SUMMARY
Termination of protein synthesis on the ribosome is catalyzed by release factors (RFs), which share a conserved glycine-glycine-glutamine (GGQ) motif. The glutamine residue is methylated in vivo, but a mechanistic understanding of its contribution to hydrolysis is lacking. Here we show that the modification, apart from increasing the overall rate of termination on all dipeptides, substantially increases the rate of peptide release on a subset of amino acids. In the presence of unmethylated RFs, we measure rates of hydrolysis that are exceptionally slow on proline and glycine residues and ~two orders of magnitude faster in the presence of the methylated factors. Structures of 70S ribosomes bound to methylated RF1 and RF2 reveal that the glutamine side-chain methylation packs against 23S rRNA nucleotide 2451 stabilizing the GGQ motif and placing the side-chain amide of the glutamine towards tRNA. These data provide a framework to understanding how release factor modifications impact termination.
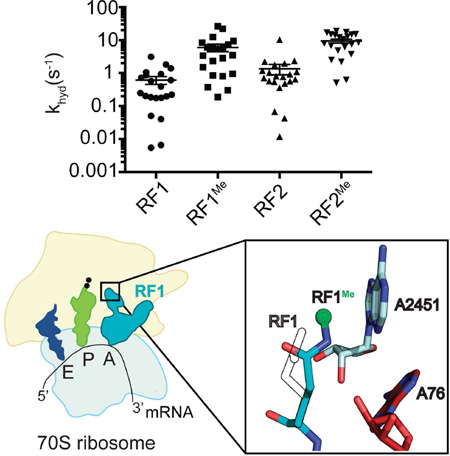
Graphical abstract
eTOC SUMMARY
Methylation of release factors is a universal process found in all organisms. Pierson et al. show that the modification substantially increases rates of peptide release on a subset of amino acids that are otherwise slow. Structures of the methylated factors show repositioning of the active-site glutamine in the peptidyl-transferase center.
Introduction
Protein synthesis is an intricate process that involves many coordinated events on the ribosome ensuring that the genetic code is decoded accurately and efficiently. Termination of protein synthesis occurs when one of three nearly universally conserved stop codons (UAA, UGA and UAG) enters the A site of the ribosome. Stop codons are recognized by protein factors called release factors (RF), which mediate the hydrolytic reaction in the peptidyl transferase center (PTC) of the ribosome. Unlike eukaryotes and archaea, which have one factor (eRF1 and aRF1, respectively) that recognizes all stop codons (Konecki et al., 1977), bacteria have two very similar factors with overlapping specificities (Scolnick et al., 1968). RF1 recognizes UAG and UAA, whereas RF2 recognizes UGA and UAA.
Despite the conserved nature of the translation machinery in all domains of life, bacterial and eukaryotic RFs are unrelated and instead appear to have evolved independently (Frolova et al., 1994). The factors, however, share a universally conserved sequence motif of Gly-Gly-Gln (GGQ) (Frolova et al., 1999). This motif is critical for the hydrolytic reaction (Frolova et al., 1999) and early low-resolution structures of termination complexes revealed that the motif engages the PTC of the ribosome (Petry et al., 2005; Rawat et al., 2003). Mutation of either glycine to any other amino acid completely inhibits activity, whereas certain substitutions at the glutamine residue are only partially defective for promoting catalysis (Frolova et al., 1999; Shaw and Green, 2007). The ribosome also plays a critical role during the reaction. This is best highlighted by early observations showing that binding of a cognate deacylated tRNA to the A site accelerates the hydrolysis of the peptidyl-tRNA (Vogel et al., 1969). Interestingly, in the presence of a deacylated tRNA, the specificity for water as a nucleophile is compromised akin to what has been observed for the RF glutamine mutants (Shaw and Green, 2007). These findings suggest that RFs contribute largely to catalysis through inducing conformational rearrangements in the PTC that expose the ester carbon for the nucleophilic attack. Consistent with this proposal, mutations of the so-called inner shell nucleotides of the PTC as well as the 2’-OH of the terminal adenosine of the peptidyl tRNA are detrimental for RF-mediated release but not for peptidyl transfer (Pavlov et al., 2009; Polacek et al., 2003; Youngman et al., 2004; Zaher et al., 2011).
Crystal structures of post-termination complexes revealed important clues about the mechanism of the reaction. The glycine residues in the GGQ motif adopt a conformation in the PTC that is incompatible with any other amino acid rationalizing earlier mutational studies (Jin et al., 2010; Korostelev et al., 2008a; Korostelev et al., 2010; Laurberg et al., 2008; Weixlbaumer et al., 2008; Zhou et al., 2012). In contrast, the role of the glutamine in catalysis has been a source of considerable debate. One group argued that the main-chain amide of the glutamine is involved in coordinating a nucleophilic water molecule (Laurberg et al., 2008). Another group argued for both the main-chain amide and the side-chain carbonyl oxygen playing a critical role in coordinating the hydrolytic water (Weixlbaumer et al., 2008). The conserved nature of the residue adds support to the side chain of glutamine contributing in some way to the mechanism.
It is worth noting that although the 3.0–3.5 Å 70S-RF structures are at resolutions typically sufficient to observe interpretable electron density of protein side chains, there is little to no electron density surrounding the unmethylated glutamine residue in all structures but the pre-termination 70S-RF2 complex and the recent post termination 70S-E. coli RF1 (Figure S1) (Jin et al., 2010; Korostelev et al., 2008a; Korostelev et al., 2010; Laurberg et al., 2008; Weixlbaumer et al., 2008; Zhou et al., 2012). Therefore, it is still ambiguous whether the glutamine side chain carbonyl or amide is directed towards the 3’-OH of the substrate tRNA A76, where it could potentially coordinate the catalytic water molecule (Shaw and Green, 2007). Recent kinetic analyses examining the pKa of the termination reaction suggested instead that a hydroxide ion could participate in the reaction with the glutamine side chain amide group donating a proton to the hydroxide to stabilize the reaction (Indrisiunaite et al., 2015).
