Abstract
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) is a metabolic disorder caused by mutations in TYMP, encoding thymidine phosphorylase (TP). In MNGIE patients, TP dysfunction produces systemic thymidine and deoxyuridine accumulation, which ultimately impairs mitochondrial DNA replication and results in mitochondrial dysfunction. To date, only allogeneic hematopoietic stem cell transplantation has demonstrated long-term clinical efficacy, but high morbidity and mortality associated with this procedure necessitate the search for safer alternatives. In a previous study, we demonstrated that hematopoietic stem cell gene therapy using a lentiviral vector containing the coding sequence of TYMP restored the biochemical homeostasis in an animal model of MNGIE. In the present follow-up study, we show that ectopic expression of TP in the hematopoietic system restores normal nucleoside levels in plasma, as well as in tissues affected in MNGIE such as small intestine, skeletal muscle, brain, and liver. Mitochondrial dNTP pool imbalances observed in liver of the animal model were also corrected by the treatment. The biochemical effects were maintained at least 20 months even with low levels of chimerism. No alterations in the blood cell counts or other toxic effects were observed in association with the lentiviral transduction or TP overexpression. These results further support the notion that gene therapy is a feasible treatment option for MNGIE.
Introduction
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) is a severe disorder caused by mutations in TYMP, the gene encoding thymidine phosphorylase (TP).1–3 This cytosolic enzyme catalyzes the phosphorolysis of the nucleosides thymidine (dThd) and deoxyuridine (dUrd) to the corresponding bases thymine and uracil, and deoxyribose-1-phosphate.4 In MNGIE patients, the lack of TP activity results in systemic dThd and dUrd overload and this excess is toxic for mitochondria.5 In vitro and in vivo studies have demonstrated that, in quiescent cells, excess of dThd results in increased mitochondrial dTTP and secondary dCTP depletion.6,7 This deoxyribonucleotide imbalance impairs mitochondrial DNA (mtDNA) replication and results in mtDNA depletion, multiple deletions, and somatic point mutations.6,8–10 MNGIE is a rare disease, with fewer than 300 patients known to be affected worldwide, although the exact incidence is unknown. Clinically, MNGIE is characterized by progressive external ophthalmoplegia, gastrointestinal dysmotility, cachexia, peripheral neuropathy, diffuse leukoencephalopathy on brain magnetic resonance imaging, and mitochondrial dysfunction. The average life expectancy of MNGIE patients is 37 years.2,11
Since MNGIE is caused by accumulation of the toxic metabolites dThd and dUrd, all therapeutic approaches have been focused on reducing them to normal levels. Allogeneic hematopoietic stem cell transplantation (Allo-HSCT) has been demonstrated to be an effective treatment in several patients.12–14 Nonetheless, allo-HSCT is limited by the difficulty of finding suitable donors and the high mortality and morbidity rates associated with this procedure, especially in MNGIE patients, who are usually in poor clinical condition when treated. Thus, allo-HSCT may be relevant only for a subset of MNGIE patients who have HLA-matched donors and are not severely medically compromised by the disease. These limitations encouraged us to investigate alternative approaches. Our previous work on a murine model of the disease has already demonstrated that gene therapy using TYMP is a feasible approach. Full normalization of biochemical abnormalities was achieved using either ex vivo transduction of a lentiviral vector targeted at hematopoietic stem cells,15 or an adeno-associated virus (AAV) vector with targeted expression at the liver.16 In the current study, we demonstrate that expression of TYMP targeted at hematopoietic tissue using a lentiviral vector is fully maintained over at least 18 months, and that the correction of biochemical derangements is sustained long-term over this entire period. These results further confirm the feasibility of gene therapy as an alternative to allo-HSCT in MNGIE patients.
Materials and Methods
Vector production, titration, and transduction
Third-generation self-inactivating (SIN) HIV-derived lentiviral vectors that express EGFP (p-sham) or EGFP and TYMP (p-TP) under the control of the hPGK promoter have been previously described.15 Viral production was performed by polyethylenimine (PEI) transient cotransfection of HEK293T with the transfer vector and the packaging plasmids pRSV REV, pMDLg RRE, and pMD VSVG as described elsewhere.15 Viral productions were titrated by transduction of HEK293T cells with serial dilutions of lentiviral vector suspensions and counting the percentage of green fluorescent cells by flow cytometry.15
Animal procedures and sample collection
All animal procedures were performed in accordance with protocols approved by our institutional review board. Tymp/Upp1 double-knockout (dKO) mice in C57b/6J genetic background6 were used in this study. Transplanted mice were routinely checked for general health status, including periodic blood cell counts. Blood samples were collected from the saphenous vein using EDTA as anticoagulant. Total and differential blood cell counts were performed in a BC-2800 Auto Hematology Analyzer (Mindray, China).
Two subgroups of mice were killed at two different timepoints (6 and 18 months after treatment) by CO2 inhalation. Intracardiac blood samples were collected and processed to obtain plasma, buffy coat, and blood cell fraction (i.e., total sedimented cells, including erithrocytes and buffy coat). Different tissues were gently washed in cold phosphate buffered saline (PBS) and immediately frozen in liquid nitrogen and stored at −80°C until analysis. Small intestine was additionally washed by flushing with PBS before freezing.
