Abstract
Lung cancer is the number one cancer in the US and worldwide. In spite of the rapid progression in personalized treatments, the overall survival rate of lung cancer patients is still suboptimal. Over the past decade, tremendous efforts have been focused on the discovery of protein biomarkers to facilitate the early detection and monitoring lung cancer progression during treatment. In addition to tumor tissues and cancer cell lines, a variety of biological material has been studied. Particularly in recent years, studies using fluid-based specimen or so-called “fluid-biopsy” specimen have progressed rapidly. Fluid specimens are relatively easier to collect than tumor tissue, and they can be repeatedly sampled during the disease progression. Glycoproteins have long been recognized to play fundamental roles in many physiological and pathological processes. In this review, we focus the discussion on recent advances of glycoproteomics, particularly in the identification of potential protein biomarkers using so-called fluid-based specimens in lung cancer. The purpose of this review is to summarize current strategies, achievements and perspectives in the field. This insight will highlight the discovery of tumor-associated glycoprotein biomarkers in lung cancer and their potential clinical applications.
Keywords: glycoproteins, lung cancer, body fluid, non-small cell lung cancer (NSCLC), glycoproteomics, protein biomarkers
1. Introduction
Lung cancer is the leading cause of cancer-related deaths in the United States and worldwide, and also accounts for 1.5 million new cases annually [1]. Lung cancer is a heterogeneous group of disease which has been divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC represents 70 to 80% of all lung cancers, of which the most dominant histological subtype is adenocarcinoma (approximately 50 to 70% of NSCLC) [2]. Approximately 40–60% of NSCLC patients present with locally advanced or metastatic disease at the time of diagnosis [3, 4]. Even when NSCLC is detected at its early stage (stage I and stage II) and a curative surgery is performed, about 30 to 40% of early stage lung cancer patients will still experience recurrent disease [3, 4]. Although targeted therapies of lung cancer have progressed rapidly, largely based upon the discovery of molecular markers such as EGFR (epidermal growth factor receptor) and KRAS (V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog) mutations, and EML4-ALK (echinoderm microtubule-associated protein-like 4 (EML4) and the anaplastic lymphoma kinase (ALK)) gene fusion [5–7], the overall progression-free survival rate of patients is still suboptimal [8–10]. To improve the survival rate and decrease mortality in lung cancer patients, the discovery of potential protein biomarkers for early detection and monitoring lung cancer progression is urgently needed.
Although research efforts have been focused on the identification of protein biomarkers in blood for early detection and monitoring disease progression in cancers, such as the discovery of glycoprotein CA19-9 for pancreatic cancer, CA125 for ovarian cancer, PSA (prostate specific antigen) for prostate cancer and CEA (carcinoembryonic antigen) for colon cancer, the study of lung cancer protein biomarker is still far from satisfactory. Tumor-associated protein biomarkers in blood are usually present as low-abundance proteins which are difficult to detect. During the protein biosynthesis, glycosylation is one of the most critical post-translation modifications [11–15]. Glycoproteins play critical roles in the regulation of cellular functions, including cell growth, differentiation and migration [13–16]. In lung cancer, glycoproteins are secreted or leaked from tumor cells into surrounding stroma and biological fluid, such as blood, sputum, airway fluid and others. Some of them are also associated with disease progression such as the development of pleural effusion when tumor metastasizes to the pleural cavity. The study of glycoprotein profiles in fluid specimens is, therefore, particularly important for understanding lung cancer development and progression as well as the discovering of candidate protein biomarkers [16–20]. These fluid samples are relatively easy to obtain and can be repeatedly sampled in patients. Recent advances in high-throughput glycoproteomics allow us for the first time to uncover hundreds of glycoproteins in biological samples [18–20]. These state-of-the-art technologies provide new platforms for characterization of the complex proteome in lung cancer.
In this review, we focus the discussion on recent advances of glycoproteomics, particularly on the identification of potential protein biomarkers using fluid-based specimens in lung cancer. The purpose of this review is to summarize current strategies, achievements and perspectives in the field. In addition to study tumor genetics, this insight will serve as another resource to understand the pathogenesis of lung cancer and to facilitate the discovery of tumor-associated protein biomarkers in lung cancer.
2. Various Technologies in Glycoprotein Detection
More than 70% of human proteins, including most secreted proteins, cell surface and intracellular proteins are glycoproteins. In eukaryotic cells, O- and N-linked glycosylations are mediated through different enzymatic and biosynthetic pathways, and they play different biological roles in cells [21–23]. For example, studies have shown that the O-linked glycosylation involves the regulation of the half-life of hormones, whereas the N-glycoproteins play important roles in cancer cell growth, differentiation, invasion and metastasis [22–23]. N-linked glycosylation also plays an important role in the regulation of the hormone-receptor binding and the bioactivity of hormones [22]. Furthermore, it is well known that glycoproteins of cancer cells express aberrant glycosylation patterns, such as increased β1-6 branching, sialylation and/or fucosylation of N-linked glycans, and truncation of O-linked glycans [13, 23–25].
Currently, several strategies, including chemical immobilization and affinity capture using lectin affinity reagents, have been developed to study glycoproteins [18, 26–29]. These approaches use different mechanisms and are complementary to each other for glycoprotein identification and enrichment. Different subsets of glycoproteins can be enriched and studied by the chemical immobilization and the lectin affinity chromatography. Glycoproteins enriched by chemistry immobilization are mostly low molecular weight proteins that function as enzymes, whereas those enriched by lectin affinity capturing are mostly high molecular weight proteins that are involved in intracellular signal transduction and cell adhesion [30].
2.1 Glycopeptides Enrichment by Chemical Capture and Immobilization SPEG (solid-phase extraction of N-linked glycopeptides)
The technique is developed by using the chemical immobilization of glycoproteins/glycopeptides on a solid support to enrich glycoproteins. It is based on the oxidation of glycans to aldehydes and subsequent reactions between aldehyde groups and hydrazide on solid support using solid-phase extraction of N-linked glycopeptides (SPEG) [26]. In the experiment, the glycans on the glycoprotein are oxidized to form aldehyde groups, then, the newly formed aldehyde reacts with hydrazide, which is attached to a solid support and forms covalent hydrazone bonds. Using SPEG, both complex N-linked and O-linked glycoproteins or glycopeptides can be conjugated to solid support via covalent bonds [26, 27]. The N-linked glycopeptides can be released by the enzyme peptide-N-glycosidase (PNGase) [26, 27]; however, O-linked glycopeptides cannot be easily released from the solid support due to the lack of specific enzymes. Chemical approaches for releasing O-linked glycosylated peptides such as alkaline β-elimination have been used with limited success [31, 32].
The advantage of SPEG is that this technique has more than 90% specificity in the identification of N-linked glycopeptides containing N-linked glycosylation sites from complex biological samples [26, 27, 33]. This method can detect approximately one to two de-glycosylated peptides on average from each glycoprotein in the peptide mixture; thus it reduces the complexity and provides an accurate analysis for low abundant glycoproteins [34]. In comparison to other enrichment methods including lectin affinity chromatography, the chemical immobilization on solid-phase extraction has a highly specific capture of glycopeptides based upon the covalent conjugation between glycans and solid support. The limitation of the method is that the associated glycans on the glycopeptides are not detected by MS. The sensitivity of the glycopeptide detection could be improved by using multiple enzymatic digestions of glycoproteins; examples include using trypsin, pepsin and thermolysin during isolation step prior to glycopeptide capturing step [26, 35, 36].
Boronic Acid Capture
Glycoprotein enrichment can also be achieved by the reaction with boronic acid [37, 38]. The principle of this method is based on the formation of stable boronic diesters by the reaction of geminal diols under basic conditions. Recently, Sparbier K et al. have successfully detected low-abundance glycoproteins in human blood samples using this method [37]. Xu Y et al. have synthesized a novel diboronic acid functionalized mesoporous silica material (FDU-12-GA) and used it for specific glycopeptide enrichment [38]. Their data has demonstrated a marked improvement of glycopeptides detection by using this method.
Hydrophilic Interaction Liquid Chromatography (HILIC)
During the glycoprotein enrichment, non-glycoprotein may interact with glycoprotein. Hagglund P et al have used a hydrophilic interaction liquid chromatography (HILIC) to reduce the non-specific interaction of proteins and the complexity of peptide/glycopeptide mixtures through depletion of hydrophobic peptides and retention of hydrophilic glycopeptides [39]. This method enables detection of glycoprotein from plasma samples using hydrophilic interaction solid-phase extraction [39]. In addition, Alvarez-Manilla G et al. have used the size-exclusion chromatography (SEC) to enrich N-linked glycopeptides. They have found a three-fold increase in the total number of glycopeptides identified in sera by LC-MS/MS [40].
2.2 Lectin Affinity Approaches
Lectins are a heterogeneous group of carbohydrate binding proteins, which selectively recognize and reversibly bind to specific glycans present on glycoproteins [41, 42]. Based on their binding specificities, various lectins are currently used for enrichment of glycoproteins or glycopeptides [43–47]. Lectins with a relatively broad specificity, such as wheat germ agglutinin (WGA) and concanavalin A (con A), are preferentially used for enriching proteins containing glycans that interact with specific lectins [44, 45].
Lectin affinity chromatography
In lectin affinity chromatography-based methods, enrichment with certain lectin is shown to be particularly useful for identifying glycoproteins or glycopeptides with particular glycan structures [41, 42]. A combination or series of lectin affinity columns may be used during the enrichment process [45–47]. Recently, a multi-lectin affinity column has been developed that allows for an almost complete enrichment of glycoproteins from biological fluids [45, 46]. In addition, lectin microcolumns have been developed for high-pressure analytical schemes. These microcolumns are directly connected on-line with reversed-phased HPLC (high-pressure liquid chromatography) to generate highly sensitive semi-automated profiling of glycoproteins [35].
