Abstract
Mesenchymal stromal cells (MSCs) are strongly immunosuppressive via producing nitric oxide (NO) and known to migrate into tumor sites to promote tumor growth, but the underlying mechanisms remain largely elusive. Here, we found that interferon alpha (IFNα)-secreting MSCs showed more dramatic inhibition effect on tumor progression than that of IFNα alone. Interestingly, IFNα-primed MSCs could also effectively suppress tumor growth. Mechanistically, we demonstrated that both IFNα and IFNβ (type I IFNs) reversed the immunosuppressive effect of MSCs on splenocyte proliferation. This effect of type I IFNs was exerted through inhibiting inducible NO synthase (iNOS) expression in IFNγ and TNFα-stimulated MSCs. Notably, only NO production was inhibited by IFNα production of other cytokines or chemokines tested was not suppressed. Furthermore, IFNα promoted the switch from signal transducer and activator of transcription 1 (Stat1) homodimers to Stat1-Stat2 heterodimers. Studies using the luciferase reporter system and chromatin immunoprecipitation assay revealed that IFNα suppressed iNOS transcription through inhibiting the binding of Stat1 to iNOS promoter. Therefore, the synergistic anti-tumor effects of type I IFNs and MSCs were achieved by inhibiting NO production. This study provides essential information for understanding the mechanisms of MSC-mediated immunosuppression and for the development of better clinical strategies using IFNs and MSCs for cancer immunotherapy.
Introduction
Interferons (IFNs) are a family of cytokines widely expressed by host cells in response to viral infections.1, 2, 3 On the basis of their structures and functions, they are classified into two main types: type I IFNs (for example, α, β, ɛ, κ, ω and δ) and type II IFN (only IFNγ).1 In addition to controlling viral infections, some type I IFNs have been used in clinical settings for treating leukemia and melanoma;4 however, their application has been limited due to their short half-life in circulation and severe side effects induced by high dosages. To overcome these limitations, various efforts have been made to find delivery vehicles that allow specific tumor targeting and controlled release strategies.
Mesenchymal stromal cells (MSCs), a heterogeneous cell population originally identified from bone marrow, are believed to be a promising stem cell population for clinical applications on account of their differentiation potential and their powerful immunosuppressive capacities. MSCs can be strongly immunosuppressive in the presence of IFNγ and TNFα5 however, the immunosuppressive effect of MSCs is plastic, depending on the tissue microenvironmental inflammation status. Our previous studies showed that following high dosages of inflammatory cytokines, mouse MSCs were immunosuppressive by producing large amount of nitric oxide (NO) and chemokines, which attract immune cells to the vicinity of MSCs. When exposed to low levels of inflammatory cytokines, MSCs failed to suppress immune responses due to insufficient NO production. However, the low levels of chemokines produced under these conditions actually enhanced immune responses through recruitment of immune cells.6 MSCs also exhibit differential responses to various inflammatory cytokines; for example, IL-17A enhances MSC-induced immunosuppression, while TGFβ reverses it.7, 8, 9, 10 In fact, in the inflammatory sites, the amount of many cytokines varies and thus further efforts are needed to define how different inflammatory cytokines regulate the immunosuppressive properties of MSCs.
MSCs can specifically migrate to inflammatory sites, such as wounds and tumors, where a variety of inflammatory cytokines exist.11, 12 MSCs from bone marrow have been shown to be an important component of the tumor microenvironment, assisting tumor escape from immunosurveillance.12 Taking advantage of their tropism for inflammatory sites, MSCs engineered to secrete IFNα or IFNβ have been employed to deliver IFNs to the tumor site.5, 13, 14 Owing to their continuous release of IFNs, these MSCs exhibited a dramatic anti-tumor effect, in an adaptive immunity-dependent manner.14 The interesting question is how type I IFNs affect the immunosuppressive property of MSCs, and whether type I IFN-secreting MSCs could have a direct role in modulating tumor growth through their immunosuppressive capacity, in addition to secreting IFNs.
In this study, we found that IFNα could not induce NO production in MSCs, even in the presence of TNFα. Unexpectedly, IFNα reversed the immunosuppressive effects of MSCs induced by IFNγ and TNFα. Further studies showed that in MSCs, IFNα decreased inducible NO synthase (iNOS) expression via promoting the switch from signal transducer and activator of transcription 1 (Stat1) homodimers to Stat1-Stat2 heterodimers and inhibiting the binding of Stat1 to iNOS promoter. On the other hand, IFNα did not affect chemokine expression in inflammatory cytokine-activated MSCs. Although MSCs alone have a little promotion on tumor growth, IFNα-secreting MSCs dramatically inhibited tumor growth, even more dramatically than high dose of recombinant IFNα, an effect that was exerted through inhibiting iNOS expression. Therefore, our study revealed the effects of IFNα on the immunosuppressive property of MSCs, providing important new concepts for designing better clinical protocols to regulate the immune response to tumors using MSCs.
Results
IFNα acts synergistically with MSCs to inhibit tumor growth
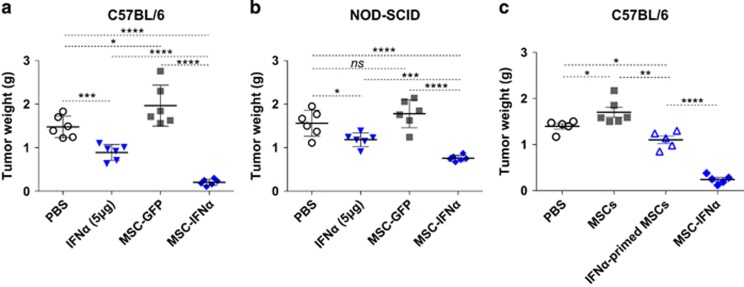
IFNα/β-secreting MSCs have been shown to be effective in treating several mouse tumor types.5, 13, 14 To demonstrate the detailed anti-tumor mechanism, we studied the effects of IFNα-secreting MSCs (MSC-IFNα) in the B16 mouse melanoma model. C57BL/6 mice were inoculated intramuscularly with a mixture of B16F0 cells with MSC-IFNα or MSC-GFP control. Previous studies of our laboratory have demonstrated that in the presence of inflammatory cytokines, including IFNγ and TNFα, in tumor microenvironment, MSCs become strongly immunosuppressive by releasing large amounts of NO.6, 15 As expected, MSC-GFP exhibited only a slight promotion in tumor growth (Figure 1a). However, this minor tumor promoting effect of MSC-GFP was diminished in NOD-SCID mice (Figure 1b). This result indicates that the host immune system is required for the observed tumor promoting effect of MSCs. On the other hand, we found that MSC-IFNα dramatically inhibited tumor growth, while IFNα protein, even at a high dosage (5 μg), inhibited tumor growth to a much lesser extent (Figure 1a). It suggests that, in addition to the direct anti-tumor effect of IFNα, MSCs also significantly contributed to the anti-tumor effect in the presence of IFNα. To directly define the anti-tumor effect of MSCs in the presence of IFNα, we primed MSCs with recombinant IFNα for 24 h. Cytokines were washed away before MSCs were intramuscularly co-injected with B16F0 melanoma cells. Interestingly, MSCs primed with IFNα also significantly inhibited tumor growth (Figure 1c). Therefore, IFNα and MSCs act in concert to inhibit tumor growth.
