Abstract
A drug’s biological half-life is defined as the time required for the human body to metabolize or eliminate 50% of the initial drug dosage. Correctly measuring the half-life of a given drug is helpful for the safe and accurate usage of the drug. In this study, we investigated which gene ontology (GO) terms and biological pathways were highly related to the determination of drug half-life. The investigated drugs, with known half-lives, were analyzed based on their enrichment scores for associated GO terms and KEGG pathways. These scores indicate which GO terms or KEGG pathways the drug targets. The feature selection method, minimum redundancy maximum relevance, was used to analyze these GO terms and KEGG pathways and to identify important GO terms and pathways, such as sodium-independent organic anion transmembrane transporter activity (GO:0015347), monoamine transmembrane transporter activity (GO:0008504), negative regulation of synaptic transmission (GO:0050805), neuroactive ligand-receptor interaction (hsa04080), serotonergic synapse (hsa04726), and linoleic acid metabolism (hsa00591), among others. This analysis confirmed our results and may show evidence for a new method in studying drug half-lives and building effective computational methods for the prediction of drug half-lives.
Introduction
A drug is any substance that contributes to the relief of various pathological symptoms, which usually induces a pharmacological change in the human body [1–3]. In pharmacology, a pharmaceutical drug or medicine is defined as the functional component that is extracted from biological material or synthesized by the modern pharmaceutical synthesis industry [4]. Drugs, such as antibiotics, have been regarded as the most effective weapons for preventing various diseases in humans and maintaining health. Once drugs are consumed, they are gradually eliminated or metabolized by a specific hepatic microsomal enzyme system [5–7]. To measure the precise amount of time that a drug is effective and control the proper drug dosage, a specific drug parameter, the drug half-life, serves to standardize the use of drugs and avoid side effects [8, 9].
In pharmacology, the drug biological half-life (usually abbreviated half-life) has been defined as the time required for the human body to metabolize or eliminate 50% of the initial value of the functional drug dosage [10]. Similarly, the plasma half-life, another relevant parameter, is defined as the time that it takes for the concentration of the drug in the blood to decrease by 50% [11]. Generally, the two parameters are not equal, but they are closely related [12]. Considering that the real half-life of a specific drug is difficult to detect and measure in most situations (except for drugs with a high tissue residual ratio such as Digitoxin), we take the plasma half-life of drugs as the reference value [13, 14].
Generally, the half-life of a specific drug is affected by six main factors, including plasma protein binding, pharmacokinetic patterns, renal/hepatic diseases, active metabolites, enterohepatic circulation and the specific distribution of the drug volume [15]. All six factors contribute to the regulation of the biotransformation and excretion of a drug, which are two core mechanisms that affect the half-life of a drug [16].
The plasma protein binding affinity has been reported to contribute to the overall metabolic flow in a drug’s transportation, function and elimination. The binding affinity extensively affects the plasma half-life of drugs, which is easily detected, reinforcing the importance of this factor in practical applications [17, 18]. For example, the drug warfarin is an anticoagulant that has a long half-life because of its high binding affinity for plasma proteins [19]. The pharmacokinetic pattern, another factor, has two main metabolic trends for common drugs, first order kinetics and zero order kinetics. According to first order kinetics, a fixed fraction of a drug will be eliminated in a given unit of time, while for zero order kinetics, a fixed amount of a drug will be excreted. The metabolic variation of the two kinetic patterns are mainly influenced by the different metabolic routes of a drug and the dosage [20]. Because of the limited metabolic ability of human bodies, most drugs follow the first pharmacological pattern of metabolism but follow the zero order kinetic pattern of metabolism for toxic doses [21].
As mentioned above, the metabolic ability of human bodies may alter the pattern and speed of specific drugs [5–7]. In humans, the liver tissue contains various hepatic microsomal enzymes and has been found to be the primary location of drug metabolism [22]. Therefore, the metabolic abilities of the hepatic microsomal enzymes may greatly affect the speed of drug metabolism and may further influence their half-lives [23]. For the elimination processes, the kidney is the junction of the urinary system and the circulating system and also affects the half-life of drugs [24, 25]. For example, the accumulation of aminoglycosides has been confirmed during diseases of the kidney [26]. Considering the functions of the liver and kidney during drug metabolism and elimination, the half-life of a drug may be greatly altered by renal or hepatic pathological conditions.
Apart from the factors above, not all drugs are in their activated states when they are absorbed by the human body. Some drugs need to be changed into an activated state (reactive form) to produce pharmacological effects [27]. For example, the half-life of aspirin is fifteen minutes, while the effective metabolic product of aspirin, salicylic acid, has a half-life as long as two hours. This illustrates the crucial role of a drug’s state during its metabolism and elimination [28, 29]. During enterohepatic circulation, such factors extend the in vivo metabolic route of drugs and may further prolong the half-life of certain drugs [30], while the volume of distribution (the ratio of the plasma concentration to the total quantity of a drug (L/kg)) reflects the overall ability to eliminate certain drugs [31]. In total, all six factors are crucial for the biotransformation and excretion of drugs, reflecting the complex regulatory mechanisms that affect their half-lives.
Based on existing experimental methods, it is difficult and time-consuming to screen and verify the proteins and biological processes that may affect the half-life of a specific drug. Most efforts made toward predicting the half-lives of drugs have been based on drug structures. Turner et al. predicted human half-lives for 20 cephalosporins based on constitutional, topological, and quantum-chemicals descriptors [32]. Arnot et al. developed two half-life prediction models in human based on molecular fragments and an automated iterative fragment selection method [33]. Lu et al. predicted elimination half-life in human by seven machine learning methods and molecular descriptors [34]. However, there are few studies investigating the biological mechanisms that may affect the half-lives of drugs.
Here, we applied a computational method to extract functional gene ontology (GO) terms and biological pathways (KEGG pathways) that may affect the half-life of a specific drug. The enrichment of GO terms and KEGG pathways was used to determine their associations with drugs with known half-life values. A popular feature selection method, minimum redundancy maximum relevance (mRMR), was employed to analyze these features and indicated a role for several important GO terms and KEGG pathways in drug metabolism. An analysis of recent publications confirmed the relevance of some of the GO terms and biological pathways that were predicted to affect drug half-life.
Materials and Methods
Materials
The terminal half-life data of 670 drugs collected by Obach et al. [35] were used in this study and are provided in S1 Table. According to their half-lives, these drugs were classified into the following five categories: (1) compounds with half-lives less than 1 h; (2) compounds with half-lives between 1 and 4 h; (3) compounds with half-lives between 4 and 12 h; (4) compounds with half-lives between 12 and 24 h; and (5) compounds with half-lives greater than 24 h. After mapping 670 drugs to their PubChem IDs and excluding those without PubChem IDs, we obtained 669 drugs. Because each drug in this study was represented by the enrichment scores of GO terms and KEGG pathways, those without these scores were discarded, resulting in 565 drugs (comprising the set S). The distribution of these 565 drugs in the aforementioned five categories is listed in Table 1.
Table 1. The distribution of drugs in five half-life categories.
| Category label | Half-life (t1/2) | Number of drugs |
|---|---|---|
| 1 | Compounds with half-lives less than 1 h | 56 |
| 2 | Compounds with half-lives between 1 and 4 h | 231 |
| 3 | Compounds with half-lives between 4 and 12 h | 154 |
| 4 | Compounds with half-lives between 12 and 24 h | 61 |
| 5 | Compounds with half-lives greater than 24 h | 63 |
Protein-chemical interactions
In this study, we investigated which GO terms and KEGG pathways were associated with effects on drug half-life. However, it was difficult to quantitatively evaluate the correlation between drugs and GO terms or KEGG pathways, which complicated further analyses. The annotated proteins for each GO term and KEGG pathway were easily obtained from public databases. Once proteins related to a specific drug were identified, the correlation between that drug and a GO term or KEGG pathway was measured using the proteins annotating to the GO term or KEGG pathway and those related to the drug.
