Abstract
The development of some familial dilated cardiomyopathies (DCM) correlates with the presence of mutations in proteins that regulate the organization and function of thin filaments in cardiac muscle cells. Harmful effects of some mutations might be caused by disruption of yet uncharacterized protein-protein interactions. We used nuclear magnetic resonance spectroscopy to localize the region of striated muscle α-tropomyosin (Tpm1.1) that interacts with leiomodin-2 (Lmod2), a member of tropomodulin (Tmod) family of actin-binding proteins. We found that 21 N-terminal residues of Tpm1.1 are involved in interactions with residues 7–41 of Lmod2. The K15N mutation in Tpm1.1, known to be associated with familial DCM, is located within the newly identified Lmod2 binding site of Tpm1.1. We studied the effect of this mutation on binding Lmod2 and Tmod1. The mutation reduced binding affinity for both Lmod2 and Tmod1, which are responsible for correct lengths of thin filaments. The effect of the K15N mutation on Tpm1.1 binding to Lmod2 and Tmod1 provides a molecular rationale for the development of familial DCM.
Keywords: leiomodin, nuclear magnetic resonance, circular dichroism, tropomodulin, dilated cardiomyopathy, intrinsically disordered regions
1. Introduction
Contraction in cardiac muscle cells results from the sliding of the overlapping thick and thin filaments in sarcomeres. The thin filaments are formed primarily by polymerized actin (F-actin) and are regulated by actin-binding proteins such as troponin and tropomyosin. The correct organization and length of the thin filaments are of crucial importance for proper-function of cardiac muscle.
The length of the muscle thin filaments is controlled by regulation of actin polymerization at the pointed (slow-growing) end of F-actin [1]. Three proteins, striated muscle α-tropomyosin, or by a recently introduced nomenclature Tpm1.1 [2], leiomodin-2 (Lmod2) and tropomodulin-1 (Tmod1), are known to associate at the pointed end and regulate the thin filament length in cardiac muscle [3, 4]. Tropomyosins are two-chained α-helical coiled coil molecules that bind along the actin filament in a head-to-tail fashion (for review see [5]). Tpm1.1 regulates actin pointed end dynamics by mediating the binding of Lmod2 and Tmod1 [3, 6] via its N-terminal region [7]. Tmod1 caps the pointed end to inhibit actin polymerization or depolymerization [8]. Lmod2 is a larger homolog of Tmod1 [9]; it is an actin filament nucleator and it elongates the thin filaments [3, 4, 10–13]. It was proposed that Tmod1 and Lmod2 work antagonistically in cardiomyocytes: Tmod1 inhibits actin polymerization at the pointed end, while Lmod2 competes for binding to the same end and allows the thin filament elongation upon Tmod1 displacement [4, 12].
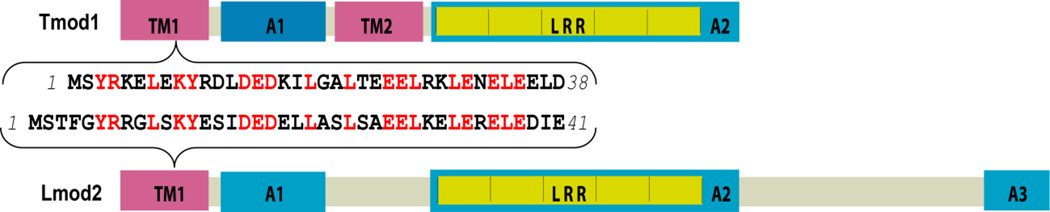
The structures of Lmod2 and Tmod1 share many commonalities [3, 4] (Fig. 1). The tropomyosin-binding site of Lmod2 is similar to the first tropomyosin-binding site (TM1) of Tmod1 (47% identity, 70% similarity) similar [3, 14]. There is high similarity between the first actin-binding site of Tmod1 (residues 58–99) and residues 60–101 of Lmod2 as well as between the LRR (leucine-rich repeat) domains of Lmod2 and Tmod1 [9]. However, despite similarity, the functionality of the sites in Lmod2 and Tmod1 may be quite different. For example, a recent report suggested that there is no N-terminal actin-binding site in Lmod2 and demonstrated that the LRR domain of Lmod2 has strong actin-nucleating effect while the LRR domain of Tmod1 has not[10]. Since Lmod2’s biological function is different from that of Tmod1 [3, 4, 10–13], a comparison of the binding sites deserves additional investigation. Understanding how Lmod2 can compete for the pointed end requires exact knowledge of binding interfaces formed between Lmod2, Tpm1.1 and actin.
Figure 1.
The schematic view of the binding sites of Tmod1 and Lmod2 molecules. TM1, TM2 tropomyosin-binding sites; A1, A2, A3 actin-binding sites; LRR leucine rich repeats. The inset shows the amino acid sequence of the tropomyosin-binding site of Lmod2 and the first tropomyosin-binding site of Tmod1. Identical residues are shown in red.
The absence of Lmod2 is lethal in mice: knockout of Lmod2 results in formation of shorter thin filaments followed by DCM [12]. Despite the active involvement of Lmod2 in sarcomere formation, no cardiomyopathy-linked mutations in this protein have been reported to date. However, detrimental mutations affecting Lmod2 function may occur in proteins such as Tpm1.1 that forms a complex with Lmod2. Such mutations would be expected to alter residues involved in the formation of protein-protein binding interfaces. Unless detailed information on the affected interface is available, understanding of the mechanistic effects of such mutations and their link to cardiomyopathy progression might remain elusive.
In this study, using nuclear magnetic resonance (NMR) spectroscopy, we determined that ~21 N-terminal amino acid residues of Tpm1.1 are involved in interactions with residues 7–41 of Lmod2. We also tested the effect of the DCM-associated Tpm1.1 mutation K15N [15], which is located within this region, on the Tpm1.1 binding to Lmod2 and Tmod1. We demonstrated that the mutation caused a drastic reduction in the affinity of Tpm1.1 for Lmod2 and Tmod1. The weakening of these interactions may explain the occurrence of DCM caused by the K15N mutation in Tpm1.1.
2. Materials & Methods
2.1. Subcloning of recombinant peptides and site-directed mutagenesis
Sequences of Tmod1 from Gallus gallus (NP_990358.1), Lmod2 from Homo sapiens (NP_997046.1) and Tpm1.1 from Homo sapiens (NP_001018005.1) were downloaded from NCBI. Deoxyoligonucleotides encoding Lmod2 and Tpm1.1 peptides, with sequences optimized for Escherichia coli expression [16], were synthesized at GenScript (Piscataway, NJ) and provided in a pUC57 vector. DNA primers for subcloning were purchased from Integrated DNA Technologies (Coralville, IA). Coding sequences for Lmod2s1 (residues 2–41) and model TM peptides were subcloned into the pET-21b(+) vector (EMD-Millipore) between NdeI and XhoI recognition sites as MFH-fusion proteins [17]. Subcloning enzymes (restriction enzymes, OneTaq DNA polymerase, T4 ligase) with pertinent solutions and buffers were from New England Biolabs (Ipswich, MA).