The limitation of the method is the non-specific binding of non-glycoproteins or non-glycopeptides onto the lectin column. Pan S. et al has conducted a study using both hydrazide chemistry immobilization and lectin affinity column for enrichment of glycoproteins in the cerebrospinal fluid (CSF) [48]. Disease-related glycoproteins in the CSF are usually low-abundance proteins; therefore, in order to comprehensive characterization of CSF proteome, they have compared the capturing specificity and capability of these two methods. In the study, they have found that the hydrazide chemical immobilization method had a higher specificity than that of the lectin affinity method. They have also found that the combination of these two methods can greatly increase the detection ability of glycoproteins in CSF.
Lectin microarray
Changes of glycan structures are hallmarks of cancer. Studies have shown that increased β1-6 branching of N-glycans resulting from the enhanced expression of N-glycan GlcNAc transferase V (GlcNAcT-V) has also been strongly correlated with metastatic potential of cancer cells [49]. This study suggests that glycoproteins and tumor-specific structural changes in glycan moieties may be used as cancer biomarkers. To investigate glycan alteration, lectin microarray technology was developed and used as an important analytical tool and a complementary technique to mass spectrometry in glycobiology [50, 51].
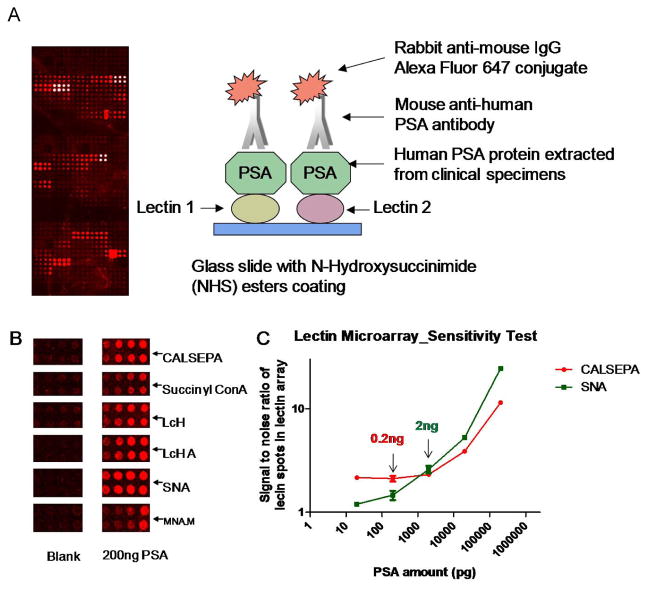
Lectin microarrays are fabricated by the immobilization of multiple lectins to a solid support using chemical reactions, such as the attachment of lysine side chain amine groups to the solid phase through epoxy-functionalized or N–hydroxysuccinimidyl derived esters; then fluorescent labeled glycoproteins are incubated with microarrays. The interactions of lectin-glycans are detected as a pattern of fluorescent intensity. In the study of Li Y et al. [Figure 1] a LLISA (lectin-linked immunosorbent assay) was used for the investigation of glycan alterations in a target glycoprotein detected by lectin microarray [52, 53]. The LLISA assay is a “sandwich” immunoassay. The target glycoprotein binds between an immobilized monoclonal antibody and the biotinylated lectin; a streptavidin SULFO TAG is then used for electrochemiluminescence detection. Previous work demonstrates that the LLISA has a detection sensitivity of 0.04–1.35 ng/mL, indicating that the assay is a sensitive investigative tool in the identification of biomarkers. [52, 53].
Figure 1.
LLISA (lectin-linked immunosorbent assay) for the detection of glycan alterations in a target glycoprotein by lectin microarray. (A) diagram of the lectin microarray for the detection of glycan in the target glycoprotein, (B) the specific binding patterns of target protein PSA to lectins on lectin microarrays, and (C) the detection sensitivity of the specific lectin-PSA interaction. SNA: Sambucus nigra agglutinin; CALSEPA: Calystege sepium agglutinin; PSA: prostatic specific antigen.
The advantage of the technique includes the high-throughput capacity and ability to analyze well-annotated clinical specimens that often have limited quantities. In addition, it requires minimal sample preparation and eliminates the step of glycan purification by protein deglycosylation. Currently, approximately 100 commercial lectins are available for lectin microarray construction [54]. However, detection sensitivity may be an issue when a small amount of the target glycoprotein is analyzed due to the weak lectin-glycan interaction. This may cause missing identification of some disease-related glycan alterations.
Multiplexed antibody array-based lectin and/or lectin microarray detection
To further improve the detection sensitivity and specificity, multiplexed techniques using antibody microarray and lectin-based glycomic analysis were developed. Chen, et al. described an immunoassay for profiling glycan structures on targeted glycoproteins [55]. In the assay, antibodies were laid onto nitrocellulose-coated slides to generate a microarray to capture target glycoproteins; glycans on target glycoprotein were then detected using biotinylated lectin followed by visualization with streptavidin-phycoerythrin. The combined use of protein microarray and lectin probes not only provides discrete glycosylation profiles of multiple target proteins, but also a specific glycosylation profile in a given protein. By using this technique, they were able to demonstrate that glycoprotein MUC-1 levels were not significantly changed in the serum of pancreatic cancer patients, but the glycan structure of the protein in cancers was significantly different than that of controls.
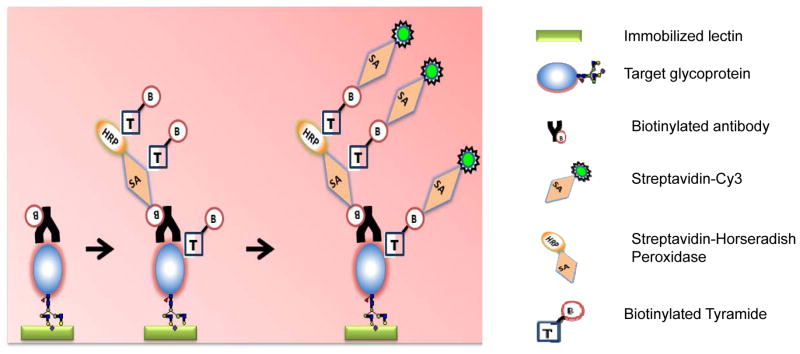
Recently a so-called antibody-overlay lectin microarray (ALM) technique was used for the detection and profiling of glycan on the target glycoprotein [56]. During this multicomplex assay, the target glycoprotein was enriched from biological samples by immunoprecipitation, and captured by lectins on the array. A glycan profile of target glycoprotein was acquired with the aid of a specific capture antibody against the target glycoprotein; then the target glycoprotein which bound to each lectin on lectin microarray was quantitatively analyzed using antibody with tyramide signal amplification [Figure 2]. The technique showed a significant improvement of the detection sensitivity of glycan. This method allows for a comprehensive and high-throughput profiling of complex glycans of the target glycoprotein [56].
Figure 2.
ALM (antibody-overlay lectin microarray) technique for the detection and profiling of glycan on the target glycoprotein. The principle of detection of a target glycoprotein on antibody-overlay lectin microarray using tyramide signal amplification (TSA). Lectin microarrays were constructed on a solid support. After incubation of the target glycoprotein, the biotinylated detection antibody was applied; then the array is incubated with streptavidin-labeled HRP (SA-HRP). The enzyme portion of the SA-HRP catalyzes localized deposition of biotin tyramide, resulting in higher levels of biotin signals that will be detected by streptavidin-labeled Cy3 (SA-Cy3).
The technique is developed to profile the glycan on a target glycoprotein, rather than to profile the whole glycoproteome; thus it may be used to characterize the glycan on a selected subset of tumor-associated glycoproteins. The limitation of such technique is the potential false positive signal caused by glycan on the capture antibodies, therefore an appropriate blocking step is necessary to reduce such false positive signals during the assay.
2.3 The isotopic glycosidase elution and labeling on lectin-column chromatography (IGEL)
Glycan structures of glycoprotein are organ-specific and correlate with disease states. In order to identify N-linked glycosylation sites between glycans and glycoproteins, Ueda K. et al. have used an approach called the isotopic glycosidase elution and labeling on lectin-column chromatography (IGEL) [57]. This technique is based on the lectin affinity chromatography and site-directed tagging of N-linked glycosylation sites by 18O during the release of N-glycopeptides with N-glycosidase [25, 29], and the combination of the IGEL method with iTRAQ stable isotope labeling [57]. They are able not only to identify N-glycosylation sites, but also to quantitatively compare glycan structures on each glycosylation site in a single LC-MS/MS analysis.
3. Type of Fluid-based Specimens
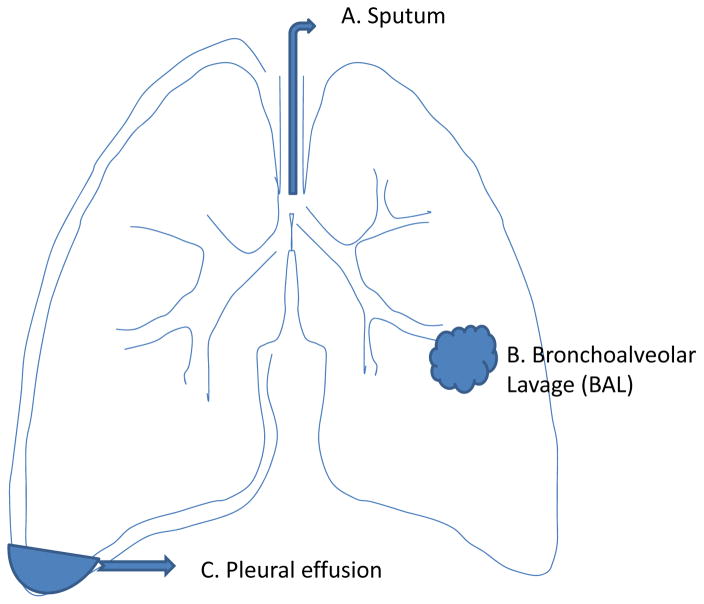
Traditionally, lung cancer cell lines, lung tumor tissue and blood samples from lung cancer patients are used for the study of protein biomarkers [20, 58–69, 112]. In recent years, other bio-fluids such as sputum, bronchoalveolar lavage (BAL) from lung airway, pleural effusion from the pleural cavity (the space between the lung and chest wall) as well as urine have been increasingly used for the study of potential biomarkers in lung diseases and cancers. These fluid-based specimens are relatively easier to collect and cause minimal risk to patients. They can be repeatedly sampled during the course of the disease. Studies of protein profile in these samples may facilitate the discovery of potential protein biomarkers for early detection or monitoring the treatment response. In addition to blood and urine specimens, Figure 3 represents the anatomical structures of the lung and the production of potential bio-fluids.