The glutamine residue of the GGQ motif in bacterial and eukaryotic RFs is N5-methylated. In E. coli, RFs are methylated by prmC (Heurgue-Hamard et al., 2002). The methylation is required for optimal growth in certain minimal media (Mora et al., 2007) and in vitro, the methylation contributes modestly to the rate of release by RF2 (Dincbas-Renqvist et al., 2000). Therefore, the coordination of water in the active site of the ribosome could be further stabilized by the methylation. Indeed, molecular dynamics studies indicate that the methylation removes the side-chain amide of the glutamine from the active site and helps in orienting the carbonyl oxygen towards the active site to coordinate the catalytic water (Trobro and Aqvist, 2007, 2009). However, beyond these models, a mechanistic understanding for the role of the modification during peptide release is missing.
Here we explore the effects of methylation on the efficiency of peptide release using a well-defined bacterial reconstituted translation system. We measured the rates of peptide release on all 20 common amino acids with unmethylated and methylated RFs. While the modification was found to increase the rate of release on all amino acids, this enhancement varies significantly depending on the identity of the amino acid. We report four structures of unmethylated and methylated E. coli RF1 or RF2 in complex with the Thermus thermophilus (Tth) ribosome. In all structures, the placement of the glutamine backbone amide is within hydrogen bonding distance to the 3’-OH of the terminal A76 of the P-site tRNA. However, in the 70S-RF1Me post-termination structure, the side-chain amide packs against A2451 of 23S rRNA and is proximal to the tRNA, both suggesting an unexpected role of the side-chain amide in stabilizing the GGQ motif.
Results
Experimental approach
To explore the full contribution of the methylation of RFs on peptide release we took advantage of our in vitro bacterial reconstituted system to generate dipeptidyl-tRNA ribosomal nascent chains (DRNCs) programmed with a UAA stop codon in the A site (Figure S2A). Twenty initiation complexes (ICs) were generated by incubating 70S ribosomes with 20 mRNAs that had the following coding sequence (AUG NNN UAA) in the presence of f[35S]-Met-tRNAfMet, initiation factors 1, 2 and 3 and GTP. We note that the NNN sequence coded for the most abundant E. coli codon for each amino acid. DRNCs were generated by incubating the initiation complexes with their corresponding ternary complexes composed of the appropriate aa-tRNA, elongation factor Tu (EF-Tu) and GTP in the presence of elongation factor G (EF-G). Following peptide-bond formation and EF-G-catalyzed translocation, the complexes were purified over sucrose cushions to remove unincorporated factors. We assessed the yield of the peptidyl-transfer reaction using an electrophoretic TLC system to follow the release reaction (Youngman et al., 2004) (Figure S2B).
Uncatalyzed hydrolysis of dipeptidyl tRNAs on and off the ribosome
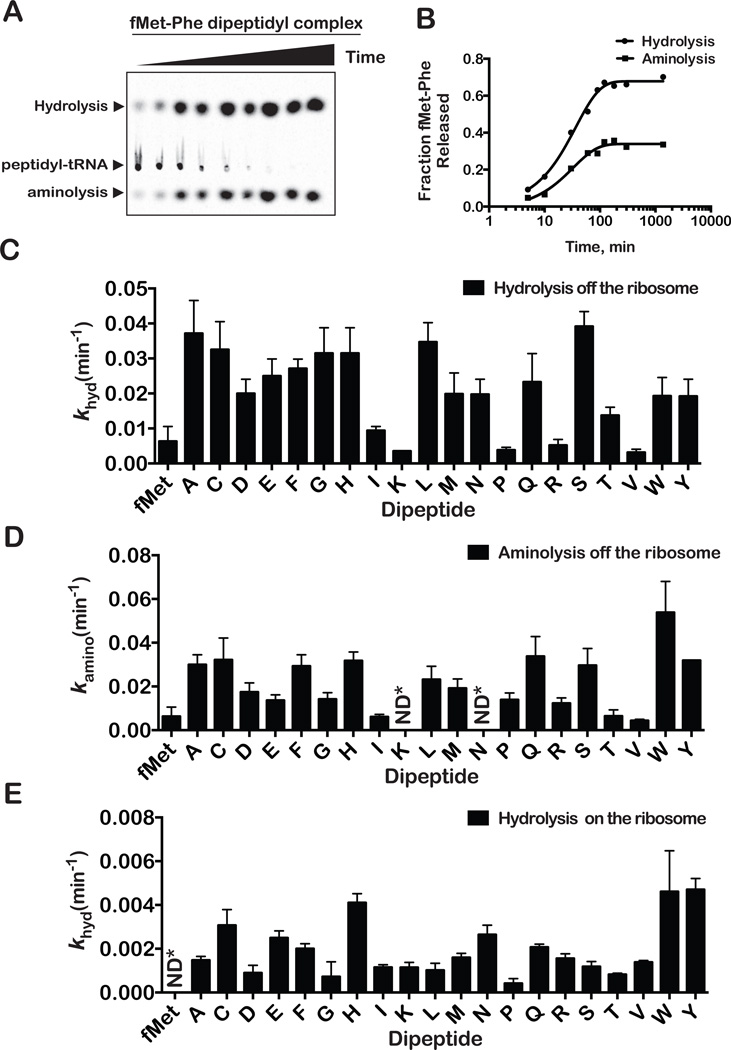
Previous experiments showed that the efficiency of nonsense suppression in E. coli is dependent on the amino acid prior to the stop codon (ultimate amino acid) (Bjornsson et al., 1996), suggesting that RF-mediated release is influenced by the identity of the C-terminal amino acid. Before exploring this hypothesis in our in vitro system, we sought to determine the rates of nonenzymatic release on all 20 dipeptidyl-tRNAs to assess if any differences observed is due to inherent chemical differences among amino acids. Nonenzymatic hydrolysis and aminolysis off the ribosome were initiated by diluting the complexes in the presence of EDTA, which facilitates splitting of the ribosomal subunits and the dissociation of the dipeptidyl-tRNA. To increase the rate of release, the pH of the dilution buffer was increased to 8.0 using Tris. Fortuitously, in these assays Tris serves as an analog for peptide bond formation, since similar to the α-amine group of aa-tRNA, its amine group attacks the peptidyl-tRNA bond to form an amide bond (Figures 1A and 1B). The rates we observed for hydrolysis and aminolysis (Figures 1C and 1D) were generally similar to one another for the same dipeptides, with