The constructs for the production of model Tpm1.1 peptides encoded an enterokinase cleavage site DDDDK to enable the MFH-tag removal. To stabilize a coiled-coil structure in solution [18], the N-terminal sequences of Tpm1.1 were fused with 15 or 18 residues of the leucine zipper domain of the yeast transcription factor GCN4 [19]. The length of the GCN4 zipper C-terminal extension was dictated by the periodicity of the coiled-coil heptad repeat. Additionally, a Gly residue was added to the N-terminus of each sequence to mimic the N-terminal acetylation of TM. The binding affinities of Tmod to the acetylated Tpm1.1 peptide and the Tpm1.1 peptide with the N-terminal glycine were shown to be similar [20]. Additionally, the Tpm1.1 peptide with the glycine binds the C-terminus of Tpm1.1 to form the overlap complex [21].
The construct for the production of Lmod2s1 included a methionine codon immediately before the sequences of interest for cyanogen bromide (CNBr) cleavage of the MFH-tag. The plasmid pET-15b-EK_C122S_His5 (entry #49048) for expression of recombinant enterokinase was obtained from the nonprofit plasmid depository Addgene. Expression, refolding and purification of the recombinant enterokinase was performed as described in [22].
αTM1a1-21Zip[K15N] was generated with PCR amplification of the plasmid encoding αTM1a1-21Zip with a complementary set of oligonucleotides and Pfu Turbo DNA Polymerase (Agilent Technologies). The template plasmid was digested with DpnI (New England Biolabs) after PCR. The reaction mixture that contained the mutated construct was transformed into E. coli MAX Efficiency® DH5α (Life Technologies) and purified. The presence of the K15N mutation was confirmed by DNA sequencing. The complementary set of oligonucleotides was synthesized by Integrated DNA Technologies (Coralville, IA) and the sequence of the sense primer was: 5’-G CAG ATG TTG AAA TTG GAC AAC GAA AAC GCC CTG GAT CC-3’. The mutated triplet is underlined. DNA sequencing for all generated constructs were performed at Genewiz, Inc. (South Plainfield, NJ).
2.2. Expression of recombinant peptides
Purified plasmids with confirmed insert sequences were used to transform E. coli BL21(DE3) cells (Life Technologies). Transformed cells were grown on LB medium overnight in the presence of 100 µg/mL ampicillin. The overnight culture was used to inoculate 200 mL of LB medium with 100 µg/mL ampicillin. Protein expression was induced with 0.1 mM IPTG after culture had reached the OD600 of 0.7–0.8. To express 15N-labeled or 15N/13C-labeled Lmod2s1, cells from an LB-ampicillin overnight culture centrifuged for 10 minutes at 4,000 g, washed, and resuspended for further growth in minimal medium with 15N-ammonium sulfate or 15N-ammonium sulfate/13C6-glucose (Cambridge Isotope Laboratories, Inc., MA), respectively, as sole sources of nitrogen and carbon [23]. The cells were harvested by centrifugation for 20 minutes at 4,000 g (Beckman Coulter JA-10 rotor) and frozen until use.
2.3. Fusion protein purification
Frozen harvested cells were thawed on ice and resuspended in 50 mM sodium phosphate buffer, pH 8.0, 8 M urea, 300 mM NaCl, 10 mM imidazole. The cell suspension was sonicated on slush ice for 10 minutes and centrifuged for 30 minutes at 16,000 rpm (Beckman Coulter JA-17 rotor) to remove cell debris. The cleared cell lysate was loaded onto Qiagen Ni-NTA resin at room temperature, washed with 50 mM sodium phosphate buffer, pH 8.0, 8 M urea, 300 mM NaCl, 20 mM imidazole and the fusion protein was eluted with 50 mM sodium phosphate buffer, pH 8.0, 8 M urea, 300 mM NaCl, 250 mM imidazole.
2.4. Recombinant Lmod2s1 peptide purification
Eluted from Ni-NTA, MFH-Lmod2s1 fusion protein was brought to 0.3 M in HCl and cleaved with a 400 molar excess of CNBr overnight. The reaction mixture was applied onto a Sep-Pak C18 cartridge from Waters (Milford, MA) and washed with 0.1% trifluoroacetic acid (TFA). The peptide and MFH-tag were eluted in 60–70% acetonitrile/0.1% TFA and lyophilized.
The MFH-tag was separated from Lmod2s1 using Ni-NTA resin (Qiagen). The lyophilized powder obtained after CNBr cleavage was reconstituted in 50 mM sodium phosphate buffer, pH 8.0, 8 M urea, 300 mM NaCl, 10 mM imidazole, and incubated with Ni-NTA resin for 1 hour at room temperature. The resin was loaded onto a column and washed with the same buffer. Flow-through and wash fractions containing the unbound Lmod2s1 peptide were collected and further purified by HPLC on a Vydac reversed-phase 218TP54 column (Grace, Columbia, MD) using a linear 1%/minute water/acetonitrile gradient in the presence of 0.1% TFA and lyophilized. The monoisotopic mass of the 15N-labeled Lmod2s1 peptide was measured at 4713.8 (theoretical 4713.3). Considering the ~0.01–0.02% error of mass determination, 15N incorporation was sufficiently uniform with the extent of labeling exceeding ~98%.
The Tmod1s1 peptide (residues 1–38) was synthesized and purified at Tufts University Core Facility (Boston, MA).
2.5. Recombinant model Tpm1.1 peptides purification
The fusion protein was dissolved in 25 mM Tris-HCl, pH 6.5, 50 mM NaCl, 2 mM CaCl2, and treated with enterokinase at room temperature for 24–72 hours. The reaction was stopped by addition of 1 mM PMSF in isopropanol, acidified, and pre-purified on a Sep-Pak C18 cartridge.
The model Tpm1.1 peptides were separated from the MFH-tag, remaining MFH-fusion proteins and products of non-specific proteolysis on the HPLC column with a 1%/minute water/acetonitrile gradient in the presence of 0.1% TFA. The peptides were lyophilized until use. The identity of all purified peptides was confirmed by MALDI mass spectrometry analysis at Washington State University Laboratory of Biotechnology & Bioanalysis 2 (Pullman, WA). All peptide concentrations were determined spectroscopically by measuring their difference spectra at 294 nm in 6 M guanidine-HCl between pH 7.0 and 12.5 using the extinction coefficient of 2357 cm−1 per 1 M tyrosine [24].
2.6. NMR spectral analysis
NMR samples were prepared in 50 mM sodium phosphate buffer, pH 6.5, 10% D2O, 0.2 mM EDTA, 0.1% sodium azide, 2X Pierce EDTA-free protease inhibitor cocktail. Two-dimensional 15N-HSQC NMR spectra were recorded at 25°C on a Varian VNMRS 600 MHz spectrometer (Agilent Technologies, Santa Clara, CA) equipped with a 5 mm triple resonance probe. The gNhsqc pulse sequence [25] found in the BioPack pulse sequence library (Agilent Technologies) was used to record the 2D 15N-HSQC spectra and extra care was given to the calibration of the water flip-back pulse to avoid water saturation and signal loss. Typically 2×128 increments were recorded with 10,000 Hz spectral width in F2 (1H) and 3700 Hz spectral width in F1 (15N). Three-dimensional HNCO, HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, and HBHA(CO)NH experiments [26] were recorded at 25°C on a Varian Inova 500 MHz spectrometer equipped with a 5 mm Nalorac Z-Spec HCNP triple resonance probe. The NMR spectra were processed using NMRPipe [27] and Felix (Felix NMR, San Diego, CA). NMRViewJ (One Moon Scientific) was used for NMR spectra visualization, peak assignment and CSI analysis [28].
