ABSTRACT
The complexity of cell-matrix adhesion convolves its roles in the development and functioning of multicellular organisms and their evolutionary tinkering. Cell-matrix adhesion is mediated by sites along the plasma membrane that anchor the actin cytoskeleton to the matrix via a large number of proteins, collectively called the integrin adhesome. Fundamental challenges for understanding how cell-matrix adhesion sites assemble and function arise from their multi-functionality, rapid dynamics, large number of components and molecular diversity. Systems biology faces these challenges in its strive to understand how the integrin adhesome gives rise to functional adhesion sites. Synthetic biology enables engineering intracellular modules and circuits with properties of interest. In this review I discuss some of the fundamental questions in systems biology of cell-matrix adhesion and how synthetic biology can help addressing them.
Keywords: building blocks, focal adhesions, reverse engineering, rewiring, self-assembly, self-organization
Background
The seed of the complexity of cell adhesion is that multi-cellular organisms develop and maintained by self-organization of their cells. Therefore cell adhesion must be a dynamic process that can respond to local cues. Cell-matrix adhesion has to sense the physical, chemical and topographical properties of the matrix as well as internal mechanical and biochemical cues. In response to these input cues, the adhesion process has to generate diverse outputs, including modulating locally the adhesion strength, signaling to the cell and modulating the matrix.1 The mechanisms by which cell-matrix adhesion achieves these functions evolved by tinkering, rather than by direct design, along the evolution of multicellular metazoan organisms with increasing complexity.2,3 These developmental, evolutionary and functional dimensions are reflected in the complexity of cell-matrix adhesion and cause the challenges in studying this process.
The founding studies of cell-matrix adhesion discovered, about 40 y ago, that this process is mediated by specific sites along the plasma membrane.4-6 Shortly later, these sites were found to be associated with actin filaments.7,8 (Fig. 1). The open question then became what are the components that mediate the association of actin filaments with the plasma membrane in cell-matrix adhesion sites.9 Integrins were identified as the transmembrane receptors that bind components of the matrix in adhesion sites.10,11 Gradually, more than hundred cytosolic proteins, collectively called the integrin adhesome, were found to be localized in cell-matrix adhesion sites.12-15 Furthermore, a complex network of regulated interactions, in which each protein can bind and affect multiple others, had been revealed.12-15 Naturally, the central question shifted from what are the components9 to how their collective actions give rise to functional cell-matrix adhesion sites.14
Figure 1.
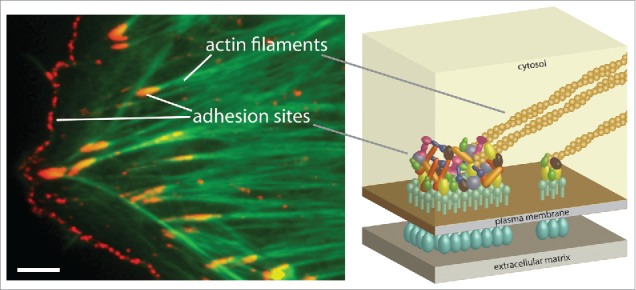
Cell-matrix adhesion sites. The image shows a porcine aortic endothelial cell labeled for phosphotyrosine (red) and actin filaments (green). Two types of cell-matrix adhesion sites are indicated, focal adhesions and focal complexes. Focal complexes are small, dot-like structures located at the cell edge while focal adhesions are bigger and ovular structures located relatively more toward the cell center. Scale bar, 5 µm. The scheme illustrates a simplified cross-section view of cell-matrix adhesion sites.
Systems biology studies how cell properties emerge from the interactions between its components. The emerging properties of the integrin adhesome are the formation of cell-matrix adhesion sites and their functions. Major challenges in studying these processes are the large number of integrin adhesome components, the molecular diversity of adhesion sites and the alternative manners by which integrin adhesome proteins can bind and regulate each other. Synthetic biology would enable, by itself, valuable bottom-up explorations of cell-matrix adhesion by reconstituting adhesion sites in minimal synthetic cells and building gradually the complexity.16 In this review I discuss a complementary path, namely – how synthetic biology can be applied to engineer intracellular modules and circuits that will facilitate studying adhesion sites in cells.
Exploring the assembly principles of cell-matrix adhesion sites
Cell-matrix adhesion sites assemble by local self-organizations of the integrin adhesome, triggered and shaped by local cues. This assembly is a multistep process along which the adhesion sites mature, change their properties, molecular content and functions.17-19 Initially, small (<0 .25 μm) and transient (< 1 min) nascent adhesions are formed at the cell edge. Nascent adhesions can evolve to bigger (˜0.5 μm) and more stable (˜1–5 min) focal complexes in response to forces applied on them from actin polymerization and its centripetal flow.18,20,21 Focal complexes can evolve to focal adhesions connected to contractile stress-fibers upon application of actomyosin contractility.1,18 Finally, fibronectin-bound fibrillar adhesions can segregate from focal adhesions and translocate toward the cell center by actomyosin contractility.22,23 Each of these types of adhesion sites has distinct functions and properties. Mainly, focal complexes are exploratory adhesions sites, focal adhesions are the main mechanical anchors and fibrillar adhesions are tension-independent sites involved in fibronectin fibrillogenesis.19
The maturation and diversification of cell-matrix adhesion sites are accompanied by changes in their internal molecular content and organization. However, the molecular events underlying these processes are still largely unknown. The primary reason for this is that the integrin adhosome is designed to form distinct structures with distinct properties. In this sense, the integrin adhesome has a higher magnitude of complexity in comparison to systems that give rise to relatively conserved multi-molecular structures within a cell, such as viral capsids,24-26 the nuclear pore complex27 and the kinetochores.28,29 The integrin adhesome can form heterogenous structures since its components interact with each other in multiple manners.14,15,30 For the sake of understanding the assembly process this property implies that there is an enormous number of potential paths by which a given adhesion site could have been assembled. Therefore, a combination of experimental and system-level analysis is required in order to decipher the assembly paths that are actually taking place, their logic and regulation.