Transduction and transplantation protocol
Eight- to 12-week-old donor mice were killed with CO2 inhalation and bone marrow (BM) was obtained from femorae, tibiae, and iliac crests. Purified murine lineage (Lin)− BM cells (Lineage Cell Depletion Kit mouse; Miltenyi Biotec, Bergisch Gladbach, Germany) were seeded at a density of 5 × 105 cells per well in 24-well plates containing 1 ml of Iscoves modified Dulbecco's medium (IMDM) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and recombinant murine growth factors (rmSCF 10 ng/μl, rmTPO 10 ng/μl, rmIL3 10 ng/μl, and rmFlt3 10 ng/μl). Lentiviral vectors p-TP and p-sham were added at an MOI of 100 and cells were incubated for 16 hr at 37°C and 5% CO2 and then washed with PBS. Transduction efficacy of Lin− selected cells, assessed by flow cytometry, ranged from 12% to 28%.
Eight- to 12-week-old Tymp/Upp1 dKO mice were tail vein injected with 3–5 × 105 transduced hematopoietic progenitors after sublethal myeloablation with total body irradiation (6 Gy, in 2 doses of 3 Gy at a dose rate of 2.24 Gy/min given at a 2 hr interval) using a cesium (Cs137) gamma-ray irradiator.
Nucleosides and TP activity determination
Plasma dThd and dUrd concentrations and blood cell TP activity were analyzed by HPLC-UV, as previously described.15 For tissue TP activity and nucleoside concentration determination, frozen samples were homogenized in lysis buffer (50 mM Tris-HCl, pH 7.2; 10 ml/l Triton X-100; 2 mM phenylmethylsulfonyl fluoride; 0.2 ml/l 2-mercaptoethanol) in a Potter homogenizer. Homogenates were centrifuged at 20,000 × g for 30 min at 4°C, and supernatants were separated into two aliquots. One aliquot was used as described elsewhere for protein determination17 and TP activity determination.18 The other aliquot was frozen until used to measure nucleosides by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Thawed supernatants were centrifuged at 20,000 × g for 10 min at 4°C to eliminate any remaining particles, and clean supernatants were deproteinized by ultrafiltration (10 kDa Amicon Ultra filters; Millipore, Darmstadt, Germany) at 14,000 × g and 4°C for 30 min. Five microliters of deproteinized homogenate was injected into an Acquity UPLC-MS/MS apparatus (Acquity UPLC-Xevo TQ Mass Spectrometer; Waters, Milford, MA) using an Acquity UPLC BEH C18 column (100 × 2.1 mm, 130 Å pore, 1.7 μm particle; Waters). The components of the sample were resolved at 0.5 ml/min through a binary gradient-elution using a saline buffer (20 mM ammonium acetate, pH 5.6) and acetonitrile as follows: 0–1.1 min, isocratic 100% saline buffer; 1.1–5 min, gradient from 0% to 13.6% acetonitrile; 5–5.1 min, gradient from 13.6% to 100% acetonitrile; 5.1–6.1 min, isocratic 100% acetonitrile; 6.1–7.2 min, isocratic 100% saline buffer. Eluate components were detected by multiple reaction monitoring, with positive electrospray for dThd (transition mass 242.8–127.1, cone voltage 10 V, collision energy 12 V) and dUrd (transition mass 228.8–113.08, cone voltage 8 V, and collision energy 12 V). Calibration curves made with aqueous standards were processed in parallel, and concentrations were obtained from interpolation of the peak areas.
Mitochondrial isolation and dNTP quantification
Liver mitochondria were isolated by differential centrifugation as previously described.9 Fourteen-month-old mice were killed by cervical dislocation and liver was homogenized in buffer A (320 mM sucrose, 1 mM EDTA, 10 mM Tris-HCl, pH 7.4). A volume (approximately 0.1–0.3 ml) of suspension with isolated mitochondria containing 0.5 mg protein was treated with trichloroacetic acid (final concentration 0.5 M) and centrifuged at 20 000 × g for 5 min at 4°C. Supernatants were neutralized with 1.5 volumes of 0.5 M tri-N-octylamine in Freon (1,1,2-trichlorotrifluoroethane) and centrifuged for 10 min at 10 000 × g at 4°C. Half the aqueous upper phase was recovered and dried under speed vacuum. Lyophillized dNTP extracts were dissolved in 125 μl of 40 mM Tris-HCl (pH 7.4) and stored at −80°C until measurement. For mitochondrial dNTP quantification, we used the polymerase-based assay previously described.9
Vector copy number
DNA was extracted from buffy coat, BM, liver, and brain with the QIAamp DNA mini kit (Qiagen, Hilden, Germany). Quantification of vector copy number was performed by qRT-PCR in the ABI PRISM 7900 sequence detection system (Applied Biosystems, Foster City, CA). Vector copy number in DNA samples from buffy coat and BM was quantified using custom-designed specific primers and TaqMan MGB probe to detect EGFP-integrated copies in the genome from p-TP or p-sham transduction, as previously described.15 Results were referred to the single-copy nuclear gene Ang1 using the predesigned TaqMan MGB gene expression assay Mm00833184_s1 (Applied Biosystems).
Flow cytometry
EGFP fluorescence was analyzed by flow cytometry. BM, spleen, and blood cells were incubated with erythrocyte lysis buffer (BD Biosciences, San Jose, CA) for 10 min at 37°C and then centrifuged for 5 min at 300 × g. Cell pellets were re-suspended in fresh medium and fluorescence was measured through a FACSCalibur flow cytometer (BD Biosciences).
Statistical analysis
Statistical analysis was performed with the SPSS 15.0 and GraphPad Prism 5 software. Tests used are indicated in the figure legends. For statistical purposes, undetectable values were considered as zero.
