SUMMARY
Culture-independent diagnostic techniques are increasingly used in clinical laboratories. They have improved turnaround times and are generally more sensitive than culture.
Their relative ease of use may increase the numbers of patients being tested.
These tests allow detection of organisms that are currently difficult or impossible to culture.
The main non-culture methods are immunoassays, which detect antibody or microbial antigen, and nucleic acid amplification testing, which detects microbial RNA or DNA.
For some infections, culture may need to be combined with these tests to determine antibiotic susceptibility.
Introduction
Traditional methods for diagnosing infection have relied largely on clinical microbiology laboratories selecting, isolating and then identifying pathogenic organisms via culture. This can be very time consuming. For some fastidious or slow-growing organisms, the delay to definitive microbiological diagnosis can stretch to weeks, while some organisms cannot be cultured at all. Other drawbacks with culture methods include problems with sensitivity, cost (resource intensive) and potential safety concerns with pathogenic organisms such as Mycobacterium tuberculosis or Coxiella burnetii (the causative organism of Q fever).
Non-culture-based diagnostic methods (see Table) can have significant advantages over traditional culture methods. For example, nucleic acid amplification testing has drastically reduced turnaround times for many routine diagnostic tests and enabled high throughput testing for multiple organisms, many of which were previously very difficult to diagnose. However, rapid changes in this area make it difficult for practitioners to keep abreast of available methods.
Table. Laboratory tests for commonly encountered infections .
| Syndrome and potential cause |
Culture | Non-culture methods | Notes | ||
|---|---|---|---|---|---|
| Serology | Antigen | NAAT | |||
| 1 = routine use | 2 = not routine use or unavailable | 3 = useful in special circumstances | |||
| Pharyngitis | |||||
|---|---|---|---|---|---|
| Group A streptococci | 1 | 3 | 1 | 2 | - |
| Epstein-Barr virus and cytomegalovirus | 2 | 1 | 2 | 2 | Epstein-Barr virus and cytomegalovirus NAAT is not useful in diagnosing acute pharyngitis in the immunocompetent patient. |
| Ocular infection | |||||
|---|---|---|---|---|---|
| Herpes simplex virus and adenovirus | 2 | 2 | 2 | 1 | - |
| Chlamydia trachomatis | 2 | 2 | 2 | 1 | - |
| Neisseria gonorrhoeae | 1 | 2 | 2 | 1 | Culture should be performed with NAAT where possible. |
| Other bacteria e.g. Bartonella henselae (granulomatous conjunctivitis) |
3 | 1 | 2 | 3 | - |
| Lower respiratory tract infection | |||||
|---|---|---|---|---|---|
| Respiratory viruses | 2 | 3 | 1 | 1 | Antigen testing on sputum is available for some pathogens, but is insensitive. PCR testing is preferred. |
| Mycoplasma pneumoniae | 2 | 1 | 2 | 1 | - |
| Chlamydia pneumoniae | 2 | 1 | 2 | 1 | - |
| Bordetella pertussis | 2 | 1 | 2 | 1 | - |
| Legionella species | 1 | 1 | 1 | 3 | Urinary antigen is only available for Legionella pneumophila serogroup 1. |
| Mycobacterium tuberculosis | 1 | 2 | 2 | 3 | - |
| Streptococcus pneumoniae | 1 | 2 | 1 | 3 | Pneumococcal urinary antigen is highly specific, but insensitive. It is useful as a rapid adjunct to culture. |
| Gastritis | |||||
|---|---|---|---|---|---|
| Helicobacter pylori | 3 | 1 | 1 | 2 | Culture is performed from gastric biopsies only. Diagnosis is most commonly made by urea breath test and faecal antigen. When an invasive procedure is performed endoscopy and biopsy urease testing can be used on tissue. |
| Gastroenteritis | |||||
|---|---|---|---|---|---|
| Bacteria (e.g. Salmonella) | 1 | 3 | 2 | 1 | - |
| Parasites | 2 | 3 | 1 | 1 | Serology is available for selected parasitic causes, e.g. Entamoeba histolytica |
| Viruses (e.g. noro, rota, adeno) | 2 | 2 | 1 | 1 | Antigen testing for these pathogens is relatively insensitive. NAAT (PCR) testing is preferred. |
| Toxigenic Clostridium difficile | 3 | 2 | 1 | 1 | - |
| Sexually transmitted infection | |||||
|---|---|---|---|---|---|
| Neisseria gonorrhoeae | 1 | 2 | 2 | 1 | Culture should be performed with NAAT testing where possible. |
| Chlamydia trachomatis | 2 | 2 | 2 | 1 | - |
| Mycoplasma genitalium | 2 | 2 | 2 | 1 | - |
| Trichomonas vaginalis | 2 | 2 | 2 | 1 | - |
| Syphilis | 2 | 1 | 2 | 1 | NAAT is useful for primary ulcers. Serology is the screening method of choice. |
| Systemic syndromes | |||||
|---|---|---|---|---|---|
| HIV | 2 | 1 | 1 | 3 | P24 antigen and NAAT may be useful in early seroconversion – discuss with laboratory. |