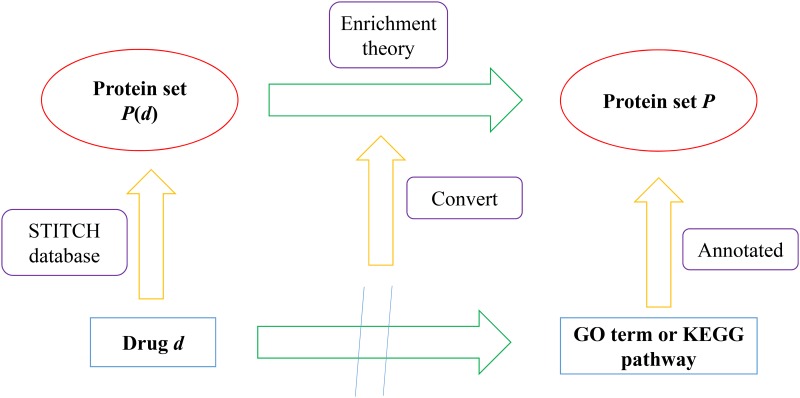
To obtain the proteins related to a specific drug, we downloaded the protein-chemical interactions from the STITCH (Search Tool for Interactions of Chemicals) database [36]. The interactions, including chemical-chemical and protein-chemical interactions, reported in STITCH are derived from experiments, databases and the literature. Thus, they can be used to determine the associations between chemicals and proteins and have been used to address several biological problems [37–43]. We extracted the protein-chemical interactions from the downloaded file ‘protein_chemical.links.v4.0.tsv.gz‘, such that chemicals were members in S and proteins were in human tissues, from which a protein set denoted by P(d) could be accessed for each drug d in S. Subsequently, the associations between one drug d and one GO term or KEGG pathway could be converted to the correlation between two protein sets, where one was P(d) and the other consisted of proteins annotated with the GO term or KEGG pathway. This idea is illustrated in Fig 1.
Fig 1. A figure illustrating how the associations between drugs and GO terms or KEGG pathways were measured.
Encoding scheme
As mentioned in Section “Protein-chemical interactions”, based on the protein-chemical interactions and the GO term or KEGG pathway protein annotations, we evaluated the associations between one drug d and one GO term or KEGG pathway by measuring the correlation between P(d) and the set consisting of proteins associated with a GO term or KEGG pathway. Here, we adopted the enrichment theory to quantify the correlation between two protein sets.
GO enrichment score
For a given drug d and one GO term GOj, let PGO denote the set consisting of proteins annotated with GOj. The GO enrichment score between d and GOj is defined as the hypergeometric test P value [44–46] of P(d) and PGO, which can be computed by:
| (1) |
where N and M denote the total number of human proteins and the number of proteins in PGO; n and m represent the number of proteins in P(d) and the number of proteins both in P(d) and PGO. The higher the score is, the stronger the correlation between drug d and GO term GOj. In total, 17,094 GO enrichment scores were calculated in this study for each drug.
KEGG enrichment score
A similar method was used to define the KEGG enrichment score, which can measure the associations between drugs and KEGG pathways. Let PKEGG denote the set consisting of proteins associated with a KEGG pathway Kj. The KEGG enrichment score between d and Kj is defined to be the hypergeometric test P value [46] of P(d) and PKEGG. Its computational formula is listed below:
| (2) |
where the parameters N and n have the same definitions as those in Eq 1, while M and m denote the number of proteins in PKEGG and the number of proteins both in P(d) and PKEGG. Similarly, a large KEGG enrichment score means a strong association between the drug and the pathway. In total, 279 KEGG pathway enrichment scores were calculated in this study for each drug.
The number of GO enrichment scores was much larger than that of the KEGG enrichment scores. Furthermore, the principles for selecting important GO terms and KEGG pathways were not the same. Thus, for each drug in dataset S, we obtained separate GO and KEGG enrichment scores resulting in the two datasets of SGO and SKEGG. Drugs in SGO were represented by 17,094 GO enrichment scores, and those in SKEGG were represented by 279 KEGG enrichment scores.
mRMR method
As described in Section “Encoding scheme”, each of the drugs in dataset S had 17,094 GO enrichment scores and 279 KEGG enrichment scores that denoted the strength of their association with a given GO term or KEGG pathway. It is obvious that not all GO terms or KEGG pathways have an equal effect on drug half-life; some of them are more important than others. To extract important GO terms and KEGG pathways, the mRMR selection method [47] was employed. This method is useful for analyzing various features and identifying the most important ones, and it has been widely used by investigators to address several biological problems [48–56]. Two excellent criteria were introduced in the mRMR method: Max-Relevance and Min-Redundancy, in which the former criterion guarantees that features with high relevance with targets can receive high ranks, and the latter one guarantees that a feature with lowest redundancies to already-selected features has priority to be selected. Two feature lists can be obtained by the mRMR method, one is called MaxRel feature list and the other is called mRMR feature list. The former list only uses the criterion of Max-Relevance, i.e., features in this list are ranked according to their relevance with targets, while the latter list uses both two criteria to rank features. It is clear that the mRMR feature list can be used to extract an optimal subspace of features for classification, while the MaxRel feature list can be adopted to access important features. Because the purpose of this study is to investigate important factors for determination of drug half-life, we only used the MaxRel feature list yielded by mRMR method. To measure the relevance between a feature f and targets, let x denote the target variable representing the drugs’ class labels, and y denote a variable representing all values under the feature f. The relevance between the target and the feature f is defined as the mutual information (MI) between x and y, which can be computed by:
| (3) |
where p(x) and p(y) are the marginal probabilities of x and y and p(x,y) is the joint probabilistic distribution of x and y. Accordingly, each feature (one GO term or KEGG pathway) was assigned an MI value, and all features were sorted by the descending order of their MI values in the MaxRel feature list. We selected the GO terms and KEGG pathways with high MI values for further analyses. The mRMR program was downloaded from http://research.janelia.org/peng/proj/mRMR.
Results and Discussion
Results of the mRMR method
As described in Section “mRMR method”, a popular feature selection method, the mRMR method, was adopted to extract important GO terms and KEGG pathways that may affect drug half-life. The mRMR program was used to produce MaxRel feature lists for SGO and SKEGG, which are provided in S2 and S3 Tables, respectively. Because our computational power was limited, we only output the first 500 features in the MaxRel feature list for GO terms.
Because GO terms or KEGG pathways with high MI values were more likely to affect drug half-life, we selected a threshold of 0.03 for the MI values of GO terms and 0.013 for the MI values of KEGG pathways. Subsequently, we obtained 23 GO terms and 18 KEGG pathways, which are listed in Tables 2 and 3, respectively. The biological characteristics and properties are analyzed extensively in the following section, producing several useful and important conclusions or suggestions for the study of drug half-lives.
Table 2. Important GO terms obtained by the mRMR method and which may be associated with different drug half-lives.
| Order | GO term ID | Name | MI value |
|---|---|---|---|
| 1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity | 0.037 |
| 2 | GO:0060033 | anatomical structure regression | 0.036 |
| 3 | GO:0050998 | nitric-oxide synthase binding | 0.036 |
| 4 | GO:0035115 | embryonic forelimb morphogenesis | 0.035 |
| 5 | GO:0046972 | histone acetyltransferase activity (H4-K16 specific) | 0.034 |
| 6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific) | 0.034 |
| 7 | GO:0043996 | histone acetyltransferase activity (H4-K8 specific) | 0.034 |
| 8 | GO:0050805 | negative regulation of synaptic transmission | 0.034 |
| 9 | GO:0042364 | water-soluble vitamin biosynthetic process | 0.032 |
| 10 | GO:0001533 | cornified envelope | 0.031 |
| 11 | GO:0008504 | monoamine transmembrane transporter activity | 0.031 |
| 12 | GO:0021853 | cerebral cortex GABAergic interneuron migration | 0.031 |
| 13 | GO:0021830 | interneuron migration from the subpallium to the cortex | 0.031 |
| 14 | GO:0021894 | cerebral cortex GABAergic interneuron development | 0.031 |
| 15 | GO:0021534 | cell proliferation in hindbrain | 0.031 |
| 16 | GO:0001965 | G-protein alpha-subunit binding | 0.03 |
| 17 | GO:1901386 | negative regulation of voltage-gated calcium channel activity | 0.03 |
| 18 | GO:0021924 | cell proliferation in external granule layer | 0.03 |
| 19 | GO:0021930 | cerebellar granule cell precursor proliferation | 0.03 |
| 20 | GO:0046341 | CDP-diacylglycerol metabolic process | 0.03 |
| 21 | GO:0019992 | diacylglycerol binding | 0.03 |
| 22 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity | 0.03 |
| 23 | GO:0090177 | establishment of planar polarity involved in neural tube closure | 0.03 |
Table 3. Important KEGG pathways obtained by the mRMR method and which may associated with different drug half-lives.