Figure 1.
IFNα and MSCs synergistically inhibit tumor growth. (a, b) B16F0 melanoma cells (1 × 106/25 μl) with or without IFNα (5 μg), MSC-GFP or MSC-IFNα (1 × 106/25 μl) were inoculated into C57BL/6 mice (a) or NOD-SCID mice (b) intramuscularly. After 12 days, tumors were excised and weighed. (c) MSCs were primed with IFNα for 24 h, and then cytokines were washed away. C57BL/6 mice were intramuscularly co-injected with IFNα-primed MSCs (1 × 106/thigh) and B16F0 melanoma cells (1 × 106/thigh). After 14 days, mice were killed and tumors were excised and weighed. All values represent means±s.d. Experiments were repeated at least twice. ns, not significant; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Distinct effect of IFNα and IFNγ in inducing NO production in MSCs
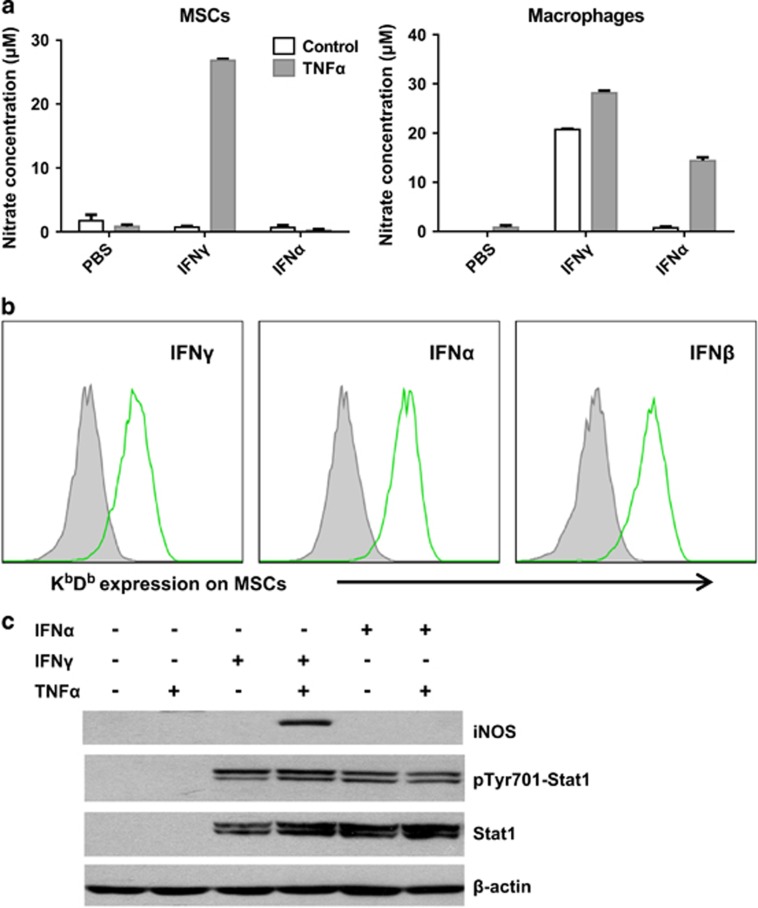
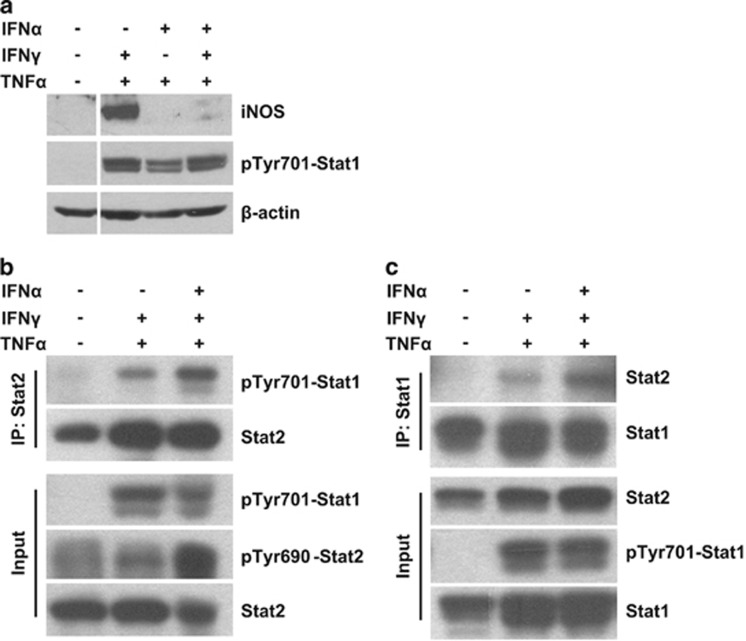
Our previous studies have shown that NO is the effector molecule that mediates the immunosuppressive property of mouse MSCs in the presence of IFNγ and TNFα.5 As IFNα shares the main components of its signaling pathway with IFNγ,2 we hypothesized that IFNα could also enable the immunosuppressive properties of MSCs. We thus checked whether IFNα could induce NO production by MSCs in combination with TNFα. Unexpectedly, we found that IFNα could not replace IFNγ in inducing NO production by MSCs (Figure 2a, left panel), even though, both IFNα and IFNγ could upregulate the expression of MHC class I molecules KbDb in MSCs (Figure 2b). Notably, both IFNα and IFNγ induced NO production in the presence of TNFα in bone marrow-derived macrophages (Figure 2a, right panel). To verify that both IFN types signal properly in MSCs, we also checked the expression and activation of Stat1, the common downstream transcription factor of the IFN signaling pathway. As shown in Figure 2c, IFNα increased and activated Stat1 to the same level as IFNγ. We also examined the expression of iNOS, which converts l-arginine into NO. Consistent with the absence of NO, iNOS protein was also not induced in MSCs upon stimulation with IFNα and TNFα (Figure 2c). Thus, IFNα did not induce an immunosuppressive effect in MSCs.
Figure 2.
IFNα could not induce NO production by MSCs. (a) MSCs and bone marrow-derived macrophages were cultured in various combinations of TNFα (10 ng/ml), IFNγ (10 ng/ml) or IFNα (2500 U/ml) for 24 h. The supernatants were collected and nitrate concentration was determined by a modified Griess reagent. Values are means±s.d. of four wells from a representative of three independent experiments. (b) MSCs were stimulated with IFNγ (10 ng/ml), IFNα (2500 U/ml) or IFNβ (2500 U/ml) for 12 h. KbDb expression was detected by flow-cytometry analysis. (c) MSCs were cultured as described in (a). The expression of Stat1, pTyr701-Stat1 and iNOS was examined by western blotting analysis. Experiments in (b) and (c) were repeated at least twice.