2.7. Circular Dichroism (CD) measurements
The CD spectra of Tpm1.1 chimera molecules at 10 µM concentration were measured between 250–195 nm at 0°C and 15°C using an Aviv model 400 spectropolarimeter (Lakewood, NJ). The binding of 40 µM Lmod2s1 and Tmod1s1 to equimolar concentrations of αTM1a1-21Zip, WT and [K15N], was assessed in 10 mM sodium phosphate pH 7.0, 100 mM NaCl in 1 mm cuvettes. The change in helical content of individual proteins and their complexes at were monitored at 222 nm as a function of temperature. Dimer formation by the two chains of the αTM1aZip model peptides is concentration-dependent and the fraction of dimers formed is greater at higher concentrations. For CD experiments, the optimal protein concentration is 0.1 mg/mL. Although at this concentration most of the Tpm1.1 peptides form a coiled-coil, it was still not high enough to obtain true thermodynamic parameters of unfolding at 222 nm. Therefore we tested several concentrations of the Tpm1.1 peptides and found ~0.4 mg/mL (40 µM for the αTM1aZip molecule) to be suitable for αTM1a1-21Zip to exhibit unfolding with two-state transition within the temperature range (0–55°C) used for the melting experiments yet not too high to measure ellipticity at 222 nm.
To extract the thermodynamic parameters, unfolding curves were fitted to the experimental data as in [29]. The dissociation constants (Kd) of the complexes were calculated from the equation: Kd=exp(−ΔΔG/RT), where ΔΔG is the difference between free energy of the folding of the complex and the folding of the Tpm1.1 peptide alone, R is the universal gas constant, and T is the absolute temperature. All measurements were done in duplicates. Statistical analysis was done using SigmaPlot 12.
Chimera software [30] was used to visualize the crystal structure of Tpm1.1 (PDB# 1IC2).
3. Results
3.1. Design of Tpm1.1 and Lmod2 peptides
We studied interactions between Tpm1.1 and Lmod2 using peptides representing their putative binding sites. The use of peptides rather than full-length proteins exploits the modular nature of tropomyosin, Lmod and Tmod interactions and alleviates technical difficulties associated with the full-length proteins. This approach has provided insights into tropomyosin interactions with Tmod and Lmod [14, 20, 31–37]. We prepared peptides corresponding to N-terminal residues 2–41 of Lmod2 (Lmod2s1), which contain the tropomyosin-binding site (Fig. 1). For NMR studies we prepared 15N-labeled and 15N13C-Lmod2s1. Model Tpm1.1 peptides of varying lengths (Fig. 2) forming a coiled coil were designed similarly to αTM1aZip described by [18].
Figure 2.
Sequences of the model Tpm1.1 peptides. The residues of αTM1a encoded by exon 1a of the Tpm1 gene are in red and are underlined. The number of residues of 1a is shown on left. A Gly residue (shown with g) was added to the N-terminus of each peptide to mimic acetylation. The C-terminal GCN4 sequence is shown in black. The positions a and d of the heptad repeat of coiled-coil molecules are listed under the corresponding residues.
3.2. Localization of regions forming the binding interface in the complex between Lmod2 and Tpm1.1
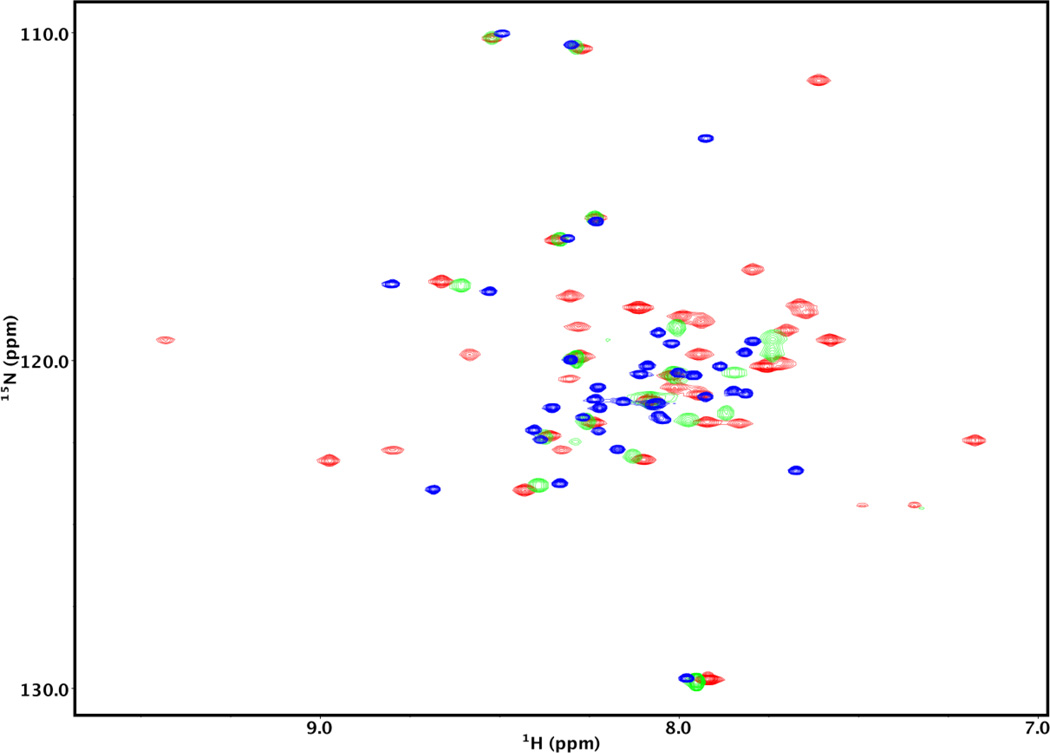
A 2D 15N-HSQC spectrum (see Fig. 3, blue) of the unbound 15N-labeled Lmod2s1 is characterized by a comparatively low resonance peak dispersion, i.e. ~1.2 ppm in the proton dimension, typical of unfolded or only partly folded proteins [38]. An addition of an excess of the unlabeled model Tpm1.1 peptide αTM1a1-14Zip increased peak dispersion indicating formation of a molecular complex. This peptide was shown to contain the binding site for Tmod1 [20].
Figure 3.
2D 15N-HSQC spectra of 15N-labeled Lmod2s1 in the presence and absence of αTM1a1-14Zip. The spectra were recorded on a Varian VNMRS 600 MHz spectrometer. The concentration of Lmod2s1 was 0.22 mM. The blue spectrum was recorded in the absence of αTM1a1-14Zip. The green and red spectra were obtained in the presence of 0.12 mM and 0.24 mM αTM1a1-14Zip, respectively.
To establish the stoichiometry of the complex, we followed the spectral changes with increasing concentrations of αTM1a1-14Zip at 25°C. When approximately one half mol of the αTM1a1-14Zip molecule per mol of the Lmod2s1peptide was added, about half of the cross-peaks broadened out beyond the detection limit (Fig. 3, green). This is typical of a system undergoing an exchange on a µs-ms time scale where a considerable portion of Lmod2s1 is in an unbound state and in equilibrium with the bound complex. Upon addition of approximately one mol of αTM1a1-14Zip per mol of Lmod2s1, the missing cross-peaks appeared (Fig. 3, red), and their position remained virtually unchanged with further increase in αTM1a1-14Zip concentration. The resonance peaks of 15N-Lmod2s1 bound to αTM1a1-14Zip were broader than those of unbound 15N-Lmod2s1, and the proton resonance dispersion increased to ~2.3 ppm indicating formation of a well-defined three-dimensional structure. These observations are consistent with formation of a complex between one two-chained coiled coil molecule of αTM1a1-14Zip and one molecule of Lmod2s1. Our results agree with the reported 1:1 stoichiometry for a complex formed between the αTM1a1-14Zip molecule and a Tmod1 fragment [20].