| Order | KEGG pathway ID | Name | MI value |
|---|---|---|---|
| 1 | hsa04080 | Neuroactive ligand-receptor interaction | 0.026 |
| 2 | hsa00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 0.024 |
| 3 | hsa05322 | Systemic lupus erythematosus | 0.02 |
| 4 | hsa04726 | Serotonergic synapse | 0.018 |
| 5 | hsa00591 | Linoleic acid metabolism | 0.017 |
| 6 | hsa05213 | Endometrial cancer | 0.016 |
| 7 | hsa00531 | Glycosaminoglycan degradation | 0.016 |
| 8 | hsa04146 | Peroxisome | 0.016 |
| 9 | hsa00100 | Steroid biosynthesis | 0.015 |
| 10 | hsa00603 | Glycosphingolipid biosynthesis—globo serie | 0.015 |
| 11 | hsa04530 | Tight junction | 0.014 |
| 12 | hsa04666 | Fc gamma R-mediated phagocytosis | 0.014 |
| 13 | hsa00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 0.013 |
| 14 | hsa04610 | Complement and coagulation cascades | 0.013 |
| 15 | hsa00240 | Pyrimidine metabolism | 0.013 |
| 16 | hsa04020 | Calcium signaling pathway | 0.013 |
| 17 | hsa04725 | Cholinergic synapse | 0.013 |
| 18 | hsa00280 | Valine, leucine and isoleucine degradation | 0.013 |
Analysis of important GO terms and KEGG terms for drug half-life
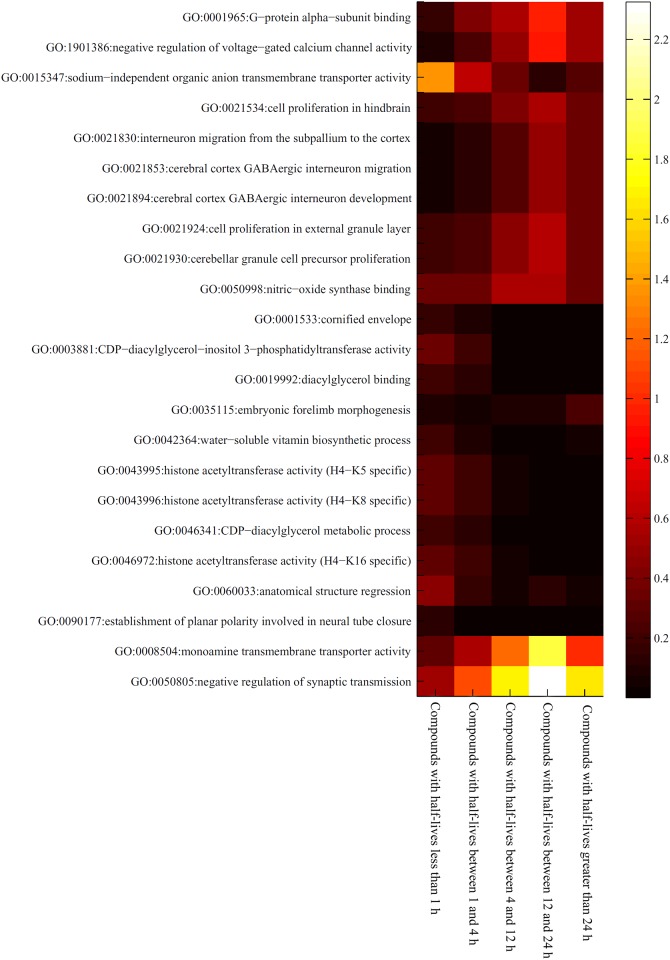
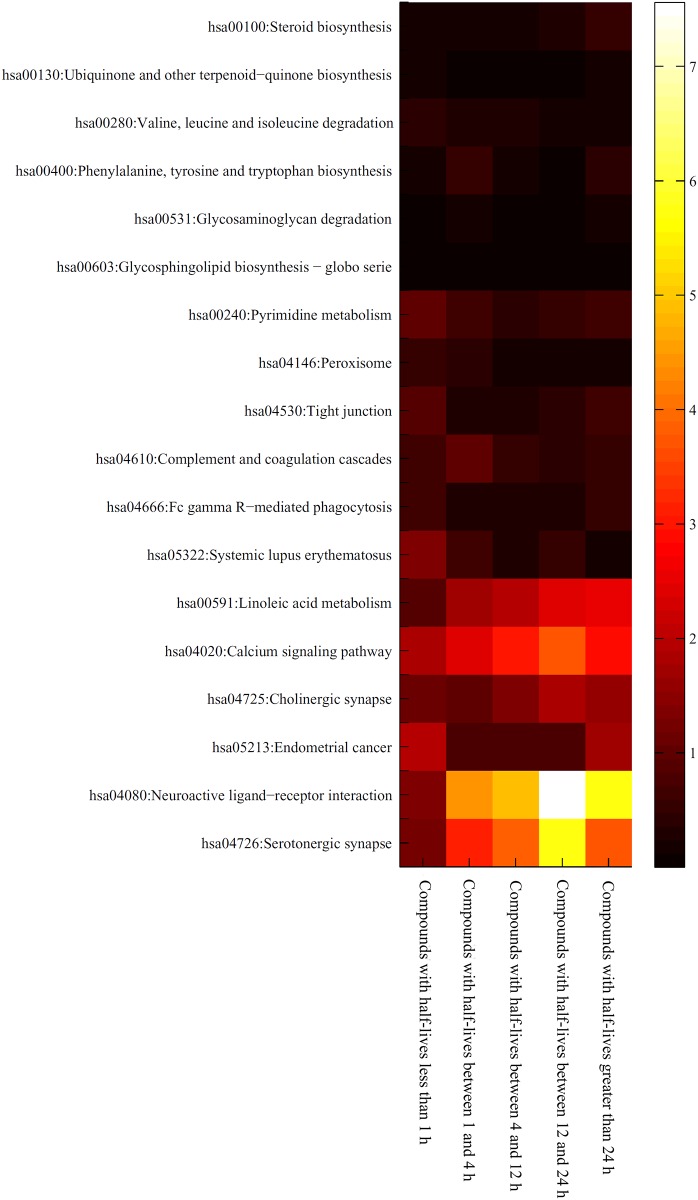
As mentioned in Section “Results of the mRMR method”, 23 GO terms and 18 KEGG pathways were identified that may have an effect on the half-lives of drugs. However, it is difficult to analyze these GO terms and KEGG pathways because drugs with different half-lives received different enrichment scores, even those drugs belonging to the same half-life category. To obtain a better understanding of the associations between a GO term or KEGG pathway and a half-life category, we calculated a “level value” for each GO term or KEGG pathway and each category. The level value was the mean of the enrichment scores for drugs in the category under the GO term or KEGG pathway. The level values for GO terms and KEGG pathways are provided in S4 and S5 Tables, respectively. In addition, for easier visualization, we plotted two heat maps of these values, one is for GO terms (shown in Fig 2), and the other is for KEGG pathways (shown in Fig 3). It can be observed that some GO terms and KEGG pathways are strongly associated with certain half-life categories. Table 4 lists the related GO terms and KEGG pathways for each half-life category. These are discussed in detail in the following sections.
Fig 2. Heat map of the level values for important GO terms in five drug half-life categories.
The rows represent GO terms and the columns represent the drug half-life categories.
Fig 3. Heat map of the level values for important KEGG pathways in five categories of drug half-life categories.
The rows represent KEGG pathways and the columns represent drug half-life categories.