Type I IFNs reverse the immunosuppressive effect of MSCs
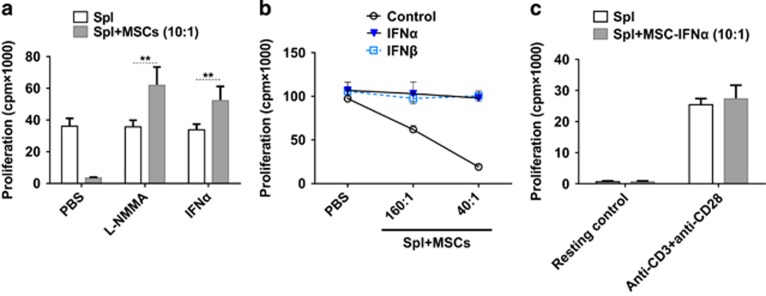
To investigate the effect of type I IFNs on cytokine-induced, MSC-mediated immunosuppression, recombinant IFNα was added to a coculture system of MSCs with splenocytes activated by anti-CD3 and anti-CD28. Surprisingly, the addition of IFNα completely reversed the immunosuppressive effect of MSCs on activated splenocytes (Figure 3a). It is noteworthy that the proliferation of activated splenocytes was not affected by IFNα, indicating the reversion of splenocyte proliferation in coculture system was not due to changes in T cells but through modulating the immunosuppressive effect of MSCs. IFNβ, another member of type I IFNs, showed a similar effect as IFNα (Figure 3b). To verify the effect of type I IFNs on MSC-mediated immunosuppression, we further tested the immunosuppressive effects of IFNα-secreting MSCs.14 MSC-IFNα cells were cocultured with resting or activated splenocytes. Interestingly, unlike wild-type MSCs in Figure 3a, MSC-IFNα lost the ability to inhibit the proliferation of activated splenocytes (Figure 3c). These results showed that the immunosuppressive effect of MSCs could be abolished by IFNα.
Figure 3.
Type I IFNs reverse the immunosuppression of MSCs. (a) MSCs were cocultured with fresh splenocytes plus anti-CD3 and anti-CD28 with or without l-NMMA (1 mm) or IFNα (2500 U/ml) for 48 h. Cell proliferation was evaluated by 3H-Tdr incorporation assay. (b, c) MSCs were plated at different concentrations and coculture with fresh splenocytes (1 × 106/well) plus anti-CD3 and anti-CD28. Cell proliferation was assayed after 48 h. Proliferation values are means±s.d. of four wells from a representative of two independent experiments. **P<0.01.
Differentiated MSCs are reported to have impaired immunosuppressive properties.16 As various cytokines in the microenvironment of the MSC niche are critical for MSC lineage commitment,17, 18 it is possible that the reversion of MSC immunosuppression by IFNα is due to inducing MSC differentiation. To test this possibility, we treated MSCs with IFNγ and TNFα or IFNγ, TNFα and IFNα for 24 h. Total RNA was collected and the expression of osteoblast marker genes or adipocyte marker genes was quantitated by real-time PCR. We found that the expression levels of osteoblast markers alpha-1 type I collagen (COL1A1), Osterix, Runx2 and adipocyte markers C/EBPβ, Leptin and Adiponectin were not affected by IFNα (Supplementary Figure S1A). To further confirm this observation, we stained osteoblasts or adipocytes with Alizarin Red S or Oil Red O after treated with IFNα for 72 h. Our results showed that IFNα alone or together with IFNγ and TNFα did not significantly promote osteogenic differentiation or adipogenic differentiation of MSCs at 72 h (Supplementary Figure S1B). Therefore, lineage differentiation was not involved in the reversion of MSC immunosuppression by IFNα.
IFNα inhibits IFNγ and TNFα-induced iNOS expression in MSCs
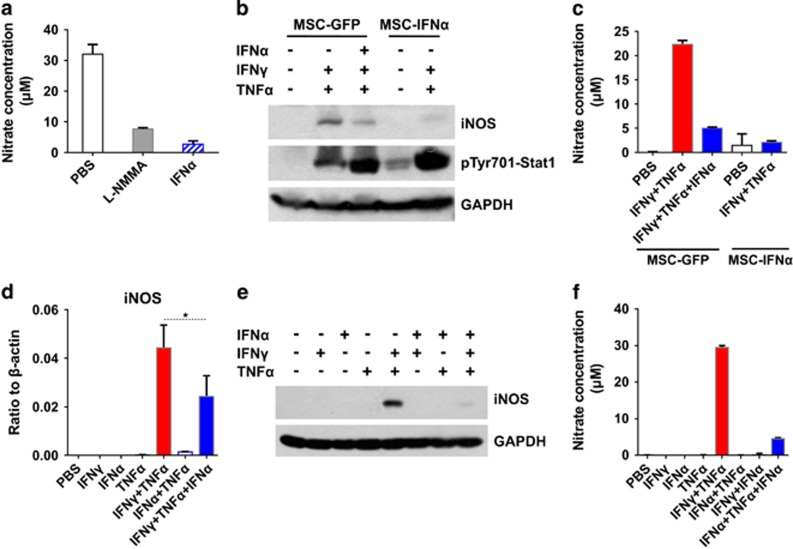
As NO is the key effector molecule of mouse MSC-mediated immunosuppression,5 we examined the production of NO by MSCs in the supernatant of the coculture system by quantifying nitrate concentration. We found that IFNα effectively inhibited NO production (Figure 4a). To test whether MSC-IFNα cells are responsive to NO induction, MSC-IFNα was cultured with IFNγ and TNFα and total protein and supernatant were collected. Compared with MSC-GFP, MSC-IFNα expressed significantly less iNOS and produced less NO in the presence of IFNγ and TNFα (Figures 4b and c). On the contrary, iNOS and NO were dramatically reduced in MSC-GFP when stimulated with IFNα in addition to IFNγ and TNFα (Figures 4b and c). To verify these results, recombinant IFNα was included in the culture medium in addition to IFNγ and TNFα. The addition of IFNα dramatically inhibited the expression of iNOS at the mRNA level and almost completely at the protein level (Figures 4d–f). We also determined whether IFNα inhibits iNOS expression by bone marrow-derived macrophages. We found that IFNα induced iNOS expression by macrophages in the presence of TNFα (Supplementary Figure S2), which was consistent with the observation in Figure 2a and previous reports.19 Furthermore, IFNα did not inhibit IFNγ and TNFα-induced iNOS expression by macrophages (Supplementary Figure S2). These results demonstrated that IFNα inhibited IFNγ and TNFα-induced iNOS expression in MSCs but not in macrophages.
Figure 4.