one exception. Aminolysis rates were higher than hydrolysis rates for the fMet-Trp dipeptide. Apart from fMet-Ile, fMet-Lys, fMet-Pro, fMet-Arg, fMet-Thr and fMet-Val, which displayed overall reduced rates, the rates of hydrolysis/aminolysis were overall similar among the 20 dipeptides. The reduced rate for fMet-Pro can be easily rationalized given the unique structure of proline. Similarly, it appears that having a positive side chain stabilizes the ester bond of the dipeptidyl tRNA for lysine and arginine. The remaining fMet-Ile, fMet-Thr and fMet-Val all share the common feature of having a branched β-carbon, but beyond that no discernable similarities are obvious.
Figure 1. Nonenzymatic dipeptide hydrolysis and aminolysis.
(A) Representative electrophoretic TLC of a time course of nonenzymatic hydrolysis and aminolysis from free fMet-Phe peptidyl tRNA.
(B) Quantification of data in A. Fractional radioactivity corresponding to the released product was plotted against time and fit to one-phase exponential association equation.
(C) Dipeptide release rates calculated for hydrolysis of free peptidyl-tRNA.
(D) Dipeptide release rates calculated for aminolysis of free peptidyl-tRNA. Note the general similarity between hydrolysis and aminolysis rates.
(E) Dipeptide release rates calculated for nonenzymatic hydrolysis of ribosomal peptidyl-tRNA. The error bars represent the standard error obtained from the nonlinear regression. Most of the reactions were done in duplicates and the deviation was less than 20 % between the observed rates.
The environment in the PTC of the ribosome is very different from that in solution and is likely to affect the accessibility/orientation of the attacking water. We measured the nonenzymatic hydrolysis rates on the ribosome in the absence of release factor. Hydrolysis rates on the ribosome were significantly slower – 2 to 30 fold, but on average tenfold - than those determined off the ribosome (Figure 1E). Similar to the solution rates of hydrolysis, fMet-Pro exhibited the slowest rates on the ribosome. Apart from this similarity, we observe no correlation between the nonenzymatic solution rates to rates on the ribosome (Figure S3A), suggesting that the environment in the PTC has an effect on the rate of hydrolysis. We note, however, that the nonenzymatic rates on the ribosome, except for proline, do not vary significantly relative to what we observe for those measured off the ribosome (< fourfold, Figures 1D and 1E). Thus, it appears that the environment in the PTC provides some uniformity to the termination process.
GGQ N5-Methylation significantly increases the rate of peptide release on proline and glycine terminating peptides
Having established that the nonenzymatic rates of release do not vary significantly among the 20 amino acids, we set out to measure the RF-mediated rates of peptide release. Our experiments were motivated by previous in vivo studies, which suggested that peptide release is particularly inefficient on peptides terminating with proline and glycine residues (Bjornsson et al., 1996). In agreement with these studies, we measured greatly reduced rates of RF1-mediated peptide release for fMet-Pro and fMet-Gly of 0.0050 s−1 and 0.013 s−1 (Figure 2A), respectively. Similarly for the RF2 reaction we measured slow rates of 0.020 s−1 and 0.035 s−1 (Figure 2B). These rates are almost two orders of magnitude slower than those previously reported rate for fMet-Ala (Freistroffer et al., 2000) (Figures 2C and 2D). Furthermore, similar to what has been observed for other amino acids (Freistroffer et al., 2000), the addition of RF3 has no effect on RF1-and RF2-mediated peptide release on proline and glycine residues (Figure S4 and data not shown). As a result, our measurements suggest that peptide release is likely to be a major bottleneck for gene expression on proline- and glycine-terminating peptides. In direct contrast to this notion, an E. coli proteome-wide analysis shows that these residues are not underrepresented at the C-terminus (Figure S4C). Therefore, we wondered if the above rates we measured in vitro do not truly represent the in vivo efficiency of peptide release and that some other factor might be contributing to peptide release.