Figure 3.
Anatomical structures of the lung and the production of potential fluid-based samples. (A) Sputum is a fluid secreted by bronchial epithelial cells from lower airways. It is usually collected as spontaneous and/or induced sputum. (B) Numerous proteins are secreted or leaked into the alveolar space from lung parenchymal cells. Proteins in the alveolar space can be recovered by bronchoalveolar lavage (BAL) using bronchoscopy. (C) The pleural cavity is a space between the lung and chest wall. A large amount of fluid, known as pleural effusion, can accumulate in the pleural cavity in a variety of diseases including lung cancers, particularly when the tumor metastases to the pleural cavity.
3.1 Blood and/or Serum
Blood and/or serum are well known specimens for the study of potential biomarkers in cancer patients [18, 19, 23, 24, 70, 71]. The most abundant protein in blood is serum albumin, which accounts for approximately 50% of total serum proteins. In addition, it contains a large amount of immunoglobulins and heterogeneous tissue proteins (as leakage from a variety of tissues). The complexity of the serum composition and the extraordinarily dynamic range of protein concentrations, which can differ by more than 10 orders of magnitude, may cause technical difficulties in the analysis of the blood proteome, since potential protein biomarkers are those of low abundant proteins and usually range in a concentration of ng/ml [72]. Several recent studies have successfully analyzed the profile of glycoproteins using serum samples from cancer patients [57, 73–76].
By using hydrazide chemistry on solid-phase extraction, Zeng X. et al. have studied pooled sera from NSCLC patients. After hydrazide chemistry enrichment and high resolution LC-MS/MS analysis, they have found that twenty-two proteins are differentially expressed in NSCLC patients [73]. In the study, they are able to further verify three glycoproteins: α-1-antichymotrypsin, insulin-like growth factor-binding protein and lipocalin-type prostaglandin D synthase; using commercially available enzyme-linked immunosorbent assay (ELISA) kits [73]. They have also performed a hierarchical clustering analysis of glycoprotein expression between different cancer types. Their data indicate that the identified glycoprotein biomarkers may be used in the separation of NSCLC from controls.
Hongsachart P. et al. have used WGA lectin affinity enrichment followed by co-immunoprecipitation, in-gel electrophoresis and MS analysis to identify serum biomarkers [74]. In this study, they have analyzed ten serum samples from stage II/III lung adenocarcinoma patients and found that twenty-seven glycoproteins are up-regulated and twelve are down-regulated in comparison to the healthy controls. The three up-regulated proteins (adiponectin, cerulolasmin and glycosylphosphatidyl-inositol-80) and two down-regulated glycoproteins (cyclin H and proto-oncogene tyrosine-protein kinase Fyn) are verified by Western blot analysis. These data highlight the potential utility of glycoprotein biomarkers in the early detection of lung cancers.
Ueda K. et al have studied eight serum samples from lung adenocarcinoma patients and found that six peptides have shown more than two-fold affinity to ConA lectin, whereas two peptides have shown less than 0.5-fold affinity to ConA lectin. Among these glycoproteins, TIMP1 (metalloproteinase inhibitor 1) has shown an up-regulated modification of high-mannose motif in stage IV lung cancer in comparison to stage I/II lung cancer and controls [57]. In addition to profiling serum glycoproteomes using lectin chromatography described above, Ueda K et al. have also performed SELDI-TOF MS (surface-enhanced laser desorption/ionization-time-of-flight mass spectrometry) analysis using lectin-coupled ProteinChip (Jacalin or SNA lectins) and found a higher frequency of loss of SNA binding in serum apolipoprotein C-III, indicating a cancer-associated aberrant glycosylation in NSCLC patients [75].
Tsai HY et al. have studied the alteration of glycans in sera of lung cancer patients using 2-DE, Western blotting with lectin staining, and MALDI (matrix-assisted laser desorption/ionization) MS and MS/MS approaches [76]. They have found that the fucosylated haptoglobin significantly increased in serum of lung cancer patients when compared to the normal controls. Further analysis has shown that the alteration of glycan in the cancer serum samples is primarily on the trisialylated triantennary N-glycan of haptoglobin [76]. Their data have suggested that specific glycans may be associated with certain subtype of NSCLC. In addition to glycoproteins, aberrant glycan may be used as a potential biomarker to monitor lung cancer progression.
In summary, these signature studies of glycoproteins provide initial evidence for the discovery of prognostic and predictive biomarkers in lung cancers.
3.2 Sputum
Sputum is a fluid secreted from lower airways. Two types of sputum, spontaneous and induced sputum, are usually collected for study of lung diseases. Spontaneous expectorated sputum is easy to collect; however, it may contain saliva from oral cavity. In order to reduce oral and upper respiratory airway contamination, induced sputum is preferentially used for the study of lung diseases. Induced sputum can be obtained after saline inhalation with a nebulizer; the procedure is usually performed in a clinic setting.
Although current MS techniques have an improved accuracy and throughput ability, the proteomic profiles of sputum have not been well studied, particularly in lung cancer. Only a few proteomic studies using this biological fluid have been published. More than 250 proteins have been identified in the sputum [77]. Gray RD, et al., showed that several proteins such as calgranulins and Clara cell secretory protein in the induced sputum were differentially expressed in patients with cystic fibrosis, asthma, COPD (chronic obstructive pulmonary disease) and bronchiectasis [78]. More recently, Nicholas BL et al., used proteomics (2-DE and tandem MS) and immunoassays (Western Blotting and ELISA) to study and compare proteins in induced sputum from COPD patients and healthy smoker controls [79]. In the study, 15 differentially expressed proteins were identified by MS/MS and subsequently validated by immunoassays; in particular, two protein biomarkers, lipocalin and alipoprotein A1, were found to be significantly reduced in patients with COPD when compared with healthy smokers [79]. Terracciano R, et al. studied the peptide profile in the induced sputum by using mesoporous silica beads (MSBs) SPE coupled to MALDI-TOF (matrix assisted laser desorption/ionization-time of flight) MS [80]. They detected more than 400 peptide peaks and found that several of them, such as human α-defensins (human neutrophil peptide (HNP)1, HNP2, HNP3) and three C-terminal amidated peptides, one of which was phosphorylated on serine, were differentially present among patients with asthma, chronic obstructive pulmonary disease and healthy controls [80].
While a number of protein biomarkers have been identified in CF (cystic fibrosis), COPD and other inflammatory airway diseases, it is quite evident that the low-abundance protein components of induced sputum could be used as biomarkers in lung diseases. However, the study of protein profile, particularly glycoproteins in sputum from lung cancer is still in its early stages. The presence of abundant, high-molecular-weight and highly charged mucin proteins may interfere with the analysis unless such disturbing proteins are removed from the sample before glycoproteomics. Other limitations of using sputum include: (1) difficulty in obtaining healthy control samples, and (2) lack of general standardization of sample collection. Despite all these apparent drawbacks, the profile of glycoproteome in sputum may provide important information in this field.
3.3 Bronchoalveolar Lavage (BAL) Fluid
Normal lung parenchyma contains several types of epithelial cells including bronchial epithelium, mucin producing cells, alveolar type I and type II pneumocytes, vascular structures and stromal cells [81]. Numerous proteins are either secreted or leaked into the alveolar space by these cells. Proteins in the alveolar space and stroma can be recovered by bronchoalveolar lavage (BAL) [82–85]. BAL fluid is collected by an endobrochoscopy. During the procedure, 10 to 20 cc of saline is used to wash the lesional area through the endobronchoscopy, and the fluid is then collected. By using this approach, airway glycoproteins can be recovered from a large area of lung parenchyma. This is especially important in the study of preinvasive and early cancer since these lesions may not have visible histological changes under bronchoscopy. It is also an important method to study peripherally-located lung cancers (particularly adenocarcinomas) since adenocarcinomas typically arise from lung parenchyma away from main bronchus and may not be reached by bronchoscopic biopsy needles.
The protein levels in BAL directly reflect the physiological or pathological status of the lung. The proteomic analysis of BAL specimens has been applied to the study of a variety of benign lung diseases, such as asthma and interstitial lung disease [82, 83]. Thus, proteomic analyses, particularly glycoproteomic approaches, can characterize the complex alveolar microenvironment and provide important insights into the pathogenesis of lung diseases and cancers, which leading to the identification of tumor-associated glycoprotein biomarkers.
Recently, we studied the protein profile in the BAL fluid from lung cancer patients [Li QK and Zhang H’s manuscript in preparation]. Among identified proteins, we also identified a subset of glycoproteins which was differentially expressed in BAL samples of lung cancer patients, compared to benign lung disease controls (Figure 4). Our study demonstrates a highly specific identification of N-glycoprotein. This finding is particularly important because it provides initial evidence that profiling glycoprotein in BAL could lead to the discovery of cancer-specific protein biomarkers.
Figure 4.