Table 4. The relationship between different drug half-lives and GO terms and KEGG pathways discovered in this study.
| Half-life (t1/2) | Related GO terms | Related KEGG pathways |
|---|---|---|
| Compounds with half-lives less than 1 h | GO: 0046972, GO: 0043995, GO:0015347 | hsa05322 |
| Compounds with half-lives between 1 and 4 h | --- | hsa00400, hsa00531, hsa04610 |
| Compounds with half-lives between 4 and 12 h | GO: 0050998 | --- |
| Compounds with half-lives between 12 and 24 h | GO: 0050805, GO: 0008504, GO: 1901386, GO: 0021924 | hsa04080, hsa04726 |
| Compounds with half-lives greater than 24 h | GO: 0035115 | hsa00591, hsa00100 |
GO terms and KEGG pathways related to compounds with half-lives less than 1 h
Nearly all the GO terms and KEGG pathways contributed to the metabolism and elimination of compounds with half-lives less than 1 h. Considering that both the intake and excretion of drugs require a significant amount of time, drugs that have a half-life less than 1 hour either have a rapid biotransformation process or lack one altogether. GO terms (GO: 0046972, GO: 0043995) that contributed to histone trans-acetylation processes were found to participate in the metabolism of drugs with half-lives less than 1 hour. Histone acetylation and deacetylation processes have been reported to be regulated by specific activators and inhibitors [57, 58]. Most of the drugs that were associated with these two GO terms have been reported to have half-lives less than 1 h. For example, experimental data from rats indicated that the antitumor drug TSA has a half-life of approximately 6 minutes and a 50 μM dosage is completely inactivated within 40 minutes, thus validating our classification and prediction [59]. Additionally, some of the drugs with half-lives less than 1 hour are enriched for sodium−independent organic anion transmembrane transporter activity (GO:0015347), as shown in Fig 2. According to recent publications, drugs that are associated with this biological process have very short half-lives [60, 61]. Niacin and Alprostadil are two classical drugs that are associated with this process and both of their half-lives are short (20–45 minutes for Niacin and approximately 42 seconds for Alprostadil (PGE1)) [61–63]. KEGG pathways enriched in drugs with half-lives less than one hour included the systemic lupus erythematosus pathway (hsa05322). Drugs used to treat systemic lupus erythematosus such as prednisone, also have a very short half-life (plasma half-life of less than 1 hour) [64], which is consistent with our results.
GO terms and KEGG pathways related to compounds with half-lives between 1 and 4 h
Unlike drugs with half-lives less than 1 hour, fewer GO terms and KEGG pathways were associated with drugs that have half-lives between 1 and 4 hours. These drugs were enriched in only 3 KEGG pathways (hsa00400, hsa00531 and hsa04610) and no GO terms. The KEGG pathway hsa00400 describes phenylalanine, tyrosine and tryptophan biosynthesis. Tetrahydrobiopterin (modified as sapropterin dihydrochloride in drugs) is mainly applied for the treatment of specific diseases such as tetrahydrobiopterin deficiency and neurotransmitter related disorders in the nervous system [65, 66]. The half-life of orally administered sapropterin has been reported to be 4 hours, which is consistent with our prediction [67]. The KEGG pathway hsa00531 describes the glycosaminoglycan degradation biological process. This process is associated with drugs such as chondroitin sulfate, elosulfase alfa and nadroparin [68–70]. Chondroitin sulfate reduces the fat in the blood stream [71, 72]. Considering its chemical nature, a sulfated glycosaminoglycan with a half-life of less than 4 hours, the metabolism and elimination processes of this drug likely involve the predicted KEGG pathway [73, 74]. The KEGG pathway for complement and coagulation cascades (hsa04610) also functions for intercellular substances, which suggests that this metabolic route may be utilized by related drugs. The coagulation factor VIIa is a significant component of the coagulation cascades and definitely participates in the predicted KEGG pathway [75]. This and similar drugs have a half-life of exactly 3.5 hours (between 1 h and 4 h), thereby confirming our prediction of the involvement of this KEGG pathway [76, 77].
GO terms and KEGG pathways related to compounds with half-lives between 4 and 12 h
Only one GO term (GO:0050998) was enriched for compounds with a half-life between 4–12 hours. GO:0050998 describes nitric−oxide synthase binding activity. Nitric oxide itself is a drug with a half-life of a few seconds in the blood [78]. However, a group of drugs that contribute to the synthesis of nitric-oxide and may participate with the synthase binding associated pathways have been confirmed to have half-lives between 4 and 12 hours [79]. For example, NXN-188 has been confirmed to be associated with the nitric-oxide synthase binding processes and has a corresponding half-life of more than four hours (8–10 hours). Thus, this result also verifies our prediction and classification [80, 81] for this drug. No KEGG pathways were enriched for drugs with half-lives of 4–12 hours.
GO terms and KEGG pathways related to compounds with half-lives between 12 and 24 h
Compared to compounds with half-lives between 4 and 12 hours, more GO terms and KEGG pathways were enriched for drugs with half-lives between 12 and 24 hours. As shown in Fig 2, we identified two GO terms (GO: 0050805; GO: 0008504) that may be related to compounds with half-lives between 12 and 24 hours. A negative regulation of synaptic transmission (GO: 0050805) has been shown to be regulated by various drugs such as imipramine, alfentanil and anileridine. Imipramine is a common antidepressant drug that has a half-life of 20 hours, which agrees with our prediction. [82, 83]. The GO term GO: 0008504 refers to monoamine transmembrane transporter activity. The drug transdermal selegiline is involved in this process and has been reported to have a half-life of 18–25 hours, indicating that our prediction was accurate [84, 85]. The term GO: 1901386 is associated with voltage-gated calcium channels, which are targeted by various drugs [86–88]. Flecainide is a crucial antiarrhythmic drug that regulates the voltage-gated calcium channel and has a specific half-life of 12–27 hours in normal pathological conditions [89–91]. Another GO term, GO: 0021924, refers to cell proliferation in the external granule layer, which has been reported to be targeted by the drug methylazoxymethanol. This drug has a half-life of 12 hours in solution [92, 93]. Several enriched KEGG pathways were associated with drugs in the 12–24 hour half-life category. Hsa04080 describes neuroactive ligand-receptor interactions, which are targeted by drugs such as hydroxyzine [94]. The half-life of hydroxyzine has been reported to be as short as 3 hours; however, the hydroxyzine derivative pamoate has been shown to have a half-life of approximately 20 hours [95, 96]. From the heat maps in Fig 3, it is evident that compounds with half-lives greater than 1 hour (especially compounds with half-lives between 12 and 24 hours) are enriched in this KEGG pathway. The serotonergic synapse pathway (hsa04726) is affected by trimipramine, an important antihistamine and sedative with an exact half-life of 23–24 hours [97, 98]. Fig 3 also shows that compounds with half-lives between 12 and 24 hours are enriched in this KEGG pathway, which supports the accuracy and efficacy of our prediction.
GO terms and KEGG pathways related to compounds with half-lives greater than 24 h
Some drugs have very long half-lives that are greater than 24 hours. Only one GO term has been predicted to be associated with such long-acting drugs. The term GO: 0035115 refers to embryonic forelimb morphogenesis. It is well known that the Hedgehog pathway contributes to normal developmental processes in the human embryo. Therefore, Hedgehog pathway inhibitors such as GDC-0449 and AAG are strongly associated with this GO term [99, 100]. Based on recent publications, the half-lives of these drugs are longer than a day (more than 7 days and 5 days for GDC-0449 and AAG, respectively) [100, 101]. Unlike the single enriched GO term, more KEGG pathways were associated with the metabolism of drugs whose half-lives were greater than 24 hours. Linoleic acid metabolism (hsa00591) is a crucial pathway for fat metabolism that is used in our daily lives [102]. This process is targeted by two derivatives of linoleic acid, di-homo-gamma-linolenic acid and alpha-linolenic acid. The half-lives of both these linoleic acid derivatives are greater than 24 hours (more than 60 hours for both di-homo-gamma-linolenic acid and alpha-linolenic acid) [103, 104]. Another similar metabolic process, steroid biosynthesis (hsa00100), was also enriched for drugs with half-lives greater than 24 hours. Pathways for steroid synthesis have been reported to be associated with rheumatoid arthritis. The drug aurothioglucose, which has been used to treat rheumatoid arthritis, has a half-life of 3–27 days, which is consistent with both our prediction and classification [105, 106].
Conclusions
This study used the mRMR method to investigate the important GO terms and KEGG pathways that may affect a drug’s half-life. The GO terms and KEGG pathways identified may provide new insights for studying drug half-life and help us build effective prediction models for drug half-lives. We hope that this study can promote pharmacological studies of the drug metabolism mechanism and expand the understanding of half-life-associated biological processes. In future, we will make our efforts in the following two points: (1) Effective models for prediction of drug half-life using some advanced machine learning algorithms [107, 108] can be built based on the extracted GO terms and KEGG pathways; (2) Refined half-life analysis of drugs on certain disease using abundant known information of this disease, such as disease-related target proteins, disease-related microRNA [109, 110], etc.