IFNα inhibits NO production by MSCs. (a) Supernatants from Figure 3a were collected for determining the concentration of nitrate. (b, c) MSC-IFNα and MSC-GFP were stimulated with combinations of IFNγ, TNFα and IFNα for 24 h. Total protein and supernatant were collected. The expression of pTyr701-Stat1 and iNOS was examined by western blotting analysis (b). Nitrate concentration was determined by Griess assay (c). (d–f) MSCs were stimulated with various combinations of IFNγ (10 ng/ml), TNFα (10 ng/ml) and IFNα (2500 U/ml) for 24 h. Total RNA (d, n=3), protein (e) and supernatants (f) were collected. Nitrate concentration and iNOS expression were determined. Nitrate values are means±s.d. of four wells from a representative of at least three independent experiments. *P<0.05.
IFNα does not affect the production of cytokines or chemokines by MSCs activated by IFNγ and TNFα
As IFNα can inhibit inflammatory cytokine-induced NO production in MSCs, we investigated whether it can also affect the expression of cytokines and chemokines. This is important because we have reported that chemokines are critically involved in MSC-mediated immunosuppression through recruiting immune cells to the vicinity of MSCs, so that labile NO could effectively inhibit the proliferation and functions of the immune cells.5 We employed microbead-based multiplex assay as previously described.20 Surprisingly, none of the cytokines or chemokines that are induced by IFNγ and TNFα was affected by IFNα, except some upregulation of IL-6 (Supplementary Figure S3). Thus, the effect of IFNα on MSCs in an inflammatory microenvironment is exerted through downregulation of NO production.
IFNα promotes the switch from Stat1 homodimers to Stat1-Stat2 heterodimers
Type I and type II IFNs have been shown to have many overlapping biological functions such as their antiviral activities and induction of the expression of major histocompatibility molecules.3 Interestingly, a recent study demonstrated that IFNα and IFNβ inhibit IFNγ-mediated macrophage activation through downregulating IFNγ receptors.21 Although this is unlikely to be true for MSCs, considering our observation that IFNα could increase the expression of IL-6 induced by IFNγ and TNFα, we performed a microarray analysis. Our data showed that the expression of IFNγ receptors was not affected by IFNα in MSCs, with or without stimulation of IFNγ and TNFα (Supplementary Figure S4A). Consistent with previous reports, the expression of MHC class I molecules H2-D1 and H2-K1 could be further increased by IFNα. In addition, no changes in IFNγ receptors were observed by flow-cytometric analysis (Supplementary Figure S4B). Therefore, the anti-tumor effect of type I IFNs is not exerted through altering the expression of IFNγ receptors.
It has been demonstrated that iNOS expression is regulated by the Stat1 and NF-κB signaling pathways.19, 22, 23 One possible mechanism of IFNα-mediated inhibition of iNOS expression is by affecting TNFα-mediated NF-κB signaling. As the phosphorylation and degradation of IκBα is required for NF-κB nuclear translocation,24 we examined the expression pattern and phosphorylation level of IκBα by western blotting analysis. We found that the IκBα levels and phosphorylation in IFNγ and TNFα-stimulated MSCs were not affected by IFNα (Supplementary Figure S5A). We also examined the nuclear distribution of NF-κB and found that addition of IFNα did not change the distribution of phosphorylated p65 in nuclei after IFNγ and TNFα stimulation (Supplementary Figure S5B). In addition, we further assessed the DNA binding activity of NF-κB using sequence-specific oligonucleotide agarose beads.25 Again, IFNα did not significantly affect the binding activity of p65 (Supplementary Figure S5C). Therefore, the NF-κB signaling pathway is not involved in IFNα-mediated inhibition of iNOS expression.
The other mechanism through which IFNα inhibits iNOS expression is through Stat1 signaling. We first examined the phosphorylation of Stat1 but found it also does not decrease upon IFNα treatment in the presence of IFNγ and TNFα (Figure 5a). IFNγ induces tyrosine phosphorylation of Stat1 and promotes the formation of Stat1 homodimers (gamma-activated factor).2 It has been shown that the binding of Stat1 homodimers to gamma-activated sequences (GAS) in the iNOS gene promoter is necessary for iNOS expression.23, 26 However, IFNα mainly promotes the formation of Stat1-Stat2 heterodimers (interact with IRF9 to form heterotrimeric complex interferon-stimulated gene factor 3, or ISGF3), and forms Stat1 homodimers to a minor extent.2, 27 ISGF3 initiates transcription by specifically binding to IFN-stimulated response element. It is possible that IFNα-induced Stat1 heterodimers compete with Stat1 homodimers for phosphorylated Stat1 and inhibit the Stat1 binding to GAS sites of iNOS promoter.28 Therefore, we checked the effect of IFNα on the formation of Stat1-Stat2 heterodimers. Co-immunoprecipitation results showed that IFNα increased the formation of Stat1-Stat2 heterodimers (Figures 5b and c). As the level of Stat1 phosphorylation was not significantly altered by IFNα, it indicated that the formation of Stat1 homodimers was inhibited after IFNα treatment. These data demonstrated that IFNα-induced formation of Stat1-Stat2 heterodimers competes with the formation of Stat1 homodimers and may affect the binding activity of Stat1 homodimers to iNOS promoter.
Figure 5.
IFNα promotes the switch from Stat1 homodimers to Stat1-Stat2 heterodimers. (a) MSCs were stimulated with combinations of IFNγ, TNFα and IFNα for 24 h. Total protein was collected and the expression of pTyr701-Stat1 and iNOS was examined by western blotting analysis. (b) MSCs were treated as in (a) for 6 h and total protein was collected. Stat2 antibody was used in the co-immunoprecipitation assay to determine the IFNα-induced Stat1-Stat2 heterodimer formation. (c) MSCs were treated as in (b) for 6 h and Stat1 antibody was used to perform the co-immunoprecipitation (Co-IP) assay. All experiments were repeated twice.