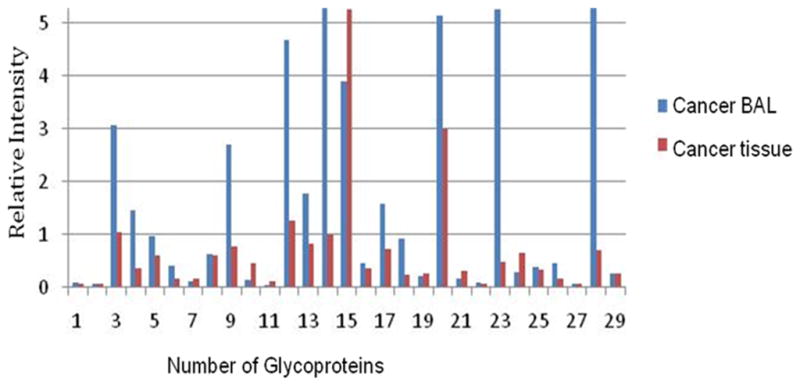
Detection of glycoproteins in BAL specimens of lung adenocarcinoma. Tumor-associated glycoproteins were identified in BAL specimens compared to tumor tissue in lung adenocarcinoma.
3.4 Pleural Effusion
The surface of the lung is covered by a thin layer of pleura, which is frequently involved in lung cancer during tumor progression. The pleura are lined by a single layer of mesothelial cells, which covers the surface of lung (visceral surface) and inner surface of the chest wall (parietal surface). The pleural cavity is formed between these two layers of cells. Normally, the pleural cavity contains only a small amount of fluid to lubricate the visceral and parietal surfaces as they move against each other during respiration. The protein composition in pleural effusion is similar to that of plasma [81]. In the presence of variety of diseases, particularly when lung cancer metastasizes to the pleura, a large amount of fluid known as pleural effusion can accumulate in the pleural cavity. This fluid accumulation is due to the increased leakage of protein and/or decreased reabsorption of fluid [81]. Pleural effusion has been considered as a biological specimen with an enrichment of tumor-derived proteins in lung cancers [86, 87]. In addition to providing diagnostic information, proteomic analysis may also provide an important insight into tumor-related biomarkers during lung cancer progression [86, 87]. Other advantages of using pleural effusion are: (a) ease of procurement by the thoracentesis, (b) minimal risk for the patient, (c) the large quantity of sample, and (d) the ability to repeatedly procure sample during disease progression for studying progressive biomarkers.
Soltermann A. et al. have applied the hydrazide chemistry capture and LC-MS/MS approach in analyzing five cases of pleural effusions from metastatic lung adenocarcinoma patients [87]. Three lung-specific proteins: mucin 5B, thyroid transcript factor 1 (TTF1) and surfactant protein A were present in their samples. Six tumor-progression or metastasis-associated glycoproteins, including CA-125, CD44, CD166, lysosomal-associated membrane glycoprotein 2 (LAMP-2), multimerin 2, and periostin were also identified [87]. This study indicated the potential utility of pleural effusion in the discovery of progression biomarkers.
In another recent study using SDS-PAGE and LC-MS/MS, four candidate proteins, α-2-HS-glycoprotein, angiogenin, cystatin-C and insulin-like growth factor-binding protein 2 were identified [88]. Among them, levels of α-2-HS-glycoprotein and insulin-like growth factor-binding protein 2 were increased in pleural effusions of lung cancer patients [88]. Although this study did not use glycoproteomics, the finding of α-2-HS-glycoprotein in malignant pleural effusion suggests that glycoproteins could be used as biomarkers in the monitoring of disease progression.
3.5 Urine
Urine is formed in the kidney by ultrafiltration of the blood to eliminate waste products such as urea and metabolites from the circulatory system [81]. In a normal adult, the protein excretion is less than 100 mg/L when urine output is about 1.5 L/day. The amount of protein in the urine may be affected by many factors such as age, gender, medication, nutritional status and many diseases; thus, both protein compositions and concentrations in the urine not only reflect the physiopathological status of the kidney/urinary system, but also reflect the status of many systems.
The urinary proteome has been analyzed by many proteomic techniques, including two-dimensional gel electrophoresis (2-DE), MS and surface-enhanced laser desorption/ionization time-of-flight (SELDI-TOF)-MS [89–91]. More than 1,500 protein peaks have been identified in the human urinary proteome [89]. Of the total urinary proteome, 70% of proteins are derived from the kidney and urinary tract while the remaining 30% of proteins represent those filtered from blood by the glomerulus. Therefore, the urinary protein may contain information from the urinary tract and other distant organs via blood and glomerular filtration. The analysis of the urinary proteome might therefore allow the identification of biomarkers for systemic diseases [90]. Indeed, urinary protein markers have been identified in COPD of the lung [92], lung cancers [93–96], prostate cancer [97], bladder cancers and a few others [90].
In the study of Boutin M et al. two proteins, desmosine (DES) and isodesmosine (IDES), were identified in the urine from COPD patients using nanoflow liquid chromatography tandem mass spectrometry with multiple reaction monitoring [92]. This method enables them to quantify DES and IDES in 50 μL of urine and provides a detection limit of 0.10 ng/mL. They analyzed a cohort of 40 urine specimens from four groups of individuals: including COPD rapid decliners, slow decliners, healthy smokers and healthy nonsmokers. Among these patients, they found that a statistically significant decrease in the level of urinary DES and IDES in COPD rapid decliner patients, compared to healthy nonsmoker controls and COPD slow decliner patients. Their data indicated that urinary DES and IDES levels may be used to monitor disease progression in COPD patients.
Tantipaiboonwong P, et al studied pooled urinary samples from five NSCLCs by using 2-DE and MALDI-MS and MS/MS analysis [93]. They found three proteins were decreased and six proteins were increased in NSCLCs in comparison to healthy controls. Among these proteins, glycoprotein of CD59 was decreased in the lung cancer patients. Li YY and Zhang Y, et al. identified 18 proteins from urine samples of 8 NSCLC patients using nano-HPLC-chip-MS/MS [94, 95]. Among them, leucine-rich α-2-glycoprotein (LRG1) was further identified in the urine of NSCLC patients and lung tumor tissues by Western blotting and immunohistochemistry [95]. In a recent study, Carrola J, et al. analyzed urinary metabolic signature of 71 lung cancer patients and 54 controls [96]. In the study, they combined analytic techniques of nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry together with multivariate statistics to unveil metabonomics in the urine of lung cancer patients. They found that levels of six metabolites including N-acetylglutamine were either elevated or decreased in urines of lung cancer patients in comparison to controls. Although these studies did not use glycoproteomics, the identification of glycoproteins and/or their metabolites in the urine suggests that glycoproteins in urine may be potential biomarkers for diagnosis of invasive lung cancers.
The advantage of studying urine samples is that they can be obtained noninvasively and repeatedly during the disease progression. However, human urine has a very low concentration of proteins while having a high concentration of salts, metabolic wastes and small molecules that could interfere with proteomic analysis [89–93]. It is crucial to use an effective protocol to eliminate interferences and to concentrate urinary proteins [89–92]. There are several protocols that can be employed to concentrate urinary proteins, such as centrifugal filtration, lyophilization, precipitation, and ultracentrifugation [89–96]. With current advanced proteomic techniques, these initial studies of urinary proteome may provide clinically relevant information regarding lung cancer diagnosis.
4. Potential Application of Glycoprotein Biomarkers
Glycoproteomics presents itself as a prominent technology in the field of cancer research. Using glycoproteomics in the study of lung cancer, however, is just emerging, and only a few studies have been published from initial studies. To facilitate the new era of personalized medicine, the profile of glycoprotein in lung cancer may play an important role. Glycoproteomic analysis can further define glycoprotein signature related to early detecton (cancer versus benign disease), prognosis (likelihood of cure or risk of progression and metastasis), and prediction (probability of response to therapy) in lung cancers.
4.1 Detection of Precancerous Lesion and Early Cancers
Studies have shown that lung cancer is the result of accumulations of genotypic and phenotypic abnormalities, and only a minority of preinvasive lung lesions progresses to invasive cancer [98–100]. These preinvasive lesions can be subtyped into the mild, moderate and severe dysplasia, and carcinoma in situ in squamous cell lung cancer. Studies using serial bronchoscopic biopsies have suggested that 3.5% of mild or moderate dysplasias progressed to severe dysplasia, 37% of severe dysplasias progressed to carcinomas in situ, and 50% of carcinomas in situ progressed to invasive carcinoma within a two- to three-year period [100]. Currently, several clinical trials to treat these patients with bronchial epithelium dysplasia using chemoprevention have shown the regression of the lesion [100]. The study of protein signature during the cancer progression may lead to the discovery of tumor-associated biomarkers. The development a quantitative measurement of probability of having lung cancer based on the glycoprotein profile of the bronchial epithelium may have important clinical implications. The identification of preinvasive lesion with a high risk of progression based on glycoprotein profile can also improve the early detection of lung cancers.
In a study of fresh frozen lung adenocarcinoma and patient-matched normal lung tissue, Rho JH et al. used a comprehensive glycoproteomic enrichment by lectins of ConA, WGA and amylase inhibitor-like protein (AIL), then analyzed glycoproteins by 2-D PAGE and MS/MS approaches [101, 102]. They analyzed both N-glycoproteins (by ConA- and WGA-capture) and O-glycoproteins (by AIL-capture) in sixteen lung adenocarcinomas and matched controls [102]. They found that eight glycoproteins are up-regulated, including α1-antitrypsin, fructose-bisphosphate aldolase A, annexin A1, calreticulin, α-enolase, protein disulfide isomerase A1, proteasome subunit β type1, and mitochondrial superoxide dismutase. In comparison, seven glycoproteins are down-regulated, including annexin A3, carbonic anhydrase 2, fetuin A, hemoglobin subunit β, peroxiredoxin-2, vimentin and receptor for advanced glycosylation end products. They also found a high level of mannose glycan structures on fetuin A protein in cancers, but not in normal tissue [102]. In addition, they also identified that transgelin is overexpressed in the stromal compartment whereas transgelin-2 is overexpressed in lung cancer tissue [101].