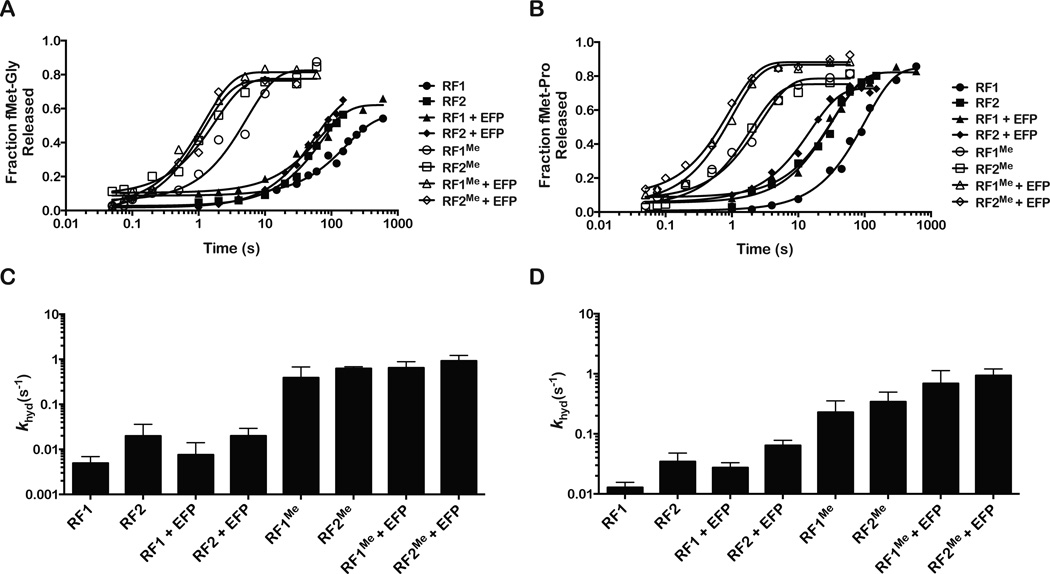
Figure 2. RF-mediated peptide release of fMet-glycyl and fMet-prolyl dipeptidyl tRNAs.
(A) Representative time courses of fMet-Gly and (B) fMet-Pro release in the presence of the indicated factors. For both dipeptides, only the addition of methylated RF1 or RF2 results in a significant increase in the rate of peptide release.
(C) fMet-Gly and (D) fMet-Pro dipeptide release rates under the indicated conditions. Error bars represent standard error of measurement calculated from at least three replicates.
Recently, it has been shown that during peptide-bond formation the elongation factor P (EF-P) increases the rate of peptide-bond formation on ribosomes stalled on dipropyl motifs (Doerfel et al., 2013; Ude et al., 2013). As a result we sought to determine whether the factor could also increase the rate of release of peptides having Cterminal proline and glycine residues. Lys34 of E. coli EF-P is modified post-translationally through the addition of a hydroxylated lysine residue by three enzymes (Peil et al., 2012). We generated modified EF-P by co-expressing the modifying enzymes (Doerfel et al., 2013) (Figure S5A), which resulted in high levels of modified protein as judged by LC MS/MS (Figure S5B). We further examined the function of the EF-P variants – unmodified, lysylated and hydroxylysylated - by assessing their effect on the yield of poly-proline peptides (Figure S5C). Having established that our modified EF-P is active, we next compared the release rates of fMet-Pro and fMet-Gly dipeptides from DRNC in the presence and absence of EF-P. Incubation with EF-P had little to no effect on the release rate of fMet-Pro and fMet-Gly regardless of the RF added (Figure 3 and S5D). These observations suggest that in contrast to peptide-bond formation EF-P does not significantly promote peptide release on proline and glycine residues.
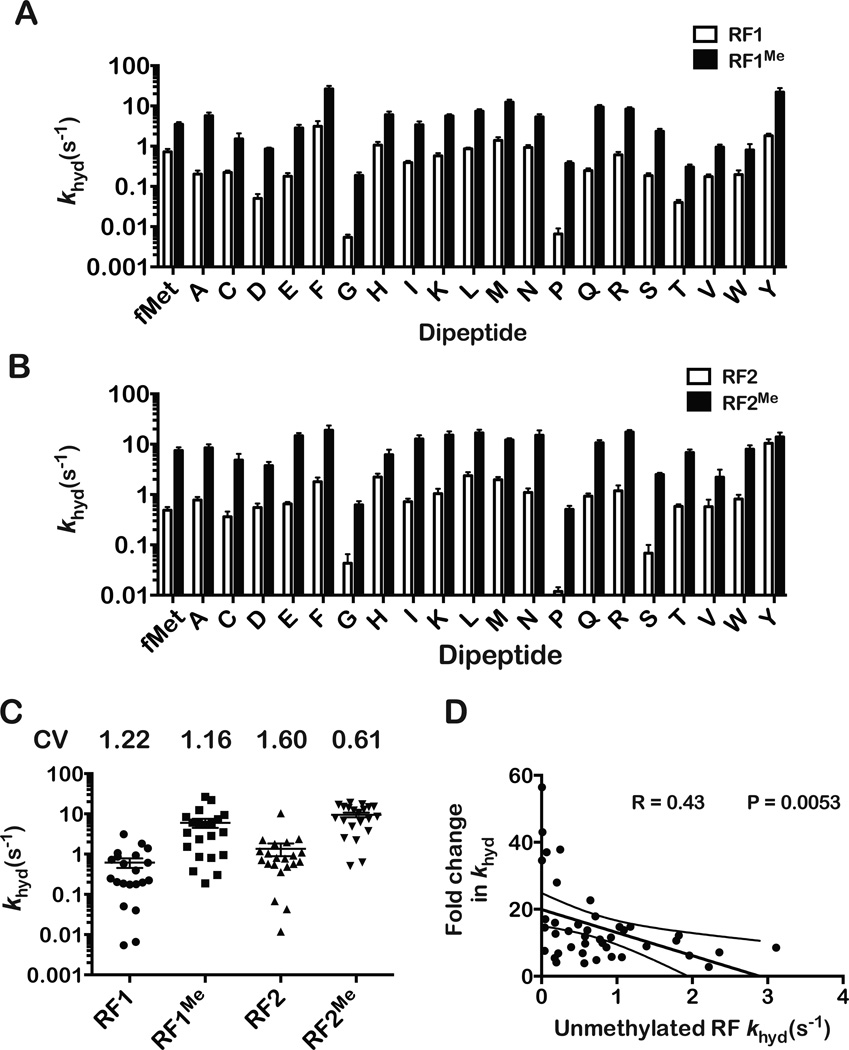
Figure 3. Effect of RF methylation on dipeptide release rate.
