Abstract
Intraperitoneal administration of hypertonic saline to the rat supraoptic nucleus (SON) increases the expression of several immediate early genes (IEG) and the vasopressin gene. These increases have usually been attributed to action of the cyclic-AMP Response Element Binding Protein (CREB). In this paper, we study the role of CREB in these events in vivo by delivering a potent dominant-negative form of CREB, known as A-CREB, to the rat SON through the use of an adeno-associated viral (AAV) vector. Preliminary experiments on HEK 293 cells in vitro showed that the A-CREB vector that we used completely eliminated CREB-induced c-fos expression. We stereotaxically injected this AAV-A-CREB into one SON and a control AAV into the contralateral SON of the same rat. Two weeks following these injections we injected hypertonic saline intraperitoneally into the rat. Using this paradigm, we could measure the relative effects of inhibiting CREB on the induced expression of c-fos, ngfi-a, ngfi-b, and vasopressin genes in the A-CREB AAV injected SON versus the control AAV injected SON in the same rat. We found only a small (20%) decrease of c-fos expression and a 30% decrease of ngfi-b expression in the presence of the A-CREB. There were no significant changes in expression found in the other IEGs nor in vasopressin that were produced by the A-CREB. This suggests that CREB may play only a minor role in the expression of IEGs and vasopressin in the osmotically activated SON in vivo.
Keywords: C-FOS, CREB, A-CREB, AAV, SON, Vasopressin
1. Introduction
Intraperitoneal (i.p.) administration of hypertonic saline increases the expression of the immediate early genes (IEG), c-fos, ngfi-a, ngfi-b, along with the expression of the vasopressin (VP) gene in the supraoptic nucleus (SON) of rats (Kawasaki et al., 2005). The presence of a cyclic adenosine 3, 5-monophosphate (cAMP) response element (CRE) sequence in the promoter region of all of these genes (Boutillier et al., 1992; DeFranco et al., 1993; Inaoka et al., 2008; Yoshida et al., 2006) suggests that the increase in expression is mediated by the CRE binding protein (CREB).
CREB is a transcription factor that has been implicated in the transcriptional regulation of many genes (Sheng et al., 1990) and is involved in a myriad of cellular responses that are involved in nervous system development, learning and memory, addiction, and neuroprotection from disease among others (Lonze and Ginty, 2002). CREB is activated in response to a vast array of physiological stimuli together with the closely related members of the same bZIP superfamily of transcription factors such as cAMP response element modulator (CREM) and activating transcription factor 1 (ATF-1).
Efforts to study CREB function in vivo have been made by the use of traditional knockout models, but success has been limited since CREB−/−mice don’t survive after birth largely because of respiratory distress (Lonze and Ginty, 2002; Mayr and Montminy, 2001). Although CREM−/−mice do survive to adulthood, the males are sterile, they exhibit disrupted circadian rhythms, and have altered behavioral patterns (Mayr and Montminy, 2001). Moreover, compensatory action by other CREB family members in CREM- or CREB-knockout mice prevents expression of a knockout phenotype (Mayr and Montminy, 2001). These drawbacks have led to the development of alternative transgenic (Carlezon et al., 1998; Herzig et al., 2001; Jancic et al., 2009; Lee et al., 2009) and viral vector (Barrot et al., 2005; Warburton et al., 2005; Yuan et al., 2003) strategies using mutant CREBs to perturb CREB function in vivo.
In the present study we stereotaxically injected A-CREB, a potent dominant negative inhibitor of CREB that disrupts the DNA binding of CREB and CREB family members (Ahn et al., 1998), into the rat SON using an adeno-associated virus (AAV) vector. A-CREB has an acidic amphipathic patch that is thought to mimic DNA in its binding to the N-terminal of the wild-type CREB leucine zipper. Experiments by Ahn et al. (1998) showed that A-CREB is able to completely and specifically inhibit CREB DNA binding. We injected an A-CREB bearing AAV vector in order to determine the role of CREB and CREB family members on the expression of c-fos, ngfi-a, ngfi-b, nr4a2, and vasopressin (VP) in this brain region. The effectiveness of using AAV to target molecules to the rat SON was recently demonstrated (Doherty et al., 2011). Since the acute, systemic administration of hypertonic saline causes rapid increases in expression of several of these genes, (Kawasaki et al., 2005) which are presumed to be primarily mediated by the transcription factor CREB, we expected that blocking CREB in vivo would significantly inhibit their increases in expression during hyperosmotic stimulation.
2. Results
2.1. Localization of the AAV to the SON
A schematic diagram of the stereotaxic injection paradigm is illustrated in Fig. 2a, which shows a coronal section of a rat brain with a syringe lowered unilaterally to the SON. Figs. 2b and c show results from control experiments in which the SON was injected with the CMV-EGFP virus and subsequently double-labeled with IHC using antibodies against both EGFP and Neurophysin (an identifying marker for the magnocellular neurons). As shown in Fig. 2d, the virus (EGFP-ir) colocalized with nearly all of the Neurophysin-positive magnocellular neurons of the SON.
Fig. 2.
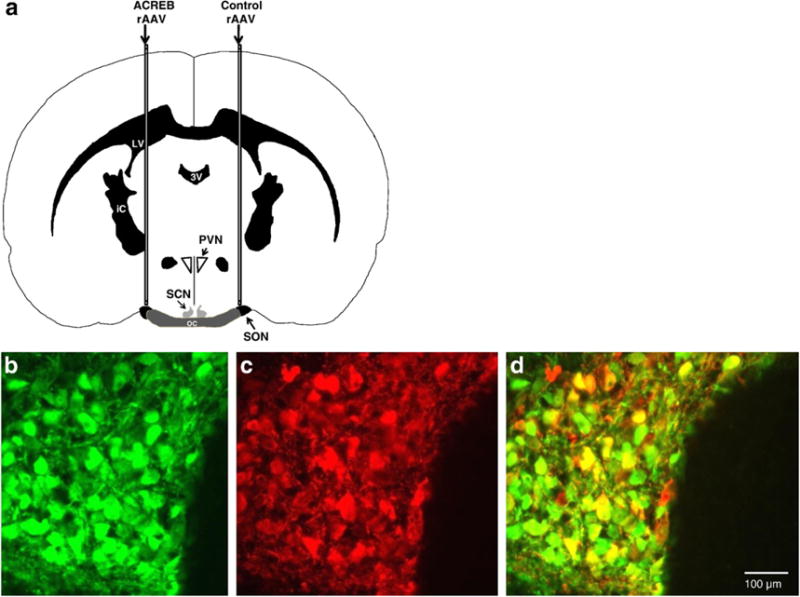
(a) Schematic drawing of cannula placements used to inject the AAVs into the rat SON. The A-CREB AAV was injected over one SON, and the control AAV was injected over the contralateral SON. Hence, each rat contained its own control SON. Abbreviations: SON = supraoptic nucleus; SCN = suprachiasmatic nucleus; PVN = paraventricular nucleus. The images in panels b–d are from the same SON that had been injected with an AAV that expressed EGFP driven by a CMV promoter. (b) Illustrates the expression of EGFP as green immunofluorescence in SON cells that were successfully transduced by the CMV-EGFP AAV. OC = optic chiasm (c) Illustrates red immunofluorescent labeling of neurophysin in both oxytocin and vasopressin magnocellular neurons in the same SON. (d) Shows the merged overlay of green (b) and red (c) fluorescence demonstrating that the AAV successfully transduced most of the magnocellular neurons in the SON and robustly expressed EGFP in virtually all of them.