4.2 Monitoring Cancer Progression (Prognostic Biomarkers)
The progression of lung cancer is a multi-step process. It is believed that tumor at a later clinical stage is more aggressive than a tumor at an early stage; therefore, tumor at different stages may express a unique subset of proteins that can be used in monitoring tumor progression. If a biomarker is correlated with the natural history of the disease and reflects the tumor progression, such as invasiveness or metastatic potential independent of the therapeutic intervention, it is defined as a prognostic biomarker. On the other hand, a biomarker is defined as predictive if the efficacy of a specific therapy on the tumor can be predicted by using the biomarker [103]. The most commonly used strategy to identify such potential biomarkers is to compare protein expression from different stages of tumor tissue or in the blood from patients responding or not responding to a certain treatment. An alternative approach is to study protein expression within different stages of tumors and then correlate them with patients’ clinical survival rates. Glycoproteomics is particular useful in discovering prognostic biomarkers since it allows not only for evaluation of differential expression of glycoproteins, but also for identification of potential changes of glycosylation sites during tumor progression.
Our recent study has shown that one subtype of lung adenocarcinoma, papillary adenocarcinoma of the lung, has a high level of NRF2 (nuclear erythroid-2 related factor 2) and p53 protein expression, and presents a poor clinical prognosis, in comparison to the conventional lung adenocarcinomas [104].
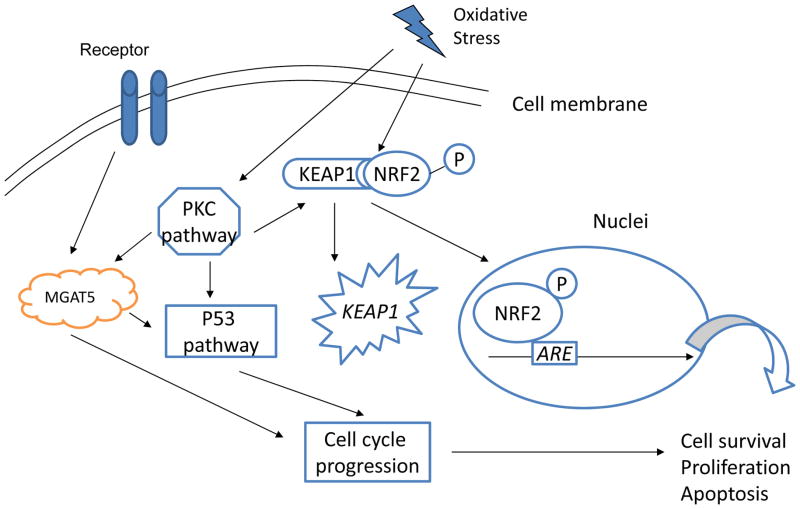
NRF2 is a transcription factor that regulates the expression of genes encoding antioxidants and xenobiotic detoxification enzymes. The elevated levels of NRF2 have been shown to relate to cancer cell survival and potential protections against chemotherapeutic agents [105]. The mechanism that regulates this increased NRF2 expression is complex. In some cases, it is related to mutations of the KEAP1 (Kelch-like ECH-associated protein) gene, which prevents binding of the KEAP1 protein to NRF2. This causes NRF2 accumulation in the nucleus where it can affect another gene, ARE (antioxidant response element) expression (Figure 5). In this complex oxidative stress signaling pathway, MGAT5 (Mannosyl (α-1,6-)-glycoprotein β-1,6,-N-acetyl-glucosaminyltransferase) gene also plays an important role. This gene encodes mannosyl (α-1,6-)-glycoprotein β-1,6-N-acetyl-glucosaminyltransferase, which regulates the synthesis of protein-bound oligosaccharides on the cell surface. In the mouse model, cell-cycle progression (p53 pathway) is dependent on Mgat5/N-glycan interaction. The alteration of cellular N-glycoproteins causes significant changes of cellular adhesion and migration [106]. These data indicate that oxidative stress and MGAT5 pathways may interact in the regulation of cell growth. The study of cellular glycoprotein in different stages of the cell cycle may help us understand mechanisms in lung cancer progression and the development of drug resistance.
Figure 5.
Oxidative stress intracellular signaling pathways in lung cancer. NRF2 (nuclear erythroid-2 related factor 2) is a transcription factor that regulates the expression of genes encoding antioxidants and xenobiotic detoxification enzymes. The elevated levels of NRF2 have been shown to regulate cancer cell survival and potentially protect against chemotherapeutic agents. The mechanism that regulates increased NRF2 expression involves KEAP1 (Kelch-like ECH-associated protein) and ARE (antioxidant response element) expression. In the network, MGAT5 (Mannosyl (α-1,6-)-glycoprotein β-1,6,-N-accetyl-glucosaminyltransferase) gene encodes mannosyl (α-1,6-)-glycoprotein β-1,6-N-acetyl-glucosaminyltransferase, which regulates the synthesis of protein-bound oligosaccharides on the cell surface. Mgat5/N-glycan interaction is involved in cell-cycle progression (p53 pathway).
4.3 Monitoring the Therapy Response
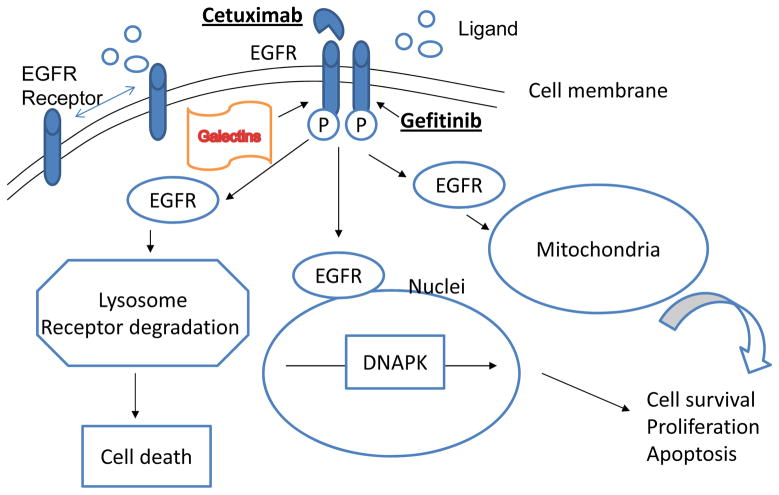
In lung cancers, the EGFR mutation is associated with a 70–80% response rate to tyrosine-kinase inhibitors (TKIs) therapy and a longer progression-free survival rate in patients [5–7, 107]. Although the growth factor receptors are N-glycosylated transmembrane proteins, their biological functions are partially regulated by intracellular endogenous lectins, such as galectins [108]. Galectins can cross-link growth factor receptor glycoproteins and regulate the distribution of receptors on the cell surface. Galectin-3 has been shown to bind to N-glycans of EGFR and limit its distribution on the plasma membrane [109–111]. Furthermore, the mutation with the deletion of extracellular domain of EGFR can cause a loss of four out of twelve N-glycan sites on the receptors of tumor cells. This deletional mutation induces receptor dimerization and signaling [109–111]. These data indicate that the interaction of N-glycan and receptor protein may regulate the distribution and residency of the growth receptor on the cell membrane in addition to receptor mutations (Figure 6). This is a poorly understood area. The study of the functional role of glycoprotein has the potential of understanding drug resistance mechanisms in lung cancer, particularly among patients who are treated with EGFR inhibitors. The analysis of glycoproteins in lung cancer cells may in fact provide highlights in the field.
Figure 6.
EGFR signaling pathways in lung cancer. In lung cancers, the EGFR mutation is associated with a 70–80% response rate to tyrosine-kinase inhibitors (TKIs) therapy, such as cetuximab and gefitinib. Intracellular endogenous lectins, such as galectin-3, have been shown to bind to N-glycans of EGFR and limit its distribution on the plasma membrane. Furthermore, the deletion of extracellular domain of EGFR can cause a loss of four out of twelve N-glycan sites on the receptor in tumor cells. This deletional mutation induces receptor dimerization and signaling. The interaction of lectin and receptor protein may regulate the distribution and residency of the growth receptor on the cell membrane. It may involve in the development of TKI drug resistance in lung cancer. DNAPK: DNA-activated protein kinase.
5. Perspectives
Although many protein biomarkers are currently published with continued exponential growth rate in the research field, they still lack full validations for both the diagnosis and monitoring progression of lung cancer. Despite years of extensive research attempting to identify and validate candidate protein biomarkers, the number of FDA-approved biomarkers is still limited, and there is still a lack of effective diagnostic and prognostic protein markers in lung cancer. The challenge in clinical proteomics is to obtain sufficient, clinically relevant, and well-characterized patient material for proteomics analysis and validation. The most common clinical samples available to study protein biomarkers include, but are not limited to, blood/plasma, induced sputum, bronchoalveolar lavage (BAL), pleural effusion, urine and tumor tissues. It is absolutely essential to use clinically relevant samples from well-characterized patients to further understand the biology of lung cancer and to discover biomarkers.
In glycoproteomic biomarker discovery, it is also important to use clinical materials in both the discovery process and subsequent validation phase. Despite the rapid progress in glycoproteomics, the workflow in analysis of clinical samples hinders the necessary throughput in large scale studies. Therefore, improvement of analytic throughput ability is needed for study of a large scale of patient cohort. In addition, technologies still need to be further improved in terms of accuracy and sensitivity in measurements of clinical material. This is evident from the serum glycoproteomic studies where the analysis of dynamic range and glycoproteome coverage needs to be more efficient, particularly in large scale of patient cohort. Finally, the potential success of glycoproteomic biomarker study largely depends on the quality and availability of patient cohort. This requires large number of carefully selected patient cohort to determine the potential utility of the candidate biomarker. Clinical validation of potential glycoprotein biomarkers must be conducted in a fashion to avoid the occurrence of false positive results. For each candidate biomarker, robust and reproducible assay needs to be developed and used in the validation phase.