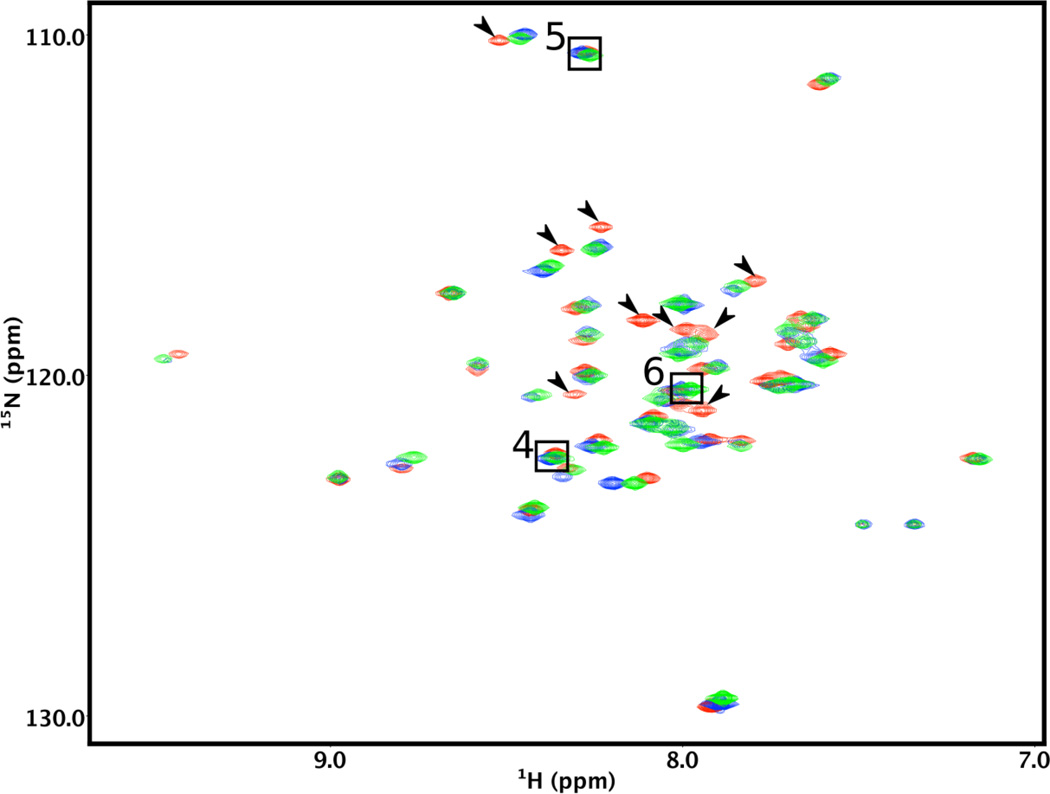
Our next step was to confirm that αTM1a1-14Zip includes all the residues interacting with Lmod2s1. We recorded 2D 15N-HSQC spectra for four NMR samples each containing 15N-Lmod2s1 mixed with a small stoichiometric excess (10–20%) of unlabeled model Tpm1.1 peptides of increasing lengths (Fig. 2). We tested Lmod2s1 interactions with αTM1a1-14Zip, αTM1a1-21Zip, αTM1a1-28Zip, or αTM1a1-52Zip at 25°C. We observed that positions of the 2D 15N-HSQC cross-peaks in the spectra with αTM1a1-21Zip, αTM1a1-28Zip, or αTM1a1-52Zip were very similar. Surprisingly, in the 2D 15N-HSQC spectrum of 15N-Lmod2s1 with αTM1a1-14Zip, many resonance peaks were markedly shifted from positions detected for the complexes with longer model Tpm1.1 peptides. An overlay of the HSQC spectra recorded in the presence of αTM1a1-14Zip, αTM1a1-21Zip or αTM1a1-28Zip is shown in Fig. 4. We also recorded the same spectra at 10°C and discovered a practically identical pattern of chemical shifts. These results suggest that all Tpm1.1 residues interacting with Lmod2s1 are present in αTM1a1-21Zip and longer peptides, whereas some are missing in αTM1a1-14Zip.
Figure 4.
Comparison of the Lmod2s1 NMR spectra in the presence of Tpm1.1 model peptides of varying lengths. The 2D 15N-HSQC spectra of 15N-labeled Lmod2s1 were recorded in the presence of a >20% stoichiometric excess of αTM1a1-14Zip (red), αTM1a1-21Zip (green), or αTM1a1-28Zip (blue). Arrows show the cross-peaks of the Lmod2s1/αTM1a1-14Zip complex that are markedly shifted with respect to those of the Lmod2s1/αTM1a1-28Zip complex. The cross-peaks of residues 4–6 are not affected by binding and are shown in boxes.
Using a uniformly 15N/13C-labeled Lmod2s1, we assigned the amide 1H/15N, carbonyl 13C', 13Cα, 13Cβ, 1Hα, and 1Hβ resonances in the unbound state of the peptide (Fig. S1). We identified and assigned all of the Lmod2s1 amide cross-peaks with the exception of Thr3. Chemical shift analysis indicated that residues Glu20-Ser24 and Ala27-Arg35 of unbound Lmod2s1 sample a considerable population of an α-helical conformation (for details, see Fig. 5). Comparison of the 2D 15N-HSQC NMR spectra of the unbound and bound 15N-labeled Lmod2s1 revealed that the cross-peaks corresponding to Phe4-Tyr6 experience little chemical shift changes upon binding to the Tpm1.1 model peptides suggesting that residues Ser2-Tyr6 of Lmod2 may not interact with Tpm1.1.
Figure 5.
Chemical shift index analysis of the unbound 15N/13C-Lmod2s1. The sequence of the peptide is displayed on top. Assigned atoms are displayed as green boxes. Chemical shifts consistent with α-helical or β-sheet structure are displayed as bars with red or blue circles, respectively. The position of two α-helices is shown schematically at the bottom of the figure.
From our NMR experiments we conclude that the binding interface between Lmod2s1 and the coiled coil αTM1a is formed by residues Arg7-Glu41 of Lmod2 and the 21 N-terminal residues of Tpm1.1.
3.3. αTM1a1-21Zip binds more strongly to Lmod2s1 than Tmod1s1
The dissociation constants (Kd) of Lmod2s1 and Tmod1s1 (res. 1–38 of Tmod1) for Tpm1.1 were previously determined using αTM1a1-14Zip [20, 32, 37]. Since the Lmod2 binding site on Tpm1.1 is longer than 14 residues, as described above, Kd was determined for the complexes between αTM1a1-21Zip and Lmod2s1 to see if the extension of the Tpm1.1 model peptide would increase the binding affinities of Lmod2 for Tpm1.1 as a consequence of increasing the number of interacting residues. For comparison, we also determined Kd for the complex formed by αTM1a1-21Zip and Tmod1s1.
