Abstract
Induced fit or protein flexibility can make a given structure less useful for docking and/or scoring. The 2015 Drug Design Data Resource (D3R) Grand Challenge provided a unique opportunity to prospectively test optimal strategies for virtual screening in these type of targets: heat shock protein 90 (HSP90), a protein with multiple ligand-induced binding modes; and, mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4), a kinase with a large flexible pocket. Using previously known co-crystal structures, we tested predictions from methods that keep the receptor structure fixed and used (a) multiple receptor/ligand co-crystals as binding templates for minimization or docking (“close”), (b) methods that align or dock to a single receptor (“cross”), and (c) a hybrid approach that chose from multiple bound ligands as initial templates for minimization to a single receptor (“min-cross”). Pose prediction using our “close” models resulted in average ligand RMSDs of 0.32 Å and 1.6 Å for HSP90 and MAP4K4, respectively, the most accurate models of the community-wide challenge. On the other hand, affinity ranking using our “cross” methods performed well overall despite the fact that a fixed receptor cannot model ligand-induced structural changes,. In addition, “close” methods that leverage the co-crystals of the different binding modes of HSP90 also predicted the best affinity ranking. Our studies suggest that analysis of changes on the receptor structure upon ligand binding can help select an optimal virtual screening strategy.
Keywords: drug discovery, virtual screening, D3R, induced fit, affinity ranking, pose prediction
Introduction
Major challenges in virtual screening are the inadequate scoring functions to evaluate the affinity of docked poses, and the difficulty to predict ligand induced flexibility observed in many important therapeutic targets [1-5]. To evaluate improvements in this area, the Drug Design Data Resource (D3R) developed the 2015 Grand Challenge, a community-wide experiment for researchers around the world to prospectively test docking and scoring methodologies against blinded data from two targets: heat shock protein 90 (HSP90), a protein that binds following an induced fit mechanism [6], i.e., the unbound or apo structure undergoes significant structural rearrangements upon ligand binding; and, mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4), a kinase with a large pocket that includes sizable flexible loops [7].
The most commonly used scoring functions can basically be classified into three types as Kitchen et al. summarized [1]: force-field-based scoring (e.g., D-Score [8], G-Score [8], GOLD [9], AutoDock [10], DOCK [11], Glide [12], SIE [13]), empirical scoring (e.g., LUDI [14, 15], F-Score [16], ChemScore [17], SCORE [18], Fresno [19], X-SCORE [20], AutoDock Vina [21]) and knowledge-based scoring (e.g., DrugScore [22], SMoG [23]). In the 2010 Community Structure-Activity Resource (CSAR) Exercise, Carlson and collaborators analyzed the performance of different scoring functions on the CSAR-NRC data set [5, 24]. The results indicated that most of the scoring functions had comparable performance (R2=0.3-0.4) and the best R2 were achieved by AutoDock and AutoDock Vina (R2=0.55) [5]. Despite the poor performance of scoring, many docking methods did well in predicting poses within 2.0 Å of the crystal conformation [3, 5].
Over the last few years, the Camacho lab has steadily built novel platforms for drug discovery, from predictions of druggable sites [25], to pharmacophore-based interactive virtual screening technologies that search billion size libraries in seconds [26]. We also developed Smina [27], a version of AutoDock Vina specially optimized to support high-throughput minimization and scoring. Based on our current implementation in AnchorQuery [28], Smina can minimize 10,000 compounds into a fixed receptor in about 10 seconds (details will be published elsewhere), the same time scale required for docking a single compound to a flexible receptor [1]. More recently, we have shifted our attention to improving our virtual screening pipeline [26-28]. We participated in the 2013/14 CSAR challenge that involved rank-ordering compounds to homology models of the receptors with a given protein primary sequence, identifying close-to-native bound conformations out of a set of decoy poses, and rank-ordering the affinity of sets of congeneric compounds to a given protein. Our predictions were among the best in the field [29, 30]. We showed that the most significant contribution to a meaningful enrichment of native-like models was the identification of the best receptor structure for docking and scoring. In particular, we showed that ranking a set of 31 congeneric compounds cross-docked to the tRNA (m1G37) methyltransferase (TRMD) structure with the largest pocket resulted in an impressive R2= 0.67, whereas other receptor structures yielded R2 ~ 0.
Here, we report our participation in the 2015 D3R Grand Challenge, where we performed a comprehensive analysis of different strategies for predicting docking poses and ranking affinities for two highly flexible targets: HSP90 and MAP4K4. These strategies included methods that utilize all available receptor/ligand co-crystals (“close”), all available ligands and a single holo-receptor structure (“min-cross”) and only a single receptor/ligand co-crystal (“cross”). As in the 2013/14 CSAR competition [29], we found that the method that predicted the best docking poses was not the same as the ones that predicted the best ranking of active compounds. Similarly, different methods were shown to predict the optimal ranking of active compounds for HSP90 and MAP4K4, i.e., “close” and “cross”, respectively. Inspection of the type of flexibility exhibited by each target, i.e., induced fit versus large flexible pocket, suggests guiding principles for selecting the optimal virtual screening for flexible targets. We note that these findings are strongly supported by the fact that our prospective pose predictions and affinity rankings for HSP90 to the 2015 D3R Grand Challenge were the best in the community-wide experiment.