In summary, glycoproteomics presents itself as a prominent technology in the field of lung cancer research. The recent advances of glycoproteomics technology clearly facilitate the discovery of novel biomarkers in lung cancer. The advances of glycoproteomics have significantly increased our knowledge in the field of lung cancer biology. It also promotes the potential clinical utility of the technique and biomarkers in the targeted therapy of lung cancer. Further improvement of both the workflow and validation process are needed in the identification of lung cancer glycoprotein biomarkers.
Acknowledgments
This work is partially supported by Drs. Ji and Li Family Foundation (QKL), and the federal fund from the National Institutes of Health/the National Cancer Institute/Early Detection Research Network grant (NIH/NCI/EDRN) U01CA152813 (HZ), NCI/Clinical Proteomics Tumor Analysis Consortium (NIH/NCI/CPTAC) U24CA160036, and by National Heart Lung and Blood Institute/Programs of Excellence in Glycosciences (NIH/NHLBI/PEG) P01HL107153. Authors thank Ms. Lynne Sakowski for her proofreading.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Global Cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon: IACR Press; 2003. World Health Organization classification of tumours. [Google Scholar]
- 3.Travis WD IASLC Staging Committee. Reporting lung cancer pathology specimens. Histopathology; Impact of the anticipated 7th Edition TNM classification based on recommendations of the IASLC Staging Committee; 2009. pp. 3–11. Review. [DOI] [PubMed] [Google Scholar]
- 4.Harpole DH, Herndon JE, Young WG, Wolfe WG, Sabiston DC. Stage I non-small cell lung cancer. Cancer. 1995;76:787–796. doi: 10.1002/1097-0142(19950901)76:5<787::aid-cncr2820760512>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasent S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–945. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 6.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, Chan WK, Li HN, Liu CC, Singh S, Chen WJ, Chen JJW, Yang PC. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 7.Coate LE, John T, Tsao MS, Shepherd FA. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10:1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- 8.National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swensen SJ, Jett JR, Hartman TE, Mandrekar SJ, Hillman SL, Sykes AM, Aughenbaugh GL, Bungum AO, Allen KL. CT screening for lung cancer: Five-year prospective experience. Radiology. 2005;235:259–265. doi: 10.1148/radiol.2351041662. [DOI] [PubMed] [Google Scholar]
- 10.Klabunde CN, Marcus PM, Silvestri GA, Han PK, Richards TB, Yuan G, Marcus SE, Vernon SW. U.S. primary care physicians’ lung cancer screening beliefs and recommendations. Am J Prev Med. 2010;39:411–420. doi: 10.1016/j.amepre.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 12.Wong CH. Protein glycosylation: new challenges and opportunities. J Org Chem. 2005;70:4219–4225. doi: 10.1021/jo050278f. [DOI] [PubMed] [Google Scholar]
- 13.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 14.Potapenko IO, Haakensen VD, Lüders T, Helland A, Bukholm I, Sørlie T, Kristensen VN, Lingjaerde OC, Børresen-Dale AL. Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Mol Oncol. 2010;4:98–118. doi: 10.1016/j.molonc.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 16.Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 17.Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clini Chem. 2010;56:223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y, Zhang H. Glycoproteomics and clinical applications. Proteomics Clin Appl. 2010;4:124–132. doi: 10.1002/prca.200900161. [DOI] [PubMed] [Google Scholar]
- 19.Kam RKT, Poon TCW. The potentials of glycomics in biomarker discovery. Clin Proteom. 2008;4:67–79. [Google Scholar]
- 20.Li QK, Gabrielson E, Zhang H. Application of glycoproteomics for the discovery of biomarkers in lung cancer. Proteomics: Clinical App. 2012;5–6:244–256. doi: 10.1002/prca.201100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fares F. The role of O-linked and N-linked oligosaccharides on the structure-function of glycoprotein hormones: development of agonists and antagonists. Biochim Biophys Acta. 2006;1760:560–567. doi: 10.1016/j.bbagen.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 23.Tong L, Baskaran G, Jones MB, Rhee JK, Yarema KJ. Glycosylation changes as markers for the diagnosis and treatment of human disease. Biotechnol Genet Eng Rev. 2003;20:199–244. doi: 10.1080/02648725.2003.10648044. [DOI] [PubMed] [Google Scholar]
- 24.Powlesland AS, Hitchen PG, Parry S, Graham SA, Barrio MM, Elola MT, Mordoh J, Dell A, Drickamer K, Taylor ME. Targeted glycoproteomic identification of cancer cell glycosylation. Glycobiology. 2009;19:899–909. doi: 10.1093/glycob/cwp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda K, Katagiri T, Shimada T, Irie S, Sato TA, Nakamura Y, Daigo Y. Comparative profiling of serum glycoproteome by sequential purification of glycoproteins and 2-nitrobenzensulfenyl (NBS) stable isotope labeling: a new approach for the novel biomarker discovery for cancer. J Proteome Res. 2007;6:3475–3483. doi: 10.1021/pr070103h. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 27.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid phase extraction of N-linked glycopeptides. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun B, Ranish JA, Utleg AG, White JT, Yan X, Lin B, Hood L. Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol Cell Proteomics. 2007;6:141–149. doi: 10.1074/mcp.T600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 30.Lee A, Kolarich D, Haynes PA, Jensen PH, Baker MS, Packer NH. Rat liver membrane glycoproteome: enrichment by phase partitioning and glycoprotein capture. J Proteome Res. 2009;8:770–781. doi: 10.1021/pr800910w. [DOI] [PubMed] [Google Scholar]
- 31.Zauner G, Koeleman CA, Deelder AM, Wuhrer M. Mass spectrometric O-glycan analysis after combined O-glycan release by beta-elimination and 1-phenyl-3-methyl-5-pyrazolone labeling. Biochim Biophys Acta. 2012;1820:1420–1428. doi: 10.1016/j.bbagen.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Y, Guo Z, Cai Z. Combination of beta-elimination and liquid chromatography/quadrupole time-of-flight mass spectrometry for the determination of O-glycosylation sites. Talanta. 2009;78:358–363. doi: 10.1016/j.talanta.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 33.Tian Y, Kelly-Spratt KS, Kemp CJ, Zhang H. Identification of glycoproteins from mouse skin tumors and plasma. Clin Proteom. 2008;4:117–136. doi: 10.1007/s12014-008-9014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, Smith RD, Kemp CJ, Aebersold R. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Madera M, Mechref Y, Klouckova I, Novotny MV. Semiautomated high-sensitivity profiling of human blood serum glycoproteins through lectin preconcentration and multidimensional chromatography/tandem mass spectrometry. J Proteome Res. 2006;5:2348–2363. doi: 10.1021/pr060169x. [DOI] [PubMed] [Google Scholar]
- 36.Chen R, Jiang X, Sun D, Han G. Wang F, Ye M, Wang L, Zou H, Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 37.Sparbier K, Wenzel T, Kostrzewa M. Exploring the binding profiles of ConA, boronic acid and WGA by MALDITOF/TOF MS and magnetic particles. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;840:29–36. doi: 10.1016/j.jchromb.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Wu Z, Zhang L, Lu H, Yang P, Webley PA, Zhao D. Highly specific enrichment of glycopeptides using boronic acid-functionalized mesoporous silica. Anal Chem. 2009;81:503–508. doi: 10.1021/ac801912t. [DOI] [PubMed] [Google Scholar]
- 39.Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- 40.Alvarez-Manilla G, Atwood J, Guo Y, Warren NL, Orlando R, Pierce M. Tools for glycoproteomic analysis: size exclusion chromatography facilitates identification of tryptic glycopeptides with N-linked glycosylation sites. J Proteome Res. 2006;5:701–708. doi: 10.1021/pr050275j. [DOI] [PubMed] [Google Scholar]
- 41.Hirabayashi J, Kuno A, Tateno H. Lectin-based structural glycomics: a practical approach to complex glycans. Electrophoresis. 2011;32:1118–1128. doi: 10.1002/elps.201000650. [DOI] [PubMed] [Google Scholar]
- 42.Kuno A, Matsuda A, Ikehara Y, Narimatsu H, Hirabayashi J. Differential glycan profiling by lectin microarray targeting tissue specimens. Methods Enzymol. 2010;478:165–179. doi: 10.1016/S0076-6879(10)78007-1. [DOI] [PubMed] [Google Scholar]
- 43.Cummings RD, Kornfeld S. Fractionation of asparagines linked oligosaccharides by serial lectin-Agarose affinity chromatography. A rapid, sensitive, and specific technique. J Biol Chem. 1982;257:11235–11240. [PubMed] [Google Scholar]
- 44.Wiener MC, van Hoek AN. A lectin screening method for membrane glycoproteins: application to the human CHIP28 water channel (AQP-1) Anal Biochem. 1996;241:267–268. doi: 10.1006/abio.1996.0411. [DOI] [PubMed] [Google Scholar]
