Abstract
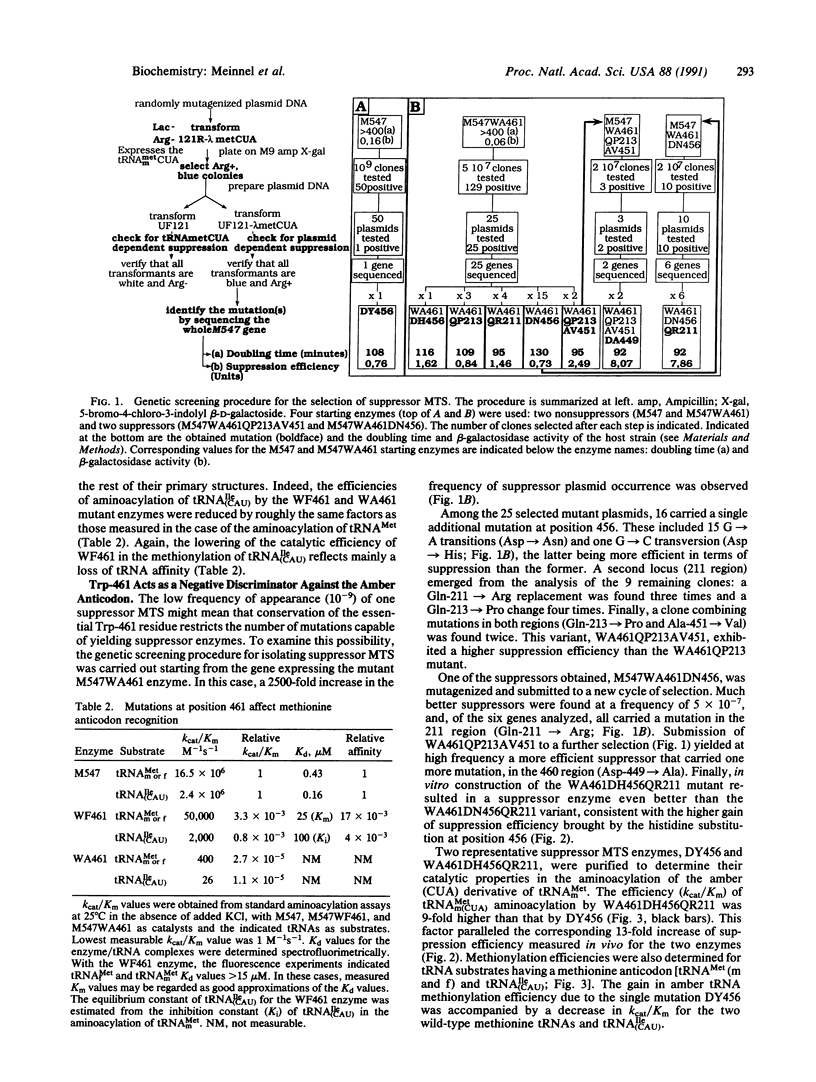
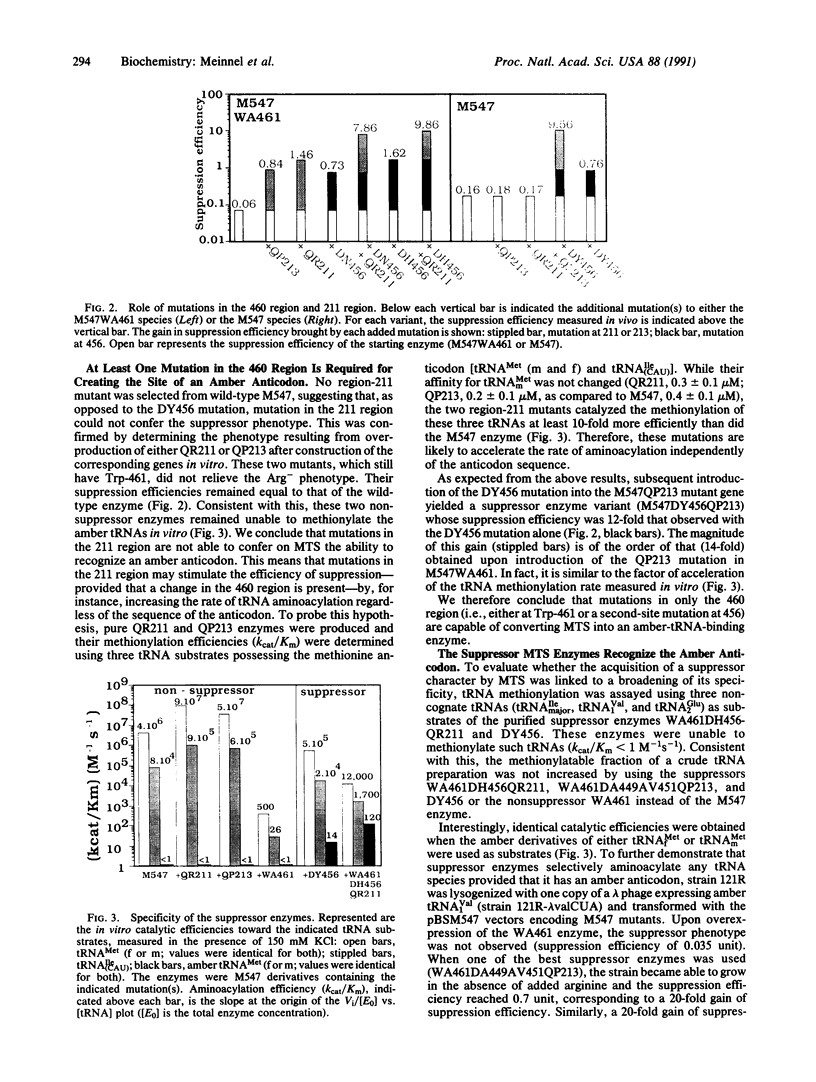
Accurate aminoacylation of a tRNA by Escherichia coli methionyl-tRNA synthetase (MTS) is specified by the CAU anticodon. A genetic screening procedure was designed to isolate MTS mutants able to aminoacylate a methionine amber tRNA (CUA anticodon). Selected suppressor MTS enzymes all possess one or several mutations in the vicinity of Trp-461, a residue that is the major contributor to the stability of complexes formed with tRNAs having the cognate CAU anticodon. Analysis of catalytic properties of purified suppressor enzymes shows that they have acquired an additional specificity toward the amber anticodon without complete disruption of the methionine anticodon site. It is concluded that both positive and negative discrimination toward the binding of tRNA anticodon sequences is restricted to a limited region of the synthetase, residues 451-467.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanquet S., Iwatsubo M., Waller J. P. The mechanism of action of methionyl-tRNA synthetase from Escherichia coli. 1. Fluorescence studies on tRNAMet binding as a function of ligands, ions and pH. Eur J Biochem. 1973 Jul 2;36(1):213–226. doi: 10.1111/j.1432-1033.1973.tb02903.x. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Ghosh G., Pelka H., Schulman L. H. Identification of the tRNA anticodon recognition site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1990 Mar 6;29(9):2220–2225. doi: 10.1021/bi00461a003. [DOI] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Blanquet S., Lederer F. Methionyl-tRNA synthetase from Escherichia coli: primary structure at the binding site for the 3'-end of tRNAfMet. Biochemistry. 1985 Feb 26;24(5):1175–1180. doi: 10.1021/bi00326a018. [DOI] [PubMed] [Google Scholar]
- Inokuchi H., Hoben P., Yamao F., Ozeki H., Söll D. Transfer RNA mischarging mediated by a mutant Escherichia coli glutaminyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5076–5080. doi: 10.1073/pnas.81.16.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence F., Blanquet S., Poiret M., Robert-Gero M., Waller J. P. The mechanism of action of methionyl-tRNA synthetase. 3. Ion requirements and kinetic parameters of the ATP-PPi exchange and methionine-transfer reactions catalyzed by the native and trypsin-modified enzymes. Eur J Biochem. 1973 Jul 2;36(1):234–243. doi: 10.1111/j.1432-1033.1973.tb02905.x. [DOI] [PubMed] [Google Scholar]