An important intermediate layer in the assembly path of adhesion sites from individual integrin adhesome proteins is the formation of soluble multi-protein building blocks in the cytosol31 (Fig. 2). These building blocks are combinatorially diversified, implying that each protein could be recruited to adhesion sites as being embedded in different types of protein complexes. Therefore, in order to understand the assembly of adhesion sites we need to find out what are the repertoire and design principles of the cytosolic building blocks for adhesion sites and how their recruitment to these sites is performed and regulated (Fig. 2).
Figure 2.
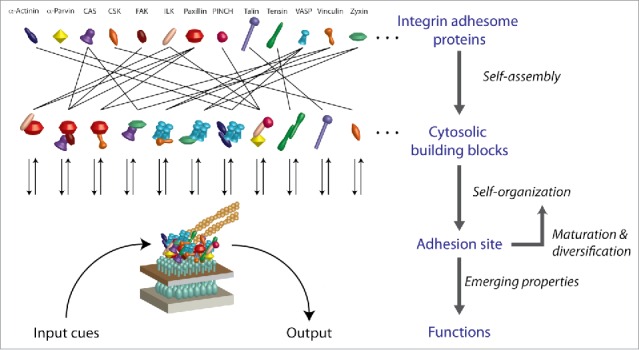
Layers of complexity along the self-organization of integrin adhesome proteins to cell-matrix adhesion sites. The integrin adhesome is self-assembled in the cytosol as diverse protein complexes, some of which serve as multi-protein building blocks for cell-matrix adhesion sites. Adhesion sites recruit these building blocks, plausibly in a specifically regulated manner. Each adhesion site matures by self-organization that affects the types of building blocks it recruits. The functions of an adhesion site emerge from the local interactions between its constituents as well as from their regulated exchange with the cytosolic pool.
Synthetic biology can help interrogating the assembly of adhesion sites by rewiring their cytosolic building blocks (Fig. 3). This rewiring could be performed by designing switchable interactions between proteins that normally do not bind each other at all or between proteins that bind each other only in adhesion sites. Acute induction of protein interactions within cells can be achieved by small, cell permeable, chemical inducers of dimerizerization (CIDs)32-41 or optogenetics42-51 (Fig. 3A). The time scales of such induced interactions are seconds to minutes, which are faster than, or comparable to, the time scale of focal adhesions assembly and maturation. The acuteness of the rewiring is important as it allows observing their direct effects on the assembly process, before indirect effects on the cellular state start taking place. Moreover, the acute induction of interaction between two ectopically expressed proteins, fused to dimerization domains, enables to detect its effects also in the presence of endogenous copies of these proteins. Photocleavable or photocaged CIDs allow for localized control of the target interaction.52-54 Such a spatial control enables rewiring the pool of cytoslic building blocks around single focal adhesions and hence to get a sensitive internal comparison of the outcomes with other focal adhesions within the same cell. Important consideration with CIDs and optogenetically induced interactions is whether the protein complex that is mediated via dimerization of fused domains preserves the relevant functionalities of the proteins and of the native complex, if such exists. Another useful rewiring of building blocks is inhibition of native interactions between proteins in the cytosol, thereby breaking down the corresponding multi-protein building blocks (Fig. 3A). Acute inhibition of protein interactions can be achieved by peptides mimicking binding domains55 or small molecule inhibitors.56,57 Genetic manipulations enable rewiring the building blocks for adhesion sites in a variety of manners, including mutations that change binding affinities,58 fusion of proteins,59,60 splitting a protein to its modules and swapping of binding domains59 (Fig. 3A). Since genetic perturbations are slow, the observed effects on adhesion sites could be due to global changes in the cellular state, like reduced spreading and overall mechanical tension. Still, due to their flexibility and generic applicability, genetic rewiring of building blocks would provide a useful complementary approach to the acute rewiring.
Figure 3.
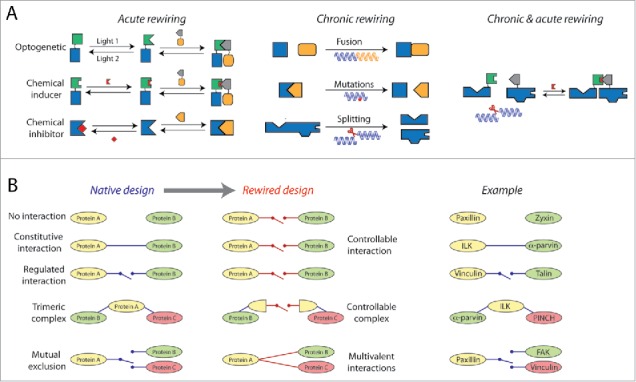
Rewiring the cytosolic pool of building blocks for studying its design principles and the assembly of adhesion sites. (A) Multi-protein building blocks can be rewired using optogentics, CIDs and genetic approaches. (B) Left, main types of building blocks rewiring based on the change from the native design of the interactions to their rewired design. Right, corresponding examples of integrin adhesome proteins that are natively wired as shown on the left.