Methods
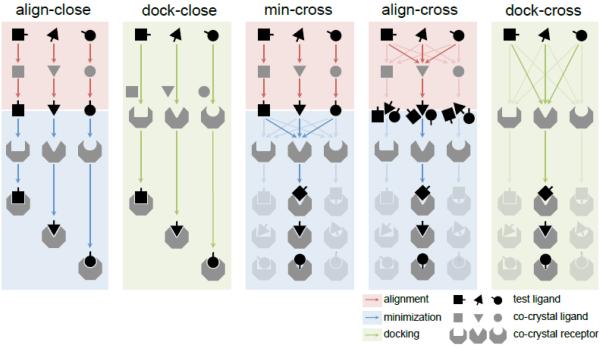
We tested the performance of five major methods (Fig. 1) on both pose and affinity predictions. Several variants of the methods were also applied to special cases, which will be discussed later in the specific challenges.
Figure 1. “Close” and “cross” methods for affinity ranking and pose prediction.
Align-close and dock-close methods minimize and dock to the “closest” receptor for each compound. Min-cross, align-cross and dock-cross methods minimize and dock to all available receptors and select “optimal” receptor based on available experimental data (see Methods). This is shown in the figure by the greyed-out shapes in the “cross” methods that ultimately select one optimal receptor. Red blocks and arrows correspond to alignment, blue blocks and arrows correspond to minimization, green blocks and arrows correspond to docking.
The methods used the following applications that are freely available for academic research. Structure preparation: all receptor structures were superimposed using the “align” command in PyMOL 1.7 [31]. Conformer generation: For structural alignment, 20 conformers were generated using Omega2 [32] with default settings. Chemical similarity: Babel 2.3.2 [33] was used with fingerprint 3 (FP3) to identify the most similar or “closest” compound among known ligands. The co-crystal receptor corresponding to the “closest” compound is referred to as “closest” receptor. Conformer alignment: Structural alignments were performed using Open3DALIGN 2.282 [34]. Minimization: Aligned conformers are minimized to a given receptor using Smina [27] with default settings. Docking: Compounds were docked with Smina with default parameters and AutoDock Vina [21] scoring function. A reference compound was used to define the docking box. The Vina-predicted energy was used to select the best ranked docked pose.
Align-close method
(a) Conformers were generated for each compound in the test set. (b) The “closest” compound among known bound ligands was identified. (c) Conformers were aligned to the “closest” compound. (d) Aligned conformers were minimized to the “closest” receptor. (e) The best Vina score was used to predict affinity for the compound.
Dock-close method
(a) The “closest” compound among known bound ligands was identified. (b) Compounds were docked to the “closest” receptor using “closest” ligand as reference to define docking box. (c) The best Vina score was used to predict affinity for the compound.
Min-cross method
(a) Conformers were generated for each compound. (b) The “closest” compound was identified. (c) Conformers were aligned to the “closest” compound. (d) The aligned conformers were minimized to all known bound receptors. (e) The best Vina score to each receptor was used to predict affinity. (f) Optimal receptor for virtual screening is selected (see below).
Align-cross method
(a) Conformers were generated for each compound. (b) Conformers were aligned to every known bound ligand. (d) Aligned conformers were minimized to the corresponding bound receptor. (e) The best Vina score among conformers was used to predict affinity to each receptor structure. (f) Optimal receptor for virtual screening is selected (see below).
Dock-cross method
(a) Compounds were docked to every known bound receptor using its bound ligand as reference. (b) The best Vina score to each receptor was used to predict affinity. (c) Optimal receptor for virtual screening is selected (see below).
These five methods can be grouped by receptor selection. The optimal receptor for “cross” methods (min-cross, align-cross and dock-cross) was chosen by comparing the Vina scores for each receptor with experimental data (IC50, see Supplementary Table 1 and 2). We calculated Spearman's rank correlation coefficient (Spearman ρ) and coefficient of determination (R2) to select the optimal receptor that performs the best for affinity ranking in our training set. Similarly, we compared the best-scored poses - for each receptor with the crystal poses to generate the ligand root-mean-square deviation (RMSD), and computed the percentage of poses that have a RMSD less than 2 Å to select the optimal receptor for pose prediction. For testing data, we use the best-performing receptor in the training data set to rank affinity and predict poses. For “close” methods (align-close and dock-close), there is no optimal receptor, but multiple receptor/ligand co-crystals are used for predictions.
Results
HSP90 Challenge
Challenge
(1) Predict binding modes of six HSP90 compounds. (2) Predict affinity ranking of P=180 HSP90 compounds, among this set, 33 unidentified compounds were said to have no inhibition. (3) Predict relative/absolute free energy of three small sets of compounds. Analyses of the 180 compounds show that they all fall into three chemical scaffolds (aminopyrimidines, benzimidazolones and benzophenone-like, Fig. 2C-2E. Upper panels show scaffolds, and lower panels show examples). Two unpublished structures, 4YKR and 4YKY, were provided as examples of benzimidazolones and benzophenone-like compound binding.
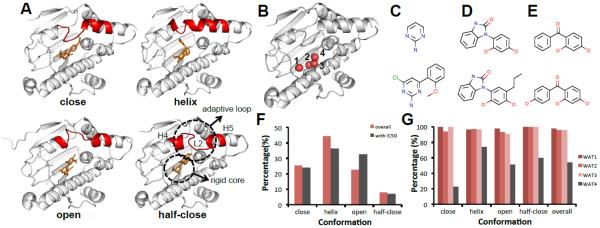
Figure 2. HSP90 adopts at least four ligand-induced binding modes.