IFNα inhibits NO production via decreasing the binding of Stat1 to iNOS promoter
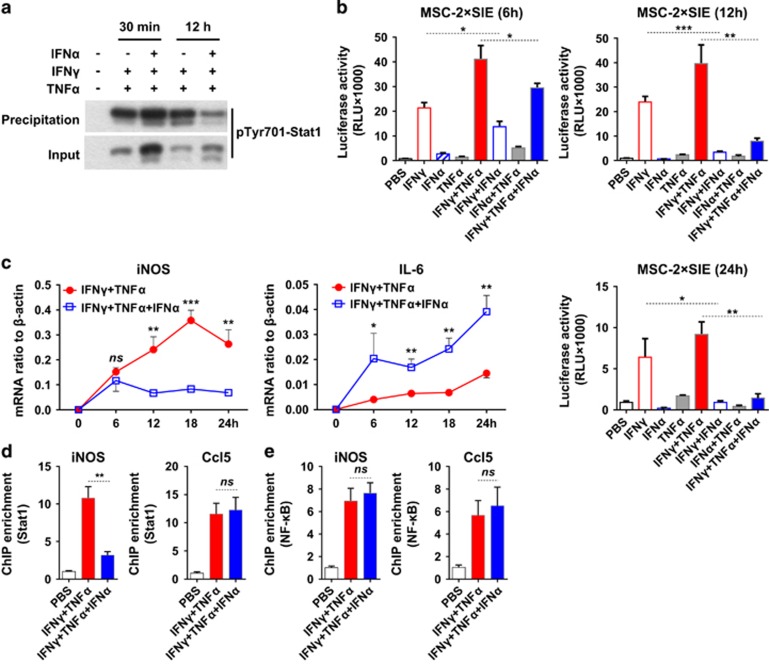
To further define the mechanism of IFNα-induced iNOS inhibition, we detected the DNA binding activity of Stat1 homodimers by GAS-containing oligonucleotide agarose beads. At different time points, we found that the DNA binding activity of Stat1 homodimers was induced by IFNγ and TNFα however, further addition of IFNα significantly inhibited the binding activity (Figure 6a). This observation could be explained by the decreased formation of Stat1 homodimers after IFNα treatment. To further verify this observation, we employed a luciferase reporter plasmid that contains GAS sites within the SIE (c-sis inducible element) as reported.25 MSCs transfected with SIE-luciferase reporter were cultured with various combinations of IFNγ, TNFα or IFNα. The transcription activity of Stat1 was quantified by luciferase assay at 6, 12 and 24 h. Consistent with the results of the GAS-containing oligonucleotide agarose beads affinity precipitation assay, the IFNγ-induced luciferase activity was slightly inhibited by IFNα starting at 6 h and significantly inhibited at 12 and 24 h (Figure 6b). This IFNα-mediated inhibition of luciferase activity was also examined for correlation with iNOS expression. We found that the inhibition in iNOS mRNA expression also started at 6 h. However, consistent with results in Supplementary Figure S3, the expression of IL-6 was not inhibited by IFNα, indicating that IFNα-induced iNOS inhibition is gene-specific rather than a systemic effect (Figure 6c).
Figure 6.
IFNα inhibits NO production via decreasing the binding activity of Stat1 homodimers to iNOS promoter. (a) MSCs were stimulated as previously for 30 min or 12 h. Total proteins were collected. Stat1 was precipitated by sequence-specific oligonucleotide agarose beads. The precipitants were determined by western blotting analysis. Total proteins were inputs. (b) MSCs were transfected with 2 × SIE-luciferase plasmids (Stat1 transcription activity reporter plasmid). Transfected MSCs were stimulated with various combinations of IFNγ, TNFα and IFNα for indicated time and analyzed by luciferase assay kit. Luciferase activity was measured as relative light units (RLU). (c) MSCs were treated with IFNγ and TNFα or IFNγ, TNFα and IFNα for indicated time and total RNA was collected. The expression pattern of IL-6 and iNOS was determined by real-time PCR. (d, e) MSCs were treated with IFNγ and TNFα or IFNγ, TNFα and IFNα for 12 h and chromatin was immunoprecipitated for ChIP assay following the manufacturer's instructions. The enrichment of Stat1 homodimers (d) and NF-κB (e) binding to respective promoters was determined by real-time PCR (relative to PBS control). Values are mean±s.d. of three replicates. All experiments were repeated at least twice. ns, not significant; *P<0.05; **P<0.01; ***P<0.001.
To provide direct evidence that IFNα inhibits the binding of Stat1 homodimers to the GAS sites of iNOS promoter, we performed the chromatin immunoprecipitation (ChIP) assay. We found that the binding of Stat1 to iNOS promoter was significantly decreased upon the addition of IFNα, while its binding activity to Ccl5, another gene known to be activated by both IFNγ and IFNα, was not affected (Figure 6d). There only exists GAS sites but not IFN-stimulated response element sites in the iNOS promoter.23, 26 However, there are both GAS sites and IFN-stimulated response element sites in the Ccl5 promoter.27, 29 We have shown that IFNα dominantly induces Stat1-Stat2 heterodimers, which compete with IFNγ-induced Stat1 homodimers for phosphorylated Stat1 and suppress the transcription activity of GAS-containing promoters. The difference between iNOS and Ccl5 of the transcription factor binding sites in promoters may explain the variation of regulatory effects by IFNα. On the other hand, the binding of NF-κB to the promoters of either iNOS or Ccl5 was not affected by IFNα treatment (Figure 6e). Therefore, IFNα-induced switch from Stat1 homodimers to Stat1-Stat2 heterodimers explains the impaired binding of Stat1 to iNOS promoter and the inhibition of NO production in MSCs.
Discussion
MSCs hold great promise for clinical applications in the treatment of various diseases based on their multi-lineage differentiation potential and immunosuppressive properties. MSCs have also been reported to migrate to tumors. Owing to this tumor tropism property, MSCs have been engineered to express anti-tumor factors (including type I IFN, TNF-related apoptosis-inducing ligand and interleukin-12), and engineered MSCs have shown dramatic anti-tumor effects.30, 31, 32 However, in the tumor microenvironment, the potent immunosuppressive effects of MSCs also promote tumor progression. Therefore, detailed mechanistic investigations, especially on the immunosuppressive property of MSCs, are expected to accelerate the development of these novel anti-tumor strategies. In this study, we examined the mechanism of IFNα-mediated anti-tumor activities. We found that IFNα-secreting MSCs could reverse the immunosuppressive effect of MSCs through inhibiting Stat1 binding to the iNOS promoter. Thus, our study provides novel insights of the anti-tumor activity of IFNα-secreting MSCs.
Our previous study demonstrated that IFNγ is required for the production of NO by MSCs.5 As it has been shown that the NF-κB and Stat1 signaling pathways are required for the induction of iNOS expression, we performed detailed analysis of the effect of IFNα on these signaling processes. We found that activation of the NF-κB pathway, as monitored by either IκBα or phosphor-p65 levels, was not affected by IFNα. Although Stat1 phosphorylation was not inhibited by IFNα treatment, the binding of Stat1 homodimers to iNOS promoter was inhibited. Stat1 activates iNOS transcription through acting as homodimers. Importantly, IFNα mainly promotes the formation of Stat1-Stat2 heterodimers, and forms Stat1 homodimers to a minor extent. Therefore, IFNα-induced Stat1-Stat2 heterodimers compete with Stat1 homodimers and inhibit the binding activity of Stat1 to iNOS promoter in MSCs. On the other hand, the possibility that an unidentified gene product was involved in IFNα-induced NO inhibition still exist. It may affect the binding activity of Stat1 homodimers to GAS sites and inhibit iNOS transcription. In addition, modifications of Stat1, such as acetylation33, 34, 35 and methylation,36 may also regulate the binding activity of Stat1 to iNOS promoter. Therefore, further studies are needed to decipher the inter-regulation of type I and type II IFN signaling in MSCs.