To determine Kd values, temperature-dependent unfolding of αTM1a1-21Zip, alone and in a complex with Lmod2s1 or Tmod1s1, was monitored by CD at 222 nm. The thermodynamic parameters of unfolding of the complexes were determined from the melting curves and then used to calculate the Kd values [29].
In agreement with our earlier reports [14, 32], both Lmod2s1 and Tmod1s1 were mostly disordered with an estimated α-helical content between 20–25%. Typical for intrinsically disordered proteins, there was no cooperativity in their unfolding (Fig.7a), while the melting curve of αTM1a1-21Zip was cooperative, due to denaturation of the coiled coil.
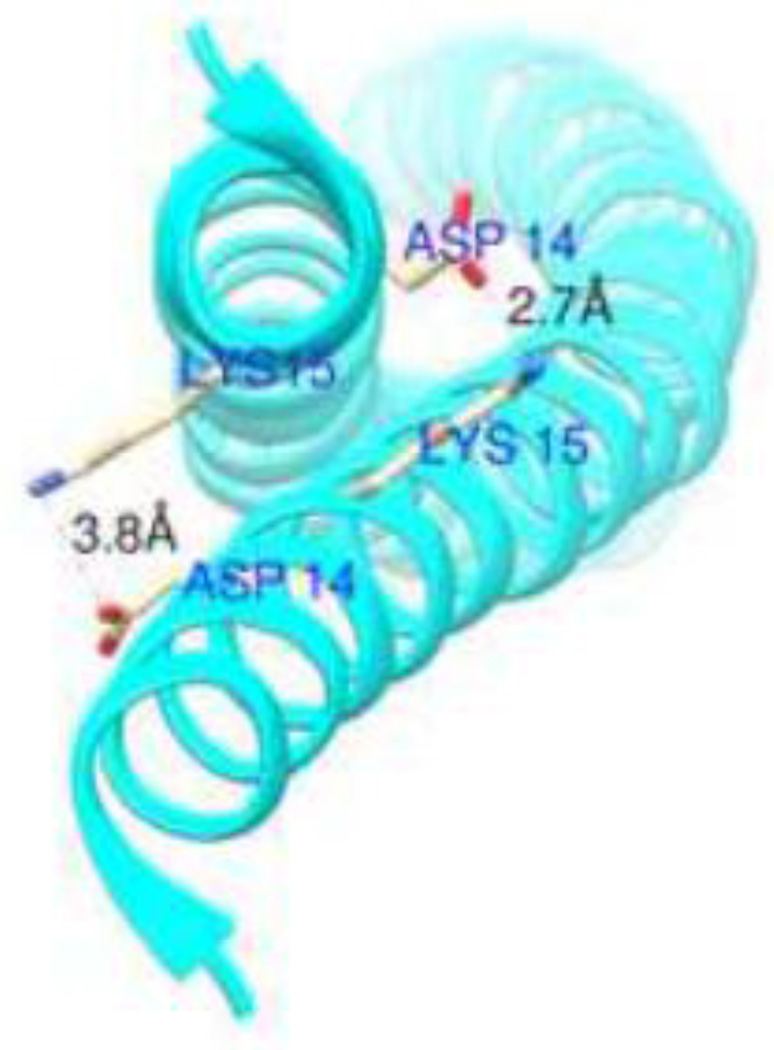
Figure 7.
Crystal structure of Tpm1.1 (PDB #1IC2) demonstrating distances between Asp14 and Lys15. Chimera software was used to calculate the distances.
The sums of the individual unfolding curves of either Lmod2s1 or Tmod1s1 and the αTM1a1-21Zip were different than the unfolding curves of the Lmod2s1/αTM1a1-21Zip and Tmod1s1/αTM1a1-21Zip complexes (Fig. 6a). The molar residue ellipticities of the Lmod2s1/αTM1a1-21Zip and Tmod1s1/αTM1a1-21Zip complexes are more negative than the sums of the individual peptides, indicating that both Lmod2s1 and Tmod1s1 form a complex with the αTM1a1-21Zip and become more folded. The melting temperatures (Tm), or the midpoints of their transition state, and the calculated Kd values for the complexes are presented in Table 1.
Figure 6.
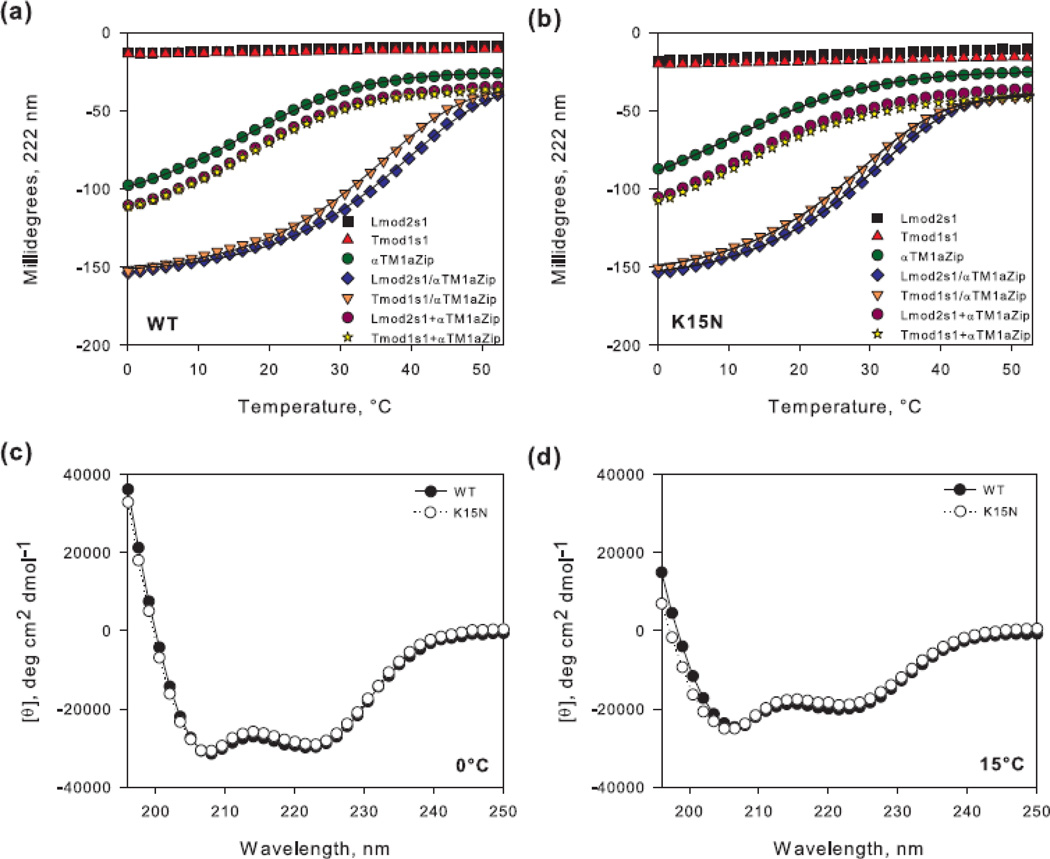
Representative unfolding curves of peptide complexes and CD spectra of αTM1a1-21Zip. Binding of 40 µM Lmod2s1 or Tmod1s1 to equimolar concentration of αTM1a1-21Zip (a) wild type (WT) and (b) [K15N] was assayed using CD spectroscopy. The graphs illustrate the unfolding curves of the individual peptides (Lmod2s1, Tmod1s1, αTM1a1-21Zip), the mixtures (Lmod2s1/αTM1a1-21Zip, Tmod1s1/αTM1a1-21Zip) and the sums of the individual unfolding curves (Lmod2s1+αTM1a1-21Zip, Tmod1s1+αTM1a1-21Zip). The lines show fitted curves to the experimental data. Spectra of αTM1a1-21Zip and αTM1a1-21Zip[K15N] at (c) 0°C and (d) 15°C.