Rewiring of cytosolic building blocks could have different informative effects on cell-matrix adhesion sites, on the cytosolic pool of integrin adhesome complexes and on other cellular phenotypes such as migration and cell-cell adhesion (Fig. 3B). Forcing two integrin adhesome proteins that do not have binding sites to each other to form a complex in the cytosol may potentially disrupt the layered structural organization of focal adhesions,61 if the proteins are normally positioned distally from each other (e.g. paxillin and zyxin61). Inducing an interaction between proteins that have binding domains to each other, but do not interact in the cytosol (e.g., talin and vinculin31), will help to study the role of the native regulation of this interaction in the assembly of adhesion sites. For example, the interaction of talin with vinculin in focal adhesions depends on the activation of talin by mechanical stretching62-64 and of vinculin by interaction with phosphatidyl-inositol-4-5-bisphosphate on the plasma membrane.65 Importantly, there is no release of talin-vinculin complexes from focal adhesions, plausibly in order to retain the checkpoint of mechanical tension and memebranal proximity for the assembly of these structures.31 Therefore, triggering the formation of a talin-vinculin building block in the cytosol may hamper the regulation of focal adhesions assembly and their mechanosensitivity. Similarly, bypassing the native regulation of an interaction between components of adhesion sites can disrupt or halt their maturation and diversification, for example by preventing the segregation of fibrillar adhesions from focal adhesions.
An intruiging question is how the integrin adhesome retains its switchability to form large adhesion sites while avoiding spontaneous nucleation and assembly of aberrant complexes in the cytosol. Considering the multiple binding domains that integrin adhesome proteins have to each other, the system is plausibly confining the undesirable assembly of large complexes in the cytosol by a network-level design of dependencies between interactions, i.e. mutual exclusion and allosteric regulations, as well as by controlling the lifetime of each interaction.66 Inducing regulated interactions, or rewiring the logical dependencies between protein interactions by genetically adding or swapping binding domains, will help to understand the robust switchability of the cytosolic pool of the building blocks for adhesion sites.
Studying how protein networks in adhesion sites function
Cell-matrix adhesion sites should not be conceived as devices that first assemble and only then are ready to perform their functions. In fact, the assembly, maturation and diversification of adhesion sites are by themselves also the means by which many of their functions are been carried out. For example, the dynamic segregation of fibrillar adhesions from focal adhesions is not only that way by which fibrillar adhesions are formed but also how they reorganize the fibronectin in the matrix.19,22,23 Therefore, the questions how protein networks in adhesion sites perform their functions and how adhesion sites assemble and mature are largely inseparable.
The first challenge in studying how adhesion sites perform their functions starts already with identifying and defining rigorously these functions (Fig. 4A). In contrast to discrete cell fate decisions (e.g. to divide or to differentiate) or phenotypes that can be described by a confined set of parameters (e.g., migration speed, persistency, etc.), cell adhesion can potentially integrate enormous types of inputs and consequently change quantitatively many aspects of its state. For example, cell-matrix adhesion sites can sense the rigidity of the matrix19,23,67 as well as spatial gradients in this rigidity68,69 – two distinct functions that require their own mechanistic designs. Once a function of adhesion sites is sharply defined at a phenomenological level, then the questions what are the involved components and how they give rise to this function can be posed (Fig. 4A).
Figure 4.
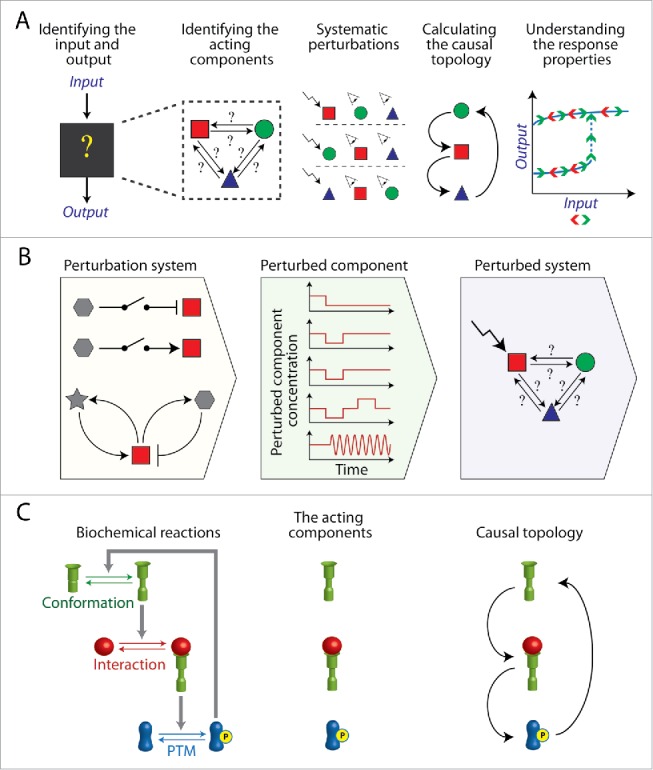
Engineering synthetic circuits for reverse engineering intracellular biochemical systems. (A) Generic pipeline of reverse engineering a protein network, starting from defining its inputs and outputs and ending up with resolving its causal topology, which explains its response properties. Experimentally, reverse engineering requires systematic perturbations of the system components and measuring their effects on each other. (B) Synthetic biology can be applied to construct intracellular circuits enabling generic, acute and flexible perturbations of the system components. (C) Left, an example of a biochemical system in which the states of the acting (i.e., active) components are defined by their conformation, interaction or post-translational modifications (PTM). The causal topology describes how the level of each acting component affects the level of the others acting components. Accordingly, perturbations applied for reverse engineering should alter the levels of the acting components either by converting their states or by depleting them.