The cell-type specificity of IFNα-mediated iNOS inhibition is also very interesting. In addition to Stat1 homodimers and NF-κB, other transcription factors have also been reported to be involved in the regulation of iNOS transcription, such as AP-1 and C/EBPβ.37, 38 Therefore, the different activation status of related signaling pathways between MSCs and macrophages can be a reason for the cell type-specific regulation of iNOS by IFNα. Indeed, our data showed that IFNγ alone is sufficient to induce iNOS expression in bone marrow-derived macrophages. Although IFNα alone did not effectively induce iNOS expression in macrophages, it gained this property in the presence of TNFα (Figure 2a and Supplementary Figure S2). These data clearly demonstrated that the activation status of transcription factors required for iNOS transcription is different between MSCs and macrophages. Notably, type I IFNs have been reported to activate p38 MAP kinase, which further activates AP-1.39, 40, 41, 42 It is possible that the expression or regulation of AP-1 is different in different types of cells, which result in the cell type-specific control of iNOS transcription. Similarly, the status of C/EBPβ and other transcription factors involved in the regulation of iNOS expression may also be different between MSCs and macrophages. Moreover, different epigenetic modifications of iNOS promoter between MSCs and macrophages may also be a reason for its cell type-specific regulation.43, 44, 45 More efforts are needed to address the cell type-specific regulation of type I and type II IFNs.
For the last decade, the immunosuppressive properties of MSCs have attracted intensive studies due to their potential clinical applications for immune-related diseases. MSCs have been showed to possess significant therapeutic effects in numerous animal disease models (including graft-versus-host disease, experimental autoimmune encephalomyelitis, inflammatory bowel disease and diabetes) and in clinical settings (including graft-versus-host disease, Crohn's disease and systemic lupus erythematosus), mediated through regulation of immune responses.46 Our previous studies showed that NO and chemokines are key factors for the immunosuppressive effect of mouse MSCs. The amount of NO is critical for the immunosuppressive capacity of MSCs.8, 15 Immunosuppressive capabilities of tumor-infiltrated MSCs counteract anti-tumor immunity within the tumor microenvironment and thus promote tumor growth.12 Studies have shown that MSCs primed with inflammatory cytokines lead to a strong therapeutic effect on ConA-induced acute hepatitis.8 On the other hand, our laboratory has demonstrated that iNOS-deficient MSCs could inhibit tumor growth through promoting immunity in a chemokine-dependent manner.6 We also found that IFNα inhibits NO production, but not that of chemokines we tested (Figures 4e and f and Supplementary Figure S3). Thus, not surprisingly, IFNα enhanced splenocyte proliferation (Figures 3a and b) and IFNα-primed MSCs inhibited tumor growth significantly (Figure 1c). Therefore, in the presence of IFNα, the absence of NO allows the chemokine-producing MSCs to enhance immune responses and exert anti-tumor effects.
Their tumor tropism, low immunogenicity and easy expansion make MSCs an ideal delivery vehicle for anti-tumor factors.47 However, the exact effect of MSCs on tumor growth is still not fully understood. Some studies demonstrated that MSCs promote tumor growth mainly through their immunosuppressive effect;12, 48, 49, 50 while others showed that MSCs showed no effect on tumor growth.51, 52, 53, 54 These variations in the outcomes could be due to tumor models employed, source of MSCs, MSC administration routes and schedules, and the dose of MSCs given. In this study, we co-injected IFNα-secreting MSCs together with B16F0 melanoma cells and demonstrated a strong anti-tumor effect. We also administered IFNα-primed MSCs to B16 tumor bearing mice and found that these primed MSCs lost NO producing ability and inhibited tumor growth significantly. To better mimic the properties of tumor microenvironment, lymphoma-derived MSCs12 were also studied, and we showed that IFNα could also inhibit NO production by lymphoma-derived MSCs (Supplementary Figure S6). Our study indicates that IFNα can exert its anti-tumor effects through altering the immune status of the tumor microenvironment.
In summary, we demonstrated that, in addition to the established anti-tumor effect, IFNα could also promote anti-tumor immunity through abolishing immunosuppressive effect of MSCs. Additionally, MSCs can also be used as a delivery vehicle to provide sustained IFNα to the tumor microenvironment for tumor therapy. Further investigations to finetune this system will lead to better clinical strategies for MSC-based tumor therapy.
Materials and methods
Mice
C57BL/6 and NOD-SCID mice were purchased from the SLAC Laboratory Animal of Chinese Academy of Science (Shanghai, China). Mice were housed in specific pathogen-free facility of Shanghai Jiao Tong University School of Medicine. Animals were matched for age and gender in each experiment. All studies were approved by the Institutional Animal Care and Use Committee of the Institute of Health Sciences, Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences.
Reagents
The following antibodies were used in flow cytometry: PE anti-mouse H-2Kb/H-2Db, PE anti-mouse IFNγR β chain (Biolegend, San Diego, CA, USA) and PE anti-mouse CD119 (IFNγ Receptor 1) (eBioscience, San Diego, CA, USA). Antibodies used in western blotting analysis were iNOS, pTyr701-Stat1, total Stat1, total Stat2, pSer536-p65, total p65 pSer32-IκBα, total IκBα, β-actin, GAPDH (Cell Signaling Technology, Danvers, MA, USA); pTyr690-Stat2 (Abcam, Cambridge, MA, USA) and LaminB (Epitomics, Burlingame, CA, USA). Recombinant mouse IFNα, IFNβ, IFNγ and TNFα were from R&D Systems (Minneapolis, MN, USA). l-NMMA was from Sigma-Aldrich (St Louis, MO, USA).
Cells
MSCs were derived from tibia and femur bone marrow of 6- to 8-week-old mice according to a protocol previously described in our laboratory.5, 55 MSC-GFP and MSC-IFNα cells were derived by transducing with lentivirus encoding green fluorescent protein alone or together with IFNα as previously described.14 The production and function of IFNα was examined by ELISA or detecting H-2Kb expression in MSCs.14
Proliferation assay
Fresh splenocytes were derived from 6- to 8-week-old C57BL/6 mice. MSCs were cocultured with splenocytes activated with anti-CD3 and anti-CD28 in RPMI-1640 medium. Cell proliferation was assayed by uptake of 3H-thymidine (3H-Tdr; Shanghai Institute of Applied Physics, Chinese Academy of Sciences, Shanghai, China). 3H-thymidine (0.5 mCi) was added to examine the cell proliferation at 42 h. Six hours later, the coculture was terminated by freezing. Incorporated 3H-Tdr was determined using a Wallac Microbeta scintillation counter (Perkin-Elmer, Waltham, MA, USA).