2.2. Effectiveness of in vitro paradigm and primer pair sensitivity
The graphs in Fig. 3 indicate that the rat neuroblastoma cells that were stimulated with forskolin had lower Ct values (more expression) than the control cells for c-fos, ngfi-a, ngfi-b, and nr4a2, demonstrating that all the primer pairs successfully detected the changes and that all the listed immediate early genes increased in vitro when the cells were stimulated.
Fig. 3.
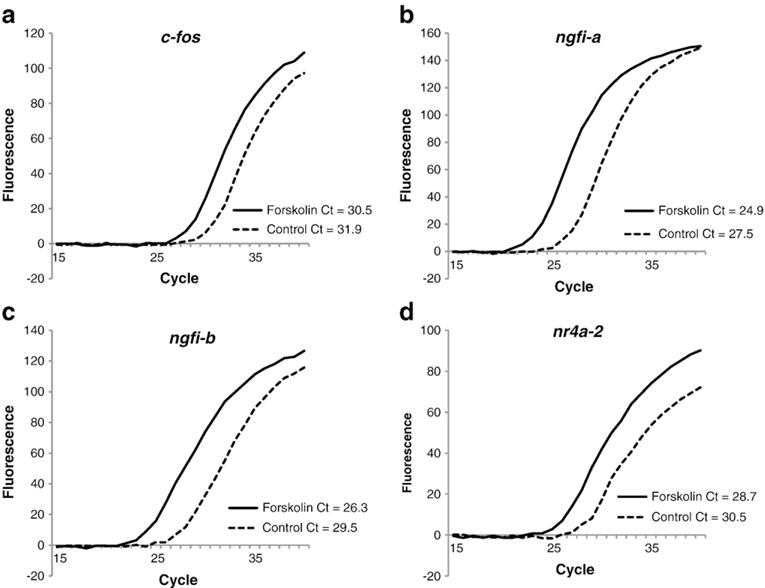
Validation of the quantitative RT-PCR assays of the immediate early genes: c-fos (a), ngfi-a (b), ngfi-b (c), nr4a-2 (d) after forskolin stimulation. The figure depicts fluorescent growth curves and cycle threshold values for each of these immediate-early genes in cultured rat neuroblastoma (B35) cells that were stimulated with forskolin (solid line) versus those that were not (dotted line). The Cfos, NGFIA, NGFIB, and NR4A2 primers are shown in Table 1. In each case, there was an increase (lower Ct value) in the IEG mRNA in the forskolin-stimulated cultures as compared with the mRNA in non-stimulated cultures.
2.3. Test of A-CREB’s effectiveness to inhibit c-fos mRNA induction in vitro
Fig. 4 shows a graph comparing the fluorescent growth curves of c-fos mRNA in PCR assays of HEK-293 t cells that were stimulated with forskolin (20 μM, 30 min) after being transduced with either the A-CREB (rAAV-CMV-A-CREB-T2A-EGFP) or control (rAAV-CMV-EGFP) AAVs (3.0 μl of each virus). A second forskolin-treated control group was included that was not transduced with AAV. The resultant Ct values were 28, 28, and 32 for the no virus control, AAV-EGFP control, and A-CREB groups, respectively. The four cycle difference in the Ct value for c-fos between the A-CREB and the control virus indicates that expression of c-fos is reduced about 16-fold in the presence of the A-CREB virus relative to the other two control groups, thereby validating the efficacy of our rAAV-CMV-A-CREB-T2A-EGFP vector.
Fig. 4.
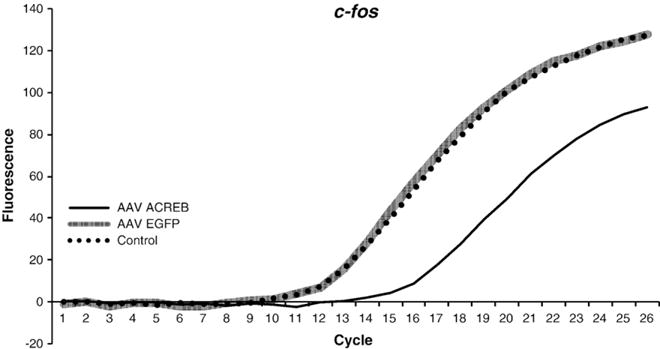
Efficacy of the A-CREB-T2A-EGFP construct to inhibit induced c-fos expression in vitro. The graph shows fluorescent growth curves and cycle threshold values for c-fos mRNA in HEK-293 t cells under control conditions and after stimulation with forskolin. Cells were either transduced with A-CREB-T2A-EGFP AAV, EGFP-AAV, or not transduced with any virus (control). Human c-fos primer was used in the qPCR assay (see Table 1). Note that there was a large reduction in c-fos mRNA (higher Ct value) in the cells that were transduced with A-CREB AAV relative to those that were transduced with the EGFP-only control AAV. The non-transduced control cells and those that were transduced with the EGFP-only AAV exhibited similar fluorescent growth curves.
2.4. Evidence for partial inhibition of Fos protein expression in the presence of A-CREB after acute hyperosmotic stimulation
The A-CREB AAV injected into the rat SON partially inhibited Fos expression. This is illustrated in the images in Fig. 5, which show IHC data using Fos antibody. The low-magnification representative section in Fig. 5a shows that Fos induced by salt loading of the rat (see Section 4.6 for salt loading methods) is reduced in the A-CREB treated (left) SON as compared to the control (right) SON (see arrows in Fig. 5a). Data from a second rat (total experiments, n=6), are illustrated at higher magnification in Figs. 5b and c which similarly show that the control SON (Fig. 5c) has both brighter and more numerous Fos immunoreactive cells than the left, A-CREB-injected SON (Fig. 5b). These data are consistent with the expectation that CREB is involved in the acute hyperosmotic stimulation of Fos expression in the SON.
Fig. 5.
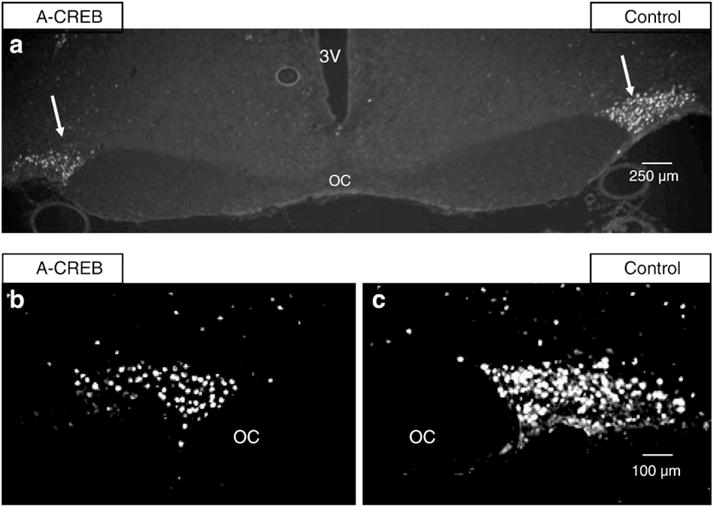
Immunohistochemical staining for c-Fos protein in the SONs of rats in which the left SON was injected with the AAV containing A-CREB and the right SON with a control AAV. (a) Fos immunoreactivity. Both left and right SONs (arrows) are demonstrated here at low power (4× magnification). Note that there is more c-Fos immunofluorescence in the control SON after an acute salt loading stimulus. The 3rd ventricle is labeled 3 V, the optic chiasm, OC. (b and c) Illustrate higher magnification views (10×) of Fos immunofluorescence in A-CREB AAV treated and control AAV SONs in a different rat. The SONs are outlined by white lines. (b) Shows Fos immunoreactivity in the A-CREB treated SON versus the contralateral control SON (c) after an acute salt loading stimulus. This image demonstrates, in an additional rat, that c-Fos immunoreactivity is reduced in the A-CREB treated SON relative to the contralateral, control SON. See Table 2 for related changes in mRNA for all the intermediate genes under these experimental conditions.