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31371335, 61303099), Shanghai Sailing Program and The Youth Innovation Promotion Association of Chinese Academy of Sciences (CAS) (2016245).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
National Natural Science Foundation of China (31371335, 61303099), Shanghai Sailing Program and The Youth Innovation Promotion Association of Chinese Academy of Sciences (CAS) (2016245). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Miyasato K. [The definition of drug dependence]. Nihon rinsho Japanese journal of clinical medicine. 2010;68(8):1431–6. . [PubMed] [Google Scholar]
- 2.Johne A, Gerloff T, Mai I. [Clinical drug investigations. Definition of terms]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2005;48(4):397–400. 10.1007/s00103-005-1012-y . [DOI] [PubMed] [Google Scholar]
- 3.Wang RI. The definition and scope of clinical pharmacology. Jama. 1980;243(19):1901–2. . [PubMed] [Google Scholar]
- 4.Modell W. Symposium on clinical drug evaluation and human pharmacology. Introduction. Clinical pharmacology: a definition. Clinical pharmacology and therapeutics. 1962;3:235–8. . [PubMed] [Google Scholar]
- 5.Orton TC, Parker GL. The effect of hypolipidemic agents on the hepatic microsomal drug-metabolizing enzyme system of the rat. Induction of cytochrome(s) P-450 with specificity toward terminal hydroxylation of lauric acid. Drug metabolism and disposition: the biological fate of chemicals. 1982;10(2):110–5. . [PubMed] [Google Scholar]
- 6.Down WH, Chasseaud LF, Grundy RK. Effect of silybin on the hepatic microsomal drug-metabolising enzyme system in the rat. Arzneimittelforschung. 1974;24(12):1986–8. . [PubMed] [Google Scholar]
- 7.Hino Y, Imai Y, Sato R. Induction by phenobarbital of hepatic microsomal drug-metabolizing enzyme system in partially hepatectomized rats. J Biochem. 1974;76(4):735–44. . [PubMed] [Google Scholar]
- 8.Berezhkovskiy LM. Prediction of drug terminal half-life and terminal volume of distribution after intravenous dosing based on drug clearance, steady-state volume of distribution, and physiological parameters of the body. J Pharm Sci. 2013;102(2):761–71. 10.1002/jps.23396 . [DOI] [PubMed] [Google Scholar]
- 9.Moriyama B, Falade-Nwulia O, Leung J, Penzak SR, C JJ, Huang X, et al. Prolonged half-life of voriconazole in a CYP2C19 homozygous poor metabolizer receiving vincristine chemotherapy: avoiding a serious adverse drug interaction. Mycoses. 2011;54(6):e877–9. 10.1111/j.1439-0507.2011.02016.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebo H. Definition of Half-Life of Drug. Drug Intel Clin Phar. 1973;7(8):357-. WOS:A1973Q486400007. [Google Scholar]
- 11.Hindmarsh PC, Charmandari E. Variation in absorption and half-life of hydrocortisone influence plasma cortisol concentrations. Clin Endocrinol. 2015;82(4):557–61. 10.1111/cen.12653. WOS:000350982900014. [DOI] [PubMed] [Google Scholar]
- 12.Johnson KE, Makanji Y, Temple-Smith P, Kelly EK, Barton PA, Al-Musawi SL, et al. Biological activity and in vivo half-life of pro-activin A in male rats. Mol Cell Endocrinol. 2016;422:84–92. 10.1016/j.mce.2015.12.007 . [DOI] [PubMed] [Google Scholar]
- 13.Bohmer T, Roseth A. Prolonged digitoxin half-life in very elderly patients. Age Ageing. 1998;27(2):222–4. . [DOI] [PubMed] [Google Scholar]
- 14.Bluschke V, Viana AP. Digitoxin plasma half-life in the dog after administration of toxic doses. Arzneimittelforschung. 1976;26(4):591–2. . [PubMed] [Google Scholar]
- 15.Mahdi AJ, Obaji SG, Collins PW. Role of enhanced half-life factor VIII and IX in the treatment of haemophilia. Br J Haematol. 2015;169(6):768–76. 10.1111/bjh.13360 . [DOI] [PubMed] [Google Scholar]
- 16.Kontermann RE. Strategies for extended serum half-life of protein therapeutics. Curr Opin Biotechnol. 2011;22(6):868–76. 10.1016/j.copbio.2011.06.012 . [DOI] [PubMed] [Google Scholar]
- 17.Malm M, Bass T, Gudmundsdotter L, Lord M, Frejd FY, Stahl S, et al. Engineering of a bispecific affibody molecule towards HER2 and HER3 by addition of an albumin-binding domain allows for affinity purification and in vivo half-life extension. Biotechnol J. 2014;9(9):1215–22. 10.1002/biot.201400009 . [DOI] [PubMed] [Google Scholar]
- 18.Lindgren J, Refai E, Zaitsev SV, Abrahmsen L, Berggren PO, Karlstrom AE. A GLP-1 receptor agonist conjugated to an albumin-binding domain for extended half-life. Biopolymers. 2014;102(3):252–9. 10.1002/bip.22474 . [DOI] [PubMed] [Google Scholar]
- 19.Yan JH, Meyers D, Lee Z, Danis K, Neelakantham S, Majumdar T, et al. Pharmacokinetic and pharmacodynamic drug-drug interaction assessment between pradigastat and digoxin or warfarin. Journal of Clinical Pharmacology. 2014;54(7):800–8. 10.1002/jcph.285. WOS:000337627100009. [DOI] [PubMed] [Google Scholar]
- 20.Gabrielsson J, Meibohm B, Weiner D. Pattern Recognition in Pharmacokinetic Data Analysis. Aaps J. 2016;18(1):47–63. 10.1208/s12248-015-9817-6. WOS:000367529900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hojo R, Stern S, Zareba G, Markowski VP, Cox C, Kost JT, et al. Sexually dimorphic behavioral responses to prenatal dioxin exposure. Environmental Health Perspectives. 2002;110(3):247–54. WOS:000174311300023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustafsson F, Foster AJ, Sarda S, Bridgland-Taylor MH, Kenna JG. A Correlation Between the In Vitro Drug Toxicity of Drugs to Cell Lines That Express Human P450s and Their Propensity to Cause Liver Injury in Humans. Toxicological Sciences. 2014;137(1):189–211. 10.1093/toxsci/kft223. WOS:000329133400019. [DOI] [PubMed] [Google Scholar]
- 23.Megaraj V, Ding XX, Fang C, Kovalchuk N, Zhu Y, Zhang QY. Role of Hepatic and Intestinal P450 Enzymes in the Metabolic Activation of the Colon Carcinogen Azoxymethane in Mice. Chemical Research in Toxicology. 2014;27(4):656–62. 10.1021/tx4004769. WOS:000334902100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YN, Wang SH, Li T, Wang Q, Tu W, Cai K, et al. Shiga toxin type 2 (Stx2), a potential agent of bioterrorism, has a short distribution and a long elimination half-life, and induces kidney and thymus lesions in rats. Archives of toxicology. 2011;85(9):1133–40. 10.1007/s00204-010-0639-0. WOS:000294169200012. [DOI] [PubMed] [Google Scholar]
- 25.Keller F. Elimination Half-life, Time of Fractional Effect Duration and Administration Interval for Sitagliptin in Patients with Kidney Failure. N-S Arch Pharmacol. 2011;383:7-. WOS:000288573100026. [Google Scholar]
- 26.Worboys PD, Wong SL, Barriere SL. Pharmacokinetics of intravenous telavancin in healthy subjects with varying degrees of renal impairment. European Journal of Clinical Pharmacology. 2015;71(6):707–14. 10.1007/s00228-015-1847-6. WOS:000354401500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou K, Khokhar JY, Zhao B, Tyndale RF. First demonstration that brain CYP2D-mediated opiate metabolic activation alters analgesia in vivo. Biochemical Pharmacology. 2013;85(12):1848–55. 10.1016/j.bcp.2013.04.014. WOS:000320429200014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trnavsky K, Zachar M. Correlation of Serum Aspirin Esterase-Activity and Half-Life of Salicylic-Acid. Agents Actions. 1975;5(5):549–52. 10.1007/Bf01972693. WOS:A1975BE30600070. [DOI] [PubMed] [Google Scholar]