| Ross River virus and Barmah Forest virus | 2 | 1 | 2 | 2 | - |
| Rickettsia and Q fever | 2 | 1 | 2 | 3 | Some laboratories may offer NAAT. But nucleic acid is usually only detectable early in acute infection (first 7 days). |
| Leptospirosis | 3 | 1 | 2 | 3 | Leptospira can be cultured, but requires direct inoculation from blood into special media. |
| Viral exanthems | |||||
|---|---|---|---|---|---|
| Parvovirus, measles, mumps, rubella | 2 | 1 | 2 | 1 | NAAT is useful for measles (urine and blood), mumps (buccal swab) and rubella (pharyngeal swab). |
| Fever in the returned traveller | |||||
|---|---|---|---|---|---|
| Malaria | 2 | 2 | 1 | 3 | - |
| Dengue | 2 | 1 | 1 | 1 | - |
| Salmonella typhi | 1 | 3 | 2 | 2 | Culture is the mainstay of diagnosis. Serology can be of some use in retrospective diagnosis. |
NAAT nucleic acid amplification testing
PCR polymerase chain reaction
Microscopy
Light microscopy is the oldest non-culture-based diagnostic method in microbiology. Its use can be enhanced using various staining techniques. For example, calcofluor white is used to detect fungal hyphae of dermatophytes that may take up to three weeks to culture. Despite being challenged by recent advances in molecular techniques, microscopy remains a central tool in laboratories. It is relatively cheap and results can be generated within minutes of receiving a sample. Microscopy can also provide important ancillary information about the likelihood of clinical infection, such as the presence, nature and differential of inflammatory cells in a specimen. Microscopy can also be highly specific for some infections, and it is the diagnostic gold standard for detecting parasitic pathogens of the blood (e.g. malaria) or gastrointestinal tract (e.g. giardiasis).
Microscopy has several clear drawbacks. Even for parasitic enteropathogens the sensitivity of a single specimen is poor,1 and for most common bacterial and fungal infections it is neither sensitive nor specific.
Microscopy is labour intensive and requires highly skilled scientists for optimal diagnostic performance. For most pathogens, microscopy is best used as an adjunct to traditional culture or molecular methods.
Immunoassays
Immunoassays use antibodies to detect either antibody or antigen in a patient’s sample (usually serum but also nasopharyngeal swabs, throat swabs and urine).
Testing for antibodies
Antibody immunoassays – usually referred to as serology – have the particular advantage over other non-culture diagnostic methods in their ability to retrospectively diagnose infection long after viable microorganisms or recoverable nucleic acid have disappeared. Other advantages include a high degree of specificity where seroconversion has occurred, fast turnaround times and improved safety compared to culture methods for some organisms (e.g. Coxiella burnetii). They can also rule out acute infection based on serological evidence of previous exposure and immunity.
However, immunoassays have a number of disadvantages. Most serological diagnoses rely on the early detection of specific IgM at the time of acute infection, with subsequent seroconversion for specific IgG. There are several pitfalls to this approach. First, during an acute infection serology may be negative as the patient has not yet generated an antibody response. Second, cross reactions with unrelated IgM can occur. Although specific IgM is classically detectable for six weeks to three months following acute infection, it occasionally persists for months to years, or may reappear as an anamnestic response due to another infection. This response is particularly common for IgM against Toxoplasma gondii, and when diagnosing the arbovirus infections such as Barmah Forest virus and Ross River virus, and may lead to false positive results and spurious diagnoses. Such errors can be reduced by measuring acute- and convalescent-phase antibody concentrations to look for changes in response to infection. This is the preferred method for definitive serological diagnosis but obviously slows down the time to confirm diagnosis. Seroconversion often takes two weeks or more to occur.
The sensitivity of serological diagnosis can be reduced by a variety of factors, including age and immunodeficiency. Serology is only useful for diagnosis when there is a clear relationship between antibody concentrations and infection. It is less useful for infections where antibodies may persist but do not provide protection against repeat infection or reactivation, such as herpes simplex, cytomegalovirus and varicella zoster virus, or for infections caused by commensal organisms.