A. Four conformations of HSP90 ligand-induced binding pocket based on the nearby adaptive loop (L2, between H4 and H5 [43]): close (2WI5), helix (4EFU), open (3RLR), half-close (3B28) (white cartoon: HSP90, red cartoon: flexible loop, orange ticks: small molecules) B. Four waters in the binding pockets labeled from 1 to 4 (white cartoon: HSP90, red sphere: water molecules) C. Aminopyrimidine scaffold and compound (2XDX) D. Benzimidazolone scaffold and compound (4YKR) E. Benzophenone-like scaffold and compound (4YKR) F. Histogram of binding modes among the N=181 known co-crystal structures and I=69 structures with IC50 data. (N: number of co-crystals, I: number of co-crystal with IC50 data) G. Histogram of conservation frequency of water molecule in Fig. 2B shows that three crystal waters are 100% conserved.
Binding Pocket Analysis
There are N=179 PDB plus two unpublished HSP90 structures bound to small molecules, with I=69 of them having known IC50 (from BindingDB [35], BindingMOAD [36] and PDBBind [37], Supplemental Table 1). We superimposed all the known receptors to the receptor structure in 4YKR. Interestingly, a distal loop (L2 between H4 and H5, Fig. 2A) is very adaptive upon different ligand binding. Basically, all co-crystal structures can be grouped into four distinct conformations based on the adaptive loop (red cartoon in Fig. 2A): close, helix, open and half-close (a conformation between open and close). The histograms of these binding modes in the whole dataset and sub-dataset with IC50 are shown in Fig. 2F. The core binding pocket is quite rigid and stable, and four crystal water molecules are observed to participate in ligand binding (Fig. 2B). Three waters are highly conserved despite the different adaptive loop conformations (Fig. 2G). These analyses suggest that the ligand-binding pocket of HSP90 consists of a rigid core part with a conserved water-mediated interacting network and a ligand-dependent adaptive loop. Therefore, when preparing models for docking and alignment/minimization, we kept conserved water molecules as part of the receptors.
Methods
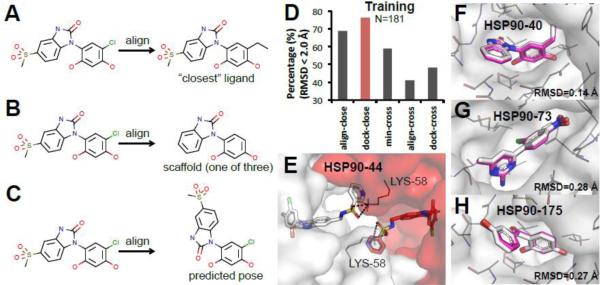
We applied the five methods listed in Fig. 1 (i.e., align-close, dock-close, min-cross, align-cross, and dock-cross) for both pose prediction and affinity ranking. For affinity ranking, we also devised several variations of the previous methods as potential improvements for ligand alignment, and others. (a) min-cross-scaffold and align-close-scaffold methods: Given the limited set of scaffolds that presumably capture the core ligand interactions, for min-cross and align-close methods we aligned the test compounds to the three scaffolds shown in Fig. 2C-2E (see, e.g., Fig. 3B) instead of the chemically “closest” compounds as in Fig. 3A. (b) min-cross-pose and align-close-pose methods: Instead of using as templates ligand structures from co-crystals, we use the actual predicted poses by “close” methods as templates for alignment in min-cross and align-close methods (see, e.g., Fig. 3C). (c) dock-close-filter and align-close-filter: We also used the aforementioned predicted poses for manually selecting inactive compounds in testing set. We then overruled the Vina score and moved this set of compounds to the bottom of the affinity ranking for the two methods that had best performance in training set. (d) HSP90 score 1-4: We used machine learning and forward selection methodologies to develop four HSP90-specific scoring functions from the set of energy terms available in Smina [27] (see Supplemental Table 2 for the selected parameters and weights). A training dataset was constructed by cross-docking the I=69 compounds with published IC50 data to crystal structure 4EFU (optimal receptor for dock-cross method) with the default Smina settings. HSP90 score 1 and 2 functions were trained on active compounds (measured by Spearman ρ), while HSP90 score 3 and 4 were trained to maximize the discrimination of active versus decoy compounds which were obtained from the HSP90 dataset in the DUD-E database [38] (measured by AUC). (e) 3DQSAR-align-pose and 3DQSAR-dock-pose: The relatively large amount of binding data made quantitative structure–activity relationship (QSAR) possible. Using Open3DQSAR 2.3 [39], we trained 3DQSAR models with the 69 HSP90 structures with IC50 data. We applied the trained models to the predicted poses in the testing set from “close” methods.
Figure 3. “Close” methods predicted high-accuracy poses for six HSP90 compounds.
A-C. Examples of different alignment methods in HSP90 challenge. A. A compound is aligned to the “closest” compound. B. A compound is aligned to one of the three scaffolds. C. A compound is aligned to the predicted pose. D. Performance of pose prediction using different methods in the training set (N: number of co-crystals) E. Lysine 58 from another HSP90 stabilizes the conformation of the extending functional group of HSP90-44. (white and red stand for two different HSP90 monomers. sticks: HSP90-44; meshes: HSP90; lines: lysine 58 of HSP90; black dash: hydrogen bonds) F-H. Best predicted poses are aligned to the co-crystal structures. F. HSP90-40 is an example of aminopyrimidines (RMSD 0.14 Å). G. HSP90-73 is an example of benzimidazolones (RMSD 0.28 Å). H. HSP90-175 is an example of benzophenone-like compounds (RMSD 0.27 Å). (white sticks: crystal pose of the compounds; magenta sticks: best pose prediction of the compounds; white meshes: HSP90 surface; grey lines: HSP90 residues close to the binding groove).