To understand the principles by which a protein network performs a certain function we need to find out how its components affect each other. A complete quantitative description of such a network of effects would include the spatial concentrations of all the components, all the biochemical reactions between them and the kinetic constants of these reactions. However obtaining such complete and quantitative information is fundamentally difficult, even for small intracellular biochemical systems. Instead, the causal topology of the protein network, which indicates how the active state of each component affects the active state of the other components, can be reverse engineered70,71 (Fig. 4A). The causal topology of a network indicates its possible dynamics and what underlie its response properties to input cues.71 Hence reverse engineering can illuminate the principles of how a protein network performs its functions. Experimentally, reverse engineering requires systematic perturbations of the active state of the network components (i.e., the acting components) and quantifying the changes in the active states of the others.71 The results are then processed to account for indirect effects between components that are mediated by other components in the considered network.70,71
Reverse engineering of protein networks in cell-matrix adhesion sites encounters several challenges. Due to the large number of integrin adhesome components and the fuzzy boundaries of the network12,13,15 it is unfeasible to define a priory the conclusive set of proteins that are relevant to a certain function. The dense network of interactions between integrin adhesome components lack a clear modularity, making it problematic to investigate it part by part. The pre-assembly of integrin adhesome proteins to multi-protein building blocks,31 as well as the multiple phosphorylation states possible for many proteins,72 increase drastically the number of distinct biochemical species in the system. Many integrin adhesome components have several activities that are regulated separately, therefore the same protein may have more than one type of active state. Finally, the molecular heterogeneity of cell-matrix adhesion sites and their rapid dynamics imply that changes in their response to perturbations should be monitored in individual adhesion sites and with high temporal resolution. Due to these reasons, effective reverse engineering of cell-matrix adhesion sites requires generic approaches for systematic acute perturbations of their components in cells.
Synthetic biology can facilitate reverse engineering of cell-matrix adhesion sites by constructing switchable interactions and circuits allowing for systematic acute perturbations of integrin adhesome components (Fig. 4B). CIDs and optogenetics can be implemented to induce an interaction between a target protein and a dimerization domain ectopically expressed on the surface of a distal compartment,73 such as the mitochondria.74 Thus, upon triggering of the interaction by an addition of the CID or by light, the target protein will be recruited to the mitochondria and thereby depleted from the cytosolic pool. Reversible CIDs systems32,75 and optogenetics42,43 enable to deplete and then to replete the target protein in time scales of seconds to minutes. Such reversible perturbations are valuable for reverse engineering as they allow assessing if the investigated system did not change qualitatively its properties upon each perturbation. Since depletion of adhesion site components can cause their gradual disassembly, reversible perturbations that are faster than the disassembly process will enable to check if components respond directly to the perturbed components or are indirectly affected by the destruction of the site. Reversible perturbations are also valuable to assess directly if the system exhibits hysteresis or irreversibility. For example, depletion and repletion of tyrosine phosphomimetic mutants of paxillin76,77 and FAK,78 as active input components, can reveal hysteresis or irreversibility in their phenotypic outputs, including focal adhesions turnover, fibrillar adhesions segregation, cell protrusion and cell migration.
Reverse engineering of intracellular biochemical networks is still a developing research field in systems biology.79-81 The number of possible causal topologies increases exponentially with the size of the considered protein network. Therefore, a major challenge is reverse engineering correctly the causal connections between proteins in large networks in the presence of noise and effects from other cellular components. The more different perturbations are applied on a studied network more information about its causal topology can be derived. Along this line, an emerging exploratory direction in the reverse engineering of intracellular systems is the application of repetitive perturbations.82-85 Reversible CIDs systems,32 as well as optogenetics with time varying light inputs,42,43 can generate such perturbations by repetitive cycles of depletion and repletion of the target component (Fig. 4B).
A quantum leap in our capabilities to reverse engineer large intracellular systems would be achieved by generic approaches for systematic, large scale, acute perturbations of protein activities in cells. Synthetic biology can be applied to construct inert intracellular circuits that would interfere with the cellular system only by perturbing one of its components86 (Fig. 4B). Such synthetic circuits can be designed to generate oscillatory perturbations.84,85,87-96 Currently, synthetic oscillatory circuits in mammalian cells are based mainly on gene networks and therefore generate low frequencies.89,90,94,97 However, with the gradual expansion of the synthetic biology toolbox toward controlling protein activities,98 the engineering of fast, protein-network based, oscillators in mammalian cells should become more feasible.91 Generating cell lines stably expressing the synthetic circuit components would provide a modular and generic platform in which only the target protein, fused to an effector binding domain and a fluorescent protein, should be ectopically expressed.
Activation of the integrin adhesome proteins can be based on conformational change, interactions or post-translational modifications (Fig. 4C). The assembly of multi-protein building blocks in the cytosol reduces the specificity of the perturbations of the acting (i.e. active) components. Depleting a protein by recruiting it to a distal compartment will deplete it in all of its states, including the protein complexes embedding it. Therefore, ultimately it would be highly beneficial to be able designing synthetic switchable interactions depending not only on external triggers but also on the states of the proteins themselves. Such conditional switchable interactions would allow the targeting of specific protein complexes, phosphorylation states and conformations. Recent studies about engineering allosteric proteins98,99 could pioneer approaches for designing synthetic state-dependent switchable protein interactions.
Conclusions
Several features make cell-matrix adhesion sites among the most complex self-organizing intracellular devices. Therefore their study requires, and inspires, novel concepts and methods. Synthetic biology can synergize with systems biology by designing intracellular modules and circuits facilitating informative interferences with the integrin adhesome. Rewiring of building blocks in the cytosol can help interrogating the assembly logic of cell-matrix adhesion sites. Synthetic circuits for acute perturbations of integrin adhesome components would facilitate reverse engineering the mechanisms by which protein networks in adhesion sites perform their functions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Dedicated to the memory of Uri Zamir.