2.5. Quantification of the effects of A-CREB on immediate early and VP gene expression in the SON
QRT-PCR was used to measure the inhibitory effects of A-CREB on the mRNA levels of IEGs relative to the control (contralateral) side of each rat brain after an acute salt loading stimulus. Table 2 shows the average Ct values of four immediate-early genes (nr4a2, ngfi-b, ngfi-a, c-fos) from the A-CREB treated SONs compared to the control SONs in each animal. Vasopressin hnRNA and vasopressin mRNA were also measured. The Ct values for vasopressin mRNA were used to normalize the data since the Ct values for vasopressin mRNA neither vary during acute osmotic stimulation (Kawasaki et al., 2009) nor do they vary in the presence or absence of A-CREB (Table 2). In this regard, it is interesting that the expression of vasopressin in the SON has recently been shown to be independent of c-fos (Arima et al., 2010). Both c-fos and ngfi-b had significantly higher Ct values on the A-CREB treated side, indicating that A-CREB inhibited the expression of these genes (p=0.03, n=7, for both). After normalization to VP mRNA using the comparative Ct method (Pfaffl, 2001) we calculate that A-CREB inhibited c-fos mRNA by 20% and ngfi-b by 30% (Table 2). Nr4a2 also had a greater Ct value on the A-CREB side (indicating lower expression), however, these results were not statistically significant (p=0.08, n=7). The apparent reduced expression of VP hnRNA on the A-CREB side was not statistically significant either (p=0.254, n=8).
Table 2.
Effects of ACREB AAV versus control AAV injections into the rat SON on immediate-early gene and vasopressin (VP) gene expression.
| c-fos (Ct) |
nr4a2 (Ct) |
ngfi-b (Ct) |
ngfi-a (Ct) |
VPhnRNA (Ct) |
VPmRNA (Ct) |
|
|---|---|---|---|---|---|---|
| ACREB | *24.2±0.26 | 25.5±0.17 | *23.8±0.37 | 22.5±0.28 | 30.8±0.62 | 18.3±0.45 |
| Control | 23.9±0.23 | 25.2±0.28 | 23.3±0.32 | 22.5±0.25 | 30.5±0.46 | 18.3±0.39 |
| P-value | 0.03 (n=8) | 0.08 (n=7) | 0.03 (n=7) | 0.42 (n=7) | 0.25 (n=8) | 0.46 (n=8) |
| Fold Change | 0.8 ** | 0.8 | 0.7 ** | 1.0 | 0.9 | – |
Data expressed as mean+/−SEM, p≤0.05 represents statistical significance.
Fold change of 0.8 for cfos and 0.7 for NGFI-B represent inhibitions of 20% and 30%, respectively, of A-CREB treated versus control SON.
3. Discussion
CREB target genes, which differ widely in their functional roles and mechanisms, all have in common the presence of a CREB binding site, the CRE sequence, in their promoter region. One of the most recognized of these target genes is the prototypic IEG, c-fos, whose transcription is rapidly activated by a variety of stimuli but peaks and returns to baseline within 1 h of the stimulus onset (Greenberg and Ziff, 1984; Sheng and Greenberg, 1990). C-fos was one of the first identified CREB target genes; its transcription was shown to be regulated by the CRE consensus site in the promoter region and to respond to stimuli that activate CREB (Lonze and Ginty, 2002; Sassone-Corsi et al., 1988). Since those early studies, however, hundreds of additional genes have been defined as having a CRE consensus site in their promoter as well as having transcription mediated by CREB. Some of the other CREB target IEGs include NURR1 (NR4A2) and Epidermal growth factor (NGFI-A) (Mayr and Montminy, 2001), and Nur77 (i.e. NGFI-B) (Fass et al., 2003).
Many of these CREB targets were first identified in vitro by using PC12 cells. Sheng et al. (1990) began studying the involvement of CREB on c-fos transcription by creating mutations in the CRE sequence of the promoter region in PC12 cells. In subsequent studies (Ginty et al., 1994; Sheng et al., 1991), a mutant of CREB (mCREB) was created by replacing Ala for Ser-133 in order to prevent phosphorylation of CREB at this position. This was used to show that c-fos transcription in activated PC12 cells also involved CREB. Ahn et al. (1998) created a different, smaller sized dominant negative CREB called A-CREB, which is a CREB leucine zipper-domain with an acidic amphipathic extension on the N terminus that disrupts DNA binding activity of CREB and CREB family factors (e.g., CREM, ATF-1). A-CREB was also used in PC12 cells and was shown to inhibit c-fos expression by blocking CREB that was induced by cAMP. The authors found, however, that A-CREB only partly inhibited Ca+2 and nerve growth factor (NGF) mediated c-fos transcription, suggesting a separate converging mechanism. Fass et al. (2003) confirmed these findings in vitro using A-CREB, and showed the same effects in the transcription of NGFI-B and other IEGs.
Thus, the CREB-c-fos pathway has been extensively studied in the literature and CREB has been identified as one of the main regulators of c-fos transcription (Fisch et al., 1989; Ginty et al., 1994; Gonzalez and Montminy, 1989; Lonze and Ginty, 2002; Mayr and Montminy, 2001; Sassone-Corsi et al., 1988). The above studies, as well as other studies in the literature, investigated the mechanisms of c-fos transcription in vitro. Our in vitro experiments on rat neuroblastoma cells also supported a robust role for CREB in forskolin-induced c-fos expression (see Fig. 4). Studies using various mammalian cell lines have described the importance of other elements within the c-fos promoter on transcription of c-fos. The cAMP Responsive Element (CRE) and, to a lesser extent, the Serum Responsive Element (SRE) are both thought to be important in mediating c-fos transcription in vitro (Ahn et al., 1998; Eto et al., 2010; Ginty et al., 1994; Hill and Treisman, 1995; Rivera et al., 1990, 1993; Sheng et al., 1991; Simon et al., 2005; Treisman, 1995; Zhang et al., 2008). However, less is known about the relative contributions of CREB and SRE to regulate c-fos, in vivo. The findings reported here shed light on this issue and lend credence to the idea that results found in vitro may not be recapitulated in vivo.
We were surprised to find that our injections of the A-CREB AAV into the SON only produced a modest reduction of c-fos expression (Table 2). A 20% reduction in c-fos expression was observed in the A-CREB treated SON relative to the control SON. We also found that another IEG known to contain a CRE binding sequence in its promoter region, NGFI-B, also had a small but significantly reduced expression in the presence of A-CREB (Table 2). Several other IEGs such as NGFI-A, NR4A-2, and the vasopressin gene, did not change in expression (Table 2).