Results
Low levels of molecular chimerism resulted in high TP activity in the hematopoietic tissues
Our previous studies using a lentiviral vector containing a copy of the human TYMP gene targeted at the hematopoietic cells in a TP-deficient mouse model showed that ectopic TP restoration in blood cells reduced dThd and dUrd concentrations to levels observed in wild-type (wt) mice.15 Here, we have analyzed blood cell fraction and other hematopoietic tissues, including BM and spleen. Six months after treatment, sustained increases in TP activity were observed in blood cells (ranging 0.9–30 nmol Thy/hr/mg prot), BM (19–242 nmol Thy/hr/mg prot), and spleen (11–152 nmol Thy/hr/mg prot), whereas TP activity was undetectable in untreated and p-sham vector-treated dKO mice. TP activities of mice treated with the p-TP vector were clearly higher than those observed in wt mice (Fig. 1).
Figure 1.
TP activity in hematopoietic tissues. TP activity in blood, BM, and spleen of wild type (WT), nontreated (KO), p-sham-treated (SHAM), and p-TP-treated (TP) Tymp/Upp1 dKO mice 6 months after treatment. To facilitate matching the results, each mouse in the TP-treated group is identified with the same symbol in Figs. 1–4. Horizontal lines indicate medians. BM, bone marrow; KO, knockout; dKO, double-knockout; TP, thymidine phosphorylase.
Molecular chimerism was assessed by flow cytometry (expressed as the percentage of EGFP+ cells), and by quantitative real-time PCR (quantifying EGFP copy number) six months after treatment (Fig. 2). Flow cytometry analysis revealed low molecular chimerism ratios in TP-treated mice, ranging from 2% to 15% (white blood cells), 2% to 28% (BM cells), and 1% to 11% (spleen). Higher chimerism ratios, although widely variable, were observed when assessed as EGFP copy number per cell, which ranged from 0.07 to 0.40 in white blood cells, and 0.15 to 1 in BM cells. Percentages of EGFP+ cells were higher in mice treated with p-sham than in the p-TP-treated counterparts, but the EGFP copy number per cell was similar in both groups, suggesting that the sham construct expressed EGFP more efficiently than the TP construct. A significant correlation between chimerism levels (% EGFP+ cells) and TP activity was found only in the BM (Supplementary Fig. S1a; Supplementary Data are available online at www.liebertpub.com/hum).
Figure 2.
Molecular chimerism. Molecular chimerism measured through flow cytometry as percentage of green fluorescent cells (a) and lentiviral integrations per cell measured by qRT-PCR, using an EGFP-specific probe referred to the single-copy mouse gene angiogenin-1 (b) in white blood cells, BM, and spleen of sham (n = 10) and TP-treated (n = 10) dKO mice 6 months after treatment. In order to facilitate matching the results, each mouse in the TP-treated group is identified with the same symbol. Horizontal lines indicate median. Statistical analyses were performed with the nonparametric Mann–Whitney U-test (*p < 0.05, **p < 0.01).
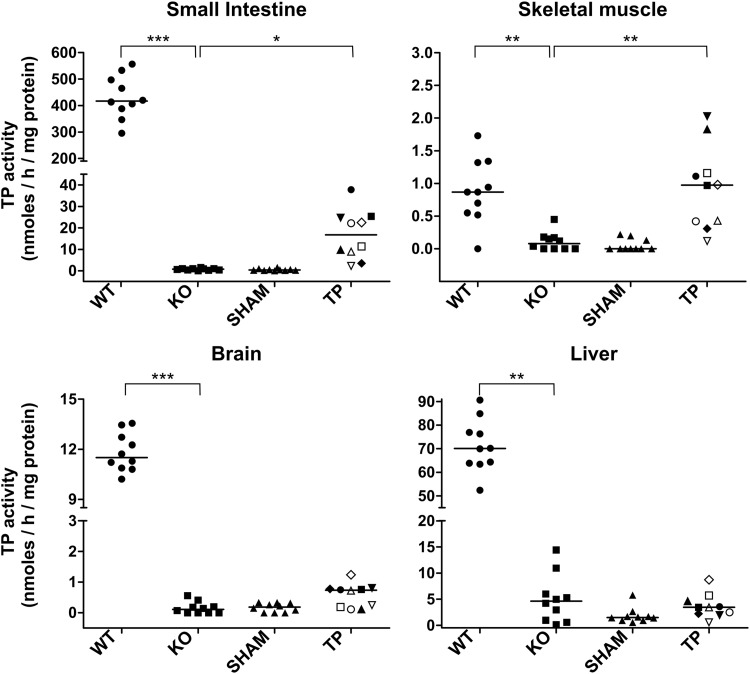
To explore the possibility of TP activity restoration in nonhematopoietic tissues as a consequence of resident cells of hematopoietic origin, we measured TP activity in organs that are usually affected in MNGIE patients (Fig. 3). In wt mice, the highest TP activity was found in small intestine (median 417 nmol Thy/hr/mg prot), and the lowest activity was found in skeletal muscle (median 0.9 nmol Thy/hr/mg prot), among the analyzed tissues. Similar values had been reported elsewhere.19 TP activity was not restored in liver, and was barely increased in brain and small intestine in treated animals; TP activity was fully restored in skeletal muscle, although the significance of this finding is limited because wt TP activity levels in this tissue are close to the lower limit of quantification of the method (0.5 nmol Thy/hr/mg prot). Interestingly, in TP-treated animals, moderate but significant correlations were observed between TP activities in small intestine, BM, blood cells, and spleen (Supplementary Fig. S1b).
Figure 3.