References
- [1].Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol 2009; 10:21-33; PMID:19197329; http://dx.doi.org/ 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- [2].Hynes RO. The evolution of metazoan extracellular matrix. J Cell Biol 2012; 196:671-9; PMID:22431747; http://dx.doi.org/ 10.1083/jcb.201109041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zaidel-Bar R. Evolution of complexity in the integrin adhesome. J Cell Biol 2009; 186:317-21; PMID:19667126; http://dx.doi.org/ 10.1083/jcb.200811067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Izzard CS, Lochner LR. Cell-to-substrate contacts in living fibroblasts: an interference reflexion study with an evaluation of the technique. J Cell Sci 1976; 21:129-59; PMID:932106 [DOI] [PubMed] [Google Scholar]
- [5].Curtis AS. The Mechanism of Adhesion of Cells to Glass. A Study by Interference Reflection Microscopy. J Cell Biol 1964; 20:199-215; PMID:14126869; http://dx.doi.org/ 10.1083/jcb.20.2.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Abercrombie M, Dunn GA. Adhesions of fibroblasts to substratum during contact inhibition observed by interference reflection microscopy. Exp Cell Res 1975; 92:57-62; PMID:1169157; http://dx.doi.org/ 10.1016/0014-4827(75)90636-9 [DOI] [PubMed] [Google Scholar]
- [7].Wehland J, Osborn M, Weber K. Cell-to-substratum contacts in living cells: a direct correlation between interference-reflexion and indirect-immunofluorescence microscopy using antibodies against actin and α-actinin. J Cell Sci 1979; 37:257-73; PMID:383732 [DOI] [PubMed] [Google Scholar]
- [8].Heath JP, Dunn GA. Cell to substratum contacts of chick fibroblasts and their relation to the microfilament system. A correlated interference-reflexion and high-voltage electron-microscope study. J Cell Sci 1978; 29:197-212; PMID:564353 [DOI] [PubMed] [Google Scholar]
- [9].Mangeat P, Burridge K. Actin-membrane interaction in fibroblasts: what proteins are involved in this association? J Cell Biol 1984; 99:95s-103s; PMID:6430913; http://dx.doi.org/ 10.1083/jcb.99.1.95s [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tamkun JW, DeSimone DW, Fonda D, Patel RS, Buck C, Horwitz AF, Hynes RO. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell 1986; 46:271-82; PMID:3487386; http://dx.doi.org/ 10.1016/0092-8674(86)90744-0 [DOI] [PubMed] [Google Scholar]
- [11].Schwartz MA, Schaller MD, Ginsberg MH. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol 1995; 11:549-99; PMID:8689569; http://dx.doi.org/ 10.1146/annurev.cb.11.110195.003001 [DOI] [PubMed] [Google Scholar]
- [12].Geiger T, Zaidel-Bar R. Opening the floodgates: proteomics and the integrin adhesome. Curr Opin Cell Biol 2012; 24:562-8; PMID:22728062; http://dx.doi.org/ 10.1016/j.ceb.2012.05.004 [DOI] [PubMed] [Google Scholar]
- [13].Horton ER, Byron A, Askari JA, Ng DH, Millon-Fremillon A, Robertson J, Koper EJ, Paul NR, Warwood S, Knight D, et al.. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol 2015; 17(12):1577-87; PMID:26479319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zaidel-Bar R, Geiger B. The switchable integrin adhesome. J Cell Sci 2010; 123:1385-8; PMID:20410370; http://dx.doi.org/ 10.1242/jcs.066183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol 2007; 9:858-67; PMID:17671451; http://dx.doi.org/ 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Frohnmayer JP, Bruggemann D, Eberhard C, Neubauer S, Mollenhauer C, Boehm H, Kessler H, Geiger B, Spatz JP. Minimal Synthetic Cells to Study Integrin-Mediated Adhesion. Angew Chem Int Ed Engl 2015; 54:12472-8; PMID:26257266; http://dx.doi.org/ 10.1002/anie.201503184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zaidel-Bar R, Cohen M, Addadi L, Geiger B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem Soc Trans 2004; 32:416-20; PMID:15157150; http://dx.doi.org/ 10.1042/bst0320416 [DOI] [PubMed] [Google Scholar]
- [18].Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol 2010; 26:315-33; PMID:19575647; http://dx.doi.org/ 10.1146/annurev.cellbio.011209.122036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci 2001; 114:3583-90; PMID:11707510 [DOI] [PubMed] [Google Scholar]
- [20].Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol 2011; 3:a005033; PMID:21441590; http://dx.doi.org/ 10.1101/cshperspect.a005033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ross TD, Coon BG, Yun S, Baeyens N, Tanaka K, Ouyang M, Schwartz MA. Integrins in mechanotransduction. Curr Opin Cell Biol 2013; 25:613-8; PMID:23797029; http://dx.doi.org/ 10.1016/j.ceb.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pankov R, Cukierman E, Katz BZ, Matsumoto K, Lin DC, Lin S, Hahn C, Yamada KM. Integrin dynamics and matrix assembly: tensin-dependent translocation of α(5)β(1) integrins promotes early fibronectin fibrillogenesis. J Cell Biol 2000; 148:1075-90; PMID:10704455; http://dx.doi.org/ 10.1083/jcb.148.5.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zamir E, Katz M, Posen Y, Erez N, Yamada KM, Katz BZ, Lin S, Lin DC, Bershadsky A, Kam Z, et al.. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat Cell Biol 2000; 2:191-6; PMID:10783236; http://dx.doi.org/ 10.1038/35008607 [DOI] [PubMed] [Google Scholar]
- [24].Zlotnick A. Theoretical aspects of virus capsid assembly. J Mol Recognit 2005; 18:479-90; PMID:16193532; http://dx.doi.org/ 10.1002/jmr.754 [DOI] [PubMed] [Google Scholar]