There have been several lines of evidence suggesting that cAMP increases VP expression mediated by CREB, predominantly from in vitro studies. Using cell lines, these studies reported that upregulating cAMP (via forskolin or other methods) led to increased VP gene expression (Iwasaki et al., 1997; Kuwahara et al., 2003). Moreover, Iwasaki et al. (1997) showed that the dominant negative CREB, KCREB, could partially block this increased expression. However, a recent in vivo study by Chiappini et al. (2011) showed that while VP production was impaired in the PVN of CREB knockout mice, it was not impaired in the SON of these mice. Their conclusion is consistent with our result that blocking CREB in the SON in vivo does not change vasopressin expression. In addition, these data further support the idea that CREB’s functions in vitro are not necessarily the same as those found in vivo.
There are several possible explanations for the modest inhibitory effect of A-CREB on c-fos expression in vivo in the SON, as compared to its potent inhibition seen in vitro in other systems. The first possibility is that the A-CREB expression in the SON was too low to fully compete with the endogenous CREB in the SON neurons. Although this cannot be absolutely discounted, it is unlikely. After the virus was stereotaxically injected into the rat SON, we were able to clearly visualize the IHC fluorescence from the A-CREB-T2A-EGFP construct in all the neurons in the sectioned SON (data not shown). This indicated that the AAV was able to enter and express A-CREB as well as the EGFP within virtually all of the cells of the SON. In addition, the IHC studies of Fos protein in the SON show that the A-CREB-treated SON had substantially lower Fos protein present than the control SON (Fig. 5). The IHC data in Fig. 5 support the view that the A-CREB vector was able to enter the SON cells to a large enough extent as to be able to inhibit Fos protein expression. Furthermore, the A-CREB/CREB heterodimer is 3300-fold more stable than CREB alone (Ahn et al., 1998), and it is likely that the A-CREB construct used here will effectively interfere with CREB binding to the CRE site in the SON in vivo. Finally, other studies using A-CREB viral vectors injected into rat brain (Warburton et al., 2005), as well as A-CREB use in transgenic animals (Jancic et al., 2009; Lee et al., 2009), have shown effective expression and action of the dominant negative A-CREB construct in rodent neurons in vivo.
Another possible explanation is suggested by the data of Yuan et al. (2010) who reported that acute hyperosmotic stimulation causes peak Fos protein expression at 45 min in astrocytes and at 90 min in neurons in the SON. It is known that c-fos mRNA expression in response to hypertonic saline peaks around 30 min in the SON (Kawasaki et al. 2005), and in view of the Yuan et al. (2010) data it is likely that at the time we did our qPCR assays of c-fos mRNA (30 min after the i.p. salt injection) increases in both astrocytic and neuronal c-fos mRNA in the SON were being measured. Since the viral vector (AAV-6) that we used to transduce the SON only targets neurons it is likely that the modest A-CREB mediated reduction of c-fos mRNA of 20% that we observed reflected only the inhibition in the magnocellular neurons, and that the presence of the uninhibited, enhanced astrocytic c-fos mRNA in the SON led to an underestimate of the A-CREB inhibition.
Finally, another possible explanation of the modest inhibitory effect of ACREB in the SON is that the regulation of c-fos in vivo in the SON in response to hyperosmotic stimuli differs from that of forskolin stimulated c-fos regulation in most cell lines in vitro. If we safely assume that there is an effective level of A-CREB expression in the SON neurons, and given that there is only a 20% reduction of c-fos expression in vivo, this suggests that full induction of c-fos expression in the rat SON may occur via alternative pathways. The canonical cAMP-CREB-c-fos pathway may make only a minor contribution. There are several locations on the c-fos promoter region that indicate c-fos can respond to molecules other than CREB, e.g. serum growth factors to the SIE region (SIF-inducible element, where SIF is an acronym for sis-inducible factor, Coulon et al., 2010). In fact, the serum response element (SRE) region in the c-fos promoter was found to be an important inducer of c-fos expression under dehydrating hyperosmotic stress (Cahill et al., 1996; Gille et al., 1995; Soh et al., 1999). This 20 base pair (bp) region is located 310 bp upstream of the c-fos gene origin and initiates transcription when the serum response factor (SRF) binds to this site in response to serum and purified growth factors. In some cell lines extracellular-signal-regulated kinase (ERK) and mitogen-and-stress-activated kinase (MSK) are recruited to the SRE promoter site and can induce c-fos transcription in response to hyperosmotic stress (Nielsen et al., 2008; Zhang et al., 2008).
Therefore, activation at the SRE site in the c-fos promoter region in the rat SON may contribute heavily to c-fos expression in response to hyperosmotic stress. Further investigations are needed to fully elucidate the roles of SRE and other possible signal-transduction and transcription factor molecules in regulating c-fos expression in the SON in vivo. It is expected that the AAV injection paradigm described in this paper will provide a useful approach for this endeavor.
4. Experimental procedures
4.1. Animals
Adult male Sprague–Dawley rats (270 g–370 g) obtained from Charles River Laboratories (Wilmington, MA) were maintained under normal laboratory conditions (temperature: 21–23 °C, 12 h light–dark cycles with light on at 6:00 AM) with access to unlimited food and drinking water. Following surgical procedures, rats were caged individually. All procedures were carried out in accordance with the National Institutes of Health (NIH) guidelines on the care and use of animals and according to an animal study protocol approved by the NINDS Animal Care and Use Committee.
4.2. A-CREB plasmid preparation
The CMV500 A-CREB plasmid (Ahn et al., 1998) was generously provided by C. Vinson (NIH, NCI). Preparation of the A-CREB virus required the use of an AAV precursor plasmid (modified pFastBac, Invitrogen) containing the CMV promoter and a dual reporter system of EGFP followed by a T2A sequence, followed by the sequence for mCherry. A WPRE sequence was placed just downstream of the mCherry. The A-CREB fragment was cloned into the plasmid in place of mCherry and just after the T2A sequence. The use of the T2A peptide allows for the same promoter to be used to drive the expression of multiple proteins (Chinnasamy et al., 2006; Szymczak et al., 2004; Yan et al., 2010). A diagram of the final construct can be seen in Fig. 1a.
Fig. 1.
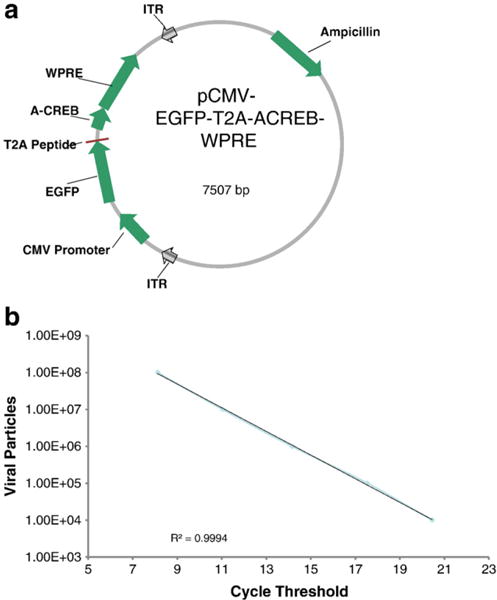
(a) Illustration of the plasmid used to construct the A-CREB AAV. In addition to the A-CREB, the construct contains an EGFP reporter separated by a T2A peptide, and therefore equimolar expression of the A-CREB and EGFP would be expected in each transduced cell. The expression of these genes in the AAV vector is driven by a CMV promoter. (b) Determination of virus titer was done by serially diluting the purified AAVs and subsequently measuring the corresponding amount of the EGFP moiety by quantitative real time PCR. The cycle threshold (Ct) values for the AAVs were compared to standard curves such as the standard dilution curve shown here (see Experimental procedure for details). In general, we injected AAVs with titers around 5×1012 vg/ml.