TP activity in different tissues. TP activity in liver, brain, small intestine, and gastrocnemius muscle from wt (WT), nontreated (KO), p-sham-treated (SHAM), and p-TP-treated (TP) Tymp/Upp1 dKO mice 6 months after treatment. Horizontal lines indicate median. In order to facilitate matching the results, each mouse in the TP-treated group is identified with the same symbol. Statistical analysis were performed with the nonparametric Dunn's multiple comparison test (*p < 0.05, **p < 0.01, ***p < 0.001).
Systemic nucleoside reduction
Six months after treatment, plasma dThd and dUrd concentrations were significantly reduced in all dKO mice treated with p-TP (p ≤ 0.001), although not all mice reached wt levels. In 60% of the animals, plasma dThd and dUrd concentrations decreased to wt levels or below (Fig. 4) (wt, dThd range 1.3–4.2 μM, dUrd range 1.7–5.1 μM; TP-treated dKO, dThd range 0.2–6.9 μM, dUrd range 1.8–6.9 μM) (Fig. 4). Reduction of dThd and dUrd levels was also observed in all tissues analyzed (Fig. 4). The reductions were more pronounced in spleen and brain (where dThd and dUrd levels reached concentrations similar to wt levels), but were only partial in liver, skeletal muscle, and small intestine. The sum of dThd and dUrd levels in plasma and all tissues analyzed (except small intestine) negatively correlated with TP activity in BM, spleen, and blood cells (Supplementary Table S1). However, when we compared nucleoside levels (dThd+dUrd) versus TP activity within a given tissue, only blood (r = −0.927; p < 0.001, Spearman) and spleen (r = −0.912; p < 0.001) showed significant correlations between these two variables, whereas no correlations were found in the other tissues (brain, liver, skeletal muscle, and small intestine), indicating that the degree of the nucleoside reduction directly depends on the TP activity provided by the hematopoietic cells.
Figure 4.
Systemic nucleoside clearance. dThd and dUrd concentration in plasma, spleen, brain, liver, skeletal muscle, and small intestine from wt (WT, n = 10), nontreated (KO, n = 10), p-sham-treated (SHAM, n = 10), and p-TP-treated (TP, n = 10) Tymp/Upp1 dKO mice 6 months after treatment. Horizontal lines indicate median. In order to facilitate matching the results, each mouse in the TP-treated group is identified with the same symbol. Statistical analyses were performed with the parametric Dunnett's multiple comparison test (*p < 0.05, **p < 0.01, ***p < 0.001).
The effect of the treatment is sustained long-term
Eighteen months after transplantation, the molecular chimerism in TP-treated mice (assessed as the percentage of EGFP+ cells) ranged from 3.1% to 50% in BM and from 0.4% to 68.1% in blood cells. The number of lentiviral integrations per cell quantified by qRT-PCR in BM and blood cells was less than one copy per cell in all animals (Table 1). TP activity in these tissues exceeded wt levels in 3 of the 5 animals treated (mice #3, #4, and #5) and correlated with the percentage of EGFP+ cells (Table 1). Plasma and liver nucleoside concentrations were also reduced to normal levels or below in these animals. In the two remaining TP-treated animals (mice #1 and #2), TP activity in blood cells and BM was negligible, correlating with the low levels of molecular chimerism. Consistently, plasma nucleoside levels remained high in these two animals. Surprisingly, liver dThd levels in one of these mice (#1) were in the range of wt animals.
Table 1.
Summary of long-term molecular and biochemical variables in treated mice
| Molecular chimerism (% EGFP+) | Copy number per cell | TP activitya | dThd levelsb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | N | ID | Age (weeks) | Blood cells | BM | Blood cells | BM | Blood cells | BM | Liver | Plasma | Liver |
| WT | 9 | — | 84.5 (84–85) | ND | ND | ND | ND | 0.08 (0.02–0.12) | 39.0 (21.5–69.9) | 51.4 (27.2–71.5) | 2.5 (0.9–4.5) | 29 (11–70) |
| KO | 9 | — | 85.6 (82–89) | ND | ND | ND | ND | Und | 1.4 (Und −5.1) | 2.0 (0.5–5.8) | 26.3 (6.4–171.4) | 245 (95–1356) |
| SHAM | 7 | — | 84.6 (80–86) | 15.5 (0.4–25.9) | 9.4 (1.4–15.4) | 0.51 (0.16–0.85) | 0.27 (0.14–0.34) | Und | 1.5 (Und −3.5) | 7.8 (0.8–25.6) | 10.1 (3.0–14.8) | 134 (79–272) |
| TP | 5 | 1 | 85 | 0.7 (1.7)c | 3.4 | 0.19 | 0.16 | 0.03 | 3.3 | 2.4 | 7.0 | 57 |
| 2 | 85 | 0.4 (0.5)c | 3.1 | 0.06 | 0.21 | 0.02 | 6.0 | 3.7 | 13.0 | 99 | ||
| 3 | 85 | 3.9 (10.1)c | 10.8 | 0.09 | 0.2 | 6.1 | 94.6 | 3.8 | 2.0 | 62 | ||
| 4 | 85 | 10.0 (8.8)c | 17.9 | 0.22 | 0.37 | 15.5 | 237.8 | 6.6 | ND | 23 | ||
| 5 | 76 | 68.1 (19.2)c | 50.0 | 0.89 | 0.87 | 90.4 | 824.3 | 31.3 | ND | 8 | ||
TP activity is measured as nmol Thy/hr/mg protein. Values are shown as mean (range).
dThd concentration in μM in plasma (nmol/mg prot in liver). Values are shown as mean (range).
Molecular chimerism 18 months after transplantation (in parentheses, molecular chimerism 1 month after transplantation).