- 29.Miners JO, Grgurinovich N, Whitehead AG, Robson RA, Birkett DJ. Influence of Gender and Oral-Contraceptive Steroids on the Metabolism of Salicylic-Acid and Acetylsalicylic-Acid. British Journal of Clinical Pharmacology. 1986;22(2):135–42. WOS:A1986D773500003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marklund M, Stromberg EA, Laerke HN, Knudsen KEB, Kamal-Eldin A, Hooker AC, et al. Simultaneous Pharmacokinetic Modeling of Alkylresorcinols and Their Main Metabolites Indicates Dual Absorption Mechanisms and Enterohepatic Elimination in Humans. Journal of Nutrition. 2014;144(11):1674–80. 10.3945/jn.114.196220. WOS:000343681400002. [DOI] [PubMed] [Google Scholar]
- 31.Mathijssen NCJ, Masereeuw R, Holme PA, van Kraaij MGJ, Laros-van Gorkom BAP, Peyvandi F, et al. Increased volume of distribution for recombinant activated factor VII and longer plasma-derived factor VII half-life may explain their long lasting prophylactic effect. Thrombosis research. 2013;132(2):256–62. 10.1016/j.thromres.2013.05.027. WOS:000324059600034. [DOI] [PubMed] [Google Scholar]
- 32.Turner JV, Maddalena DJ, Cutler DJ, Agatonovic-Kustrin S. Multiple pharmacokinetic parameter prediction for a series of cephalosporins. J Pharm Sci. 2003;92(3):552–9. Epub 2003/02/15. 10.1002/jps.10314 . [DOI] [PubMed] [Google Scholar]
- 33.Arnot JA, Brown TN, Wania F. Estimating screening-level organic chemical half-lives in humans. Environ Sci Technol. 2014;48(1):723–30. Epub 2013/12/05. 10.1021/es4029414 . [DOI] [PubMed] [Google Scholar]
- 34.Lu J, Lu D, Zhang X, Bi Y, Cheng K, Zheng M, et al. Estimation of elimination half-lives of organic chemicals in humans using gradient boosting machine. Biochimica et Biophysica Acta (BBA)—General Subjects. 2016. 10.1016/j.bbagen.2016.05.019 [DOI] [PubMed] [Google Scholar]
- 35.Obach RS, Lombardo F, Waters NJ. Trend analysis of a database of intravenous pharmacokinetic parameters in humans for 670 drug compounds. Drug metabolism and disposition: the biological fate of chemicals. 2008;36(7):1385–405. Epub 2008/04/23. 10.1124/dmd.108.020479 . [DOI] [PubMed] [Google Scholar]
- 36.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Research. 2008;36(Database issue):D684–8. Epub 2007/12/18. 10.1093/nar/gkm795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaal W, Hammerling U, Gustafsson MG, Spjuth O. Automated QuantMap for rapid quantitative molecular network topology analysis. Bioinformatics. 2013;29(18):2369–70. 10.1093/bioinformatics/btt390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, Lu J, Zhang N, Huang T, Cai YD. A hybrid method for prediction and repositioning of drug Anatomical Therapeutic Chemical classes. Mol Biosyst. 2014;10(4):868–77. 10.1039/c3mb70490d . [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Vogt I, Haque T, Campillos M. HitPick: a web server for hit identification and target prediction of chemical screenings. Bioinformatics. 2013;29(15):1910–2. 10.1093/bioinformatics/btt303 [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Zeng WM, Cai YD, Feng KY, Chou KC. Predicting Anatomical Therapeutic Chemical (ATC) Classification of Drugs by Integrating Chemical-Chemical Interactions and Similarities. PLoS ONE. 2012;7(4):e35254 10.1371/journal.pone.0035254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu LL, Chen C, Huang T, Cai YD, Chou KC. Predicting Biological Functions of Compounds Based on Chemical-Chemical Interactions. PLoS ONE. 2011;6(12):e29491 10.1371/journal.pone.0029491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen L, Lu J, Huang T, Yin J, Wei L, Cai YD. Finding candidate drugs for hepatitis C based on chemical-chemical and chemical-protein interactions. PLoS One. 2014;9(9):e107767 10.1371/journal.pone.0107767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J, Chen L, Yin J, Huang T, Bi Y, Kong XY, et al. Identification of new candidate drugs for lung cancer using chemical-chemical interactions,chemical-protein interactions and a K-means clustering algorithm. Journal of Biomolecular Structure & Dynamics. 2016;34(4):906–17. 10.1080/07391102.2015.1060161 [DOI] [PubMed] [Google Scholar]
- 44.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8(1):R3 Epub 2007/01/06. 10.1186/gb-2007-8-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Li BQ, Feng KY. Predicting Biological Functions of Protein Complexes Using Graphic and Functional Features. Current Bioinformatics. 2013;8(5):545–51. ISI:000325754000004. [Google Scholar]
- 46.Huang T, Zhang J, Xu ZP, Hu LL, Chen L, Shao JL, et al. Deciphering the effects of gene deletion on yeast longevity using network and machine learning approaches. Biochimie. 2012;94(4):1017–25. Epub 2012/01/14. 10.1016/j.biochi.2011.12.024 . [DOI] [PubMed] [Google Scholar]
- 47.Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2005;27(8):1226–38. 10.1109/TPAMI.2005.159 [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Ding C, Li T. Gene selection algorithm by combining reliefF and mRMR. BMC genomics. 2008;9(Suppl 2):S27 10.1186/1471-2164-9-S2-S27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Shi X, Kong X, Zeng Z, Cai YD. Identifying protein complexes using hybrid properties. J Proteome Res. 2009;8(11):5212–8. Epub 2009/09/22. 10.1021/pr900554a . [DOI] [PubMed] [Google Scholar]
- 50.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. Journal of bioinformatics and computational biology. 2005;3(02):185–205. [DOI] [PubMed] [Google Scholar]
- 51.Chen L, Chu C, Huang T, Kong X, Cai Y-D. Prediction and analysis of cell-penetrating peptides using pseudo-amino acid composition and random forest models. Amino acids. 2015;47(7):1485–93. 10.1007/s00726-015-1974-5 [DOI] [PubMed] [Google Scholar]
- 52.Mohabatkar H, Mohammad Beigi M, Esmaeili A. Prediction of GABAA receptor proteins using the concept of Chou's pseudo-amino acid composition and support vector machine. Journal of Theoretical Biology. 2011;281(1):18–23. 10.1016/j.jtbi.2011.04.017 [DOI] [PubMed] [Google Scholar]
- 53.Chen L, Chu C, Feng K. Predicting the types of metabolic pathway of compounds using molecular fragments and sequential minimal optimization. Combinatorial Chemistry & High Throughput Screening. 2016;19(2):136–43. [DOI] [PubMed] [Google Scholar]
- 54.Mohabatkar H, Beigi MM, Abdolahi K, Mohsenzadeh S. Prediction of allergenic proteins by means of the concept of Chou's pseudo amino acid composition and a machine learning approach. Med Chem. 2013;9(1):133–7. Epub 2012/08/31. MC-EPUB-20120817-1 [pii]. . [DOI] [PubMed] [Google Scholar]
- 55.Li Z, Zhou X, Dai Z, Zou X. Classification of G-protein coupled receptors based on support vector machine with maximum relevance minimum redundancy and genetic algorithm. BMC bioinformatics. 2010;11(1):325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang PW, Chen L, Huang T, Zhang N, Kong XY, Cai YD. Classifying ten types of major cancers based on reverse phase protein array profiles. PLoS ONE. 2015;10(3):e0123147 Epub 2015/03/31. 10.1371/journal.pone.0123147 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, et al. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. Journal of Biological Chemistry. 2001;276(41):38307–19. WOS:000171526500079. 10.1074/jbc.M100290200 [DOI] [PubMed] [Google Scholar]
- 58.Evertts AG, Zee BM, DiMaggio PA, Gonzales-Cope M, Coller HA, Garcia BA. Quantitative Dynamics of the Link between Cellular Metabolism and Histone Acetylation. Journal of Biological Chemistry. 2013;288(17):12142–51. 10.1074/jbc.M112.428318. WOS:000318157600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elaut G, Laus G, Alexandre E, Richert L, Bachellier P, Tourwe D, et al. A metabolic screening study of trichostatin A (TSA) and TSA-like histone deacetylase inhibitors in rat and human primary hepatocyte cultures. Journal of Pharmacology and Experimental Therapeutics. 2007;321(1):400–8. 10.1124/jpet.106.116202. WOS:000244989600044. [DOI] [PubMed] [Google Scholar]