4.3. Virus preparation
Adeno-associated viruses (AAVs) were prepared according to the methods of Urabe et al. (2002). Briefly, DH10Bac cells (Invitrogen, cat# 10361-012) were transformed with donor AAV plasmids (either the A-CREB or the control EGFP) and plated on appropriate selection media containing tetracycline, kanamycin, gentamicin, X-gal and IPTG. If the donor plasmid is integrated into a composite bacmid, the resulting colony appears white and can be isolated and amplified for the next step.
Insect cells (Sf9, Gibco, cat# 11496-015) were plated on a 6-well plate at a density of 2×106 cells/ml and were then transfected with the appropriate bacmid. The transfection reactions were prepared in ~500 μl of Grace’s Medium (Gibco, cat# 11595) using 5 μg/well of bacmid and 5 μl/well of Cellfectin transfection reagent (Invitrogen, cat# 10362-010). Sf9 cells were transfected with the bacmid by the addition of ~250 μl of transfection reaction mixture to the well and the cells were maintained at 27 °C for 72–96 h. This resulted in bacculovirus P1 concentrations of ~1×107–9 viral genomes/ml (vg/ml) as assayed by quantitative PCR (F: 5′-ATGAAGCA-CAGTGTCCACGGTC-3′; R: 5′-GGTGAAGCGGCAGAATAACAATC-3′). An amplification step was then performed whereby the supernatant containing the P1 bacculovirus was added to 45–50 ml of Sf9 cells gently shaking in a 125 ml Erlenmeyer flask (Corning, cat# 431143) at 115 RPM and maintained 27.5 °C for 48–72 h. This resulted in bacculovirus concentrations of ~1×1011–12 vg/ml.
To make the injectible AAVs, the A-CREB or control EGFP bacculovirus was added along with a second AAV-6 packaging bacculovirus (generous gift from R. Kotin, NIH, NHLBI) to another 45–50 ml flask of Sf9 cells containing 2×106 cells/ml. The AAV-6 serotype was chosen because it was found to be among the most efficient in transducing magnocellular SON neurons in preliminary experiments in which we tested AAV-2, AAV-4, AAV-5, AAV-6, and AAV-8 (unpublished data). The bacculoviruses were added so that their final ‘virus:cell’ ratios were 3–10:1; controlling this ratio and keeping it relatively low appeared to result in higher AAV titers. Packaging reactions were performed over ~96 h until Sf9 cell viability was reduced to less than 50%, and then the contents of the flasks were poured into a 50 ml Falcon tube and centrifuged such that all of the insect cells were pelleted. The supernatant was then returned to the original flask, the pellet resuspended in 5 ml of sterile PBS and subjected to 3 freeze–thaw cycles, before another centrifugation (3500×g, 10 min). The supernatant was then returned to the flask, to which was added MgCl2 to a final concentration of 2 mM and 500 units of Benzonase (Sigma, cat# E1014-25KU). The flask was then maintained at 37 °C for 30 min, after which polyethylene glycol (Fluka, cat# 83271) was added to a final concentration of 2%. The flask was gently shaken at 4 °C overnight.
The contents of the flask were then poured into a 50 ml falcon tube and centrifuged for 15 min at 3500×g. The supernatant was discarded and the pellet was resuspended in 10– 12 ml of CsCl2 solution having a refractive index of 1.372. This suspension was then ultracentrifuged at 38 k RPM for 36 h at 25 °C and the resulting gradients were collected in 1.5 ml aliquots. The fractions having a refractive index between 1.378 and 1.368 (approximately four 1.5 ml fractions) were kept and pooled together in a centrifugal filter device (Millipore, cat# UFC910008) having a MW cutoff of 100 kDa. The filter devices were centrifuged at 3000×g for 20 min at 25 °C and, following the first centrifuge, 2 ml of PBS with 1 mM MgCl2 was added to the upper reservoir in order to dialyze out the CsCl2. Following 3 additions of PBS w/MgCl2, the resulting virus (usually ~20–100 μl) was placed in aliquots for storage at −80 °C.
4.4. Titering procedures
Determination of virus titer was done as indicated in Urabe et al. (2002). QPCR was performed on the concentrated AAVs to determine their titers. Briefly, we serially diluted the viruses (both AAVs and bacculoviruses) and subsequently measured the corresponding concentrations through quantitative real time PCR (Smartcycler, Cepheid, Sunnyvale, CA). In addition to 1ul of template, primers for EGFP (to measure AAV titers) or the bacculovirus backbone (to measure bacculovirus titers) were added to the SYBR green QPCR master mix (Applied Biosystems, Foster City, CA). The cycle threshold (Ct) values for the viruses were compared to standard curves. Fig. 1b demonstrates a typical standard dilution curve that was used to titer both the A-CREB and EGFP virus titers. The AAV titer was determined by comparison with a standard curve made using pFBOT100 plasmid DNA and using the following primers: EGFP F-ACCCTCGTGACCACCCTGAC and EGFP R-ACCTTGATGCCGTTCTTCTGC which amplify a 130 bp fragment of the EGFP gene. Viral samples were usually diluted 1:10,000 and approximately 2 μl of sample was used for the qPCR. To illustrate the calculation, a Ct measurement of 14 would correspond to 1×106 viral particles according to the graph or 1×1010 viral particles when taking into account the dilution factor. Since 2 μl was used in the sample this would be equivalent to 5×1012 vg/ml. PCR conditions were the same as for the rBV titers. AAV titers are expressed as viral genome/ml (vg/ml).
4.5. Stereotaxic surgery
Rats were anesthetized with 1–5% isoflurane (Baxter) in air using a Stoelting gas anesthesia adaptor for stereotaxic instruments (David Kopf Instruments, Tujunga, CA, Calbiochem Cat# 05-23-0101). The rats were placed in the stereotaxic instrument and the hair on top of the scalp was shaved. The scalp was sterilized by two rounds of application of betadine, followed by a 50:50 betadine/ethanol mixture, followed by 70% ethanol. A 15–20 mm incision was made using a size 15 scalpel starting rostral to bregma and ending just caudal to lambda. Ensuring a flat skull position was done by determination that both bregma and lambda were on the same horizontal plane. Two holes were drilled into the skull directly dorsal to the SON in order to lower the 30-guage syringe needles just over the SON bilaterally (Shahar et al., 2004). The stereotaxic coordinates were determined using the Paxinos and Watson (1986) atlas and were: 1.3 mm posterior to bregma, 1.8 mm lateral on each side, and 8.8 mm (for rats between 270 and 320 g) or 8.9 mm (for rats between 321 and 370 g) dorsal to ventral.