BM, bone marrow; KO, knockout; ND, not determined; TP, thymidine phosphorylase; Und, undetectable (dThd <0.45 μM; blood cells TP activity <0.02; BM and liver TP activity <0.5); WT, wild type.
Molecular chimerism in blood cells did not decline in old animals; mice #1 and #2 displayed very low levels of chimerism from the first month after treatment, which were maintained 18 months after the treatment (20 months of age); chimerism was maintained in mouse #4, and increased in mouse #5. In mouse #3, chimerism decreased from 10% to 4%, but was sufficient to maintain TP activity in blood and low nucleoside levels in plasma (Table 1).
Effect on survival
Kaplan–Meier analysis showed that untreated wt and dKO mice had no differences in survival, which was reduced in the treated groups regardless of whether they received sham- or TP-corrected cells (mean of survival for TP and sham groups was 70 and 77 weeks, respectively, and 91 weeks for wt and untreated dKO mice) (Fig. 5a and Supplementary Table S2). These results indicate that the treatment is associated with an increase in mortality, but not because of the effect of the transgene, as survival rates were similar in animals treated with p-sham and p-TP. To elucidate if the higher mortality was associated with the transplantation procedure, we studied an additional group of animals transplanted with nontransduced total BM cells from syngeneic wt donors, following the conditioning and transplantation protocols used for the animals treated with p-sham and p-TP. The results showed that mice transplanted with nontransduced BM survived for an average of 75 weeks, similar to that observed in p-sham-transduced or p-TP-transduced transplanted animals (Fig. 5a and Supplementary Table S2). This suggests that the reduction of lifespan is related to the transplantation procedure rather than to the lentiviral transduction or the TP overexpression.
Figure 5.
Survival curve and blood cell counts. (a) Kaplan–Meier survival representation of wt (n = 111), nontreated (n = 181), p-sham-treated (n = 21), p-TP-treated (n = 22), and BM-transplanted (n = 9) dKO mice. (b) Blood cell counts in wt (n = 9), nontreated (n = 10), p-sham-treated (n = 7), and p-TP-treated (n = 5) dKO mice 18 months after treatment (20-month-old mice). Box plots represent the median (horizontal line), interquartile range (box), and minimum and maximum (whiskers), except outliers, which are depicted as dots.
To rule out undesired hematopoietic toxicity attributable to the lentiviral transduction and/or ectopic TP expression, blood cell counts were performed periodically and at the end of the study. No differences in white blood cell counts were observed among groups (Fig. 5b).
dNTP homeostasis
In vitro and in vivo models of MNGIE show that dThd overload results in mitochondrial dNTP pool imbalances characterized by increased dTTP and secondary dCTP depletion.6,8,9 We measured mitochondrial dNTPs in livers of 14-month-old treated and untreated mice (Fig. 6). Mitochondrial dCTP level was significantly lower in untreated and p-sham-treated KO animals, as compared with the wt values. Surprisingly, dTTP levels did not significantly increase in untreated dKO mice, but clearly did in mice treated with p-sham, and the same pattern was observed for mitochondrial dGTP. This enhanced biochemical effect observed in the mice treated with p-sham coincides with a more pronounced nucleoside accumulation in these mice (Supplementary Fig. S2). When compared with mice treated with p-sham, mitochondrial dCTP, dTTP, and dGTP levels were restored to wt values in the group treated with the TP-vector. Results obtained for mitochondrial dATP were unreliable because they were very close to the lower limit of quantification or undetectable (data not shown).
Figure 6.
Liver mitochondrial dNTP concentrations. Mitochondrial dNTP concentration in liver from wt (WT, n = 10), nontreated (KO, n = 10), p-sham-treated (SHAM, n = 7), and p-TP-treated (TP, n = 5) dKO mice 12 months after treatment. Box plots represent the median (horizontal line), interquartile range (box), and minimum and maximum (whiskers), except outliers, which are depicted as open circles. Asterisks indicate significant differences with WT (*p < 0.05; **p < 0.001; ***p < 0.0001, Student's t-test).
Discussion
All therapeutic approaches for MNGIE have been focused on reducing systemic dThd and dUrd overload, and most have shown biochemical efficacy, though in some cases, such as hemodialysis, the reductions are transient and suboptimal.20–23 Eznyme replacement through erythrocyte-entrapped TP administration has shown to improve the health in MNGIE patients, although the biochemical correction was only partial.21,24 Only allo-HSCT has been demonstrated to be metabolically and clinically effective and associated with objective evidence of health improvement in the long-term.13,14 However, this procedure has produced high morbidity and mortality rates in MNGIE patients.13
In recent years, gene therapy has emerged as a promising alternative for MNGIE patients. Our previous work has demonstrated that ex vivo hematopoietic stem cell gene therapy (HSCGT) using a lentiviral vector was able to provide ectopic TP activity in blood cells for at least six months after transplantation of sublethally irradiated Tymp/Upp1 dKO mice. Lentiviral-mediated TP expression restored normal dThd and dUrd homeostasis in this mouse model one month after treatment.15 In this follow-up study, we expand our previous results that support two important concepts. First, that TP activity provided by lentiviral HSCGT as well as its effects on systemic nucleoside homeostasis (i.e., restoration of dThd and dUrd concentrations to wt levels in blood and tissues) are maintained throughout the lifespan of treated mice. Although the mortality rate of the animals was significantly increased by the transplantation procedure, which limited the number of mice available for postmortem studies at advanced age, we were still able to study biochemically and molecularly 5 animals 18 months after treatment. None lost the gene-modified cells or the transgene expression. Two animals had low levels of chimerism, negligible TP activity, and increased nucleoside levels in blood 18 months after treatment, but those mice already had very low chimerism early after treatment. For unknown reasons, ex vivo transduction and/or engraftment after transplant were not efficient in these two animals. A primary graft failure related to the quality or the viability of the BM cells transplanted or the presence of an immune response to the transgene products cannot be ruled out. On the other hand, the 3 animals with moderate chimerism levels immediately after treatment maintained relatively good levels of chimerism and TP activity 18 months after the treatment. Overall, these results indicate that, when transduction and engraftment are successful (i.e., moderate levels of chimerism achieved after transplant), TP function is maintained and is effective in lowering nucleosides over the lifespan of the mice. Other preclinical studies in larger model animals have demonstrated that lentiviral HSCGT is stable during six years after transplantation.25 Furthermore, there are running clinical trials using lentiviral HSCGT for X-linked adrenoleukodystrophy and Wiskott–Aldrich syndrome that report long-term therapeutic effect for more than three years.26,27 These observations support the notion that lentiviral HSCGT may provide stable TP expression in MNGIE patients.