- [25].Baschek JE, HC RK, Schwarz US. Stochastic dynamics of virus capsid formation: direct versus hierarchical self-assembly. BMC Biophys 2012; 5:22; PMID:23244740; http://dx.doi.org/ 10.1186/2046-1682-5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dokland T. Freedom and restraint: themes in virus capsid assembly. Structure 2000; 8:R157-62; PMID:10997898; http://dx.doi.org/ 10.1016/S0969-2126(00)00181-7 [DOI] [PubMed] [Google Scholar]
- [27].Kabachinski G, Schwartz TU. The nuclear pore complex–structure and function at a glance. J Cell Sci 2015; 128:423-9; PMID:26046137; http://dx.doi.org/ 10.1242/jcs.083246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gascoigne KE, Cheeseman IM. Kinetochore assembly: if you build it, they will come. Curr Opin Cell Biol 2011; 23:102-8; PMID:20702077; http://dx.doi.org/ 10.1016/j.ceb.2010.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J 2009; 28:2511-31; PMID:19629042; http://dx.doi.org/ 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zamir E, Geiger B. Components of cell-matrix adhesions. J Cell Sci 2001; 114:3577-9; PMID:11707509 [DOI] [PubMed] [Google Scholar]
- [31].Hoffmann JE, Fermin Y, Stricker RL, Ickstadt K, Zamir E. Symmetric exchange of multi-protein building blocks between stationary focal adhesions and the cytosol. Elife 2014; 3:e02257; PMID:24894463; http://dx.doi.org/ 10.7554/eLife.02257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Voss S, Klewer L, Wu YW. Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells. Curr Opin Chem Biol 2015; 28:194-201; PMID:26431673; http://dx.doi.org/ 10.1016/j.cbpa.2015.09.003 [DOI] [PubMed] [Google Scholar]
- [33].Peterson-Kaufman KJ, Carlson CD, Rodriguez-Martinez JA, Ansari AZ. Nucleating the assembly of macromolecular complexes. Chembiochem 2010; 11:1955-62; PMID: 20812316; http://dx.doi.org/ 10.1002/cbic.201000255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Miyamoto T, DeRose R, Suarez A, Ueno T, Chen M, Sun TP, Wolfgang MJ, Mukherjee C, Meyers DJ, Inoue T. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat Chem Biol 2012; 8:465-70; PMID:22446836; http://dx.doi.org/ 10.1038/nchembio.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lin YC, Nihongaki Y, Liu TY, Razavi S, Sato M, Inoue T. Rapidly reversible manipulation of molecular activity with dual chemical dimerizers. Angew Chem Int Ed Engl 2013; 52:6450-4; PMID:23649661; http://dx.doi.org/ 10.1002/anie.201301219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Erhart D, Zimmermann M, Jacques O, Wittwer MB, Ernst B, Constable E, Zvelebil M, Beaufils F, Wymann MP. Chemical development of intracellular protein heterodimerizers. Chem Biol 2013; 20:549-57; PMID:23601644; http://dx.doi.org/ 10.1016/j.chembiol.2013.03.010 [DOI] [PubMed] [Google Scholar]
- [37].DeRose R, Miyamoto T, Inoue T. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch 2013; 465:409-17; PMID:23299847; http://dx.doi.org/ 10.1007/s00424-012-1208-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Controlling protein association and subcellular localization with a synthetic ligand that induces heterodimerization of proteins. Proc Natl Acad Sci U S A 1996; 93:4604-7; PMID:8643450; http://dx.doi.org/ 10.1073/pnas.93.10.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Amara JF, Clackson T, Rivera VM, Guo T, Keenan T, Natesan S, Pollock R, Yang W, Courage NL, Holt DA, et al.. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proc Natl Acad Sci U S A 1997; 94:10618-23; PMID:9380684; http://dx.doi.org/ 10.1073/pnas.94.20.10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Stein V, Alexandrov K. Synthetic protein switches: design principles and applications. Trends Biotechnol 2015; 33:101-10; PMID:25535088; http://dx.doi.org/ 10.1016/j.tibtech.2014.11.010 [DOI] [PubMed] [Google Scholar]
- [41].Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 2006; 126:995-1004; PMID:16959577; http://dx.doi.org/ 10.1016/j.cell.2006.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang K, Cui B. Optogenetic control of intracellular signaling pathways. Trends Biotechnol 2015; 33:92-100; PMID:25529484; http://dx.doi.org/ 10.1016/j.tibtech.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 2013; 155:1422-34; PMID:24315106; http://dx.doi.org/ 10.1016/j.cell.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol 2014; 15:551-8; PMID:25027655; http://dx.doi.org/ 10.1038/nrm3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pathak GP, Vrana JD, Tucker CL. Optogenetic control of cell function using engineered photoreceptors. Biol Cell 2013; 105:59-72; PMID:23157573; http://dx.doi.org/ 10.1111/boc.201200056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Muller K, Weber W. Optogenetic tools for mammalian systems. Mol Biosyst 2013; 9:596-608; PMID:23412367; http://dx.doi.org/ 10.1039/c3mb25590e [DOI] [PubMed] [Google Scholar]
- [47].Kim B, Lin MZ. Optobiology: optical control of biological processes via protein engineering. Biochem Soc Trans 2013; 41:1183-8; PMID:24059506; http://dx.doi.org/ 10.1042/BST20130150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kianianmomeni A. UVB-based optogenetic tools. Trends Biotechnol 2015; 33:59-61; PMID:24985334; http://dx.doi.org/ 10.1016/j.tibtech.2014.06.004 [DOI] [PubMed] [Google Scholar]
- [49].Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009; 461:997-1001; PMID:19749742; http://dx.doi.org/ 10.1038/nature08446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Schmidt D, Cho YK. Natural photoreceptors and their application to synthetic biology. Trends Biotechnol 2015; 33:80-91; PMID:25466878; http://dx.doi.org/ 10.1016/j.tibtech.2014.10.007 [DOI] [PubMed] [Google Scholar]