(A), (B) Comparison of dipeptide release rates for (A) RF1 and RF1Me and (B) RF2 and RF2Me. Note the near universal rate increase for methylated RFs. The error bars represent the standard error obtained from the nonlinear regression. For most dipeptides, reactions were carried out at least in duplicates and the observed variation was less than 20 % between the rates.
(C) Variation in dipeptide release rates for each release factor. Each data point corresponds to the release rate of a different dipeptidyl ribosome complex. Numbers above plot indicate the Coefficient of Variation (CV) for the release rates for each RF.
(D) Negative correlation between release rate for unmethylated RF and increase in release rate due to GGQ N5-methylation, consistent with the notion that methylation has a more significant effect on slow release reactions.
As discussed earlier, in E. coli the glutamine residue of the GGQ motif is N5-methylated by prmC (Heurgue-Hamard et al., 2002). Our experiments, so far, have been conducted with overexpressed release factors (Figure S5E), which exhaust prmC and as a result are unmethylated (Figure S5F). To address the effect of methylation on peptide release of fMet-Pro and fMet-Gly, we generated methylated RFs by coexpressing the methyltransferase prmC alongside the His-tagged RF. Using this strategy we obtained fully methylated RF1 (99.8%) and 75% methylated RF2 as assessed by LC MS/MS (Figure S5F and S5G). Although we expected the methylation to stimulate peptide release (Dincbas-Renqvist et al., 2000), we were surprised by its extent of stimulation on these particular dipeptides. Methylation of RF1 increased the rate of peptide release by ~70 fold and 30 fold for glycine and proline, respectively (Figure 2). Likewise, for RF2, methylation increased the rate of peptide release by 30 and 10 fold for glycine and proline, respectively (Figure 2). Similar to the unmethylated factors, the addition of RF3 had minimal effects on RF2Me-mediated release (Figure S4B). In contrast, the addition of EF-P to the methylated factors further increased the rate of release by 2 to 5 fold bringing the rates of peptide release to ~ 1 s−1 (Figure 2 and S5D). Collectively these findings suggest that methylation is critical for peptide release on a subset of amino acids.
Effects of GGQ N5-Methylation on peptide release on all amino acids
Our data on peptide release with glycine- and proline-terminating peptides suggest that methylation of release factor may play a larger role in catalysis than previously appreciated (Dincbas-Renqvist et al., 2000) and is likely to be specific to certain amino acids. To test this, we measured the khyd values for the remaining 18 amino acids by incubating our DRNCs with saturating concentrations of RF1 and RF2. In agreement with previous in vivo studies (Bjornsson et al., 1996), release rates were significantly influenced by the identity of the terminal amino acid. The rates vary by more than 500-fold for RF1- and by more than 800-fold for RF2-mediated release (Figures 3A and 3B). We note that although rates of peptide release by both factors on glycine and proline were the obvious outliers, rates on other amino acids were also relatively slow. This variation in the rates of peptide release is in direct contrast to what we observe for the nonenzymatic reactions, for which we measure a maximum of tenfold difference in the rate of hydrolysis (Figure 1E). These data suggest that the release factor significantly alters the environment in the PTC and perhaps alters the reaction pathway. In agreement with this proposal, rates for RF1-mediated release do not correlate well to those of nonenzymatic release (R = 0.332, Figure S3B). RF2-mediated release correlates a little better to nonenzymatic release (R=0.562, Figure S3C). This is consistent with recent data from our laboratory showing that while RF1 and RF2 share many similarities, under certain conditions the factors interact differently with the ribosome (Petropoulos et al., 2014). Nevertheless, statistically significant correlation (R=0.544, p = 0.01) was observed between RF1 and RF2 khyd values (Figure S3D). In summary, our observations suggest that in the absence of methylation peptide release varies significantly and could potentially affect gene expression.
This disparity in the rates of peptide release appears to be alleviated by the methylation of the factors. GGQ N5-methylation increased release rates for both factors (Figure 3A and 3B). Similar to our earlier analysis, we observe little to no correlation between the nonenzymatic rates and those of RFMe-mediated release (Figure S3E and S3F). We do observe, however, significant correlation between rates of release for RF1Me and RF2Me (R = 0.651, p = 0.0014; Figure S3G). Perhaps more interesting was the observation of a correlation between the effect of RF methylation on dipeptide release and the release rate observed for the unmethylated RF (Figure 3D). DRNCs that exhibited slow dipeptide release rates for RF1 and RF2 generally exhibited disproportionately high release rates for RF1Me and RF2Me, while those that exhibited high release rates for RF1 and RF2 exhibited more modest increases for RF1Me and RF2Me. For instance, methylation of RF2 increased the rate of fMet-Ser release by nearly 40 fold (0.07 s−1 to 2.5 s−1), but had little effect on the rate of fMet-Tyr release (10 s−1 to 14 s−1). In addition, dipeptide release rates varied less between DRNCs for RF1Me- and RF2Me-mediated release than for RF1 and RF2, (Figure 3C). The coefficient of variation (CV) for dipeptide hydrolysis rates was 1.215 for RF1, 1.164 for RF1Me, 1.599 for RF2 and 0.613 for RF2Me. This is also visible in the condensed vertical spread of data points for RF1Me and RF2Me. These observations suggest that the GGQ N5-methylation plays an important role in tuning the environment of the PTC during peptide release to ensure that different peptidyl-tRNA substrates are released with somewhat similar efficiency.