Table 1.
Melting temperatures (Tm) and dissociation constants (Kd) of the peptide complexes.
We found that the affinity of Tmod1s1 for αTM1a1-21Zip is nearly identical to that for αTM1a1-14Zip [37], whereas the affinity of Lmod2s1 for αTM1a1-21Zip is ~two-fold higher than what we previously measured for the Lmod2s1/αTM1a1-14Zip complex [32]. In other words, increasing the length of the Tpm1.1 peptide increased binding affinity of Lmod2s1, but had no effect on the binding of Tmod1s1.
3.4. The K15N mutation decreases the stability of αTM1a1-21Zip and its affinity for Lmod2s1 and Tmod1s1
The K15N mutation in Tpm1.1 is a cause of dilated cardiomyopathy [15]. Lys15 is within the newly determined site for binding Lmod2. To establish if the mutation affects the secondary structure of the Tpm1.1 peptide, we compared CD spectra of wild type (WT) and K15N mutant αTM1a1-21Zip molecules (αTM1a1-21Zip[K15N]).
The CD spectra of 10 µM WT and mutant Tpm1.1 molecules measured at 0°C and 15°C (Fig. 6c,d) were typical for α-helical proteins [39], with minima near 222 and 208 nm. However, a difference between the CD spectra for the WT and mutant Tpm1.1 molecules became evident at 15°C. The random coil content in the mutant was somewhat higher than that of WT, since the interception point of the mutant’s spectrum with the x-axis shifted to lower wavelengths. The more disordered structure of αTM1a1-21Zip[K15N] at a higher temperature is indicative of decreased stability and therefore weaker dimer formation. This conclusion was supported by comparison of melting curves for αTM1a1-21Zip and αTM1a1-21Zip[K15N] (Fig. 6a,b). The mutation caused ~6°C decrease in the Tm (Table 1).
To test the effect of the K15N mutation on the affinities of complexes between αTM1a1-21Zip and Lmod2s1 or Tmod1s1, we repeated the melting experiments described in the previous section with αTM1a1-21Zip[K15N] (Fig. 6b). The mutation destabilized the complexes and reduced the Tm values. The decreases in Tm suggest formation of weaker and less folded complexes for the K15N mutant versus WT Tpm1.1. We calculated Kds from the melting curves with the mutated Tpm1.1 fragment (Table 1) and found that the K15N mutation reduces the affinities of Lmod2s1 and Tmod1s1 for αTM1a1-21Zip approximately six- and four-fold, respectively (P<0.001, One Way ANOVA).
4. Discussion
Dilated cardiomyopathy (DCM) is the most frequent type of cardiomyopathy [40, 41]. It is characterized by the enlargement of the ventricular chamber and systolic dysfunction that can result in heart failure. Mutations in over 30 genes were shown to be linked to familial DCM; many of the mutations are found in genes that encode for proteins involved in sarcomere assembly and muscle contraction [15, 42]. Several cardiomyopathy-related mutations identified in Tpm1.1 still lack molecular explanations [43, 44]. Some of these mutations are located in proximity to the sites of interaction with Lmod2 and Tmod1, suggesting a molecular basis for the DCM disorders [43]. Establishing details of molecular interactions between Tpm1.1, Lmod2 and Tmod1 may shed light on molecular mechanisms behind the development of these cardiomyopathies.
It was previously shown that Tmod1 binds to the 14 N-terminal amino acids of Tpm1.1 [20]. Our NMR results demonstrated that the amino acid sequence that forms the binding site for Lmod2 on Tpm1.1 is seven residues longer, encompassing the first 21 residues of Tpm1.1. Our CD experiments also support this finding; a two-fold increase in the affinity of Lmod2s1 for αTM1a1-21Zip was observed compared to that for αTM1a1-14Zip. Regardless of a large degree of sequence similarity between Lmod2s1 and Tmod1s1 (Fig. 1), a difference in their Kd values suggest that their mode of binding to Tpm1.1 may not be identical. These differences are further corroborated by the fact that the unbound Lmod2s1 has a tendency to form two α-helices spanning residues Glu20-Ser24 and Ala27-Arg35. For comparison, formation of a single α-helix localized to residues 24–35 (aligning with residues 27–38 of Lmod2) was reported for the N-terminal fragment (res. 1–92) of unbound Tmod1 [14]. It is not yet clear to what extent these variations in unbound secondary structures affect the mode of Lmod2 and Tmod1 binding. Solving the three-dimensional structure of the Lmod2s1/αTM1a1-21Zip and Tmod1s1/αTM1a1-21Zip complexes will allow us to answer this question.
K15N mutation in Tpm1.1 is associated with familial DCM [15] but the underlying molecular basis for the physiological consequences of this mutation is unknown [43]. Our CD spectroscopy analysis indicates that the stability of αTM1a1-21Zip decreases when the K15N mutation is present. Lys15 is at position a of the heptad repeat of Tpm1.1 and it is highly conserved [45]. Residues in position a are generally hydrophobic and they form the core of the coiled coils in tropomyosins [46]. In the Tpm1.1 crystal structure (PDB #1IC2) Lys15 is charged and its side-chain is exposed to avoid disruption of the coiled-coil hydrophobic core [47]. Lys15 and Asp14 located on the neighboring helices form salt linkages (Fig. 7). Asn has a shorter sidechain than Lys and is not charged. Therefore, with the introduction of Asn instead of Lys, the ion pair would not be formed and the decreased stability of αTM1a1-21Zip may be explained by this destabilization of the coiled coil.
The K15N mutation resulted in approximately six- and four-fold decreases in the affinities of αTM1a1-21Zip for Lmod2s1 and Tmod1s1, respectively. The mutation is outside the previously proposed Tmod-binding site on Tpm1.1 but it still decreased the affinity of Tmod1s1. However, our CD spectroscopy results with Tmod1s1 and the WT αTM1a1-21Zip support the length of the Tmod-binding site on Tpm1.1, which encompasses the 14 N-terminal residues. The change of Tmod1’s affinity for αTM1a1-21Zip[K15N] may be explained by long-range effects of the mutation. Lys15 is located within the binding interface of Tpm1.1 for Lmod2s1 according to our NMR data and it may form direct contacts with Lmod2s1 in the complex. In addition to the structural changes in mutated Tpm1.1, changing Lys to Asn would affect putative direct molecular interactions between Tpm1.1 and Lmod2s1. Consistently, the magnitude of reduction in the affinity of Lmod2s1 for the mutant αTM1a1-21Zip was greater than with Tmod1s1.
There are two possible reasons for the pathogenic effect of the K15N mutation. Tropomyosin is an important regulator of the actin-nucleating ability of Lmod2 [4, 13] and recruits Tmod1 and Lmod2 to the pointed end [3, 6, 13, 36]. The mutation in Tpm1.1 may reduce the localization of Lmod2 and Tmod1 to the pointed end of thin filaments due to the decreased binding affinity for Tpm1.1. This alone may lead to formation of thin filaments with improper lengths and result in cardiomyopathies since reduced levels of Lmod2 and Tmod1 in cardiomyocytes disrupts myofibril formation [3, 48].