- 45.Bunkenborg J, Pilch BJ, Podtelejnikov AV, Wisniewski JR. Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics. 2004;4:454–465. doi: 10.1002/pmic.200300556. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto K, Tsuji T, Osawa T. Analysis of asparagines linked oligosaccharides by sequential lectin-affinity chromatography. Methods Mol Biol. 1998;76:35–51. doi: 10.1385/0-89603-355-4:35. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Wu SL, Hancock WS. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap--Fourier transform mass spectrometry. Glycobiology. 2006;16:514–523. doi: 10.1093/glycob/cwj091. [DOI] [PubMed] [Google Scholar]
- 48.Pan S, Wang Y, Quinn JF, Peskind ER, Waichunas D, Wimberger JT, Jin J, Li JG, Zhu D, Pan C, Zhang J. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- 49.Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med. 2000;6:306–312. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- 50.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Evanescent-field fluorescence assisted lectin microarray: a new strategy for glycan profiling. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 51.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycolpatterns. Chembiochem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Tao SC, Bova GS, Liu AY, Chan DW, Zhu H, Zhang H. Detection and verification of glycosylation patterns of glycoproteins from clinical specimens using lectin microarrays and lectin-based immunosorbent assays. Anal Chem. 2011;83:8509–8516. doi: 10.1021/ac201452f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meany DL, Zhang Z, Sokoll LJ, Zhang H, Chan DW. Glycoproteomics for Prostate Cancer Detection: Changes in Serum PSA Glycosylation Patterns. J Proteome Res. 2009;8:613–619. doi: 10.1021/pr8007539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tao SC, Li Y, Zhou J, Qian J, Schnaar RL, Zhang Y, Goldstein IJ, Zhu H, Schneck JP. Lectin microarrays identify cell-specific and functionally significant cell surface glycan markers. Glycobiology. 2008;18:761–769. doi: 10.1093/glycob/cwn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 56.Meany DL, Hackler L, Jr, Zhang H, Chan DW. Tyramide signal amplification for antibody-overlay lectin microarray: a strategy to improve the sensitivity of targeted glycan profiling. J Proteome Res. 2011;10:1425–1414. doi: 10.1021/pr1010873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueda K, Takami S, Saichi N, Daigo Y, Ishikawa N, Kohno N, Katsumata M, Yamane A, Ota M, Sato TA, Nakamura Y, Nakagawa H. Development of serum glycoproteomic profiling technique; simultaneous identification of glycosylation sites and site-specific quantification of glycan structure changes. Mol Cell Proteomics. 2010;9:1819–1828. doi: 10.1074/mcp.2010/000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanagisawa K, Shyr Y, Xu BJ, Massion PP, Larsen PH, White BC, Roberts JR, Edgerton M, Gonzalez A, Nadaf S, Moore JH, Caprioli RM, Carbone DP. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362:433–439. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 59.Buhrens RI, Amelung JT, Reymond MA, Beshay M. Protein expression in human non-small cell lung cancer: a systematic database. Pathobiology. 2009;76:277–285. doi: 10.1159/000245893. [DOI] [PubMed] [Google Scholar]
- 60.Lehtio J, De Petris L. Lung cancer proteomics, clinical and technological considerations. J Proteome. 2010;73:1851–1863. doi: 10.1016/j.jprot.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 61.Vegvari A, Marko-Varga G. Clinical protein science and bioanalytical mass spectrometry with an emphasis on lung cancer. Chem Rev. 2010;110:3278–3298. doi: 10.1021/cr100011x. [DOI] [PubMed] [Google Scholar]
- 62.Tian T, Hao J, Xu A, Hao J, Luo C, Liu C, Huang L, Xiao X, He D. Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis. Cancer Sci. 2007;98:1265–1274. doi: 10.1111/j.1349-7006.2007.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao H, Zhang Z, Xiao Z, Chen Y, Li C, Zhang P, Li M, Liu Y, Guan Y, Yu Y, Chen Z. Identification of metastasis associated proteins in human lung squamous carcinoma using two-dimensional difference gel electrophoresis and laser capture microdissection. Lung Cancer. 2009;65:41–48. doi: 10.1016/j.lungcan.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 64.Rahman SM, Shyr Y, Yildiz PB, Gonzalez AL, Li H, Zhang X, Chaurand P, Yanagisawa K, Slovis BS, Miller RF, Ninan M, Miller YE, Franklin WA, Caprioli RM, Carbone DP, Massion PP. Proteomic patterns of preinvasive bronchial lesions. Am J Respir Crit Care Med. 2005;172:1556–1562. doi: 10.1164/rccm.200502-274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Au JS, Cho WC, Yip TT, Law SC. Proteomic approach to biomarker discovery in cancer tissue from lung adenocarcinoma among nonsmoking Chinese women in Hong Kong. Cancer Investig. 2008;26:128–135. doi: 10.1080/07357900701788031. [DOI] [PubMed] [Google Scholar]
- 66.Campa MJ, Wang MZ, Howard B, Fitzgerald MC, Patz EF., Jr Protein expression profiling identifies macrophage migration inhibitory factor and cyclophilin a as potential molecular targets in non-small cell lung cancer. Cancer Res. 2003;63:1652–1656. [PubMed] [Google Scholar]
- 67.Kawamura T, Nomura M, Tojo H, Fujii K, Hamasaki H, Mikami S, Bando Y, Kato H, Nishimura T. Proteomic analysis of laser-microdissected paraffin-embedded tissues: (1) stage-related protein candidates upon non-metastatic lung adenocarcinoma. J Proteome. 2010;73:1089–1099. doi: 10.1016/j.jprot.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 68.Nishimura T, Nomura M, Tojo H, Hamasaki H, Fukuda T, Fujii K, Mikami S, Bando Y, Kato H. Proteomic analysis of laser-microdissected paraffin-embedded tissues: (2) MRM assay for stage-related proteins upon non-metastatic lung adenocarcinoma. J Proteome. 2010;73:1100–1110. doi: 10.1016/j.jprot.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Oshita F, Morita A, Ito H, Kameda Y, Tsuchiya E, Asai S, Miyagi Y. Proteomic screening of completely resected tumors in relation to survival in patients with stage I non-small cell lung cancer. Onco Rep. 2010;24:637–645. doi: 10.3892/or_00000902. [DOI] [PubMed] [Google Scholar]
- 70.Zhang H, Chan DW. Cancer biomarker discovery in plasma using a tissue-targeted proteomic approach. Cancer Epidemiol Biomarkers Prev. 2007;16:1915–1917. doi: 10.1158/1055-9965.EPI-07-0420. [DOI] [PubMed] [Google Scholar]
- 71.Li Y, Sokoll LJ, Barker PE, Zhang H, Chan DW. Mass spectrometric identification of proteotypic peptides from clinically used tumor markers. Clin Proteom. 2008;4:58–66. [Google Scholar]
- 72.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 73.Zeng X, Hood BL, Sun M, Conrads TP, Day RS, Weissfeld JL, Siegfried JM, Bigbee WL. Lung cancer serum biomarker discovery using glycoprotein capture and liquid chromatography mass spectrometry. J Proteome Res. 2010;9:6440–6449. doi: 10.1021/pr100696n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hongsachart P, Huang-Liu R, Sinchaikul S, Pan FM, Phutrakul S, Chuang YM, Chen ST. Glycoproteomic analysis of WGA-bound glycoprotein biomarkers in sera from patients with lung adenocarcinoma. Electrophoresis. 2009;30:1206–1220. doi: 10.1002/elps.200800405. [DOI] [PubMed] [Google Scholar]
- 75.Ueda K, Fukase Y, Katagiri T, Ishikawa N, Irie S, Sato TA, Ito H, Nakayama H, Miyagi Y, Tsuchiya E, Kohno N, Shiwa M, Nakamura Y, Daigo Y. Targeted serum glycoproteomics for the discovery of lung cancer-associated glycosylation disorders using lectin-coupled ProteinChip arrays. Proteomics. 2009;9:2182–2192. doi: 10.1002/pmic.200800374. [DOI] [PubMed] [Google Scholar]
- 76.Tsai HY, Boonyapranai K, Sriyam S, Yu CJ, Wu SW, Khoo KH, Phutrakul S, Chen ST. Glycoproteomics analysis to identify a glycoform on haptoglobin associated with lung cancer. Proteomics. 2011;11:2162–2170. doi: 10.1002/pmic.201000319. [DOI] [PubMed] [Google Scholar]
- 77.Nicholas B, Skipp P, Mould R, Rennard S, Davies DE, O’Connor CD, Djukanovi R. Shotgun proteomic analysis of human-induced sputum. Proteomics. 2006;6:4390–4401. doi: 10.1002/pmic.200600011. [DOI] [PubMed] [Google Scholar]
- 78.Gray RD, MacGregor G, Noble D, Imrie M, Dewar M, Boyd AC, Innes JA, Porteous DJ, Greening AP. Sputum proteomics in inflammatory and suppurative respiratory diseases. Am J Respir Crit Care Med. 2008;178:444–452. doi: 10.1164/rccm.200703-409OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicholas BL, Skipp P, Barton S, Singh D, Bagmane D, Mould R, Angco G, Ward J, Guha-Niyogi B, Wilson S, Howarth P, Davies DE, Rennard S, O’Connor CD, Djukanovic R. Identification of lipocalin and apolipoprotein A1 as biomarkers of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;181:1049–1060. doi: 10.1164/rccm.200906-0857OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Terracciano R, Preianò M, Palladino GP, Carpagnano GE, Barbaro MP, Pelaia G, Savino R, Maselli R. Peptidome profiling of induced sputum by mesoporous silica beads and MALDI-TOF MS for non-invasive biomarker discovery of chronic inflammatory lung diseases. Proteomics. 2011;11:3402–3414. doi: 10.1002/pmic.201000828. [DOI] [PubMed] [Google Scholar]