Phase 1: Pose prediction results
Retrospective study of known ligands demonstrated that dock-close and align-close methods predicted the most accurate poses. For the analysis shown in Fig. 3D the co-crystal of each ligand was first removed from the dataset, and poses were then predicted based on the remaining co-crystal structures in the training set. Given the large dataset of available co-crystal structures, our results reflect the empirical observation that crystallographic information is superior to any computational model. Hence, we were able to predict high-accuracy poses for all six testing compounds. We took the top five poses predicted by “close” methods (sorted by Vina score), and submitted the best models. The mean RMSD for the first ranked and best pose were 0.46 Å and 0.32 Å, respectively. Fig. 3F-3H show an example of the best-predicted poses of each scaffold. The predicted pose for HSP90-44 had a flexible group sticking out of the binding site. We used molecular dynamics to predict the most likely conformation, yet the co-crystal shows that this group is stabilized by Lys58 from the second HSP90 monomer in the dimer structure (Fig. 3E). When structural data is available, our results demonstrate that “close” methods are significantly better in pose prediction than “cross” methods, while dock-cross have an upper limit of about 50% success rate using a single receptor structure.
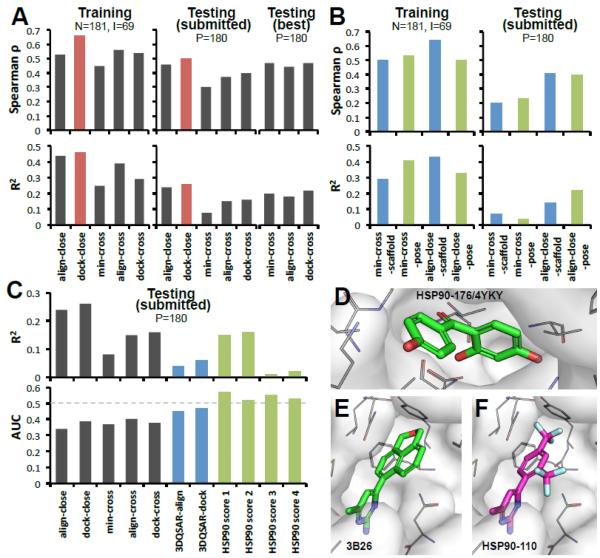
Phase 2: Affinity prediction results
The results of our predictions are summarized in Table 1. Dock-close (Spearman ρ=0.42, R2=0.26) and align-close (Spearman ρ=0.45, R2=0.24) methods have the best performance. The relative performance of the five methods is consistent between the training set and our submitted predictions (Fig. 4A). An interesting question to ask is whether for the “cross” methods, were we able to predict the optimal receptors? The answer is that our R2 analysis correctly predicted an open structure (Fig. 2A) as optimal receptor. However, in retrospect, we found that other open structures were marginally better, see Testing (best) in Fig. 4A. Thus, a receptor is only assumed to be “optimal” based on the data available. Overall, the relatively similar outcomes of “close” and “cross” methods suggest that our scoring function cannot account for the change in free energy associated with different receptor structures, and therefore ranking ligands to induced-fit targets is still limited.
Table 1.
Affinity ranking prediction results of HSP90 challenge.
| Method | Phase a | Spearman ρ | Kendall Tau b |
|---|---|---|---|
| align-close | P1, P2 | 0.45 | 0.31 |
| dock-close | P1, P2 | 0.42 | 0.29 |
| align-cross | P1, P2 | 0.33 | 0.22 |
| dock-cross | P1 | 0.37 | 0.25 |
| align-close-scaffold | P1, P2 | 0.42 | 0.3 |
| min-cross-pose | P2 | 0.26 | 0.18 |
| align-close-pose | P2 | 0.37 | 0.26 |
| align-close-filter | P1, P2 | 0.38 | 0.26 |
| dock-close-filter | P1, P2 | 0.38 | 0.26 |
| HSP90 score 1 | P1 | 0.17 | 0.12 |
| HSP90 score 2 | P1 | 0.23 | 0.16 |
| HSP90 score 3 | P1 | −0.01 | −0.01 |
| HSP90 score 4 | P1 | 0.09 | 0.06 |
| 3DQSAR-align-pose | P2 | 0.18 | 0.13 |
| 3DQSAR-dock-pose | P2 | 0.24 | 0.16 |
P1 means this method was submitted for evaluation in HSP 90 Phase 1 challenge. P2 stands for Phase 2.
Spearman ρ and Kendall Tau are from D3R result evaluation.
Figure 4. “Close” methods have better performance than “cross” methods for affinity ranking in HSP90 challenge.
A. Prediction rates on: training set, testing set submitted prospectively, and testing (best) set reassessed retrospectively. Optimal receptors for align-cross, min-cross and dock-cross were (prospectively) 3OWD, 4BQJ, 3K98 and (retrospectively) 3T10, 3RLP, 3OWD, respectively. N: number of co-crystals, I: number of co-crystal with IC50 data, P: number of compounds for prediction. B. Results of variant methods: aligning to scaffold, to predicted pose, and using human expertise to eliminate non-binders. C. Distinguishing active from 33 inactive compounds using general methods, human discrimination, 3DQSAR, and special purpose scoring functions to discriminate HSP90 ligands. The lower panel shows binding/non-binding AUC performances, and upper panel shows the corresponding affinity ranking. D-F. Examples of binding poses of inactive compounds. D. Co-crystal of inactive compound 176 (4YKY). E. Co-crystal from PDB 3B26 (unknown IC50). F. prediction for compound 110 (inactive).