Lmod2 and Tmod1 were proposed to compete for the pointed end and their competition to promote formation of sarcomeres with appropriate thin filament lengths in cardiomyocytes [4]. Another explanation of the mutant pathogenicity can be the unequal decrease in the affinities of these two proteins for Tpm1.1 with K15N mutation. The mutation can shift the balance in their competition and therefore alter their localization at the pointed end. DCM was observed in mice upon Tmod1 overexpression [49] or Lmod2 knockout [12]. Consequently, the impaired displacement of Tmod1 by Lmod2 at the pointed end of thin filaments can potentially contribute to the development of DCM.
Previously, M8R, a nemaline myopathy- [50, 51] and DCM-associated mutation in Tpm1.1 [52], was shown to cause loss of its ability to interact with Tmod1 [20]. It also destroyed the formation of the coiled coil at the N-terminal region [53], decreased actin-binding affinity by 100-fold [54, 55] and severely weakened the head-to-tail polymerization ability of Tpm1.1 [55, 56]. Similar to Met8, Lys15 is at position a of the heptad repeat (Fig. 2), however it is not within the 11 N-terminal residues shown to form the overlap junction of Tpm1.1 [57]. Therefore, the K15N mutation is not expected to have a direct effect on the head-to-tail association of Tpm1.1; and the decrease of Tmod1 and Lmod2 binding may have a major impact in the development of familial DCM caused by K15N mutation on Tpm1.1.
Nevertheless, K15N mutation in Tpm1.1 may also simultaneously affect its interaction with other binding partners. It was proposed that troponin T interacts with Lys15 of Tpm1.1 [58]. Lys15 is also positioned next to Glu16 in Tpm1.1, which is known to interact with actin [59]. K15N mutation, therefore, could be altering the interaction of Tpm1.1 with these binding partners as well. Further studies can reveal the roles of other binding partners of Tpm1.1 in the development of K15N-associated DCM.
Supplementary Material
Highlights.
21 N-terminal residues of tropomyosin (Tpm1.1) represent leiomodin-2 binding site.
Dilated cardiomyopathy-causing mutation K15N disrupts the structure of Tpm1.1.
K15N mutation decreases Tpm1.1 binding to leiomodin-2 and tropomodulin-1.
Acknowledgments
We would like to express our gratitude to Drs. Hans Brandstetter, Wofgang Skala and Peter Goettig, who created and deposited at Addgene a plasmid for high-yield recombinant enterokinase production. We thank Drs. Norma Greenfield and Sarah Hitchcock-DeGregori for helpful comments and careful reading of the manuscript. We appreciate Dr. Gerhard Munske’s help with mass spectrometry analysis. This study was supported in part by National Institutes of Health Grant GM081688 and startup funds to ASK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Littlefield RS, Fowler VM. Thin filament length regulation in striated muscle sarcomeres: pointed-end dynamics go beyond a nebulin ruler. Seminars in cell & developmental biology. 2008;19:511–519. doi: 10.1016/j.semcdb.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geeves MA, Hitchcock-DeGregori SE, Gunning PW. A systematic nomenclature for mammalian tropomyosin isoforms. Journal of muscle research and cell motility. 2015;36:147–153. doi: 10.1007/s10974-014-9389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukada T, Pappas CT, Moroz N, Antin PB, Kostyukova AS, Gregorio CC. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. Journal of cell science. 2010;123:3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. Tropomyosin - master regulator of actin filament function in the cytoskeleton. Journal of cell science. 2015;128:2965–2974. doi: 10.1242/jcs.172502. [DOI] [PubMed] [Google Scholar]
- 6.Tsukada T, Kotlyanskaya L, Huynh R, Desai B, Novak SM, Kajava AV, Gregorio CC, Kostyukova AS. Identification of residues within tropomodulin-1 responsible for its localization at the pointed ends of the actin filaments in cardiac myocytes. The Journal of biological chemistry. 2011;286:2194–2204. doi: 10.1074/jbc.M110.186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung LA, Lin JJ. Erythrocyte tropomodulin binds to the N-terminus of hTM5, a tropomyosin isoform encoded by the gamma-tropomyosin gene. Biochemical and biophysical research communications. 1994;201:627–634. doi: 10.1006/bbrc.1994.1747. [DOI] [PubMed] [Google Scholar]
- 8.Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. The Journal of cell biology. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73:127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 10.Boczkowska M, Rebowski G, Kremneva E, Lappalainen P, Dominguez R. How Leiomodin and Tropomodulin use a common fold for different actin assembly functions. Nature communications. 2015;6:8314. doi: 10.1038/ncomms9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Ni F, Kondrashkina E, Ma J, Wang Q. Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:12687–12692. doi: 10.1073/pnas.1512464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappas CT, Mayfield RM, Henderson C, Jamilpour N, Cover C, Hernandez Z, Hutchinson KR, Chu M, Nam KH, Valdez JM, Wong PK, Granzier HL, Gregorio CC. Knockout of Lmod2 results in shorter thin filaments followed by dilated cardiomyopathy and juvenile lethality. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:13573–13578. doi: 10.1073/pnas.1508273112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skwarek-Maruszewska A, Boczkowska M, Zajac AL, Kremneva E, Svitkina T, Dominguez R, Lappalainen P. Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres. Molecular biology of the cell. 2010;21:3352–3361. doi: 10.1091/mbc.E10-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostyukova AS, Hitchcock-Degregori SE, Greenfield NJ. Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein. Journal of molecular biology. 2007;372:608–618. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circulation. Cardiovascular genetics. 2010;3:155–161. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic acids research. 2007;35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolkatchev D, Plamondon J, Gingras R, Su Z, Ni F. Recombinant Production of Intrinsically Disordered Proteins for Biophysical and Structural Characterization. In: Uversky VN, Longhi S, editors. Instrumental Analysis of Intrinsically Disordered Proteins: Assessing Structure And Conformation. Hoboken, NJ: John Wiley & Sons, Inc.; 2010. [Google Scholar]
- 18.Greenfield NJ, Montelione GT, Farid RS, Hitchcock-DeGregori SE. The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies. Biochemistry. 1998;37:7834–7843. doi: 10.1021/bi973167m. [DOI] [PubMed] [Google Scholar]
- 19.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 20.Greenfield NJ, Fowler VM. Tropomyosin requires an intact N-terminal coiled coil to interact with tropomodulin. Biophysical journal. 2002;82:2580–2591. doi: 10.1016/S0006-3495(02)75600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palm T, Greenfield NJ, Hitchcock-DeGregori SE. Tropomyosin ends determine the stability and functionality of overlap and troponin T complexes. Biophysical journal. 2003;84:3181–3189. doi: 10.1016/S0006-3495(03)70042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skala W, Goettig P, Brandstetter H. Do-it-yourself histidine-tagged bovine enterokinase: a handy member of the protein engineer's toolbox. Journal of biotechnology. 2013;168:421–425. doi: 10.1016/j.jbiotec.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osborne MJ, Su Z, Sridaran V, Ni F. Efficient expression of isotopically labeled peptides for high resolution NMR studies: application to the Cdc42/Rac binding domains of virulent kinases in Candida albicans. Journal of biomolecular NMR. 2003;26:317–326. doi: 10.1023/a:1024043620972. [DOI] [PubMed] [Google Scholar]
