Abstract
Summary: Circularized Chromosome Conformation Capture followed by deep sequencing (4C-Seq) is a powerful technique to identify genome-wide partners interacting with a pre-specified genomic locus. Here, we present a computational and statistical approach to analyze 4C-Seq data generated from both enzyme digestion and sonication fragmentation-based methods. We implemented a command line software tool and a web interface called w4CSeq, which takes in the raw 4C sequencing data (FASTQ files) as input, performs automated statistical analysis and presents results in a user-friendly manner. Besides providing users with the list of candidate interacting sites/regions, w4CSeq generates figures showing genome-wide distribution of interacting regions, and sketches the enrichment of key features such as TSSs, TTSs, CpG sites and DNA replication timing around 4C sites.
Availability and Implementation: Users can establish their own web server by downloading source codes at https://github.com/WGLab/w4CSeq. Additionally, a demo web server is available at http://w4cseq.wglab.org.
Contact: kaiwang@usc.edu or wangelu@usc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
With the development of microscopy and genomic technologies, it has been shown that three-dimensional genome architecture within nucleus tremendously affects gene function and cell property (Pombo and Dillon, 2015). The underlying component of genome architecture is chromatin interactions that occur at the genome‐wide scale. The specific long-range genomic segments contacts are achieved by chromatin looping and usually mediated by transcription factors. Most of the interactions are between enhancer and promoter, leading to well-regulated gene expression patterns (Dekker, 2008). Originated from 3C (chromosome conformation capture) (Dekker et al., 2002), 4C-Seq (van de Werken et al., 2012) is specifically developed to assess the long-range DNA-DNA interactions where one of the DNA segments is the locus of interest, usually called ‘bait’ or ‘viewpoint’. Classical 4C-Seq technique uses enzyme digestion to fragment chromatin. To minimize the bias caused by enzyme efficiency and uneven distribution of enzyme sites across genome, an alternative 4C-Seq approach is developed where sonication is used as the fragmentation method (Gao et al., 2013).
It is noted that the analysis of 4C-Seq data requires extensive data manipulation and computation, thus posing a daunting challenge for most researchers. On the other hand, most of the existing analytical tools are either difficult to use or mainly focus on near-cis intra-chromosomal interactions. Besides, there is no tool available currently that can analyze 4C-Seq data generated by sonication method. That motivates us to develop a new tool to facilitate the analysis of 4C-Seq data. Here, we summarize the analysis pipelines, present a web application named w4CSeq, and show the results generated with w4CSeq by analyzing real biological datasets.
2 Methods and results
2.1 Implementation
The core w4CSeq software is implemented in Perl and R. Required external tools include BWA, SAMtools and BEDTools. Detailed descriptions of statistical models in analytical pipelines are included in Supplementary Information. w4CSeq can be installed as a local web application. The server backend is implemented in Perl CGI.
2.2 Input and output
Once the server is running, users need to upload a single-end FASTQ file for enzyme digestion-based 4C-Seq analysis and paired-end FASTQ files for sonication-based 4C-Seq analysis. A genome build needs to be specified against which the sequencing reads are mapped. Users also need to provide information on the bait region. In addition, users can specify window sizes and FDR (False Discovery Rate) threshold for statistical analysis. For enzyme-digestion-based 4C-Seq portal, users need to choose the primary restriction enzyme and provide primer sequence used in library construction. For sonication-based 4C-Seq, users need to specify the length for extension to define the ‘bait’ neighborhood for alignment purpose. Optionally, we provide a module where users can upload their own BED formatted annotation files containing the regions of interest, such as ChIP-Seq peaks.
After the analysis is done, w4CSeq outputs a link navigating to the result page that has five modules: (i) summary of parameters that users have specified; (ii) summary of metrics throughout analysis; (iii) figures illustrating: (a) the genome‐wide distribution of 4C regions, (b) distance distribution to reference genes, TSSs, TTSs and CpG islands of 4C sites compared to random, (c) the comparison of DNA replication timing of 4C regions versus the whole genome, and (d) the enrichment of user provided feature around 4C regions compared to random; (iv) files which users can download to perform their downstream analysis and (v) a link to the UCSC genome browser where users can visualize the contact map.