The experimental and control viruses were unilaterally injected over the SON in vivo by convection enhanced delivery (Szerlip et al., 2007) at a rate of 0.30 μl/min for 10 min (total injected volume=3 μl). Each rat received the experimental A-CREB virus injection on one side of the brain and the control virus injection on the contralateral side (Cetin et al., 2006). Before any immunohistochemical analysis was done, sections were studied under a fluorescent microscope to ensure in vivo targeting was accurate. Although the same stereotaxic coordinates were used in all surgical injections, occasional off-target injections did occur. Accordingly, all injections included in the analysis were verified to contain accurate targeting before proceeding with immunohistochemistry and qPCR.
Following these procedures, the incision was closed using interrupted sutures (Ethilon, Nylon Suture, Black Monofilament 4–0) and Ketoprofen was administered subdermally (5 mg/kg, diluted in 0.9% NaCl). Rats were then kept individually in cages (18′′/9′′/9′′–l/w/h) for two weeks to allow the vector to fully express.
4.6. Salt loading of rats
Rats were injected i.p. with 1 ml/100 g of either 1.5 M or 2 M NaCl, kept without access to water for 30 min, and then killed by decapitation. Magnocellular neurons of the SON synthesize Fos protein in response to injections of hypertonic saline, but not in response to injections of isotonic saline (Giovannelli et al., 1990; Kawasaki et al., 2009).
4.7. Immunohistochemistry
For experiments involving immunohistochemical assays, two weeks after the surgical injection, animals were injected with 2 M NaCl (1 ml/100 g). Two hours later, the animals were anesthetized with isoflurane and immediately perfused transcardially with 100 ml of 0.1 M PBS at 10 ml/min followed by 200 ml of 4% fresh paraformaldehyde (PFA) in 0.1 M PBS, Ph 7.4 at 10 ml/min. Brains were removed and post-fixed in PFA overnight. Brains were then cryoprotected in increasing concentrations of sucrose in 0.9% saline. Sucrose concentrations were 5%, 10%, and 15%.
Coronal sections (16 μm) were made on a cryostat (Reichert-Jung 2800; Frigocut, Heidelberg, Germany) and mounted onto coated slides (Fisher Scientific Cat# 12-550-19, Pittsburgh, PA). The sections were rinsed with 0.3% Triton-X in 0.1 M PBS for 5 min, followed by two rinses of 5 min each of PBS. The sections were incubated overnight at 4 °C followed by 2 h at room temperature with Fos antibody (sc-52, Santa Cruz Biotechnology Inc, Santa Cruz, CA) at a dilution ratio of 1:2000 in 0.1 M PBS. The slides were then rinsed 3 times with 0.1 M PBS, and then incubated in secondary antibody, Alexa Flour 594-conjugated donkey anti-rabbit prepared at a dilution of 1:500 (Molecular Probes, Eugene, OR), for two hours. Slides were then rinsed three times in 0.1 M PBS and coverslipped using Prolong Gold Antifade reagent (Molecular Probes) as a mounting medium. This procedure resulted in red Fos labeling together with native, green EGFP.
Stained sections were visualized using an epifuorescent microscope (Nikon Eclipse E400) was used. Representative images were then recorded using the accompanying camera (Q-Imaging RETIGA EXi Fast, Cooled Mono 12-bit). Optimizing the exposure time of the photograph to best display the representative images was done using Image-Pro Plus 5.1 (Media-Cybernetics, Inc) imaging software.
4.8. qRT-PCR
For experiments using qRT-PCR analyses, the rats received an intraperitoneal hypertonic 2 M NaCl solution injection (10 ml/kg) two weeks after the surgical procedure. Thirty minutes later, the rats were sacrificed under isoflurane anesthesia. The thirty minute time course was shown (Kawasaki et al., 2005) to be the duration for the examined genes to reach peak expression after hypertonic stimulation. The brains were quickly removed and placed in a 1X-Phosphate Buffer Saline (PBS) solution. A single 1 mm coronal hypothalamic slice containing the SONs, located at 0.8 to 1.8 mm in the Paxinos and Watson (1986) atlas, were made using a Jacobowitz brain slicer (Zivic Instruments, Pittsburgh, PA). The slice was then placed in a Petri dish filled with PBS and the left and right SONs were removed and placed in lysis/RNA stabilization buffer (Stratagene, La Jolla, CA) for later purification of the RNA.
The SON RNA was purified using an Absolutely RNA Miniprep Kit (Stratagene, La Jolla, CA, CAT #400800) according to the manufacturer’s protocol. To eliminate DNA contamination of the RNA, a Qiagen DNase reagent (CAT #79254) was used. Following purification, the RNA was quantified in triplicate on a spectrophotometer (NanoDrop, Wilmington, DE) and stored at −80 °C until further use. All RNA used had a 260/280 nm ratio between 1.90 and 2.10, representing minimal contamination by organic solutes.
Eight sets of primer pairs, several of which were reported previously (Kawasaki et al., 2009; Ponzio et al., 2007) were designed to measure VP hnRNA, VP mRNA, OT mRNA, c-fos mRNA, NGFI-A mRNA, NGFI-B mRNA, NR4A2 mRNA, and EGFP mRNA expression, the sequences can be seen in Table 1.
Table 1.
PCR primers used to assay immediate-early gene, vasopressin and EGFP mRNAs.
| Primers | Sequence | TA * | TM (°C) * |
|---|---|---|---|
| Rat c-fos mRNA primer pairs | 5′-AGCATGGGCTCCCCTGTCA (forward) 5′-GAGACCAGAGTGGGCTGCA (reverse) |
60 °C, 30 s | 83.4 |
| Rat NGFI-A mRNA primer pairs | 5′-GCATGCGTAATTTCAGTCGTAGTG (forward) 5′-GGCAAACTTTCTCCCACAAATG (reverse) |
60 °C, 30 s | 80.8 |
| Rat NGFI-B mRNA primer pairs | 5′-AAGATGCCGGTGACGTGCA (forward) 5′-CGGACTCTAGCAACAGGTCT (reverse) |
60 °C, 30 s | 80.5 |
| Rat NR4A2 mRNA primer pairs | 5′-AGTCTGATCAGTGCCCTCGT (forward) 5′-ATAGTCAGGGTTTGCCTGGAA (reverse) |
60 °C, 30 s | 81.7 |
| Rat VP mRNA primer pairs | 5′-TGCCTGCTACTTCCAGAACTGC (forward) 5′-AGGGGAGACACTGTCTCAGCTC (reverse) |
60 °C, 30 s | 80.7 |
| Rat OT mRNA primer pairs | 5′-AAGGCAGACTCAGGGTCG (forward) 5′-GGCATCTGCTGTAGCCCG (reverse) |
60 °C, 30 s | 84.5 |
| Rat EGFP mRNA primer pairs | 5′-GTGCCCATCCTGGTCGA (forward) 5′-CTTGCCGGTGGTGCAGA (reverse) |
60 °C, 30 s | 66.2 |
| Human c-fos mRNA primer pairs | 5′-TAGCCTCTCTTACTACCACTCA (forward) 5′-AGTGACCGTGGGAATGAAGTT (reverse) |
60 °C, 30 s | 84.2 |
TA, annealing temperature and time; TM, PCR product melting temperature.
Based on Kawasaki et al., 2009 and Ponzio et al., 2007.