Alignment is an area that can be improved particularly for large and/or low similarity compounds. Thus, we developed two variants of the above methods to test different structural alignments. First, we surmised that aligning to the scaffold would lead to better core interactions (Fig. 2C-2E). Although this was the case in our training data set, the opposite was observed for “-scaffold” methods in the testing set (Fig. 4B). In retrospect, we found that our method was able to improve some bad alignments, but it also eliminated some good ones. The latter was particularly true for benzophenone-like compounds whose structures are quite diverse. Second, using our models for the testing set as “predicted closest” templates also failed to improve the affinity ranking, observing only a minor “-pose” improvement for min-cross in the training set (Fig. 4B). The failure may come from the inadequacy of the force field to smoothly remove clashes upon minimization. In summary, neither aligning to scaffolds nor to predicted poses improved affinity ranking relative to aligning to the “closest” compound.
As a control of blind versus human predictions, we visually inspected all dock-close and align-close poses and predicted whether they were binders/active or non-binders/inactive (“-filter” methods in Table 1). Humbly, the blind methods performed better than the subjective human filtered scores. In hindsight, one problem is that a compound may bind but it might also be deemed inactive. For instance, HSP90-176 and HSP90-110 are both inactive compounds (Fig. 4D-F), yet, compound 176 binds HSP90 (4YKY), same thing happened for our binding model for HSP90-110 (Fig. 4F) that is based on a highly similar co-crystal (3B26 in Fig. 4E).
The 3DQSAR models perform poorly in affinity ranking (Table 1 and Fig. 4C). The major reason seems to be that there were no cases for aminopyrimidine scaffold among the 69 compounds with IC50 data. Therefore, when applying the models and functions to testing set, they did poorly at scoring aminopyrimidine compounds.
The HSP90 score 1-2 were trained to better rank active compounds, and their predicted ranking was similar to other “cross” methods. However, these scoring functions had a meaningful improvement in the discrimination between actives and inactives (Fig. 4C). On the other hand, the HSP90 score 3 and 4 that were specially designed solely to distinguish actives from inactives. As expected, these methods performed poorly in affinity ranking. However, training on inactive compounds from the DUD-E database did not improve the discrimination of active compounds. In hindsight, we realized that the inactive compounds in the testing set had different scaffolds than the DUD-E decoy compounds. Thus, in all likelihood the observed discrimination might be close to random. These results show how dangerous is to evaluate machine learning scoring functions without a rigorous benchmarking. Overall, these results indicate that target specific scoring functions and 3DQSAR models can do better at distinguishing active from inactive compounds than the default Vina scoring function used in the methods in Fig. 1.
MAP4K4 Challenge
Challenge
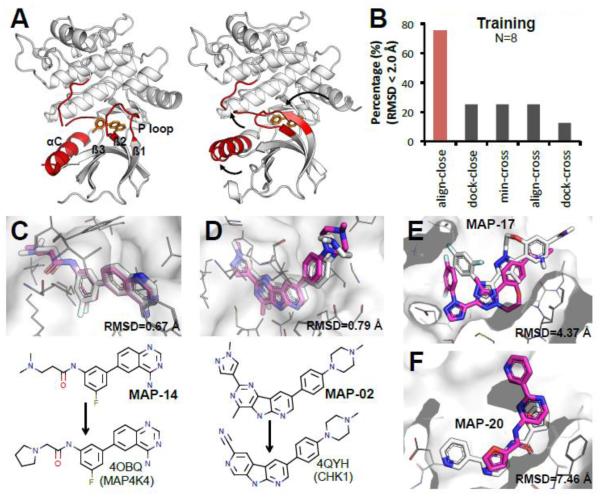
(1) Predict the binding modes of P=30 MAP4K4 compounds. (2) Predict affinity ranking of P=18 MAP4K4 compounds. The structural data available for MAP4K4 were fairly limited. Only N=8 published co-crystal structures of MAP4K4 bound to small molecules were found in the PDB, and all of them (I=8) had IC50 data (from BindingDB [35], and literature [40, 41]). MAP4K4 is a kinase with a large ATP binding pocket surrounded by several flexible regions (Fig. 5A): residues 171-to-190, residue 30-to-39 (P loop) and residue 60-to-75 (αC). Residue 171-190 is a disordered region that is not resolved in most co-crystals [42]. The P loop links two β-sheets, β1 and β2, in the binding groove, and acts as a lid to adaptively adjust conformation upon ligand binding. Helix αC is also flexible and the loop between αC and β3 is sometimes not resolved.
Figure 5. Align-close predicted the best models for 30 MAP4K4 compounds with a mean RMSD of 1.6 Å.
A. Flexible regions around the MAP4K4 binding pocket adapt to different conformations upon ligand binding. (left panel: 4OBO, right panel: 4U44, white cartoon: MAP4K4, red cartoon: flexible loop/helix, orange sticks: small molecules) B. Pose prediction performance across different methods in training set. C, D. Alignment of our best-predicted pose with the co-crystal structure. MAP-14 is an example of aligning to compound from MAP4K4 (RMSD 0.67 Å). MAP-02 is an example of aligning to the compound from other kinase (CHK1, 4QYH) (RMSD 0.79 Å). E, F. Two cases we did poor in pose prediction: MAP-17 and MAP-20. (white sticks: crystal; magenta sticks: predicted; white meshes: MAP4K4 surface; grey lines: MAP4K4 residues close to the binding groove).
Methods
We first applied the five methods described in Fig. 1 for pose prediction and affinity ranking. Scaffolds of testing compounds were quite different from the eight published ligands. Thus, we enriched our structural sampling by collecting an additional eight co-crystal structures of related kinases (Supplemental Table 1), which we only used to improve the alignment of compounds in the MAP4K4 binding pocket. Otherwise, the methods are the same as for HSP90. For affinity ranking, we also tested: (1) Min-cross-pose and align-close-pose (see description in HSP90 challenge methods); (2) Co-crystal-min: Since for Phase 2 we learned the co-crystal structures of all the testing compounds, we ranked these ligands using Vina scores after minimization; (3) Interaction-based ranking: Since the Vina scoring function does not have a directional Hydrogen bond term, we generated an alternative ranking by counting the number of intermolecular hydrogen bonds between the compound and the receptor.