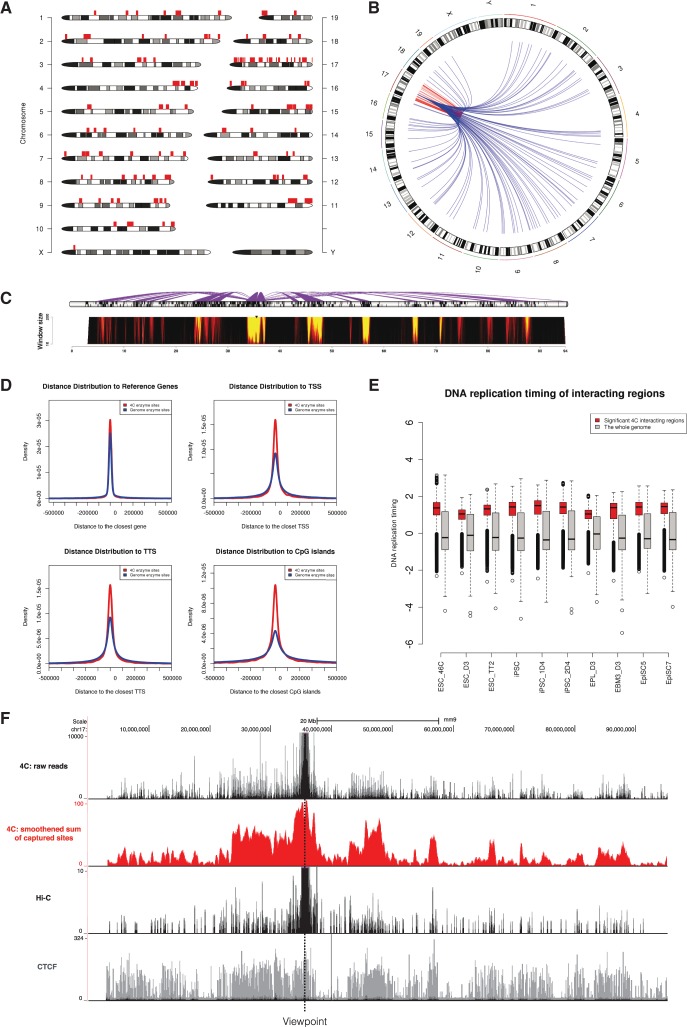
Figure 1 shows the analysis of a 4C-Seq dataset (Wei et al., 2013) aimed to investigate interaction landscape centered on a distal enhancer of Oct4 gene in mouse pluripotent stem cells. Extensive interactions are observed both in cis and in trans. Regions interacting with the Oct4 distal enhancer are mostly gene-rich regions. In addition, 4C sites are closer to annotated genes, TSSs, TTSs and CpG islands, and show an earlier DNA replication timing. This suggests the Oct4 distal enhancer interacting regions tend to be actively engaged in gene function and transcription activity. Of note, after processed by w4CSeq, the interaction landscape is smoother with clear demarcations. We also find strong correlations between the profile of 4C-Seq and that of CTCF with virtual-4C derived from Hi-C.
Fig. 1.
w4CSeq generated output for an enzyme digestion-based 4C-Seq dataset. (A) Genome wide‐distribution of 4C regions as indicated by red rectangles on top of chromosome ideogram. (B) Circos plot of genome‐wide distribution of 4C interactions as indicated by curves extended from the ‘bait’ region. Intra-chromosomal (cis) and inter-chromosomal (trans) interactions are shown in red and blue, respectively. (C) Spider plot (top) depicting contacts centered on the ‘bait’ region and domainogram (bottom) depicting interaction intensities across window size ranging from 2 to 200 restriction sites. (D) Density curve showing relative distance of 4C sites from key genomic features including reference genes, TSSs, TTSs and CpG islands compared to random. (E) Box plot showing the distribution of DNA replication timing values of 4C regions compared to the whole genome in 10 pluripotent cell lines. Early replication domains have the logarithm of replication timing ratio > 0. (F) 4C signals distributed in cis with the viewpoint. Upper black track shows the raw reads distribution with counts ranging from 0 to 10 000; Middle red track shows the running sum of captured sites in window (window size: 100 enzyme sites); Middle black track shows virtual‐4C profile extracted from Hi-C result; Lower grey track shows CTCF binding profile.
3 Conclusion
We describe w4CSeq, a web application for analyzing 4C-Seq data in an automated manner, with rich visualization functionalities. w4CSeq will be a valuable tool to unveil the functional genome organization.
Funding
This work was supported by the National Institutes of Health [HG006465 to K.W., 2R01CA13692406A1 to W.L.].
Conflict of Interest: K.W. is a board member and shareholder of Tute Genomics, Inc. The study did not involve and did not use any product from Tute Genomics.
Supplementary Material
References
- Dekker J. (2008) Gene regulation in the third dimension. Science, 319, 1793–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. et al. (2002) Capturing chromosome conformation. Science, 295, 1306–1311. [DOI] [PubMed] [Google Scholar]
- Gao F. et al. (2013) Comparative analysis of 4C-Seq data generated from enzyme-based and sonication-based methods. BMC Genomics, 14, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A., Dillon N. (2015) Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol., 16, 245–257. [DOI] [PubMed] [Google Scholar]
- van de Werken H.J. et al. (2012) 4C technology: protocols and data analysis. Methods Enzymol., 513, 89–112. [DOI] [PubMed] [Google Scholar]
- Wei Z. et al. (2013) Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell, 13, 36–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.