The second important finding of our study is that partial restoration of TP activity, irrespective of the organ or tissue where it is expressed, is sufficient to clear dThd and dUrd systemically. Indeed, the present results confirm that moderate levels of molecular chimerism in blood (around 10%) are sufficient to reach biochemical efficacy, and nucleosides are cleared from all organs even though TP activity is only present in hematopoietic cells. Therefore, the use of a moderate and well-accepted low genotoxic promoter such as hPGK, previously used in a lentiviral-based clinical trial,28 suffices to restore the nucleoside homeostasis in our model. This represents an advantage in terms of safety, because the need for stronger promoters would be associated with higher risks of genotoxicity attributable to potential transactivation of oncogenes.29
Hematopoietic progenitors transduced with the therapeutic lentiviral vector were infused in dKO mice after sublethal total body irradiation. As a result, treated animals expressed TP in hematopoietic tissues (blood cells, BM, and spleen). TP restoration was very low in other tissues; minute or moderate increases of TP activity were observed in brain, skeletal muscle, and small intestine. Blood contamination of the tissue sample likely contributed to these activities, as organs were collected without prior exsanguination. Nevertheless, we cannot rule out the contribution of tissue-resident cells of hematopoietic origin, such as macrophages or other cell types, as suggested by the fact that TP activity (expressed per mg of protein) in the small intestine is similar or even higher than that observed in blood cell extracts. Nucleoside reduction observed in skeletal muscle, brain, and liver correlated with TP activity in BM, blood cells, and spleen but not with their own TP activity, indicating that systemic dThd and dUrd clearance is mainly carried out by the system, supporting the notion that the biochemical defect in MNGIE can be repaired by molecular correction in any tissue or cell type. Furthermore, we have recently demonstrated that another gene therapy strategy using an AAV targeted at the liver provides enough TP activity to this tissue to clear systemic nucleoside accumulation.19
It should be noted that, in contrast with other tissues analyzed, reductions of nucleoside concentrations in the small intestine were only partial, and this observation coincides with our prior studies using a liver-targeted AAV to deliver TP.19 The reasons for the difficulty in clearing excess nucleoside from the intestine in the animal model are unknown. A substantial proportion of the nucleosides found in this organ could be derived from direct absorption from the diet; therefore, continuous diet supply would be hindering the total removal of these compounds by the blood stream. Efforts should be focused on understanding the specific difficulty in clearing nucleosides from small intestine, and on finding the way to improve it, as the small intestine is one of the most clinically affected organs in MNGIE.11
One of the consequences of dThd and dUrd overload in MNGIE is the imbalance of the mitochondrial dNTP pool. Observations from in vitro and in vivo studies have demonstrated that dThd excess induces dTTP expansion and dCTP depletion in mitochondria, which interferes with mtDNA replication.6,8,9 Our results here reveal a modest dNTP imbalance in liver mitochondria of dKO mice, as compared with those of their wt counterparts. In fact, this dNTP imbalance (including a dGTP expansion) is enhanced in mice treated with p-sham, coinciding with a more pronounced nucleoside accumulation in this group. Our findings recapitulate in vitro and in vivo results previously reported by others.10,30 The reasons why dNTP imbalances are more pronounced in the animals treated with p-sham are not clear. Irradiation and transplantation procedures may perturb nucleotide metabolism of the mice; however, the mice were treated at 2 months of age, and dNTPs were analyzed at 14 months, and it is unlikely that alterations in dNTP metabolism early in the life of the animals would have such long-lasting effects. In any case, the treatment with the therapeutic vector largely prevented the dNTP imbalance observed in mice treated with p-sham, which further supports the positive effects of the treatment.
One of the aims of our long-term study was to assess the effects of treatment on other phenotype traits of the animal model, as it has been previously reported that old animals develop mtDNA depletion, histological alterations in brain, and leukoencephalopathy.6 However, we did not find differences in brain and liver mtDNA copy number, brain magnetic resonance imaging, or histological changes in muscle or brain in our experimental mice. In fact, in the original report the magnitude of these phenotypic differences was small and elusive6; therefore, different conditions of the colony in our animal facilities may account for this discrepancy. The limitation of this murine model of the disease (the only one so far available) constitutes a significant issue for preclinical in vivo studies; the absence of clinical phenotype makes it difficult to evaluate whether correction of the biochemical imbalances is an appropriate demonstration of efficacy of the treatment. However, there are repeated observations that the normalization (or at least, significant reductions) of systemic dThd and dUrd levels achieved by bone marrow transplantation,13 peritoneal dialysis,23 or infusion of erythrocyte-entrapped TP24 is associated with transient or long term-maintained clinical improvement in MNGIE patients. Therefore, the demonstration that a therapy is able to restore the biochemical homeostasis in the murine model should be combined with the increasing evidence, obtained from other treatments tested in MNGIE patients, that such biochemical restoration is beneficial.