- 60.Breuer J, Breuer H. Half-Life of [4-C-14] Estradiol-17beta in-Vivo, and Metabolism of [4-C-14] Estrone and [4-C-14]Oestriol in-Vitro in Normal Subjects and in Patients with Liver Cirrhosis. Z Klin Chem Klin Bio. 1973;11(6):263–9. WOS:A1973Q163900008. [PubMed] [Google Scholar]
- 61.Chen H, Zhang Q, Li X, Zhang H, Sun Y, Yin L, et al. Single- and multiple-dose pharmacokinetics and tolerability of limaprost in healthy Chinese subjects. Clin Drug Investig. 2015;35(3):151–7. 10.1007/s40261-014-0265-3 . [DOI] [PubMed] [Google Scholar]
- 62.Basuli F, Li C, Xu B, Williams M, Wong K, Coble VL, et al. Synthesis of fluorine-18 radio-labeled serum albumins for PET blood pool imaging. Nucl Med Biol. 2015;42(3):219–25. 10.1016/j.nucmedbio.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keenan JM. Wax-matrix extended-release niacin vs inositol hexanicotinate: a comparison of wax-matrix, extended-release niacin to inositol hexanicotinate "no-flush" niacin in persons with mild to moderate dyslipidemia. J Clin Lipidol. 2013;7(1):14–23. 10.1016/j.jacl.2012.10.004 . [DOI] [PubMed] [Google Scholar]
- 64.Sagcal-Gironella ACP, Sherwin CMT, Tirona RG, Rieder MJ, Brunner HI, Vinks AA. Pharmacokinetics of Prednisolone at Steady State in Young Patients With Systemic Lupus Erythematosus on Prednisone Therapy: An Open-Label, Single-Dose Study. Clinical Therapeutics. 2011;33(10):1524–36. 10.1016/j.clinthera.2011.09.015. WOS:000297037800020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kietadisorn R, Kietselaer BL, Schmidt HHHW, Moens AL. Role of tetrahydrobiopterin (BH4) in hyperhomocysteinemia-induced endothelial dysfuction: new indication for this orphan-drug? Am J Physiol-Endoc M. 2011;300(6):E1176–E. 10.1152/ajpendo.00084.2011. WOS:000290959000025. [DOI] [PubMed] [Google Scholar]
- 66.Lee EHY, Mandell AJ. Relationships between Drug-Induced Changes in Tetrahydrobiopterin and Biogenic-Amine Concentrations in Rat-Brain. Journal of Pharmacology and Experimental Therapeutics. 1985;234(1):141–6. WOS:A1985AMB6300020. [PubMed] [Google Scholar]
- 67.Sanford M, Keating GM. Sapropterin A Review of its Use in the Treatment of Primary Hyperphenylalaninaemia. Drugs. 2009;69(4):461–76. WOS:000265588200006. 10.2165/00003495-200969040-00006 [DOI] [PubMed] [Google Scholar]
- 68.Lyseng-Williamson KA. Elosulfase Alfa: A Review of Its Use in Patients with Mucopolysaccharidosis Type IVA (Morquio A Syndrome). Biodrugs. 2014;28(5):465–75. 10.1007/s40259-014-0108-z. WOS:000344602900006. [DOI] [PubMed] [Google Scholar]
- 69.Diepstraten J, Janssen EJH, Hackeng CM, van Dongen EPA, Wiezer RJ, van Ramshorst B, et al. Population pharmacodynamic model for low molecular weight heparin nadroparin in morbidly obese and non-obese patients using anti-Xa levels as endpoint. European Journal of Clinical Pharmacology. 2015;71(1):25–34. 10.1007/s00228-014-1760-4. WOS:000347156900003. [DOI] [PubMed] [Google Scholar]
- 70.Wakao M, Obata R, Miyachi K, Kaitsubata Y, Kondo T, Sakami C, et al. Synthesis of a chondroitin sulfate disaccharide library and a GAG-binding protein interaction analysis. Bioorganic & medicinal chemistry letters. 2015;25(7):1407–11. 10.1016/j.bmcl.2015.02.054. WOS:000351688700009. [DOI] [PubMed] [Google Scholar]
- 71.Hu S, Chang Y, He M, Wang J, Wang Y, Xue C. Fucosylated chondroitin sulfate from sea cucumber improves insulin sensitivity via activation of PI3K/PKB pathway. Journal of food science. 2014;79(7):H1424–9. 10.1111/1750-3841.12465 . [DOI] [PubMed] [Google Scholar]
- 72.Hu SW, Tian YY, Chang YG, Li ZJ, Xue CH, Wang YM. Fucosylated chondroitin sulfate from sea cucumber improves glucose metabolism and activates insulin signaling in the liver of insulin-resistant mice. J Med Food. 2014;17(7):749–57. 10.1089/jmf.2013.2924 . [DOI] [PubMed] [Google Scholar]
- 73.Gomes CLR, Leao CL, Venturotti C, Barreira AL, Guimaraes G, Fonseca RJC, et al. The Protective Role of Fucosylated Chondroitin Sulfate, a Distinct Glycosaminoglycan, in a Murine Model of Streptozotocin-Induced Diabetic Nephropathy. Plos One. 2014;9(9):e106929 10.1371/journal.pone.0106929. WOS:000347993600082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao YL, Li PL, Cheng YN, Zhang XK, Sheng JZ, Wang DC, et al. Enhancing the intestinal absorption of low molecular weight chondroitin sulfate by conjugation with alpha-linolenic acid and the transport mechanism of the conjugates. Int J Pharmaceut. 2014;465(1–2):143–58. 10.1016/j.ijpharm.2014.02.009. WOS:000333675100019. [DOI] [PubMed] [Google Scholar]
- 75.Sullivan BP, Kopec AK, Joshi N, Cline H, Brown JA, Bishop SC, et al. Hepatocyte tissue factor activates the coagulation cascade in mice. Blood. 2013;121(10):1868–74. 10.1182/blood-2012-09-455436. WOS:000321751800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee AYY, Vlasuk GP. Recombinant nematode anticoagulant protein c2 and other inhibitors targeting blood coagulation factor VIIa/tissue factor. J Intern Med. 2003;254(4):313–21. 10.1046/j.1365-2796.2003.01224.x. WOS:000185349100002. [DOI] [PubMed] [Google Scholar]
- 77.Montoro JB, Altisent C, Pico M, Cabanas MJ, Vila M, Puig LL. Recombinant factor VIIa in continuous infusion during central line insertion in a child with factor VIII high-titre inhibitor (vol 4, pg 762, 1998). Haemophilia. 1999;5(5):371-. WOS:000085545800013. [DOI] [PubMed] [Google Scholar]
- 78.Vrancken K, Schroeder HJ, Longo LD, Power GG, Blood AB. Role of ceruloplasmin in nitric oxide metabolism in plasma of humans and sheep: a comparison of adults and fetuses. Am J Physiol-Reg I. 2013;305(11):R1401–R10. 10.1152/ajpregu.00266.2013. WOS:000328489300015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shafran Y, Zurgil N, Afrimzon E, Tauber Y, Sobolev M, Shainberg A, et al. Correlative Analyses of Nitric Oxide Generation Rates and Nitric Oxide Synthase Levels in Individual Cells Using a Modular Cell-Retaining Device. Analytical chemistry. 2012;84(17):7315–22. 10.1021/ac202741z. WOS:000308255400008. [DOI] [PubMed] [Google Scholar]
- 80.Bhatt DK, Gupta S, Jansen-Olesen I, Andrews JS, Olesen J. NXN-188, a selective nNOS inhibitor and a 5-HT1B/1D receptor agonist, inhibits CGRP release in preclinical migraine models. Cephalalgia. 2013;33(2):87–100. 10.1177/0333102412466967. WOS:000312550000003. [DOI] [PubMed] [Google Scholar]
- 81.Chen D, Whitcomb R, MacIntyre E, Tran V, Do ZN, Sabry J, et al. Pharmacokinetics and Pharmacodynamics of AR9281, an Inhibitor of Soluble Epoxide Hydrolase, in Single- and Multiple-Dose Studies in Healthy Human Subjects. Journal of Clinical Pharmacology. 2012;52(3):319–28. 10.1177/0091270010397049. WOS:000300755600003. [DOI] [PubMed] [Google Scholar]