Waning immunity and reinfection commonly occur with Bordetella pertussis, which causes whooping cough. The detection of IgG specific for B. pertussis toxin greater than 100 IU/mL is suggestive of acute infection, and in older children and adults this may be supported by the presence of IgA to B. pertussis toxin.
Infections for which serology remains the mainstay of diagnosis in general practice include syphilis, Epstein-Barr virus, cytomegalovirus, toxoplasmosis, parvovirus, Barmah Forest virus, Ross River virus, dengue, chikungunya and Zika virus. Historically, detection of polyclonal antibody (Monospot test) has been used to diagnose acute glandular fever. It lacks sensitivity and specificity and has generally been replaced by the detection of specific IgM/IgG to Epstein-Barr virus capsid antigen in combination with the absence of IgG to nuclear antigen which develops six weeks to three months after acute infection and remains positive lifelong.
Testing for microbial antigens
An antigen is a component of a pathogen that stimulates an immune response. Immunoassays can measure this in various sample types. Many of these tests are in current use, including urinary antigen tests for Streptococcus pneumoniae and Legionella pneumophila serogroup 1. These are useful to identify the causative organism of acute community-acquired pneumonia, and the group A streptococcal antigen test of throat swabs for bacterial pharyngitis. Other examples of useful antigen assays include cryptococcal antigen detection in serum and cerebrospinal fluid in both immunocompetent and immunocompromised patients, and galactomannan antigen which is a surrogate marker for invasive aspergillosis, usually in immunocompromised individuals.
Antigen testing can provide rapid results – the S. pneumoniae antigen test can be completed within 15 minutes. Many of these tests have very good specificity. For example, a positive group A streptococcal antigen from a throat swab can allow targeted treatment if indicated and obviate the need for culture. Unfortunately, these tests often lack sensitivity in comparison to traditional culture methods and particularly compared to nucleic acid amplification tests. Their usefulness therefore often lies in enabling rapid diagnosis, rather than excluding clinical infection.
Combined immunoassay tests
The drawbacks of using antigen or antibody assays in isolation can be overcome by combining them. Assays that include both antigen and antibody, such as dengue virus NS1 antigen with IgM/IgG, or HIV antigen/antibody screening testing, offer reduced diagnostic window periods and enhanced sensitivity and specificity. Dengue NS1 antigen detection (Fig.) in particular has allowed rapid confirmation of dengue with the ability to initiate public health interventions earlier. Its sensitivity equates to that of a polymerase chain reaction (PCR) test for dengue in the first week of illness.
Fig.
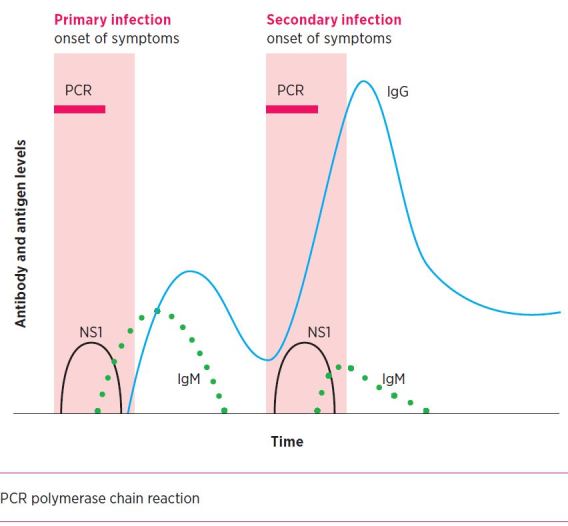
Immune response with primary and secondary dengue infections
Nucleic acid amplification testing
Nucleic acid amplification testing involves the detection of pathogen-specific DNA or RNA sequences in patient samples. There are a number of different methods – PCR is one type. Compared to traditional methods, nucleic acid amplification testing offers improved turnaround times and markedly enhanced sensitivity. These techniques are easily adapted to high-throughput testing and can allow multiple pathogen identification within a single test. Nucleic acid amplification testing is currently revolutionising areas where traditional microbiological methods have been complex, costly and time consuming, such as the diagnosis of faecal pathogens.
However, nucleic acid amplification testing is not without its challenges. Loss or changes to the target nucleic acid sequence through mutation can lead to false negative results. Furthermore, because of their extreme sensitivity, contamination can lead to false positives. While strict quality-control measures reduce this risk, nucleic acid amplification testing is not endorsed for screening in critical diagnoses such as HIV.