In addition, it has been shown that feeding the dKO animals with dThd and dUrd exacerbates the phenotype in the long-term, leading to reduced survival, body weight, and muscle strength, as well as enhanced leukoencephalopathy, reduction of smooth muscle cells, and increased fibrosis in the small intestine.30 These new findings constitute a relevant improvement and a valuable tool to test the therapy and will allow us to definitively show in this in vivo model whether this and other therapy approaches are effective improving the clinical endpoints.
The most concerning issue of HSCGT using lentiviral or other integrative vectors is the risk of insertional oncogenesis, as it was observed in initial clinical trials performed with gamma retroviral vectors.31,32 The development of less genotoxic self-inactivating (SIN) vectors33 has contributed to recent successes of HSCGT in clinical trials for X-linked SCID, β-thalassemia, Wiskott–Aldrich syndrome, metachromatic leukodystrophy, and X-linked adrenoleukodystrophy, with no adverse effects observed so far.27,28,34–37 We used a SIN vector, and no oncogenic events associated to the treatment with the sham or TP vectors have been observed over 18 months, as assessed by macroscopic survey and inspection of the living animals, more specifically by periodic complete blood cell counts, and in necropsies. In addition, these analyses indicated that blood cell differentiation was not affected by the hematopoietic TP overexpression. It is noteworthy that we recently demonstrated that, in TP-deficient mice, TYMP can act as a suicide gene in combination with capecitabine, a commonly used anticancer drug, and this would represent a valuable safeguard if MNGIE patients undergoing gene therapy were to develop tumors because of insertional oncogenesis.16
Although no specific adverse effects were caused by the use of the vectors (neither the sham nor the TP vectors), the transplantation procedure, which includes irradiation and BM transplantation, was associated with a lower survival rate. It is beyond the scope of this study to address the issue of the secondary effects of the conditioning and hematopoietic transplantation, but this limitation should be obviously considered when balancing the potential benefits and risks of HSCGT for MNGIE. In this regard, it should be noted that the gene transfer protocol can still be optimized to achieve higher transduction rates in the hematopoietic progenitors, which would allow us the use of milder myeloablative methods based on chemotherapeutic regimes currently used in autologous transplantation, reducing the toxicity of the procedure. A HSCGT clinical trial for Wiskott–Aldrich syndrome showed that reduction of flurabadine and busulfan doses to 50% and 30% of those used for standard allogeneic transplantation results in multilineage engraftment of gene-corrected cells in BM and blood.37 Our results in the murine model indicate that relatively low levels of correction that can be achieved with reduced intensity conditioning regimens currently available could also correct the metabolic abnormalities in humans. On the other hand, even if high-intensity myeloablative conditioning regimens were required (as occurs in patients with thalassemia, metachromatic leukodystrophy, or adrenoleukodystrophy),28,34,36 this toxicity could still be acceptable in comparison with that of allogeneic transplantation, which in these particular patients is extremely risky.13
Our preclinical studies have demonstrated that submyeloablative HSCGT using a SIN lentiviral vector containing a functional TYMP is able to restore nucleoside homeostasis in the animal model of MNGIE during the entire life of the mice. This should be considered in the context of the recent demonstration that the use of an AAV2/8 vector transcriptionally targeted at the liver also resulted in the long-term restoration of nucleoside homeostasis, using the same animal model.19 Based on these results and on their safe profiles, we consider the use of an AAV vector as a potential first option in MNGIE. Nevertheless, there will be a percentage of patients who would not be eligible for such strategy because the presence of liver disease, which is not uncommon in MNGIE patients,11 or the existence of anti-AAV8-neutralizing antibodies would preclude the use of this strategy.38 Hence, these patients would benefit from the HSCGT strategy studied here. In summary, gene therapy using different target tissues and vectors offers plausible alternatives to treat MNGIE, and the implementation of clinical trials to demonstrate their safety and efficacy warrants serious consideration.
Supplementary Material
Acknowledgments
This work was supported by the Instituto de Salud Carlos III (Grant PI12/00322 to R.M., co-funded with FEDER funds), the United Mitochondrial Disease Foundation (Postdoctoral Grant 12–029 to J.T.-T.), and the French Muscular Dystrophy Association–Téléthon (AFMTéléthon Postdoctoral Grant 18247 to J.T.-T.).
Author Disclosure
No competing financial interests exist.