- Leon O., Schulman L. H. tRNA recognition site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1987 Aug 25;26(17):5416–5422. doi: 10.1021/bi00391a030. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Chen Y. M., Foss K., Schneider J. Association of transfer RNA acceptor identity with a helical irregularity. Science. 1988 Dec 23;242(4886):1681–1684. doi: 10.1126/science.2462282. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- Mechulam Y., Blanquet S., Fayat G. Dual level control of the Escherichia coli pheST-himA operon expression. tRNA(Phe)-dependent attenuation and transcriptional operator-repressor control by himA and the SOS network. J Mol Biol. 1987 Oct 5;197(3):453–470. doi: 10.1016/0022-2836(87)90558-4. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Fayat G. Fast purification of a functional elongator tRNAmet expressed from a synthetic gene in vivo. Nucleic Acids Res. 1988 Aug 25;16(16):8095–8096. doi: 10.1093/nar/16.16.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellot P., Mechulam Y., Le Corre D., Blanquet S., Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol. 1989 Aug 5;208(3):429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Albertini A. M. Effects of surrounding sequence on the suppression of nonsense codons. J Mol Biol. 1983 Feb 15;164(1):59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Normanly J., Masson J. M., Kleina L. G., Abelson J., Miller J. H. Construction of two Escherichia coli amber suppressor genes: tRNAPheCUA and tRNACysCUA. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Ogden R. C., Horvath S. J., Abelson J. Changing the identity of a transfer RNA. Nature. 1986 May 15;321(6067):213–219. doi: 10.1038/321213a0. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Swanson R. N., Rould M. A., Steitz T. A., Söll D. Structural basis for misaminoacylation by mutant E. coli glutaminyl-tRNA synthetase enzymes. Science. 1989 Dec 1;246(4934):1152–1154. doi: 10.1126/science.2686030. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Sayers J. R., Schmidt W., Eckstein F. 5'-3' exonucleases in phosphorothioate-based oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1988 Feb 11;16(3):791–802. doi: 10.1093/nar/16.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Parameters for the molecular recognition of transfer RNAs. Biochemistry. 1989 Apr 4;28(7):2747–2759. doi: 10.1021/bi00433a001. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon loop size and sequence requirements for recognition of formylmethionine tRNA by methionyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6755–6759. doi: 10.1073/pnas.80.22.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Springer M., Graffe M., Dondon J., Grunberg-Manago M. tRNA-like structures and gene regulation at the translational level: a case of molecular mimicry in Escherichia coli. EMBO J. 1989 Aug;8(8):2417–2424. doi: 10.1002/j.1460-2075.1989.tb08372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]