Detection of cytokines and NO in supernatants
The levels of cytokine and chemokines in supernatants were determined by multiplexed bead immunoassay using the Luminex Technology (Bio-Plex, Bio-Rad Laboratories, Hercules, CA, USA). NO was detected using a modified Griess reagent (Sigma-Aldrich). Briefly, the mixture of supernatants (50 μl) and Griess reagent (50 μl) was incubated for 15 min in the dark at room temperature, the plate was read at 540 nm and nitrate concentrations were calculated.
Western blotting analysis
Total protein was extracted from the cell pellet with RIPA lysis buffer (Upstate, Charlottesville, VA, USA). Nuclear proteins were extracted by NE-PER Nuclear and Cytoplasmic Extraction Reagents from Thermo (Waltham, MA, USA). The proteins were boiled in SDS sample buffer for 10 min. The lysates were separated by polyacrylamide gel electrophoresis and transferred onto a 0.45-μm polyvinylidene fluoride blotting membrane (Whatman Inc., Clifton, NJ, USA). The membrane was then incubated at room temperature in a blocking solution composed of 5% skimmed milk powder dissolved in TBST (0.05% Tween-20, 10 mm Tris, pH 8.0 and 140 mm NaCl) for 1 h followed by incubation with the primary antibodies overnight at 4 °C. The membrane was washed three times in TBST (5 min each), then incubated with horseradish peroxidase-conjugated secondary antibody in the blocking solution. The blot was then exposed by ECL (Pierce, Rockford, IL, USA) after another three washes in TBST.
The binding activity of p65 and Stat1 homodimers was determined using NF-κB-specific or GAS-containing oligonucleotide agarose beads (Santa Cruz Biotechnology, SantaCruz, CA, USA). Total proteins were used for the precipitation assay according to the instruction.
For the co-immunoprecipitation assay, cells were lysed in lysis buffer containing Triton X-100, protease inhibitors, PMSF, Na3VO4 and NaF for 30 min on ice. Lysates were clarified by centrifugation at 14 000 g for 10 min. Supernatant was incubated with primary antibody with gentle rocking overnight at 4 °C. Protein A sepharose beads were added to the mixture and incubated with gentle rocking for additional 2 h at 4 °C. After washing five times with lysis buffer, beads were suspended and analyzed by western blotting analysis.
Mouse melanoma model
B16F0 mouse melanoma cells were expanded in complete Dulbecco's modified Eagle's medium (high glucose) in vitro. Each mouse was inoculated with B16F0 cells (1 × 106 in 25 μl PBS) intramuscularly on the left thigh, with or without co-injection of MSCs as indicated. Mice were observed daily and killed when tumor burden began to significantly affect mobility. The tumors were then excised and weighed.
Real-time PCR
Total RNA was isolated from cell pellets using an RNAprep pure Cell/Bacteria Kit (Tiangen Biotech, Beijing, China). First-strand cDNA synthesis was performed using the 1st strand cDNA Synthesizing Kit with random hexamer primers (Tiangen Biotech). Genes of interest were quantitated by real-time PCR. The mRNA levels of genes of interest were measured by real-time PCR (7900 HT by Applied Biosystems, Foster City, CA, USA) using SYBR Green Master Mix (TaKaRa Biotech, Dalian, China). Total amount of mRNA was normalized to endogenous β-actin mRNA. Sequences of PCR primer pairs were as follows: mouse IL-6, forward 5′-AGATAAGCTGGAGTCACAGAAGGAG-3′ and reverse 5′-CGCACTAGGTTTGCCGAGTAG-3′ mouse iNOS, forward 5′-CAGCTGGGCTGTACAAACCTT-3′ and reverse 5′-CATTGGAAGTGAAGCGTTTCG-3′ mouse β-actin, forward 5′-TTCCAGCCTTCCTTCTTGGG-3′ and reverse 5′-TGTTGGCATAGAGGTCTTTACGG-3′.
Flow-cytometry analysis
Cells were harvested and washed once with PBS. The cell pellets were then suspended in staining buffer (PBS, 3% FCS, 0.01% NaN3) at a concentration of 1 × 107 cells/ml. Cell suspension (100 μl) was incubated for 30 min on ice with either directly conjugated antibodies or biotinylated antibodies followed by streptavidin-PE for an additional 30 min on ice after washing. Cells were then washed with the staining buffer. The samples were subjected to flow-cytometric analysis using an FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA). FlowJo software was used for data analysis.
ChIP and real-time PCR detection
Chromatin from MSCs was crosslinked by formaldehyde treatment and immunoprecipitated using a Pierce Agarose ChIP kit (Thermo Fisher Scientific, Rockford, IL, USA). Purified chromatin (10 μg) was normalized to input and immunoprecipitated using 6 μg of NF-κB p65 antibody (Abcam), Stat1 antibody at 1:50 (Cell Signaling Technology) or 1 μl of rabbit IgG control. Immunoprecipitated DNA was subjected to quantitative PCR to determine the enrichment of Stat1 homodimers and NF-κB binding to respective promoters, and results were normalized to control group. Primers used for quantitative PCR were as follows: binding of Stat1 homodimers to mouse iNOS promoter, forward 5′-GGCACCATCTAACCTCAC-3′ and reverse 5′-CAGCACGTAGTCACTTCA-3′ NF-κB binding to mouse iNOS promoter, forward 5′- TGAGGATACACCACAGAGT-3′ and reverse 5′-GTGCAAGTTAGCTCATTCAT-3′ binding of Stat1 homodimers to mouse Ccl5 promoter, forward 5′-TATAGGGAGCCAGGGTAGCA-3′ and reverse 5′-GCAACAAGTGTTTGGTGTCTTT-3′ NF-κB binding to mouse Ccl5 promoter, forward 5′-AGCCAGGGTAGCAGAGGAA-3′ and reverse 5′-ATGACAGCAACAAGTGTTTGGT-3′.
Statistical analysis
The GraphPad Prism 6 software (GraphPad Software, Inc., San Diego, CA, USA) was used for the statistical analyses. Statistical significance was assessed by unpaired two-tailed Student's t-test.