Phase 1: Pose prediction results
Analysis of the training set showed that only the align-close method predicted top quality models (Fig. 5B). Contrary to HSP90 where dock-close performed very well, docking on the large kinase pocket performed very poorly. However, expanding the set of ligands to those bound to homolog kinases allowed us to find related compounds for 27 out of the 30 testing compounds, using align-close for pose prediction resulted in a mean RMSD for the first predicted poses and best poses of 2.6 Å and 1.6 Å, respectively. Figure 5C and Fig. 5D show successful examples of pose predictions with reference in MAP4K4 training set or other kinases (CHK1). Two particularly bad predictions were MAP-17 and MAP-20. For MAP-17, we predicted a binding mode similar to MAP-12 and MAP-13, however, the co-crystal showed that the compound is not as deeply buried (Fig. 5E). For MAP-20, the reference structure we used was 3FV8 from JNK3. Here, the co-crystal revealed a pose that is 180° rotated from our predicted pose (Fig. 5F). Without these two incorrect predictions, our mean RMSD was 1.1 Å. The above notwithstanding, our results produced the best overall models of D3R, and our method proved to be a robust approach for pose prediction of kinases even when there is limited available data.
Phase 2: Affinity prediction results
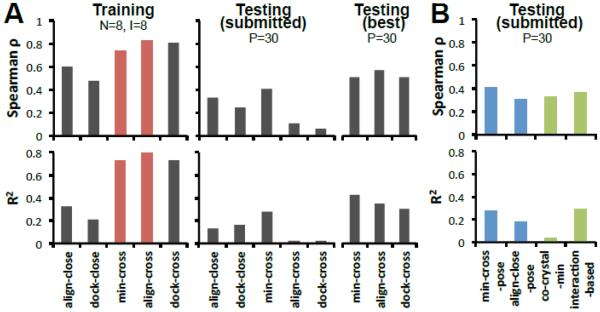
The ranking predictions of the 18 MAP4K4 compounds are summarized in Table 2. Our best submitted predictions were obtained using the min-cross and min-cross-pose methods (Spearman ρ=0.41, R2=0.28). Based on the IC50 data, we predicted 4OBP as the optimal receptor for all “cross” methods, achieving a remarkable Spearman ρ=0.8 in the training set. Of course, this prediction lacked statistical significance due to the small number of IC50’s. For Phase 2, we were given the co-crystal structures of all the compounds in the testing set but we still selected the same optimal receptor as in Phase 1. Hence, as shown in Table 2, results from Phase 1 and Phase 2 did not show significant differences for most of the methods. The two methods that improved were min-cross-pose and interaction-based methods, in which predicted poses were replaced by co-crystal poses. However, it is important to note that our retrospective analysis shows that with more affinity data our approach could have improved the affinity ranking (see Testing best Fig. 6A). In fact, just choosing MAP23 as our optimal receptor would have resulted on a Spearman ρ=0.57, comparable to the best prediction for this target in the D3R challenge.
Table 2.
Affinity ranking prediction results of MAP4K4 challenge.
| Spearman Value | Kendall Tau Value a | |||
|---|---|---|---|---|
| Method |
|
|||
| Phase 1 | Phase 2 | Phase 1 | Phase 2 | |
| align-close | 0.33 | - c | 0.22 | - |
| dock-close | 0.03 | - | 0.06 | - |
| min-cross | 0.41 | 0.41 | 0.29 | 0.29 |
| align-cross | 0.11 | 0.11 | 0.07 | 0.07 |
| dock-cross | 0.06 | 0.01 | 0.06 | -0.01 |
| min-cross-pose | 0.31 | 0.41 | 0.23 | 0.29 |
| align-close-pose | - | 0.29 | - | 0.22 |
| interaction-based | 0.30 | 0.37 | 0.22 | 0.27 |
| co-crystal-min | - | 0.33 | - | 0.33 |
The results are from D3R results evaluation.
This method is not submitted in this Phase.
Figure 6. “Cross” methods perform better than “close” methods for affinity ranking of MAP4K4 ligands.
A. Five methods in training set, submitted testing set, and retrospective best predictions for testing set. Optimal receptors for align-cross, min-cross and dock-cross were (prospectively) 4OBP, 4OBP, MAP03 and (retrospectively) MAP29, MAP16, 4U45, respectively. Overall, min-cross and align-cross performed better in our submitted predictions. N: number of co-crystals, I: number of co-crystal with IC50 data, P: number of compounds for prediction. B. Comparison of several pose related methods. (Spearman ρ and R2 are generated by comparing the Vina scores from different methods and experimental IC50 data).
Several variations of the main methods resemble “close” methods that overall did not perform as well as some “cross” methods. Figure 6B showed that min-cross-pose and align-close-pose, which in Phase 2 aligned compounds to the crystal poses as opposed to the “closest” poses, had almost the same Spearman outcomes. Consistent with our training data predicting “cross” methods superior to “close” methods, the co-crystal-min method that combined scores from different receptors performed poorly. The interaction-based method (Table 2 and Fig. 6B) showed that hydrogen bonding is important in determining the affinity of the compounds, highlighting a limitation of the Vina scoring function.