Structural insights into catalysis by RF1Me and RF2Me
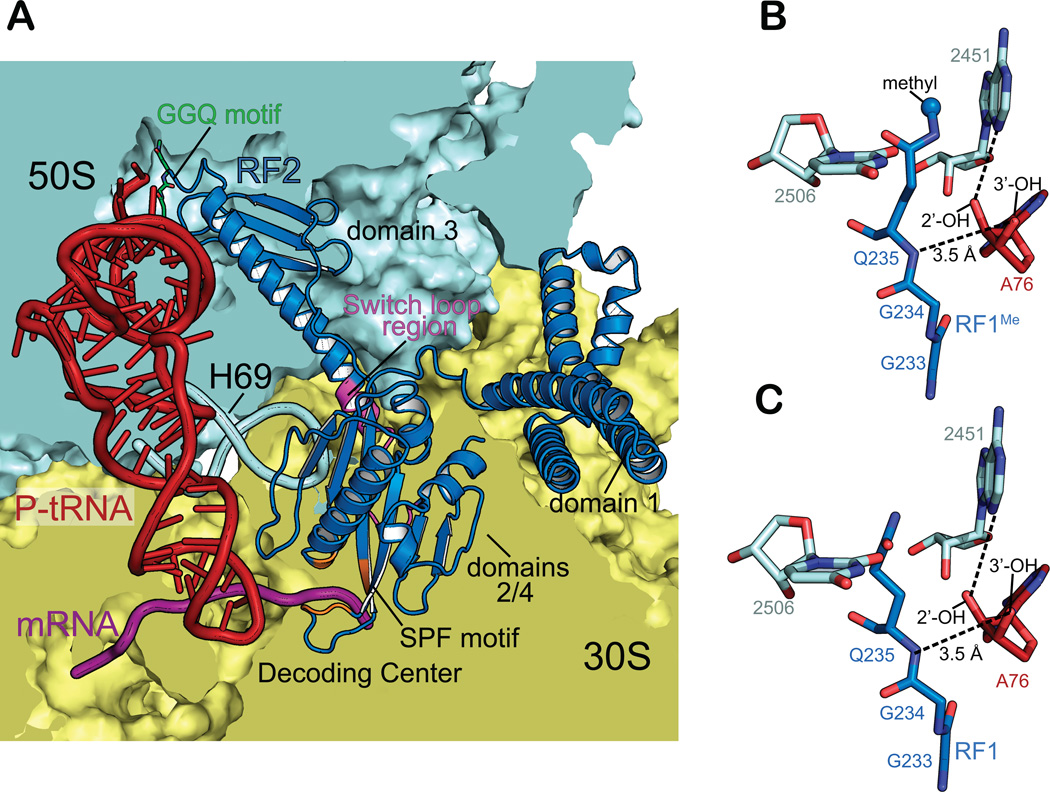
To understand the structural basis of the methylated glutamine, we next determined four structures of E. coli unmethylated and methylated RF1 and RF2 bound to the Tth 70S ribosome. In all previous structures of Tth RFs bound to the 70S, domain three closely packs against 23S rRNA nucleotides of the PTC (Figure 4A) (Jin et al., 2010; Korostelev et al., 2008a; Santos et al., 2013; Weixlbaumer et al., 2008). The universally conserved GGQ motif is located at the tip of domain three and the glutamine (Q235 in RF1, Q252 in RF2) backbone amide is located in an equivalent position at the 2’ - end of the A-site tRNA as previously reported (Figure 4B, Figure S1). (Jin et al., 2010; Korostelev et al., 2008a; Santos et al., 2013; Weixlbaumer et al., 2008). The structures of the 70S-RF1Me with and without the Q235 methylation reveal that the overall location of the backbone of the GGQ motif is unaffected by the modification. Surprisingly, in the 70S-RF1Me complex, the side-chain amide of glutamine in RF1Me is positioned facing A76 of the tRNA and packs against 23S rRNA A2451 and C2452 (Figure 4B). This positioning is clear based on our electron density maps and leaves positive difference density if rotated 180° with the side-chain carbonyl facing the tRNA instead (Figure S1). In the RF2 structure, the methylated Q252 positioning is more ambiguous likely due to the lower resolution (3.4 Å) (Figure S1). In contrast to the sidechain carbonyl, the side-chain amide has historically been proposed to play little to no role in catalysis (Weixlbaumer et al., 2008). Recent studies have suggested that the packing of the methyl group against A2451 of the 23S provides support for the glutamine side chain amide group to participate in hydrogen bonding with a hydroxide ion (Indrisiunaite et al., 2015). However future studies assessing the role of the methylated glutamine in pre-termination states and with different substrates will be required to reconcile the mechanism of termination.
Figure 4. 70S-RFMe interactions.
(A) Overview of E. coli RF2Me (blue) interactions with the Thermus thermophilus 30S (yellow) and 50S (blue) subunits programmed with P-site tRNAfMet (red) and mRNA (magenta).
(B) Interactions between RF1Me GGQ motif (blue), the peptidyl transferase center 23S rRNA residues 2506 and 2452 (cyan), and the terminal A76 of P-site tRNAfMet (red). The backbone amide of Q235 is within hydrogen bond distance to the 3’-OH of A76. The side chain methyl group (blue sphere) packs against the nucleobase of 2451 placing its side chain positioned towards A76.
(C) Interactions between RF1 GGQ motif. The back-bone amide is still within hydrogen bond distance to the 3’-OH of A76, but the glutamine side chain no longer interacts with 2451. The color scheme is the same as (B).