- 81.Kobzik L. Lung. In: Cotran RS, Kumer V, Collins T, editors. Robbins pathologic basis of disease. 7. Philadelphia: Saunders; 2004. pp. 697–755. [Google Scholar]
- 82.Magi B, Bargagli E, Bini L, Rottoli P. Proteome analysis of bronchoalveolar lavage in lung diseases. Proteomics. 2006;6:6354–6369. doi: 10.1002/pmic.200600303. [DOI] [PubMed] [Google Scholar]
- 83.Rottoli P, Bargagli E, Landi C, Magi B. Proteomic analysis in interstitial lung diseases: a review. Curr Opin Pulm Med. 2009;15:470–478. doi: 10.1097/MCP.0b013e32832ea4f2. [DOI] [PubMed] [Google Scholar]
- 84.Wattiez R, Falmagne P. Proteomics of BAL fluid. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:169–178. doi: 10.1016/j.jchromb.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 85.Plymoth A, Löfdahl CG, Ekberg-Jansson A, Dahlbäck M, Lindberg H, Fehniger TE, Marko-Varga G. Human bronchoalveolar lavage: biofluid analysis with special emphasis on sample preparation. Proteomics. 2003;3:962–972. doi: 10.1002/pmic.200300387. [DOI] [PubMed] [Google Scholar]
- 86.Tyan YC, Wu HY, Lai WW, Su WC, Liao PC. Proteomic profiling of human pleural effusion using two-dimensional nano liquid chromatography tandem mass spectrometry. J Proteome Res. 2005;4:1274–1286. doi: 10.1021/pr049746c. [DOI] [PubMed] [Google Scholar]
- 87.Soltermann A, Ossola R, Kilgus-Hawelski S, von Eckardstein A, Suter T, Aebersold R, Moch H. N-glycoprotein profiling of lung adenocarcinoma pleural effusions by shotgun proteomics. Cancer Cytopathol. 2008;114:124–133. doi: 10.1002/cncr.23349. [DOI] [PubMed] [Google Scholar]
- 88.Yu CJ, Wang CL, Wang CI, Chen CD, Dan YM, Wu CC, Wu YC, Lee IN, Tsai YH, Chang YS, Yu JS. Comprehensive proteome analysis of malignant pleural effusion for lung cancer biomarker discovery by using multidimensional protein identification technology. J Proteome Res. 2011;10:4671–4682. doi: 10.1021/pr2004743. [DOI] [PubMed] [Google Scholar]
- 89.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coon JJ, Zürbig P, Dakna M, Dominiczak AF, Decramer S, Fliser D, Frommberger M, Golovko I, Good DM, Herget-Rosenthal S, Jankowski J, Julian BA, Kellmann M, Kolch W, Massy Z, Novak J, Rossing K, Schanstra JP, Schiffer E, Theodorescu D, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clin Appl. 2008;2:964–973. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sinchaikul S, Tantipaiboonwong P, Sriyam S, Tzao C, Phutrakul S, Chen ST. Different sample preparation and detection methods for normal and lung cancer urinary proteome analysis. Methods Mol Biol. 2010;641:65–88. doi: 10.1007/978-1-60761-711-2_5. [DOI] [PubMed] [Google Scholar]
- 92.Boutin M, Berthelette C, Gervais FG, Scholand MB, Hoidal J, Leppert MF, Bateman KP, Thibault P. High-sensitivity nanoLC-MS/MS analysis of urinary desmosine and isodesmosine. Anal Chem. 81:1881–1887. doi: 10.1021/ac801745d. 20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tantipaiboonwong P, Sinchaikul S, Sriyam S, Phutrakul S, Chen ST. Different techniques for urinary protein analysis of normal and lung cancer patients. Proteomics. 2005;5:1140–9. doi: 10.1002/pmic.200401143. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y, Li Y, Qiu F, Qiu Z. Comparative analysis of the human urinary proteome by 1D SDS-PAGE and chip-HPLC-MS/MS identification of the AACT putative urinary biomarker. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:3395–401. doi: 10.1016/j.jchromb.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 95.Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 32:1976–1983. doi: 10.1002/elps.201000598. 201. [DOI] [PubMed] [Google Scholar]
- 96.Carrola J, Rocha CM, Barros AS, Gil AM, Goodfellow BJ, Carreira IM, Bernardo J, Gomes A, Sousa V, Carvalho L, Duarte IF. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J Proteome Res. 2011;10:221–230. doi: 10.1021/pr100899x. [DOI] [PubMed] [Google Scholar]
- 97.Theodorescu D, Fliser D, Wittke S, Mischak H, Krebs R, Walden M, Ross M, Eltze E, Bettendorf O, Wulfing C, Semjonow A. Pilot study of capillary electrophoresis coupled to mass spectrometry as a tool to define potential prostate cancer biomarkers in urine. Electrophoresis. 2005;26:2797–2808. doi: 10.1002/elps.200400208. [DOI] [PubMed] [Google Scholar]
- 98.Venmans BJ, van Boxem TJ, Smith EF, Postmus PE, Sutedja TG. Outcome of bronchial carcinoma in situ. Chest. 2000;117:1572–1576. doi: 10.1378/chest.117.6.1572. [DOI] [PubMed] [Google Scholar]
- 99.Vermylen P, Roufosse C, Ninane V, Sculier JP. Biology of pulmonary preneoplastic lesions. Cancer Treat Rev. 1997;23:241–262. doi: 10.1016/s0305-7372(97)90013-x. [DOI] [PubMed] [Google Scholar]
- 100.Kelloff GJ, Lippman SM, Dannenberg AJ, Sigman CC, Pearce HL, Reid BJ, Szabo E, Jordan VC, Spitz MR, Mills GB, Papadimitrakopoulou VA, Lotan R, Aggarwal BB, Bresalier RS, Kim J, Arun B, Lu KH, Thomas ME, Rhodes HE, Brewer MA, Follen M, Shin DM, Parnes HL, Siegfried JM, Evans AA, Blot WJ, Chow WH, Blount PL, Maley CC, Wang KK, Lam S, Lee JJ, Dubinett SM, Engstrom PF, Meyskens FL, Jr, O’Shaughnessy J, Hawk ET, Levin B, Nelson WG, Hong WK AACR Task Force on Cancer Prevention. Progress in chemoprevention drug development: the promise of molecular biomarkers for prevention of intraepithelial neoplasia and cnacer—a plan to move forward. Clin Cancer Res. 2006;12:3661–3697. doi: 10.1158/1078-0432.CCR-06-1104. [DOI] [PubMed] [Google Scholar]
- 101.Rho JH, Roehrl MH, Wang JY. Tissue proteomics reveals differential and compartment-specific expression of the homologs transgelin and transgelin-2 in lung adenocarcinoma and its stroma. J Proteome Res. 2009;8:5610–5618. doi: 10.1021/pr900705r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rho JH, Roehrl MH, Wang JY. Glycoproteomic analysis of human lung adenocarcinomas using glycoarrays and tandem mass spectrometry: differential expression and glycosylation patterns of vimentin and fetuin A isoforms. Protein J. 2009;28:148–160. doi: 10.1007/s10930-009-9177-0. [DOI] [PubMed] [Google Scholar]
- 103.Mischak H, Allmaier G, Apweiler R, Attwood T, Baumann M, Benigni A, Bennett SE, Bischoff R, Bongcam-Rudloff E, Capasso G, Coon JJ, D’Haese P, Dominiczak AF, Dakna M, Dihazi H, Ehrich JH, Fernandez-Llama P, Fliser D, Frokiaer J, Garin J, Girolami M, Hancock WS, Haubitz M, Hochstrasser D, Holman RR, Ioannidis JP, Jankowski J, Julian BA, Klein JB, Kolch W, Luider T, Massy Z, Mattes WB, Molina F, Monsarrat B, Novak J, Peter K, Rossing P, Sánchez-Carbayo M, Schanstra JP, Semmes OJ, Spasovski G, Theodorescu D, Thongboonkerd V, Vanholder R, Veenstra TD, Weissinger E, Yamamoto T, Vlahou A. Recommendations for biomarker identification and qualification in clinical proteomics. Sci Transl Med. 2010;2:46ps42. doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- 104.Li QK, Singh A, Biswal S, Askin F, Gabrielson E. KEAP1 gene mutations and NRF2 activation are common in pulmonary papillary adenocarcinoma. J Hum Genet. 2011;56:230–234. doi: 10.1038/jhg.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tian H, Zhang B, Di J, Jiang G, Chen F, Li H, Li L, Pei D, Zheng J. Keap1: One stone kills three birds Nrf2, IKKβ and Bcl-2/Bcl-xL. Cancer Lett. 2012;325:26–34. doi: 10.1016/j.canlet.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 106.Lau K, Partridge EA, Silvescu CI, Grigorian A, Pawling J, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–124. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 107.Munfus-McCray D, Adams C, Harada S, Askin F, Clark DP, Gabrielson E, Li QK. EGFR and KRAS mutations in metastatic lung adenocarcinomas. Human Pathol. 2011;42:1447–1453. doi: 10.1016/j.humpath.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 108.Lee RT, Lee YC. Affinity enhancement by multivalent lectin-carbohydrate interaction. Glycoconj J. 2000;17:543–551. doi: 10.1023/a:1011070425430. [DOI] [PubMed] [Google Scholar]
- 109.Fernandes H, Cohen S, Bishayee S. Glycosylation-induced conformational modification positively regulates receptor-receptor association: A study with an aberrant epidermal growth factor receptor (EGFRvIII/deltaEGFR) expressed in cancer cells. J Boil Chem. 2001;276:5375–5383. doi: 10.1074/jbc.M005599200. [DOI] [PubMed] [Google Scholar]
- 110.Lajoie P, Partrideg EA, Guay G, Goetz JG, Pawling J, Lagana A, Dennis JW, Nabi IR. Plasma membrane domain organization regulates EGFR signaling in tumor cells. J Cell Bio. 2007;179:341–356. doi: 10.1083/jcb.200611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu YC, Yen HY, Chen CY, Chen CH, Cheng PF, Juan YH, Chen CH, Khoo KH, Yu CJ, Yang PC, Hsu TL, Wong CH. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci USA. 2011;108:11332–11337. doi: 10.1073/pnas.1107385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tarro G, Perna A, Esposito C. Early diagnosis of lung cancer by detection of tumor liberated protein. J Cell Physiol. 2005;203:1–5. doi: 10.1002/jcp.20195. [DOI] [PubMed] [Google Scholar]