Discussion
The Drug Design Data Resource (D3R) 2015 Grand Challenge provided a unique platform for the prospective validation of new methods addressing two of the fundamental challenges in computer-aided drug discovery: predicting the pose of compounds bound to a non-cognate receptor (the cross-docking problem) and ranking series of congeneric compounds based on their affinity (the scoring problem). For the last several decades, the field has worked to develop solutions to these problems and many advances have been made. However, most evaluations of docking and scoring are retrospective wherein the correct answers (poses or affinities) are already known [3]. This retrospective analysis can lead to an unintentional bias to overestimate the performance of the method as the persons developing the methods may notice problems that can lead them to the circumstantial corrections of flaws in the workflow. Unfortunately, this does not reflect the real world use-case of docking and scoring methods where the correct answers are unknown until tested experimentally. This highlights the importance of prospective validation of methods as the true test of their performance since after the submission of the predictions, no further tweaking of parameters or workflows can take place.
The 2015 Grand Challenge featured two challenging and therapeutically relevant targets: HSP90 and MAP4K4. Here we presented five methods for pose prediction and affinity ranking that we employed in the challenge: align-close, dock-close, min-cross, align-cross and dock-cross. As reported, our approach of using “close” methods for pose predictions, where all available crystallographic information (mostly co-crystals) is used, yielded the most accurate poses in the community-wide experiment for both targets. Consistent with our earlier work [29], align-close was particularly robust for both flexible targets. Although dock-close predicted better-docked poses to the rather tight pocket of HSP90, the same method failed predicting good poses in the much larger binding site of MAP4K4. The reason is that docking to a large pocket naturally relies more on scoring to select among several “reasonable” poses.
Affinity ranking is still a major challenge in drug discovery, where sometimes not even the co-crystal structure helps much. Part of the problem is that often scoring functions are optimized for virtual screening, whereas calculating the true binding free energy is a multidimensional process that involve different interaction energies, water molecules, polarization effects, conformational changes and dynamics in the two components. The methods that we present here are aimed to optimize virtual screening technologies. As such we keep the receptor structure fixed for docking and/or minimization. With this constraint, we find that “cross” methods had more robust predictions than “close” methods, stressing the limitations scoring different receptor structures. Interestingly, for HSP90, dock-close had slightly better predictions than dock-cross. Analyses of the HSP90 bound structures suggest that dock-close performed better because the induced-fit binding process modifies the distal helix of the binding pocket but keeps the core motif almost intact (Fig. 2A), providing a better normalization between receptors. The latter is very different from MAP4K4 where most of the binding pocket is malleable (Fig. 5A), and the binding energy associated with those differences varies greatly.
There is still significant room for improvements. For instance, our methods use Vina scoring function, which represents the state of the art in open source scoring functions but it is still far from perfect [5]. Additionally, it is well known that experimental data coming from different assays is not well normalized. The above notwithstanding, compared with 3D-QSAR and machine learning algorithms, the structure-based methods sketched in Fig. 1 were significantly more reliable with limited data (Table 1). From a methodology point of view, we have shown that “close” methods show great power to predict poses. For affinity ranking, the choice of methods seems to be more dependent on the binding pocket features. Consistent with MAP4K4, another kinase from CSAR 2013/14 competition, SYK, had “cross” methods as the best ranking method [29], whereas for TRMD, a target with a rigid and buried pocket perhaps resembling HSP90, dock-cross performed the best [29]. These results suggest a preference of method selection in different pocket types, but the specific features that dominate the selection of the optimal method remains to be determined. In the 2013/14 CSAR competitions, we suggested that the structure with the largest binding pocket should have the best performance [29]. Similar analyses have been applied to HSP90 and MAP4K4, but no strong correlations were observed. Further research is needed to understand what are the conditions to select the optimal receptor or set of receptors that would predict the best ranking of compounds.
In presenting the results of our participation in the D3R 2015 Grand Challenge, we have validated five major methods for pose prediction, docking and scoring. These methods prospectively predicted the overall best poses for both flexible targets and best affinity ranking (Spearman ρ) for HSP90. Furthermore, these general methods can be adapted and applied in a number of different situations by modifying the scoring functions or docking protocols, improving the outcome of virtual screening experiments.
Supplementary Material
Acknowledgement
The authors thank D3R for organizing and evaluating the 2015 Grand Challenge. We are grateful to the OpenEye Scientific for providing an academic license for their software. The work is funded by National Institution of Health 2GM097082. Zhaofeng Ye also thanks the Tsinghua University-University of Pittsburgh joint Program and Chinese Scholar Council for providing the research opportunities and resources.