DISCUSSION
During termination of protein synthesis, release factors encounter a diverse set of peptidyl-tRNA substrates. As expected, for the nonenzymatic reaction this diversity of the leaving group was found to impart a significant effect on the rate of hydrolysis (Figure 1) that is likely the result of chemical differences among the 20 amino acids. However on the ribosome this variation in the rates of hydrolysis is not simply the result of intrinsic chemical differences among the leaving groups, as we observe no correlation between ribosome-independent and –dependent reactions (Figure S3A). Instead it appears that the side-chain of the C-terminal amino acid, through interactions with the active site of the ribosome, affects the environment of the PTC and hence possibly the reaction pathway.
Release factors play an active role during the course of the hydrolysis reaction and accelerate the rate of the reaction by a factor of ~105 (Figures 2 and 3). It is therefore reasonable to assume that variations in the efficiency of peptide release that we observe for the uncatalyzed reactions are irrelevant during cellular protein synthesis. Release factors, in principle, could bring about uniformity to the reaction by reshaping the environment in the PTC, which they are known to do (Jin et al., 2010; Korostelev et al., 2008a; Korostelev et al., 2010; Laurberg et al., 2008; Weixlbaumer et al., 2008; Zhou et al., 2012), and/or by altering the rate-limiting step for the reaction. As a result, we were surprised by the observations that in the presence of RF1 and RF2 the rates are much more dramatically different relative to the uncatalyzed reaction. For fMet-Pro and fMet-Gly dipeptidyl tRNAs, the rates of release were so greatly reduced (0.005–0.01 s−1) relative to other dipeptidyl tRNAs that if the rates we measure reflect the in vivo values, ribosomes terminating on these amino acids are likely to stall. Interestingly Hayes and colleagues have previously documented that transcripts having proline codons before stop codons induce ssrA tagging in E. coli indicating that termination following proline residues is inefficient and causes ribosomal stalling (Hayes et al., 2002). In E. coli, 113 and 279 genes terminate with proline and glycine, respectively, and they are in fact not underrepresented taking into account the overall amino acid frequencies (Figure S4C); unless there are mechanisms in vivo that improve termination efficiency on these amino acids, cellular fitness is likely to be impeded given the large fraction of ribosomes that would be unavailable for translation. This termination inefficiency appears to be partly alleviated by adjusting the sequence context around the C-terminal amino acid. In particular, the penultimate amino acid appears to play a critical role during the hydrolysis reaction as it affects the efficiency of stop-codon readthrough (Mottagui-Tabar et al., 1994). In agreement with these findings, proteome-wide analysis of E. coli shows that the same subset of amino acids are either forbidden or significantly underrepresented at the penultimate position for genes terminating with proline (Figure S4D). We note, however, all of our in vitro analysis conducted here was done with Met as the penultimate amino acid, which is not significantly underrepresented at this position for genes terminating with proline. Hence, it is highly unlikely that the termination inefficiencies that we document for certain peptidyl-tRNAs are the result of poor sequence context. Instead we argue that, in addition to the sequence context around the stop codon, the methylation status of release factor plays a critical role in ensuring termination proceeds uniformly irrespective of the identity of the C-terminal amino acid.
What is clear from our analysis and a previous in vivo survey of nonsense suppression (Bjornsson et al., 1996) is that the identity of the C-terminal amino acid is important for efficiency of the hydrolysis reaction. Our data suggest that the side-chain of the ultimate amino acid is likely to influence the orientation of the catalytic water. Our structures of RF1Me and RF2Me bound to 70S reveals that the backbone placement of the GGQ motif is unaffected by the methylation and preserves the previously noted key interaction of the main-chain amide interaction of the glutamine with the 3’-OH of A76 (Figures 4B and 4C). Previous models for how the methylation contributes to release argued that only the main-chain amide is responsible for coordinating the water while a second model argued that the side-chain carbonyl is also involved in coordinating the water (Korostelev et al., 2008b; Korostelev et al., 2010; Laurberg et al., 2008; Santos et al., 2013). A recent alternative hypothesis is that methylation positions the side chain amide towards the 3’-OH of A76 where it forms a hydrogen bond with a hydroxide ion (Indrisiunaite et al., 2015). Our structures show that the methyl-glutamine of the GGQ motif maintains the main chain amide interaction with the P-site tRNA. Unexpectedly, the methylation causes the side-chain carbonyl to face away from the active site and instead, positions the side-chain amide towards the P-site tRNA and A2451 of the 23S (Figure 4B). Clearly presenting the side-chain amide adjacent to the P-site tRNA must be beneficial to the hydrolysis reaction and two possibilities exist. One is that the methylation rigidifies the important glutamine to provide a tight pocket for the catalytic water regardless of the ultimate amino acid. The other possibility is that the methylation stabilizes the glutamine in the PTC to allow for the side chain amide to interact with a hydroxide ion in the reaction. Future studies aimed at the determination of pretermination state structures with different substrates will be required to reconcile this important mechanistic question.
METHODS
Detailed methods are provided in Supplemental Experimental procedures
Release Assays
Ribosome complexes were prepared as previously described (Simms et al., 2014). Rates of peptide release were determined on a Quench-Flow instrument (RQF-3 Rapid Quench-Flow, KinTek) and reactions resolved with electrophoresis on cellulose TLC plates (Youngman et al., 2004). Observed khyd values were calculated by fitting a first-order rate equation to the curve using GraphPad Prism software.
Nonenzymatic hydrolysis and aminolysis
Nonenzymatic hydrolysis on and off the ribosome were performed as previously described (Zaher et al., 2011).