References
- 1.Hirano M, Garcia-de-Yebenes J, Jones AC, et al. Mitochondrial neurogastrointestinal encephalomyopathy syndrome maps to chromosome 22q13.32-qter. Am J Hum Genet 1998;63:526–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirano M, Nishigaki Y, Marti R. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): A disease of two genomes. Neurologist 2004;10:8–17 [DOI] [PubMed] [Google Scholar]
- 3.Hirano M, Silvestri G, Blake DM, et al. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): Clinical, biochemical, and genetic features of an autosomal recessive mitochondrial disorder. Neurology 1994;44:721–727 [DOI] [PubMed] [Google Scholar]
- 4.Desgranges C, Razaka G, Rabaud M, et al. Catabolism of thymidine in human blood platelets: Purification and properties of thymidine phosphorylase. Biochim Biophys Acta 1981;654:211–218 [DOI] [PubMed] [Google Scholar]
- 5.Marti R, Nishigaki Y, Vila MR, et al. Alteration of nucleotide metabolism: A new mechanism for mitochondrial disorders. Clin Chem Lab Med 2003;41:845–851 [DOI] [PubMed] [Google Scholar]
- 6.Lopez LC, Akman HO, Garcia-Cazorla A, et al. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum Mol Genet 2009;18:714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pontarin G, Ferraro P, Valentino ML, et al. Mitochondrial DNA depletion and thymidine phosphate pool dynamics in a cellular model of mitochondrial neurogastrointestinal encephalomyopathy. J Biol Chem 2006;281:22720–22728 [DOI] [PubMed] [Google Scholar]
- 8.Ferraro P, Pontarin G, Crocco L, et al. Mitochondrial deoxynucleotide pools in quiescent fibroblasts: A possible model for mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). J Biol Chem 2005;280:24472–24480 [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Vioque E, Torres-Torronteras J, Andreu AL, et al. Limited dCTP availability accounts for mitochondrial DNA depletion in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). PLoS Genet 2011;7:e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song S, Wheeler LJ, Mathews CK. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J Biol Chem 2003;278:43893–43896 [DOI] [PubMed] [Google Scholar]
- 11.Garone C, Tadesse S, Hirano M. Clinical and genetic spectrum of mitochondrial neurogastrointestinal encephalomyopathy. Brain 2011;134:3326–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halter J, Schupbach WM, Casali C, et al. Allogeneic hematopoietic SCT as treatment option for patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): A consensus conference proposal for a standardized approach. Bone Marrow Transplant 2010;46:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halter JP, Michael W, Schupbach M, et al. Allogeneic haematopoietic stem cell transplantation for mitochondrial neurogastrointestinal encephalomyopathy. Brain 2015;138:2847–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano M, Marti R, Casali C, et al. Allogeneic stem cell transplantation corrects biochemical derangements in MNGIE. Neurology 2006;67:1458–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres-Torronteras J, Gomez A, Eixarch H, et al. Hematopoietic gene therapy restores thymidine phosphorylase activity in a cell culture and a murine model of MNGIE. Gene Ther 2011;18:795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Estevez S, Ferrer G, Torres-Torronteras J, et al. Thymidine phosphorylase is both a therapeutic and a suicide gene in a murine model of mitochondrial neurogastrointestinal encephalomyopathy. Gene Ther 2014;21:673–681 [DOI] [PubMed] [Google Scholar]
- 17.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–254 [DOI] [PubMed] [Google Scholar]
- 18.Valentino ML, Marti R, Tadesse S, et al. Thymidine and deoxyuridine accumulate in tissues of patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). FEBS Lett 2007;581:3410–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Torronteras J, Viscomi C, Cabrera-Perez R, et al. Gene therapy using a liver-targeted AAV vector restores nucleoside and nucleotide homeostasis in a murine model of MNGIE. Mol Ther 2014;22:901–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lara MC, Weiss B, Illa I, et al. Infusion of platelets transiently reduces nucleoside overload in MNGIE. Neurology 2006;67:1461–1463 [DOI] [PubMed] [Google Scholar]
- 21.Moran NF, Bain MD, Muqit MM, et al. Carrier erythrocyte entrapped thymidine phosphorylase therapy for MNGIE. Neurology 2008;71:686–688 [DOI] [PubMed] [Google Scholar]
- 22.Spinazzola A, Marti R, Nishino I, et al. Altered thymidine metabolism due to defects of thymidine phosphorylase. J Biol Chem 2002;277:4128–4133 [DOI] [PubMed] [Google Scholar]
- 23.Yavuz H, Ozel A, Christensen M, et al. Treatment of mitochondrial neurogastrointestinal encephalomyopathy with dialysis. Arch Neurol 2007;64:435–438 [DOI] [PubMed] [Google Scholar]
- 24.Bax BE, Bain MD, Scarpelli M, et al. Clinical and biochemical improvements in a patient with MNGIE following enzyme replacement. Neurology 2013;81:1269–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beard BC, Kiem HP. Canine models of gene-modified hematopoiesis. Methods Mol Biol 2009;506:341–361 [DOI] [PubMed] [Google Scholar]
- 26.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol 2012;507:187–198 [DOI] [PubMed] [Google Scholar]
- 27.Hacein-Bey Abina S, Gaspar HB, Blondeau J, et al. Outcomes following gene therapy in patients with severe Wiskott-Aldrich syndrome. JAMA 2015;313:1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biffi A, Montini E, Lorioli L, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013;341:1233158. [DOI] [PubMed] [Google Scholar]
- 29.Cesana D, Ranzani M, Volpin M, et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol Ther 2014;22:774–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Diaz B, Garone C, Barca E, et al. Deoxynucleoside stress exacerbates the phenotype of a mouse model of mitochondrial neurogastrointestinal encephalopathy. Brain 2014;137:1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 2003;348:255–256 [DOI] [PubMed] [Google Scholar]
- 32.Stein S, Ott MG, Schultze-Strasser S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med 2010;16:198–204 [DOI] [PubMed] [Google Scholar]
- 33.Montini E, Cesana D, Schmidt M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest 2009;119:964–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 2009;326:818–823 [DOI] [PubMed] [Google Scholar]
- 35.Hacein-Bey-Abina S, Pai SY, Gaspar HB, et al. A modified gamma-retrovirus vector for X-linked severe combined immunodeficiency. N Engl J Med 2014;371:1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 2010;467:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with wiskott-aldrich syndrome. Science 2013;341:1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boutin S, Monteilhet V, Veron P, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Hum Gene Ther 2010;21:704–712 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.