Reference
- [1].Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- [2].Rauh D, Klebe G, Stubbs MT. Understanding protein-ligand interactions: the price of protein flexibility. J Mol Biol. 2004;335:1325–1341. doi: 10.1016/j.jmb.2003.11.041. [DOI] [PubMed] [Google Scholar]
- [3].Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- [4].Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov Today. 2006;11:580–594. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Smith RD, Dunbar JB, Jr., Ung PM, Esposito EX, Yang CY, Wang S, Carlson HA. CSAR benchmark exercise of 2010: combined evaluation across all submitted scoring functions. J Chem Inf Model. 2011;51:2115–2131. doi: 10.1021/ci200269q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lauria A, Ippolito M, Almerico AM. Inside the Hsp90 inhibitors binding mode through induced fit docking. J Mol Graph Model. 2009;27:712–722. doi: 10.1016/j.jmgm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- [7].Wang L, Stanley M, Boggs JW, Crawford TD, Bravo BJ, Giannetti AM, Harris SF, Magnuson SR, Nonomiya J, Schmidt S, Wu P, Ye W, Gould SE, Murray LJ, Ndubaku CO, Chen H. Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors. Bioorg Med Chem Lett. 2014;24:4546–4552. doi: 10.1016/j.bmcl.2014.07.071. [DOI] [PubMed] [Google Scholar]
- [8].Kramer B, Rarey M, Lengauer T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins. 1999;37:228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- [9].Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- [10].Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- [11].Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- [12].Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- [13].Rahaman O, Estrada TP, Doren DJ, Taufer M, Brooks CL, 3rd, Armen RS. Evaluation of several two-step scoring functions based on linear interaction energy, effective ligand size, and empirical pair potentials for prediction of protein-ligand binding geometry and free energy. J Chem Inf Model. 2011;51:2047–2065. doi: 10.1021/ci1003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bohm HJ. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J Comput Aided Mol Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- [15].Bohm HJ. Prediction of binding constants of protein ligands: a fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J Comput Aided Mol Des. 1998;12:309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- [16].Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J Mol Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- [17].Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- [18].Wang R, Liu L, Lai L, Tang Y. SCORE: A new empirical method for estimating the binding affinity of a protein-ligand complex. Molecular modeling annual. 1998;4:379–394. [Google Scholar]
- [19].Rognan D, Lauemoller SL, Holm A, Buus S, Tschinke V. Predicting binding affinities of protein ligands from three-dimensional models: application to peptide binding to class I major histocompatibility proteins. J Med Chem. 1999;42:4650–4658. doi: 10.1021/jm9910775. [DOI] [PubMed] [Google Scholar]
- [20].Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- [21].Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gohlke H, Hendlich M, Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. J Mol Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- [23].DeWitte RS, Shakhnovich EI. SMoG: de Novo design method based on simple, fast, and accurate free energy estimates .1. Methodology and supporting evidence. J Am Chem Soc. 1996;118:11733–11744. [Google Scholar]
- [24].Dunbar JB, Jr., Smith RD, Yang CY, Ung PM, Lexa KW, Khazanov NA, Stuckey JA, Wang S, Carlson HA. CSAR benchmark exercise of 2010: selection of the protein-ligand complexes. J Chem Inf Model. 2011;51:2036–2046. doi: 10.1021/ci200082t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Koes DR, Camacho CJ. PocketQuery: protein-protein interaction inhibitor starting points from protein-protein interaction structure. Nucleic Acids Res. 2012;40:W387–392. doi: 10.1093/nar/gks336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Koes DR, Camacho CJ. ZINCPharmer: pharmacophore search of the ZINC database. Nucleic Acids Res. 2012;40:W409–414. doi: 10.1093/nar/gks378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Koes DR, Baumgartner MP, Camacho CJ. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J Chem Inf Model. 2013;53:1893–1904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Koes D, Khoury K, Huang Y, Wang W, Bista M, Popowicz GM, Wolf S, Holak TA, Domling A, Camacho CJ. Enabling large-scale design, synthesis and validation of small molecule protein-protein antagonists. PLoS One. 2012;7:e32839. doi: 10.1371/journal.pone.0032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Baumgartner MP, Camacho CJ. Choosing the Optimal Rigid Receptor for Docking and Scoring in the CSAR 2013/2014 Experiment. J Chem Inf Mode. 2015 doi: 10.1021/acs.jcim.5b00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Smith RD, Damm-Ganamet KL, Dunbar JB, Jr., Ahmed A, Chinnaswamy K, Delproposto JE, Kubish GM, Tinberg CE, Khare SD, Dou J, Doyle L, Stuckey JA, Baker D, Carlson HA. CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schrödinger L. The PyMOL Molecular Graphics System, Version 1.7. 4. Schrödinger, LLC; [Google Scholar]
- [32].Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model. 2010;50:572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tosco P, Balle T, Shiri F. Open3DALIGN: an open-source software aimed at unsupervised ligand alignment. J Comput Aided Mol Des. 2011;25:777–783. doi: 10.1007/s10822-011-9462-9. [DOI] [PubMed] [Google Scholar]
- [35].Chen X, Liu M, Gilson MK. BindingDB: a web-accessible molecular recognition database. Comb Chem High Throughput Screen. 2001;4:719–725. doi: 10.2174/1386207013330670. [DOI] [PubMed] [Google Scholar]
- [36].Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases) Proteins. 2005;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- [37].Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- [38].Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tosco P, Balle T. Open3DQSAR: a new open-source software aimed at high-throughput chemometric analysis of molecular interaction fields. J Mol Model. 2011;17:201–208. doi: 10.1007/s00894-010-0684-x. [DOI] [PubMed] [Google Scholar]
- [40].Ndubaku CO, Crawford TD, Chen H, Boggs JW, Drobnick J, Harris SF, Jesudason R, McNamara E, Nonomiya J, Sambrone A, Schmidt S, Smyczek T, Vitorino P, Wang L, Wu P, Yeung S, Chen J, Chen K, Ding CZ, Wang T, Xu Z, Gould SE, Murray LJ, Ye W. Structure-Based Design of GNE-495, a Potent and Selective MAP4K4 Inhibitor with Efficacy in Retinal Angiogenesis. ACS Med Chem Lett. 2015;6:913–918. doi: 10.1021/acsmedchemlett.5b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schroder P, Forster T, Kleine S, Becker C, Richters A, Ziegler S, Rauh D, Kumar K, Waldmann H. Neuritogenic militarinone-inspired 4-hydroxypyridones target the stress pathway kinase MAP4K4. Angew Chem Int Ed Engl. 2015;54:12398–12403. doi: 10.1002/anie.201501515. [DOI] [PubMed] [Google Scholar]
- [42].Patel RY, Doerksen RJ. Protein kinase-inhibitor database: structural variability of and inhibitor interactions with the protein kinase P-loop. J Proteome Res. 2010;9:4433–4442. doi: 10.1021/pr100662s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.