Structures of 70S and release factor complexes
Tth 70S ribosomes were purified, complexed with mRNA, tRNA and RFs as described (Selmer et al., 2006). Crystals were screened for diffraction at the SER-CAT 22-ID beamline and X-ray data were collected at the NE-CAT 24-IDC beamline, both at the APS, Argonne National Laboratory. The data were integrated and scaled using XDS (Kabsch, 2010), refined with PHENIX (Adams et al., 2010) and modeled using Coot (Emsley et al., 2010) (Table S1).
Supplementary Material
HIGHLIGHTS.
-
-
Efficiency of peptide release is dependent on the C-terminal amino acid identity
-
-
Methylation of release factor affects peptide release disparately depending on the substrate
-
-
Structures show the methylated glutamine repositions against the tRNA and 23S nt 2451
Acknowledgments
The authors wish to thank Joe Jez and Doug Chalker for comments on earlier version of the manuscript and members of the Zaher and Dunham laboratories for useful discussions. This work was supported by the National Institutes of Health (NIH R01GM093278 to CMD, R01GM112641 to HSZ), a Searle Scholars award (to HSZ) and a Pew Scholar in the Biomedical Sciences award (to CMD). The Donald Danforth Plant Science Center Proteomics and Mass Spectrometry Facility is supported the National Science Foundation Grant No. DBI-0922879 for acquisition of the LTQ-Velos Pro Orbitrap LC-MS/MS. The Emory Integrated Proteomics Core is supported by the Neuroscience NINDS Core Facilities (P30NS055077), the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. The X-ray crystallography datasets were collected at the NE-CAT beamlines, which are funded by the National Institute of General Medical Sciences from the NIH (P41 GM103403), and at the SERCAT beamlines. The Pilatus 6M detector on 24-ID-C beam line is funded by a NIHORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DEAC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
WP designed and performed the peptide release assays. EH designed and carried out the X-ray crystallography assays. HK performed the nonenzymatic release assays. CLS carried out the in vivo readthrough assays. All authors interpreted the results. CMD and HSZ designed the experiments and wrote the manuscript.
SUPPLEMENTAL INFORMATION
Document S1. Figures S1–S5
Table S1
Data collection and refinement statistics for the 70S-RF complexes
Accession numbers Coordinates and structure factors were deposited in the RCSB Protein Data Bank with accession codes 5CZP, 5DFE, 5J30 and 5J3C.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D-Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson A, Mottagui-Tabar S, Isaksson LA. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 1996;15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D-Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci USA. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Champ S, Engstrom A, Ehrenberg M, Buckingham RH. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002;21:769–778. doi: 10.1093/emboj/21.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrisiunaite G, Pavlov MY, Heurgue-Hamard V, Ehrenberg M. On the pH dependence of class-1 RF-dependent termination of mRNA translation. J Mol Biol. 2015;427:1848–1860. doi: 10.1016/j.jmb.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci USA. 2010;107:8593–8598. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr Sect D-Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecki DS, Aune KC, Tate W, Caskey CT. Characterization of reticulocyte release factor. J Biol Chem. 1977;252:4514–4520. [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA. 2008a;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhua J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA. 2008b;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Zhu J, Asahara H, Noller HF. Recognition of the amber UAG stop codon by release factor RF1. EMBO J. 2010;29:2577–2585. doi: 10.1038/emboj.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Mora L, Heurgue-Hamard V, de Zamaroczy M, Kervestin S, Buckingham RH. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J Biol Chem. 2007;282:35638–35645. doi: 10.1074/jbc.M706076200. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Bjornsson A, Isaksson LA. The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J. 1994;13:249–257. doi: 10.1002/j.1460-2075.1994.tb06255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Virumae K, Atkinson GC, Tenson T, Remme J, Wilson DN. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat Chem Biol. 2012;8:695–697. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- Petropoulos AD, McDonald ME, Green R, Zaher HS. Distinct roles for release factor 1 and release factor 2 in translational quality control. J Biol Chem. 2014;289:17589–17596. doi: 10.1074/jbc.M114.564989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FVt, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Polacek N, Gomez MJ, Ito K, Xiong L, Nakamura Y, Mankin A. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol Cell. 2003;11:103–112. doi: 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- Santos N, Zhu J, Donohue JP, Korostelev AA, Noller HF. Crystal structure of the 70S ribosome bound with the Q253P mutant form of release factor RF2. Structure. 2013;21:1258–1263. doi: 10.1016/j.str.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci USA. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Shaw JJ, Green R. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell. 2007;28:458–467. doi: 10.1016/j.molcel.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Hudson BH, Mosior JW, Rangwala AS, Zaher HS. An active role for the ribosome in determining the fate of oxidized mRNA. Cell reports. 2014;9:1256–1264. doi: 10.1016/j.celrep.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol Cell. 2007;27:758–766. doi: 10.1016/j.molcel.2007.06.032. [DOI] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. Mechanism of the translation termination reaction on the ribosome. Biochemistry. 2009;48:11296–11303. doi: 10.1021/bi9017297. [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Vogel Z, Zamir A, Elson D. Possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry. 1969;8:5161–5168. doi: 10.1021/bi00840a070. [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Shaw JJ, Strobel SA, Green R. The 2'-OH group of the peptidyl-tRNA stabilizes an active conformation of the ribosomal PTC. EMBO J. 2011;30:2445–2453. doi: 10.1038/emboj.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Korostelev A, Lancaster L, Noller HF. Crystal structures of 70S ribosomes bound to release factors RF1, RF2 and RF3. Curr Opin Struct Biol. 2012;22:733–742. doi: 10.1016/j.sbi.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.