A positive result reflects the presence of nucleic acid only, not viable organisms. Failure to recognise this can lead to pitfalls in interpretation of results. For example, when infection has resolved (with or without treatment), nucleic acid may persist. Re-testing at an early interval can lead to positive results and false assumptions about reinfection or failure of therapy. For this reason if re-testing for Chlamydia trachomatis is required, it is recommended at least three weeks after initial diagnosis. The same applies to other sexually transmitted infections such as Neisseria gonorrhoeae and Trichomonas vaginalis.
Problems with assay specificity can potentially lead to false positives. Early nucleic acid amplification tests for N. gonorrhoeae resulted in significant rates of false positives due to cross reactions with commensal Neisseria species.2 Most testing platforms include both C. trachomatis and N. gonorrhoeae in multiplex assays. Laboratories report both results despite the generally lower prevalence of N. gonorrhoeae. To reduce false positive results, laboratories invariably confirm initial positive N. gonorrhoeae results using a second independent assay.3,4 This increases the positive predictive value of the reported positive result.
For many pathogens, it is important to pair nucleic acid amplification testing with traditional culture methods for additional information regarding antimicrobial susceptibility, microbial virulence and epidemiology. Currently, these cannot be determined by most molecular assays.5 For example, around 33% of all notified N. gonorrhoeae cases were diagnosed by culture, allowing antimicrobial susceptibility to be performed. With the introduction of faecal bacterial enteropathogen testing (Campylobacter, Salmonella, Shigella) the same concerns around availability of isolates for typing and susceptibility are surfacing.
Nucleic acid amplification testing has limitations when applied to organisms that potentially form part of the normal human flora (either transiently or permanently). For example, Clostridium difficile may be present in the bowel without causing illness. Its detection alone does not necessarily indicate a disease state,6 and a positive result in the wrong clinical context can lead to inappropriate diagnosis and therapy.
Nucleic acid is generally robust, so amplification testing on blood and other specimens (including skin swabs, urine, genital swabs, throat swabs, nasopharyngeal swabs, tissue aspirates) are usually stable at room temperature for 24 hours. If processing is delayed, samples should be refrigerated at 4° C. Dedicated samples for nucleic acid amplification testing are desirable to reduce the risk of contamination. Swabs in bacterial transport medium (Amies and Stuarts) may be inhibitory for nucleic acid amplification testing. Dry dacron-tipped or flocked swabs are the preferred sample type. Universal swabs suitable for all types of testing will become routinely available, although it is best to liaise with the local pathology laboratory regarding the preferred specimen types.
Conclusion
Non-culture-based diagnostic methods, particularly nucleic acid amplification tests, often as multiple PCR tests in a syndromic panel, are revolutionising the modern medical microbiology laboratory. They have enabled or simplified many difficult diagnoses, improved turnaround times and been adapted to allow high-throughput testing. This area will continue to expand and may even replace many traditional culture methods in the future. The optimal use of these diagnostic tests requires recognition of their limitations and judicious use of supporting clinical and laboratory evidence (including culture-based methods). When questions arise regarding the choice or interpretation of non-culture-based diagnostics, it is advisable to liaise with your local laboratory.
Footnotes
Conflict of interest: none declared
REFERENCES
- 1.McHardy IH, Wu M, Shimizu-Cohen R, Couturier MR, Humphries RM. Detection of intestinal protozoa in the clinical laboratory. J Clin Microbiol 2014;52:712-20. 10.1128/JCM.02877-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trembizki E, Costa AM, Tabrizi SN, Whiley DM, Twin J. Opportunities and pitfalls of molecular testing for detecting sexually transmitted pathogens. Pathology 2015;47:219-26. 10.1097/PAT.0000000000000239 [DOI] [PubMed] [Google Scholar]
- 3.Chow EP, Fehler G, Read TR, Tabrizi SN, Hocking JS, Denham I, et al. Gonorrhoea notifications and nucleic acid amplification testing in a very low-prevalence Australian female population. Med J Aust 2015;202:321-3. 10.5694/mja14.00780 [DOI] [PubMed] [Google Scholar]
- 4.Whiley DM, Lahra MM, National Neisseria Network . Review of 2005 Public Health Laboratory Network Neisseria gonorrhoeae nucleic acid amplification tests guidelines. Commun Dis Intell Q Rep 2015;39:E42-5. [PubMed] [Google Scholar]
- 5.Langley G, Besser J, Iwamoto M, Lessa FC, Cronquist A, Skoff TH, et al. Effect of culture-independent diagnostic tests on future emerging infections program surveillance. Emerg Infect Dis 2015;21:1582-8. 10.3201/eid2109.150570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphries RM, Linscott AJ. Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev 2015;28:3-31. 10.1128/CMR.00073-14 [DOI] [PMC free article] [PubMed] [Google Scholar]