- [51].Pathak GP, Strickland D, Vrana JD, Tucker CL. Benchmarking of optical dimerizer systems. ACS Synth Biol 2014; 3:832-8; PMID:25350266; http://dx.doi.org/ 10.1021/sb500291r [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zimmermann M, Cal R, Janett E, Hoffmann V, Bochet CG, Constable E, Beaufils F, Wymann MP. Cell-permeant and photocleavable chemical inducer of dimerization. Angew Chem Int Ed Engl 2014; 53:4717-20; PMID:24677313; http://dx.doi.org/ 10.1002/anie.201310969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Brown KA, Zou Y, Shirvanyants D, Zhang J, Samanta S, Mantravadi PK, Dokholyan NV, Deiters A. Light-cleavable rapamycin dimer as an optical trigger for protein dimerization. Chem Commun (Camb) 2015; 51:5702-5; PMID:25716548; http://dx.doi.org/ 10.1039/C4CC09442E [DOI] [PubMed] [Google Scholar]
- [54].Ballister ER, Aonbangkhen C, Mayo AM, Lampson MA, Chenoweth DM. Localized light-induced protein dimerization in living cells using a photocaged dimerizer. Nat Commun 2014; 5:5475; PMID:25400104; http://dx.doi.org/ 10.1038/ncomms6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rubinstein M, Niv MY. Peptidic modulators of protein-protein interactions: progress and challenges in computational design. Biopolymers 2009; 91:505-13; PMID:19226619; http://dx.doi.org/ 10.1002/bip.21164 [DOI] [PubMed] [Google Scholar]
- [56].Bienstock RJ. Computational drug design targeting protein-protein interactions. Curr Pharm Des 2012; 18:1240-54; PMID:22316151; http://dx.doi.org/ 10.2174/138161212799436449 [DOI] [PubMed] [Google Scholar]
- [57].Opitz R, Muller M, Reuter C, Barone M, Soicke A, Roske Y, Piotukh K, Huy P, Beerbaum M, Wiesner B, et al.. A modular toolkit to inhibit proline-rich motif-mediated protein-protein interactions. Proc Natl Acad Sci U S A 2015; 112:5011-6; PMID:25848013; http://dx.doi.org/ 10.1073/pnas.1422054112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sharabi O, Shirian J, Shifman JM. Predicting affinity- and specificity-enhancing mutations at protein-protein interfaces. Biochem Soc Trans 2013; 41:1166-9; PMID:24059503; http://dx.doi.org/ 10.1042/BST20130121 [DOI] [PubMed] [Google Scholar]
- [59].Wang B, Barahona M, Buck M, Schumacher J. Rewiring cell signalling through chimaeric regulatory protein engineering. Biochem Soc Trans 2013; 41:1195-200; PMID:24059508; http://dx.doi.org/ 10.1042/BST20130138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yu K, Liu C, Kim BG, Lee DY. Synthetic fusion protein design and applications. Biotechnol Adv 2014; 33:155-64; PMID:25450191; http://dx.doi.org/ 10.1016/j.biotechadv.2014.11.005 [DOI] [PubMed] [Google Scholar]
- [61].Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature 2010; 468:580-4; PMID:21107430; http://dx.doi.org/ 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science 2009; 323:638-41; PMID:19179532; http://dx.doi.org/ 10.1126/science.1162912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yan J, Yao M, Goult BT, Sheetz MP. Talin dependent mechanosensitivity of cell focal adhesions. Cell Mol Bioeng 2015; 8:151-9; PMID:26097520; http://dx.doi.org/ 10.1007/s12195-014-0364-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Margadant F, Chew LL, Hu X, Yu H, Bate N, Zhang X, Sheetz M. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol 2011; 9:e1001223; PMID:22205879; http://dx.doi.org/ 10.1371/journal.pbio.1001223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature 1996; 381:531-5; PMID:8632828; http://dx.doi.org/ 10.1038/381531a0 [DOI] [PubMed] [Google Scholar]
- [66].Koster J, Zamir E, Rahmann S. Efficiently mining protein interaction dependencies from large text corpora. Integr Biol (Camb) 2012; 4:805-12; PMID:22706334; http://dx.doi.org/ 10.1039/c2ib00126h [DOI] [PubMed] [Google Scholar]
- [67].Katz BZ, Zamir E, Bershadsky A, Kam Z, Yamada KM, Geiger B. Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions. Mol Biol Cell 2000; 11:1047-60; PMID:10712519; http://dx.doi.org/ 10.1091/mbc.11.3.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 2012; 151:1513-27; PMID:23260139; http://dx.doi.org/ 10.1016/j.cell.2012.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Plotnikov SV, Waterman CM. Guiding cell migration by tugging. Curr Opin Cell Biol 2013; 25:619-26; PMID:23830911; http://dx.doi.org/ 10.1016/j.ceb.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kholodenko BN, Kiyatkin A, Bruggeman FJ, Sontag E, Westerhoff HV, Hoek JB. Untangling the wires: a strategy to trace functional interactions in signaling and gene networks. Proc Natl Acad Sci U S A 2002; 99:12841-6; PMID:12242336; http://dx.doi.org/ 10.1073/pnas.192442699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zamir E, Bastiaens PI. Reverse engineering intracellular biochemical networks. Nat Chem Biol 2008; 4:643-7; PMID:18936743; http://dx.doi.org/ 10.1038/nchembio1108-643 [DOI] [PubMed] [Google Scholar]
- [72].Robertson J, Jacquemet G, Byron A, Jones MC, Warwood S, Selley JN, Knight D, Humphries JD, Humphries MJ. Defining the phospho-adhesome through the phosphoproteomic analysis of integrin signalling. Nat Commun 2015; 6:6265; PMID:25677187; http://dx.doi.org/ 10.1038/ncomms7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods 2005; 2:415-8; PMID:15908919; http://dx.doi.org/ 10.1038/nmeth763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Robinson MS, Sahlender DA, Foster SD. Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev Cell 2010; 18:324-31; PMID:20159602; http://dx.doi.org/ 10.1016/j.devcel.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Liu P, Calderon A, Konstantinidis G, Hou J, Voss S, Chen X, Li F, Banerjee S, Hoffmann JE, Theiss C, et al.. A bioorthogonal small-molecule-switch system for controlling protein function in live cells. Angew Chem Int Ed Engl 2013; 53:10049-55; http://dx.doi.org/ 10.1002/anie.201403463 [DOI] [PubMed] [Google Scholar]
