1992; 40:71-87; PMID:NOT_FOUND; http://dx.doi.org/ 10.1016/0304-3991(92)90235-C [DOI] [Google Scholar]
- [14].Dierksen K, Typke D, Hegerl R, Baumeister W. Towards Automatic Electron Tomography .2. Implementation of Autofocus and Low-Dose Procedures. Ultramicroscopy 1993; 49:109-20; http://dx.doi.org/ 10.1016/0304-3991(93)90217-L [DOI] [Google Scholar]
- [15].Dubrovsky A, Sorrentino S, Harapin J, Sapra KT, Medalia O. Developments in cryo-electron tomography for in situ structural analysis. Arch Biochem Biophys 2015; 581:78-85; PMID:25921875. [DOI] [PubMed] [Google Scholar]
- [16].Asano S, Engel BD, Baumeister W. In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology. J Mol Biol 2016; 428:332-43; PMID:26456135; http://dx.doi.org/ 10.1016/j.jmb.2015.09.030 [DOI] [PubMed] [Google Scholar]
- [17].Patla I, Volberg T, Elad N, Hirschfeld-Warneken V, Grashoff C, Fassler R, Spatz JP, Geiger B, Medalia O. Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat Cell Biol 2010; 12:909-15; PMID:20694000; http://dx.doi.org/ 10.1038/ncb2095 [DOI] [PubMed] [Google Scholar]
- [18].Elad N, Volberg T, Patla I, Hirschfeld-Warneken V, Grashoff C, Spatz JP, Fässler R, Geiger B, Medalia O. The role of integrin-linked kinase in the molecular architecture of focal adhesions. J Cell Sci 2013; 126:4099-107; PMID:23843624; http://dx.doi.org/ 10.1242/jcs.120295 [DOI] [PubMed] [Google Scholar]
- [19].Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature 2010; 468:580-4; PMID:21107430; http://dx.doi.org/ 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 1988; 21:129-228; PMID:3043536; http://dx.doi.org/ 10.1017/S0033583500004297 [DOI] [PubMed] [Google Scholar]
- [21].White J. Arrangements of actin filaments in the cytoskeleton of human platelets. Am J Pathol 1984; 117:207-17; PMID:6149688. [PMC free article] [PubMed] [Google Scholar]
- [22].Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol 2006; 18:18-25; PMID:16337369; http://dx.doi.org/ 10.1016/j.ceb.2005.11.002 [DOI] [PubMed] [Google Scholar]
- [23].Small JV. Pushing with actin: from cells to pathogens. Biochemical Society transactions 2015; 43:84-91; PMID:25619250; http://dx.doi.org/ 10.1042/BST20140184 [DOI] [PubMed] [Google Scholar]
- [24].Wagner CL, Mascelli MA, Neblock DS, Weisman HF, Coller BS, Jordan RE. Analysis of GPIIb/IIIa receptor number by quantification of 7E3 binding to human platelets. Blood 1996; 88:907-14; PMID:8704248. [PubMed] [Google Scholar]
- [25].Amiry-Moghaddam M, Ottersen OP. Immunogold cytochemistry in neuroscience. Nat Neurosci 2013; 16:798-804; PMID:23799472; http://dx.doi.org/ 10.1038/nn.3418 [DOI] [PubMed] [Google Scholar]
- [26].Kahner BN, Kato H, Banno A, Ginsberg MH, Shattil SJ, Ye F. Kindlins, integrin activation and the regulation of talin recruitment to alphaIIbbeta3. PLoS One 2012; 7:e34056; PMID:22457811; http://dx.doi.org/ 10.1371/journal.pone.0034056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dai A, Ye F, Taylor DW, Hu G, Ginsberg MH, Taylor KA. The Structure of a Full-length Membrane-embedded Integrin Bound to a Physiological Ligand. J Biol Chem 2015; 290:27168-75; PMID:26391523; http://dx.doi.org/ 10.1074/jbc.M115.682377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim C, Ye F, Ginsberg MH. Regulation of Integrin Activation. Annu Rev Cell Dev Biol 2011; PMID:21663444 [DOI] [PubMed] [Google Scholar]
- [29].Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH. Recreation of the terminal events in physiological integrin activation. J Cell Biol 2010; 188:157-73; PMID:20048261; http://dx.doi.org/ 10.1083/jcb.200908045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med 2008; 14:325-30; PMID:18278053; http://dx.doi.org/ 10.1038/nm1722 [DOI] [PubMed] [Google Scholar]
- [31].Theodosiou M, Widmaier M, Bottcher RT, Rognoni E, Veelders M, Bharadwaj M, Lambacher A, Austen K, Müller DJ, Zent R, et al.. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife 2016; 5:pii: e10130; PMID:26821125; http://dx.doi.org/ 10.7554/eLife.10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tamir A, Sorrentino S, Motahedeh S, Shai E, Dubrovsky A, Dahan I, Eibauer M, Studt JD, Sapra KT, Varon D, et al.. The macromolecular architecture of platelet-derived microparticles. J Struct Biol 2016; 193(3):181-7; PMID:26767592. [DOI] [PubMed] [Google Scholar]
- [33].Muller DJ, Helenius J, Alsteens D, Dufrene YF. Force probing surfaces of living cells to molecular resolution. Nat Chem Biol 2009; 5:383-90; PMID:19448607; http://dx.doi.org/ 10.1038/nchembio.181 [DOI] [PubMed] [Google Scholar]
- [34].Stolz M, Gottardi R, Raiteri R, Miot S, Martin I, Imer R, Staufer U, Raducanu A, Düggelin M, Baschong W, et al.. Early detection of aging cartilage and osteoarthritis in mice and patient samples using atomic force microscopy. Nat Nanotechnol 2009; 4:186-92; PMID:19265849; http://dx.doi.org/ 10.1038/nnano.2008.410 [DOI] [PubMed] [Google Scholar]
- [35].Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, Hyotyla JT, Aebi U, Bentires-Alj M, Lim RY, et al.. The nanomechanical signature of breast cancer. Nat Nanotechnol 2012; 7:757-65; PMID:23085644; http://dx.doi.org/ 10.1038/nnano.2012.167 [DOI] [PubMed] [Google Scholar]
- [36].Radmacher M, Fritz M, Kacher CM, Cleveland JP, Hansma PK. Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophys J 1996; 70:556-67; PMID:8770233; http://dx.doi.org/ 10.1016/S0006-3495(96)79602-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Franz CM, Muller DJ. Analyzing focal adhesion structure by atomic force microscopy. J Cell Sci 2005; 118:5315-23; PMID:16263758; http://dx.doi.org/ 10.1242/jcs.02653 [DOI] [PubMed] [Google Scholar]