The qRT-PCR tests used templates of 10 or 20 ng SON-enriched RNA from dissected tissue. Each of the aforementioned primer pairs was used at a concentration of 0.6 μM in separate one-step qRT-PCR reactions (Qiagen, Valencia CA). All qRT-PCR procedures were done according to the manufacturer’s protocol and were previously described in (Kawasaki et al., 2009; Ponzio et al., 2007). Annealing temperature was set at 60 °C for all primers. Ct values were calculated by the Real-Time PCR apparatus (Step-One-Plus, Applied Biosystems, Foster City, CA). The Ct values represent the PCR cycle number at which the measured fluorescence of the indicator dye, SYBR Green, increases linearly over the background; this represents the quantity of the amplified products. All reactions were run in triplicate and an average Ct value was calculated. One additional reaction was done using an absence of reverse transcriptase enzyme (−RT) in order to control for DNA contamination in the RNA templates. Only templates that had differences of at least one thousand-fold between the +RT compared to the −RT PCRs assays, indicative of little or no DNA contamination, were included in the analysis.
4.9. Tests of primer pairs for qRT-PCR of IEGs
In addition to examining Fos via immunohistochemistry, primers pairs were designed for use in qRT-PCR quantification of c-fos transcript and several other immediate-early genes. To test the primers, rat neuroblastoma (B35, ATCC CRL 2754) cells were grown in culture DMEM+10% FBS and then stimulated by incubation in 20 μm forskolin (Tocris 1099) for 30 min to stimulate cyclic AMP-sensitive immediate early genes (see Fig. 3).
4.10. Culturing of HEK-293t cells
To investigate the efficacy of the AAV A-CREB viral vector, we transfected HEK-293t cells that were then, 48 h later, stimulated by forskolin (10 μM, 30 min) to increase intracellular cAMP and subsequently increase the expression of c-fos. The cells were grown in a cell-culture medium made of 90% Dulbecco’s Modified Eagle’s Medium (DMEM) and 10% Fetal Bovine Serum (FBS) at 37 °C.
4.11. Statistical analysis
The Ct values±standard errors of the mean (SEMs) were calculated for each experiment and a paired Student’s t-test was used to compare the Ct values, with statistical significance set at p<0.05. Ct values of EGFP mRNA were used as a measure of virus targeting in the SON for each section extracted. Samples with an EGFP Ct value greater than 25 were not included in the analysis due to poor targeting. Moreover, the Ct value of OT mRNA was used as a measure for enrichment of the SON within the tissue section. Samples with OT mRNA Ct values greater than 20 were not included in the analysis due to lack of tissue enrichment. Relative quantification of c-fos mRNA was determined by the comparative cycle threshold method using the formula 2−ΔΔCt and data were normalized to VP mRNA in each sample (Pfaffl, 2001; Ponzio et al., 2007). The fold change in c-fos mRNA in each sample was calculated as: fold change=(2.0)ΔCt between experimental side and control side of SONs (c-fos mRNA)/(2.0)ΔCt experimental side and control side of SONs (VP mRNA) and is expressed as percent change from control.
Acknowledgments
This research was supported by the Intramural Research Program of the NINDS, NIH. We would like to thank Dr. Kory Johnson (NINDS, NIH) for his advice about statistical tests, and Dr. C. Vinson (NCI, NIH) for his gift of the A-CREB plasmid.
Contributor Information
Daniel Lubelski, Email: DLubelski@gmail.com.
Todd A. Ponzio, Email: ponziot@mail.nih.gov.
References
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima H, Baler R, Aguilera G. Fos proteins are not prerequisite for osmotic induction of vasopressin transcription in supraoptic nucleus of rats. Neurosci Lett. 2010;486:5–9. doi: 10.1016/j.neulet.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrot M, Wallace DL, Bolaños CA, Graham DL, Perrotti LI, Neve RL, Chambliss H, Yin JC, Nestler EJ. Regulation of anxiety and initiation of sexual behavior by CREB in the nucleus accumbens. Proc Natl Acad Sci U S A. 2005;102:8357–8362. doi: 10.1073/pnas.0500587102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutillier AL, Barthel F, Roberts JL, Loeffler JP. Beta-adrenergic stimulation of cFOS via protein kinase A is mediated by cAMP regulatory element binding protein (CREB)-dependent and tissue-specific CREB-independent mechanisms in corticotrope cells. J Biol Chem. 1992;267:23520–23526. [PubMed] [Google Scholar]
- Cahill MA, Janknecht R, Nordheim A. Signalling pathways: jack of all cascades. Curr Biol. 1996;6:16–19. doi: 10.1016/s0960-9822(02)00410-4. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, Duman RS, Neve RL, Nestler EJ. Regulation of cocaine reward by CREB. Science. 1998;282:2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- Cetin A, Komai S, Eliava M, Seeburg PH, Osten P. Stereotaxic gene delivery in the rodent brain. Nat Protoc. 2006;1:3166–3173. doi: 10.1038/nprot.2006.450. [DOI] [PubMed] [Google Scholar]
- Chiappini F, Cunha LL, Harris JC, Hollenberg AN. Lack of cAMP-response element-binding protein 1 in the hypothalamus causes obesity. J Biol Chem. 2011;286:8094–8105. doi: 10.1074/jbc.M110.178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnasamy D, Milsom MD, Shaffer J, Neuenfeldt J, Shaaban AF, Margison GP, Fairbairn LJ, Chinnasamy N. Multicistronic lentiviral vectors containing the FMDV 2A cleavage factor demonstrate robust expression of encoded genes at limiting MOI. Virol J. 2006;3:14. doi: 10.1186/1743-422X-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon V, Chebli K, Cavelier P, Blanchard JM. A novel mouse c-fos intronic promoter that responds to CREB and AP-1 is developmentally regulated in vivo. PLoS One. 2010;5:e11235. doi: 10.1371/journal.pone.0011235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco C, Damon DH, Endoh M, Wagner JA. Nerve growth factor induces transcription of NGFIA through complex regulatory elements that are also sensitive to serum and phorbol 12-myristate 13-acetate. Mol Endocrinol. 1993;7:365–379. doi: 10.1210/mend.7.3.8483478. [DOI] [PubMed] [Google Scholar]
- Doherty FC, Schaack JB, Sladek CD. Comparison of the efficacy of four viral vectors for transducing hypothalamic magnocellular neurons in the rat supraoptic nucleus. J Neurosci Methods. 2011;197:238–248. doi: 10.1016/j.jneumeth.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto K, Hommyo A, Yonemitsu R, Abe SI. ErbB4 signals neuregulin1-stimulated cell proliferation and c-fos gene expression through phosphorylation of serum response factor by mitogen-activated protein kinase cascade. Mol Cell Biochem. 2010;339:119–125. doi: 10.1007/s11010-009-0375-z. [DOI] [PubMed] [Google Scholar]
- Fass DM, Butler JEF, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J Biol Chem. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- Fisch TM, Prywes R, Simon MC, Roeder RG. Multiple sequence elements in the c-fos promoter mediate induction by cAMP. Genes Dev. 1989;3:198–211. doi: 10.1101/gad.3.2.198. [DOI] [PubMed] [Google Scholar]