- [76].Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J Cell Sci 2007; 120:137-48; PMID:17164291; http://dx.doi.org/ 10.1242/jcs.03314 [DOI] [PubMed] [Google Scholar]
- [77].Clarke DM, Brown MC, LaLonde DP, Turner CE. Phosphorylation of actopaxin regulates cell spreading and migration. J Cell Biol 2004; 166:901-12; PMID:15353548; http://dx.doi.org/ 10.1083/jcb.200404024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Deramaudt TB, Dujardin D, Hamadi A, Noulet F, Kolli K, De Mey J, Takeda K, Rondé P. FAK phosphorylation at Tyr-925 regulates cross-talk between focal adhesion turnover and cell protrusion. Mol Biol Cell 2011; 22:964-75; PMID:21289086; http://dx.doi.org/ 10.1091/mbc.E10-08-0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].DREAM5 Consortium, Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Kellis M, Collins JJ, et al.. Wisdom of crowds for robust gene network inference. Nat Methods 2012; 9:796-804; PMID:22796662; http://dx.doi.org/ 10.1038/nmeth.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Costello JC, Heiser LM, Georgii E, Gonen M, Menden MP, Wang NJ, Bansal M, Ammad-ud-din M, Hintsanen P, Khan SA, et al.. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol 2014; 32:1202-12; PMID:24880487; http://dx.doi.org/ 10.1038/nbt.2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].De Smet R, Marchal K. Advantages and limitations of current network inference methods. Nat Rev Microbiol 2010; 8:717-29; PMID:20805835 [DOI] [PubMed] [Google Scholar]
- [82].Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 2008; 319:482-4; PMID: 18218902; http://dx.doi.org/ 10.1126/science.1151582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bennett MR, Pang WL, Ostroff NA, Baumgartner BL, Nayak S, Tsimring LS, Hasty J. Metabolic gene regulation in a dynamically changing environment. Nature 2008; 454:1119-22; PMID:18668041; http://dx.doi.org/ 10.1038/nature07211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lipan O, Wong WH. The use of oscillatory signals in the study of genetic networks. Proc Natl Acad Sci U S A 2005; 102:7063-8; PMID:15883385; http://dx.doi.org/ 10.1073/pnas.0403790102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Konopka T, Rooman M. Gene expression model (in)validation by Fourier analysis. BMC Syst Biol 2010; 4:123; PMID:20815892; http://dx.doi.org/ 10.1186/1752-0509-4-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol 2009; 10:410-22; PMID:19461664; http://dx.doi.org/ 10.1038/nrm2698 [DOI] [PubMed] [Google Scholar]
- [87].Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet 2010; 11:367-79; PMID:20395970; http://dx.doi.org/ 10.1038/nrg2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sohka T, Heins RA, Phelan RM, Greisler JM, Townsend CA, Ostermeier M. An externally tunable bacterial band-pass filter. Proc Natl Acad Sci U S A 2009; 106:10135-40; PMID:19502423; http://dx.doi.org/ 10.1073/pnas.0901246106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Tigges M, Denervaud N, Greber D, Stelling J, Fussenegger M. A synthetic low-frequency mammalian oscillator. Nucleic Acids Res 2010; 38:2702-11; PMID:20197318; http://dx.doi.org/ 10.1093/nar/gkq121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Greber D, Fussenegger M. Mammalian synthetic biology: engineering of sophisticated gene networks. J Biotechnol 2007; 130:329-45; PMID:17602777; http://dx.doi.org/ 10.1016/j.jbiotec.2007.05.014 [DOI] [PubMed] [Google Scholar]
- [91].Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol 2008; 9:981-91; PMID:18971947; http://dx.doi.org/ 10.1038/nrm2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature 2008; 456:516-9; PMID:18971928; http://dx.doi.org/ 10.1038/nature07389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature 2009; 457:309-12; PMID:19148099; http://dx.doi.org/ 10.1038/nature07616 [DOI] [PubMed] [Google Scholar]
- [94].Ye H, Fussenegger M. Synthetic therapeutic gene circuits in mammalian cells. FEBS Lett 2009; 588:2537-44; http://dx.doi.org/ 10.1016/j.febslet.2014.05.003 [DOI] [PubMed] [Google Scholar]
- [95].Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat Rev Mol Cell Biol 2014; 15:95-107; PMID:24434884; http://dx.doi.org/ 10.1038/nrm3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Del Vecchio D. Modularity, context-dependence, and insulation in engineered biological circuits. Trends Biotechnol 2015; 33:111-9; PMID:25544476; http://dx.doi.org/ 10.1016/j.tibtech.2014.11.009 [DOI] [PubMed] [Google Scholar]
- [97].Wieland M, Fussenegger M. Engineering molecular circuits using synthetic biology in mammalian cells. Annu Rev Chem Biomol Eng 2012; 3:209-34; PMID:22468602; http://dx.doi.org/ 10.1146/annurev-chembioeng-061010-114145 [DOI] [PubMed] [Google Scholar]
- [98].Raman S, Taylor N, Genuth N, Fields S, Church GM. Engineering allostery. Trends Genet 2014; 30:521-8; PMID:25306102; http://dx.doi.org/ 10.1016/j.tig.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Page MJ, Carrell CJ, Di Cera E. Engineering protein allostery: 1.05 A resolution structure and enzymatic properties of a Na+-activated trypsin. J Mol Biol 2008; 378:666-72; PMID:18377928; http://dx.doi.org/ 10.1016/j.jmb.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]