- 24.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 25.Kay LE, Keifer P, Saarinen T. Pure Absorption Gradient Enhanced Heteronuclear Single Quantum Correlation Spectroscopy with Improved Sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 26.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Res Sp. 1999;34:93–158. [Google Scholar]
- 27.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in molecular biology. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 29.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nature protocols. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Greenfield NJ, Kostyukova AS, Hitchcock-DeGregori SE. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophysical journal. 2005;88:372–383. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostyukova AS. Leiomodin/tropomyosin interactions are isoform specific. Archives of biochemistry and biophysics. 2007;465:227–230. doi: 10.1016/j.abb.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Kostyukova AS, Choy A, Rapp BA. Tropomodulin binds two tropomyosins: a novel model for actin filament capping. Biochemistry. 2006;45:12068–12075. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kostyukova AS, Hitchcock-DeGregori SE. Effect of the structure of the N terminus of tropomyosin on tropomodulin function. The Journal of biological chemistry. 2004;279:5066–5071. doi: 10.1074/jbc.M311186200. [DOI] [PubMed] [Google Scholar]
- 35.Kostyukova AS, Rapp BA, Choy A, Greenfield NJ, Hitchcock-DeGregori SE. Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry. 2005;44:4905–4910. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- 36.Moroz NA, Novak SM, Azevedo R, Colpan M, Uversky VN, Gregorio CC, Kostyukova AS. Alteration of tropomyosin-binding properties of tropomodulin-1 affects its capping ability and localization in skeletal myocytes. The Journal of biological chemistry. 2013;288:4899–4907. doi: 10.1074/jbc.M112.434522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uversky VN, Shah SP, Gritsyna Y, Hitchcock-DeGregori SE, Kostyukova AS. Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. Journal of molecular recognition : JMR. 2011;24:647–655. doi: 10.1002/jmr.1093. [DOI] [PubMed] [Google Scholar]
- 38.Yao J, Dyson HJ, Wright PE. Chemical shift dispersion and secondary structure prediction in unfolded and partly folded proteins. FEBS letters. 1997;419:285–289. doi: 10.1016/s0014-5793(97)01474-9. [DOI] [PubMed] [Google Scholar]
- 39.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nature protocols. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Camethon MR, Dai S, de Simone G, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Greenlund KJ, Hailpem SM, Heit JA, Ho PM, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, McDermott MM, Meigs JB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Rosamond WD, Sorlie PD, Stafford RS, Turan TN, Turner MB, Wong ND, Wylie-Rosett J, Comm AHAS, Subcomm SS. Heart Disease and Stroke Statistics-2011 Update A Report From the American Heart Association. Circulation. 2011;123:E18–E209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fatkin D. Familial dilated cardiomyopathy: Current challenges and future directions. Global cardiology science & practice. 2012;2012:8. doi: 10.5339/gcsp.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hershberger RE, Siegfried JD. Update 2011: clinical and genetic issues in familial dilated cardiomyopathy. Journal of the American College of Cardiology. 2011;57:1641–1649. doi: 10.1016/j.jacc.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marston S, Memo M, Messer A, Papadaki M, Nowak K, McNamara E, Ong R, El-Mezgueldi M, Li X, Lehman W. Mutations in repeating structural motifs of tropomyosin cause gain of function in skeletal muscle myopathy patients. Human molecular genetics. 2013;22:4978–4987. doi: 10.1093/hmg/ddt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Memo M, Marston S. Skeletal muscle myopathy mutations at the actin tropomyosin interface that cause gain- or loss-of-function. Journal of muscle research and cell motility. 2013;34:165–169. doi: 10.1007/s10974-013-9344-y. [DOI] [PubMed] [Google Scholar]
- 45.Barua B, Pamula MC, Hitchcock-DeGregori SE. Evolutionarily conserved surface residues constitute actin binding sites of tropomyosin. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10150–10155. doi: 10.1073/pnas.1101221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry SV. Vertebrate tropomyosin: distribution, properties and function. Journal of muscle research and cell motility. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 47.Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, Cohen C. Deciphering the design of the tropomyosin molecule. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8496–8501. doi: 10.1073/pnas.131219198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sussman MA, Baque S, Uhm CS, Daniels MP, Price RL, Simpson D, Terracio L, Kedes L. Altered expression of tropomodulin in cardiomyocytes disrupts the sarcomeric structure of myofibrils. Circulation research. 1998;82:94–105. doi: 10.1161/01.res.82.1.94. [DOI] [PubMed] [Google Scholar]
- 49.Sussman MA, Welch S, Cambon N, Klevitsky R, Hewett TE, Price R, Witt SA, Kimball TR. Myofibril degeneration caused by tropomodulin overexpression leads to dilated cardiomyopathy in juvenile mice. The Journal of clinical investigation. 1998;101:51–61. doi: 10.1172/JCI1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laing NG, Wilton SD, Akkari PA, Dorosz S, Boundy K, Kneebone C, Blumbergs P, White S, Watkins H, Love DR, et al. A mutation in the alpha tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy NEM1. Nature genetics. 1995;10:249. doi: 10.1038/ng0695-249. [DOI] [PubMed] [Google Scholar]
- 51.Tan P, Briner J, Boltshauser E, Davis MR, Wilton SD, North K, Wallgren-Pettersson C, Laing NG. Homozygosity for a nonsense mutation in the alpha-tropomyosin slow gene TPM3 in a patient with severe infantile nemaline myopathy. Neuromuscular disorders : NMD. 1999;9:573–579. doi: 10.1016/s0960-8966(99)00053-x. [DOI] [PubMed] [Google Scholar]
- 52.Lakdawala NK, Funke BH, Baxter S, Cirino AL, Roberts AE, Judge DP, Johnson N, Mendelsohn NJ, Morel C, Care M, Chung WK, Jones C, Psychogios A, Duffy E, Rehm HL, White E, Seidman JG, Seidman CE, Ho CY. Genetic testing for dilated cardiomyopathy in clinical practice. Journal of cardiac failure. 2012;18:296–303. doi: 10.1016/j.cardfail.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moraczewska J, Greenfield NJ, Liu Y, Hitchcock-DeGregori SE. Alteration of tropomyosin function and folding by a nemaline myopathy-causing mutation. Biophysical journal. 2000;79:3217–3225. doi: 10.1016/S0006-3495(00)76554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heald RW, Hitchcock-DeGregori SE. The structure of the amino terminus of tropomyosin is critical for binding to actin in the absence and presence of troponin. The Journal of biological chemistry. 1988;263:5254–5259. [PubMed] [Google Scholar]
- 55.Urbancikova M, Hitchcock-DeGregori SE. Requirement of amino-terminal modification for striated muscle alpha-tropomyosin function. The Journal of biological chemistry. 1994;269:24310–24315. [PubMed] [Google Scholar]
- 56.Cho YJ, Liu J, Hitchcock-DeGregori SE. The amino terminus of muscle tropomyosin is a major determinant for function. The Journal of biological chemistry. 1990;265:538–545. [PubMed] [Google Scholar]
- 57.Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. Journal of molecular biology. 2006;364:80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 58.Murakami K, Stewart M, Nozawa K, Tomii K, Kudou N, Igarashi N, Shirakihara Y, Wakatsuki S, Yasunaga T, Wakabayashi T. Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7200–7205. doi: 10.1073/pnas.0801950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophysical journal. 2011;100:1005–1013. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.