- Gille H, Strahl T, Shaw PE. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr Biol. 1995;5:1191–1200. doi: 10.1016/s0960-9822(95)00235-1. [DOI] [PubMed] [Google Scholar]
- Ginty DD, Bonni A, Greenberg ME. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Giovannelli L, Shiromani PJ, Jirikowski GF, Bloom FE. Oxytocin neurons in the rat hypothalamus exhibit c-fos immunoreactivity upon osmotic stress. Brain Res. 1990;531:299–303. doi: 10.1016/0006-8993(90)90789-e. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hill CS, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaoka Y, Yazawa T, Uesaka M, Mizutani T, Yamada K, Miyamoto K. Regulation of NGFI-B/Nur77 gene expression in the rat ovary and in leydig tumor cells MA-10. Mol Reprod Dev. 2008;75:931–939. doi: 10.1002/mrd.20788. [DOI] [PubMed] [Google Scholar]
- Iwasaki Y, Oiso Y, Saito H, Majzoub JA. Positive and negative regulation of the rat vasopressin gene promoter. Endocrinology. 1997;138:5266–5274. doi: 10.1210/endo.138.12.5639. [DOI] [PubMed] [Google Scholar]
- Jancic D, Lopez de Armentia M, Valor LM, Olivares R, Barco A. Inhibition of cAMP response element-binding protein reduces neuronal excitability and plasticity, and triggers neurodegeneration. Cereb Cortex. 2009;19:2535–2547. doi: 10.1093/cercor/bhp004. [DOI] [PubMed] [Google Scholar]
- Kawasaki M, Yamaguchi K, Saito J, Ozaki Y, Mera T, Hashimoto H, Fujihara H, Okimoto N, Ohnishi H, Nakamura T, Ueta Y. Expression of immediate early genes and vasopressin heteronuclear RNA in the paraventricular and supraoptic nuclei of rats after acute osmotic stimulus. J Neuroendocrinol. 2005;17:227–237. doi: 10.1111/j.1365-2826.2005.01297.x. [DOI] [PubMed] [Google Scholar]
- Kawasaki M, Ponzio TA, Yue C, Fields RL, Gainer H. Neurotransmitter regulation of c-fos and vasopressin gene expression in the rat supraoptic nucleus. Exp Neurol. 2009;219:212–222. doi: 10.1016/j.expneurol.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara S, Arima H, Banno R, Sato I, Kondo N, Oiso Y. Regulation of vasopressin gene expression by cAMP and glucocorticoids in parvocellular neurons of the paraventricular nucleus in rat hypothalamic organotypic cultures. J Neurosci. 2003;23:10231–10237. doi: 10.1523/JNEUROSCI.23-32-10231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Cao R, Choi YS, Cho HY, Rhee AD, Hah CK, Hoyt KR, Obrietan K. The CREB/CRE transcriptional pathway: protection against oxidative stress-mediated neuronal cell death. J Neurochem. 2009;108:1251–1265. doi: 10.1111/j.1471-4159.2008.05864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Nielsen MB, Christensen ST, Hoffmann EK. Effects of osmotic stress on the activity of MAPKs and PDGFR-beta-mediated signal transduction in NIH-3T3 fibroblasts. Am J Physiol Cell Physiol. 2008;294:C1046–C1055. doi: 10.1152/ajpcell.00134.2007. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic; New York: 1986. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzio TA, Yue C, Gainer H. An intron-based real-time PCR method for measuring vasopressin gene transcription. J Neurosci Methods. 2007;164:149–154. doi: 10.1016/j.jneumeth.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera VM, Sheng M, Greenberg ME. The inner core of the serum response element mediates both the rapid induction and subsequent repression of c-fos transcription following serum stimulation. Genes Dev. 1990;4:255–268. doi: 10.1101/gad.4.2.255. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Miranti CK, Misra RP, Ginty DD, Chen RH, Blenis J, Greenberg ME. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol Cell Biol. 1993;13:6260–6273. doi: 10.1128/mcb.13.10.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P, Visvader J, Ferland L, Mellon PL, Verma IM. Induction of proto-oncogene fos transcription through the adenylate cyclase pathway: characterization of a cAMP-responsive element. Genes Dev. 1988;2:1529–1538. doi: 10.1101/gad.2.12a.1529. [DOI] [PubMed] [Google Scholar]
- Shahar T, House SB, Gainer H. Neural activity protects hypothalamic magnocellular neurons against axotomy-induced programmed cell death. J Neurosci. 2004;24:6553–6562. doi: 10.1523/JNEUROSCI.0886-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- Sheng M, Thompson MA, Greenberg ME. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- Simon AR, Severgnini M, Takahashi S, Rozo L, Andrahbi B, Agyeman A, Cochran BH, Day RM, Fanburg BL. 5-HT induction of c-fos gene expression requires reactive oxygen species and Rac1 and Ras GTPases. Cell Biochem Biophys. 2005;42:263–276. doi: 10.1385/CBB:42:3:263. [DOI] [PubMed] [Google Scholar]
- Soh JW, Lee EH, Prywes R, Weinstein IB. Novel roles of specific isoforms of protein kinase C in activation of the c-fos serum response element. Mol Cell Biol. 1999;19:1313–1324. doi: 10.1128/mcb.19.2.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlip NJ, Walbridge S, Yang L, Morrison PF, Degen JW, Jarrell ST, Kouri J, Kerr PB, Kotin R, Oldfield EH, Lonser RR. Real-time imaging of convection-enhanced delivery of viruses and virus-sized particles. J Neurosurg. 2007;107:560–567. doi: 10.3171/JNS-07/09/0560. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DAA. Correction of multi-gene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urabe M, Ding C, Kotin RM. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- Warburton EC, Glover CPJ, Massey PV, Wan H, Johnson B, Bienemann A, Deuschle U, Kew JNC, Aggleton JP, Bashir ZI, Uney J, Brown MW. cAMP responsive element-binding protein phosphorylation is necessary for perirhinal long-term potentiation and recognition memory. J Neurosci. 2005;25:6296–6303. doi: 10.1523/JNEUROSCI.0506-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang H, Xu Q, Jain N, Toxavidis V, Tigges J, Yang H, Yue G, Gao W. Signal sequence is still required in genes downstream of “autocleaving” 2A peptide for secretary or membrane-anchored expression. Anal Biochem. 2010;399:144–146. doi: 10.1016/j.ab.2009.11.032. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Iwasaki Y, Asai M, Takayasu S, Taguchi T, Itoi K, Hashimoto K, Oiso Y. Identification of a functional AP1 element in the rat vasopressin gene promoter. Endocrinology. 2006;147:2850–2863. doi: 10.1210/en.2005-1222. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Harley CW, Darby-King A, Neve RL, McLean JH. Early odor preference learning in the rat: bidirectional effects of cAMP response element-binding protein (CREB) and mutant CREB support a causal role for phosphorylated CREB. J Neurosci. 2003;23:4760–4765. doi: 10.1523/JNEUROSCI.23-11-04760.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Gao B, Duan L, Jiang S, Cao R, Xiong YF, Rao ZR. Acute hyperosmotic stimulus-induced Fos expression in neurons depends on activation of astrocytes in the supraoptic nucleus of rats. J Neurosci Res. 2010;88:1364–1373. doi: 10.1002/jnr.22297. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Li L, Papadopoulou N, Hodgson G, Evans E, Galbraith M, Dear M, Vougier S, Saxton J, Shaw PE. Mitogen-induced recruitment of ERK and MSK to SRE promoter complexes by ternary complex factor Elk-1. Nucleic Acids Res. 2008;36:2594–2607. doi: 10.1093/nar/gkn099. [DOI] [PMC free article] [PubMed] [Google Scholar]