- 82.Le Noury J, Nardo JM, Healy D, Jureidini J, Raven M, Tufanaru C, et al. Restoring Study 329: efficacy and harms of paroxetine and imipramine in treatment of major depression in adolescence. Bmj-Brit Med J. 2015;351:h4320 10.1136/bmj.h4320. WOS:000361678800002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mukherjee S, Mukherjee B, Mukhopadhyay R, Naskar K, Sundar S, Dujardin JC, et al. Imipramine Is an Orally Active Drug against Both Antimony Sensitive and Resistant Leishmania donovani Clinical Isolates in Experimental Infection. PLoS neglected tropical diseases. 2012;6(12):e1987 10.1371/journal.pntd.0001987. WOS:000312910200043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Killen JD, Fortmann SP, Murphy GM, Hayward C, Fong D, Lowenthal K, et al. Failure to improve cigarette smoking abstinence with transdermal selegiline plus cognitive behavior therapy. Addiction. 2010;105(9):1660–8. 10.1111/j.1360-0443.2010.03020.x. WOS:000280668200027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frampton JE, Plosker GL. Selegiline transdermal system in major depressive disorder—Profile report (Reprinted from Drugs, vol 67, pg 257–267, 2007). Cns Drugs. 2007;21(6):521–4. 10.2165/00023210-200721060-00007. WOS:000247518000008. [DOI] [PubMed] [Google Scholar]
- 86.Catterall WA, Swanson TM. Structural Basis for Pharmacology of Voltage-Gated Sodium and Calcium Channels. Molecular Pharmacology. 2015;88(1):141–50. 10.1124/mol.114.097659. WOS:000357091800017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rahman W, Dickenson AH. Voltage gated sodium and calcium channel blockers for the treatment of chronic inflammatory pain. Neuroscience letters. 2013;557:19–26. 10.1016/j.neulet.2013.08.004. WOS:000329601300003. [DOI] [PubMed] [Google Scholar]
- 88.Naziroglu M, Sahin M, Cig B, Aykur M, Erturan I, Ugan Y. Hypericum perforatum Modulates Apoptosis and Calcium Mobilization Through Voltage-Gated and TRPM2 Calcium Channels in Neutrophil of Patients with Behcet's Disease. J Membrane Biol. 2014;247(3):253–62. 10.1007/s00232-014-9630-7. WOS:000332009700005. [DOI] [PubMed] [Google Scholar]
- 89.Lim KS, Cho JY, Jang IJ, Kim BH, Kim J, Jeon JY, et al. Pharmacokinetic interaction of flecainide and paroxetine in relation to the CYP2D6*10 allele in healthy Korean subjects. Br J Clin Pharmacol. 2008;66(5):660–6. 10.1111/j.1365-2125.2008.03267.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Conard GJ, Ober RE. Metabolism of flecainide. Am J Cardiol. 1984;53(5):41B–51B. . [DOI] [PubMed] [Google Scholar]
- 91.Sikkel MB, Collins TP, Rowlands C, Shah M, O'Gara P, Williams AJ, et al. Flecainide reduces Ca(2+) spark and wave frequency via inhibition of the sarcolemmal sodium current. Cardiovasc Res. 2013;98(2):286–96. 10.1093/cvr/cvt012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bacon E, Matsokis N, Roujansky P, Debarry J, Gombos G. Alteration of Benzodiazepine Receptors in Mouse Cerebellum Following Methylazoxymethanol Treatment during Development. Dev Brain Res. 1989;47(2):293–7. 10.1016/0165-3806(89)90185-5. WOS:A1989AB74400014. [DOI] [PubMed] [Google Scholar]
- 93.Ferrer I, Pozas E, Marti M, Blanco R, Planas AM. Methylazoxymethanol acetate-induced apoptosis in the external granule cell layer of the developing cerebellum of the rat is associated with strong c-Jun expression and formation of high molecular weight c-Jun complexes. Journal of neuropathology and experimental neurology. 1997;56(1):1–9. WOS:A1997WC09700001. [DOI] [PubMed] [Google Scholar]
- 94.Mistraletti G, Umbrello M, Sabbatini G, Miori S, Taverna M, Cerri B, et al. Melatonin reduces the need for sedation in ICU patients: a randomized controlled trial. Minerva Anestesiol. 2015;81(12):1298–310. . [PubMed] [Google Scholar]
- 95.Simons FE, Simons KJ, Becker AB, Haydey RP. Pharmacokinetics and antipruritic effects of hydroxyzine in children with atopic dermatitis. J Pediatr. 1984;104(1):123–7. . [DOI] [PubMed] [Google Scholar]
- 96.Levis S, Preat S, Beersaerts J, Dauby J, Beelen L, Baugniet V. [Pharmcological study on hydroxyzine, U.C.B. 492; a disubstituted piperazine derivative]. Arch Int Pharmacodyn Ther. 1957;109(1–2):127–42. . [PubMed] [Google Scholar]
- 97.Degen J, Wolke E, Seiberling M, Pintar P, Hoxter G, Steinhauer HB, et al. [Comparative study of the pharmacokinetics of amitriptyline oxide and trimipramine after single administration in healthy male probands and patients with renal failure]. Med Klin (Munich). 1993;88(3):129–33, 71 . [PubMed] [Google Scholar]
- 98.Iribarren C, Round AD, Peng JA, Lu M, Zaroff JG, Holve TJ, et al. Validation of a population-based method to assess drug-induced alterations in the QT interval: a self-controlled crossover study. Pharmacoepidemiol Drug Saf. 2013;22(11):1222–32. 10.1002/pds.3479 . [DOI] [PubMed] [Google Scholar]
- 99.Kim EJ, Sahai V, Abel EV, Griffith KA, Greenson JK, Takebe N, et al. Pilot clinical trial of hedgehog pathway inhibitor GDC-0449 (vismodegib) in combination with gemcitabine in patients with metastatic pancreatic adenocarcinoma. Clin Cancer Res. 2014;20(23):5937–45. 10.1158/1078-0432.CCR-14-1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Graham RA, Lum BL, Cheeti S, Jin JY, Jorga K, Von Hoff DD, et al. Pharmacokinetics of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with locally advanced or metastatic solid tumors: the role of alpha-1-acid glycoprotein binding. Clin Cancer Res. 2011;17(8):2512–20. 10.1158/1078-0432.CCR-10-2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Borel-Giraud N, Maillavin A, Vernet-Nyssen M. [Determination of alpha-1-foetoprotein in human serum by electroimmuno-diffusion on agarose (author's transl)]. Clin Chim Acta. 1976;71(2):117–27. . [DOI] [PubMed] [Google Scholar]
- 102.Saccenti E, van Duynhoven J, Jacobs DM, Smilde AK, Hoefsloot HCJ. Strategies for Individual Phenotyping of Linoleic and Arachidonic Acid Metabolism Using an Oral Glucose Tolerance Test. Plos One. 2015;10(3):e0119856 10.1371/journal.pone.0119856. WOS:000352138500136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pawlosky RJ, Hibbeln JR, Novotny JA, Salem N. Physiological compartmental analysis of alpha-linolenic acid metabolism in adult humans. Journal of lipid research. 2001;42(8):1257–65. WOS:000170272900011. [PubMed] [Google Scholar]
- 104.Catella F, Lawson JA, Fitzgerald DJ, Fitzgerald GA. Endogenous Biosynthesis of Arachidonic-Acid Epoxides in Humans—Increased Formation in Pregnancy-Induced Hypertension. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(15):5893–7. 10.1073/pnas.87.15.5893. WOS:A1990DR71400064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yarwood A, Martin P, Bowes J, Lunt M, Worthington J, Barton A, et al. Enrichment of vitamin D response elements in RA-associated loci supports a role for vitamin D in the pathogenesis of RA. Genes and immunity. 2013;14(5):325–9. 10.1038/gene.2013.23. WOS:000322325000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Raspe HH, Kindel P, Vesterling K, Kohlmann T. [Change in functional capacity and pain intensity of 81 cP patients treated with Azulfidine RA or aurothioglucose. Preliminary statistical assessment of the German multicenter study of the treatment of chronic polyarthritis with Azulfidine RA]. Z Rheumatol. 1987;46(2):71–5. . [PubMed] [Google Scholar]
- 107.Tinghuai M, Jinjuan Z, Meili T, Yuan T, Abdullah A-D, Mznah A-R, et al. Social network and tag sources based augmenting collaborative recommender system. IEICE transactions on Information and Systems. 2015;98(4):902–10. [Google Scholar]
- 108.Wen X, Shao L, Xue Y, Fang W. A rapid learning algorithm for vehicle classification. Inform Sciences. 2015;295:395–406. [Google Scholar]
- 109.Zou Q, Li J, Song L, Zeng X, Wang G. Similarity computation strategies in the microRNA-disease network: a survey. Briefings in Functional Genomics. 2016;15(1):55–64. 10.1093/bfgp/elv024 [DOI] [PubMed] [Google Scholar]
- 110.Zeng X, Zhang X, Zou Q. Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Briefings in bioinformatics. 2016;17(2):193–203. 10.1093/bib/bbv033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.