Acknowledgments
We thank Ms Fengying Li and Ms Jingjing Li for technical assistances in breeding the mice; Ms Yanyan Han, Mr Jianchang Cao, Mr Shijia Wang, Ms Qing Li and Ms Chenxi Zhang for technical assistances and helpful discussion. This work was supported by grants from the Ministry of Science and Technology of China (2015CB964400, 2012CBA01303), Scientific Innovation Project of the Chinese Academy of Science (XDA01040110, XDA01040107), the External Cooperation Program of Bureau of International Cooperation, Chinese Academy of Sciences (GJHZ201307), the Programs of National Natural Science of China (81330046, 81171653, 31570877, 31570908), grants from the National Institutes of Health of the United States of America (GM866889) and a grant to the Child Health Institute from the Robert Wood Johnson Foundation (67038).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Frasca L, Lande R. Overlapping, additive and counterregulatory effects of type II and I interferons on myeloid dendritic cell functions. ScientificWorldJournal 2011; 11: 2071–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 2005; 5: 375–386. [DOI] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol 2004; 22: 929–979. [DOI] [PubMed] [Google Scholar]
- Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem 2007; 282: 20047–20051. [DOI] [PubMed] [Google Scholar]
- Ren G, Zhang L, Zhao X, Xu G, Zhang Y, Roberts AI et al. Mesenchymal stem cell-mediated immunosuppression occurs via concerted action of chemokines and nitric oxide. Cell Stem Cell 2008; 2: 141–150. [DOI] [PubMed] [Google Scholar]
- Li W, Ren G, Huang Y, Su J, Han Y, Li J et al. Mesenchymal stem cells: a double-edged sword in regulating immune responses. Cell Death Differ 2012; 19: 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yu P, Han X, Du L, Gan J, Wang Y et al. TGF-beta promotes immune responses in the presence of mesenchymal stem cells. J Immunol 2014; 192: 103–109. [DOI] [PubMed] [Google Scholar]
- Han X, Yang Q, Lin L, Xu C, Zheng C, Chen X et al. Interleukin-17 enhances immunosuppression by mesenchymal stem cells. Cell Death Differ 2014; 21: 1758–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Su J, Roberts AI, Shou P, Rabson AB, Ren G. How mesenchymal stem cells interact with tissue immune responses. Trends Immunol 2012; 33: 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen X, Cao W, Shi Y. Plasticity of mesenchymal stem cells in immunomodulation: pathological and therapeutic implications. Nat Immunol 2014; 15: 1009–1016. [DOI] [PubMed] [Google Scholar]
- Karp JM, Leng Teo GS. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell 2009; 4: 206–216. [DOI] [PubMed] [Google Scholar]
- Ren G, Zhao X, Wang Y, Zhang X, Chen X, Xu C et al. CCR2-dependent recruitment of macrophages by tumor-educated mesenchymal stromal cells promotes tumor development and is mimicked by TNFalpha. Cell Stem Cell 2012; 11: 812–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Kumar S, Chanda D, Chen J, Mountz JD, Ponnazhagan S. Therapeutic potential of mesenchymal stem cells producing interferon-alpha in a mouse melanoma lung metastasis model. Stem Cells 2008; 26: 2332–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Lin L, Cao G, Chen Q, Shou P, Huang Y et al. Interferon-alpha-secreting mesenchymal stem cells exert potent antitumor effect in vivo. Oncogene 2014; 33: 5047–5052. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yu P, Li W, Ren G, Roberts AI, Cao W et al. p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene 2014; 33: 3830–3838. [DOI] [PubMed] [Google Scholar]
- Liu H, Kemeny DM, Heng BC, Ouyang HW, Melendez AJ, Cao T. The immunogenicity and immunomodulatory function of osteogenic cells differentiated from mesenchymal stem cells. J Immunol 2006; 176: 2864–2871. [DOI] [PubMed] [Google Scholar]
- Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ 2016; doi:10.1038/cdd.2015.168. [DOI] [PMC free article] [PubMed]
- Chen Q, Shou P, Zhang L, Xu C, Zheng C, Han Y et al. An osteopontin-integrin interaction plays a critical role in directing adipogenesis and osteogenesis by mesenchymal stem cells. Stem Cells 2014; 32: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie QW, Whisnant R, Nathan C. Promoter of the mouse gene encoding calcium-independent nitric oxide synthase confers inducibility by interferon gamma and bacterial lipopolysaccharide. J Exp Med 1993; 177: 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Liu Y, Zhao X, Zhang J, Zheng B, Yuan ZR et al. Tumor resident mesenchymal stromal cells endow naive stromal cells with tumor-promoting properties. Oncogene 2014; 33: 4016–4020. [DOI] [PubMed] [Google Scholar]
- Rayamajhi M, Humann J, Penheiter K, Andreasen K, Lenz LL. Induction of IFN-alphabeta enables Listeria monocytogenes to suppress macrophage activation by IFN-gamma. J Exp Med 2010; 207: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefano D, Maiuri MC, Iovine B, Ialenti A, Bevilacqua MA, Carnuccio R. The role of NF-kappaB, IRF-1, and STAT-1alpha transcription factors in the iNOS gene induction by gliadin and IFN-gamma in RAW 264.7 macrophages. J Mol Med (Berl) 2006; 84: 65–74. [DOI] [PubMed] [Google Scholar]
- Gao J, Morrison DC, Parmely TJ, Russell SW, Murphy WJ. An interferon-gamma-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-gamma and lipopolysaccharide. J Biol Chem 1997; 272: 1226–1230. [DOI] [PubMed] [Google Scholar]
- Kretz-Remy C, Mehlen P, Mirault ME, Arrigo AP. Inhibition of I kappa B-alpha phosphorylation and degradation and subsequent NF-kappa B activation by glutathione peroxidase overexpression. J Cell Biol 1996; 133: 1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TR, Hong YK, Wang XD, Ling MY, Dragoi AM, Chung AS et al. SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem 2002; 277: 47572–47580. [DOI] [PubMed] [Google Scholar]
- Burke SJ, Updegraff BL, Bellich RM, Goff MR, Lu D, Minkin SC Jr et al. Regulation of iNOS gene transcription by IL-1beta and IFN-gamma requires a coactivator exchange mechanism. Mol Endocrinol 2013; 27: 1724–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begitt A, Droescher M, Meyer T, Schmid CD, Baker M, Antunes F et al. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat Immunol 2014; 15: 168–176. [DOI] [PubMed] [Google Scholar]
- Ghislain JJ, Wong T, Nguyen M, Fish EN. The interferon-inducible Stat2:Stat1 heterodimer preferentially binds in vitro to a consensus element found in the promoters of a subset of interferon-stimulated genes. J Interferon Cytokine Res 2001; 21: 379–388. [DOI] [PubMed] [Google Scholar]
- Kim MO, Suh HS, Brosnan CF, Lee SC. Regulation of RANTES/CCL5 expression in human astrocytes by interleukin-1 and interferon-beta. J Neurochem 2004; 90: 297–308. [DOI] [PubMed] [Google Scholar]
- Lee ST, Jang JH, Cheong JW, Kim JS, Maemg HY, Hahn JS et al. Treatment of high-risk acute myelogenous leukaemia by myeloablative chemoradiotherapy followed by co-infusion of T cell-depleted haematopoietic stem cells and culture-expanded marrow mesenchymal stem cells from a related donor with one fully mismatched human leucocyte antigen haplotype. Br J Haematol 2002; 118: 1128–1131. [DOI] [PubMed] [Google Scholar]
- Zou W, Zheng H, He TC, Chang J, Fu YX, Fan W. LIGHT delivery to tumors by mesenchymal stem cells mobilizes an effective antitumor immune response. Cancer Res 2012; 72: 2980–2989. [DOI] [PubMed] [Google Scholar]
- Elzaouk L, Moelling K, Pavlovic J. Anti-tumor activity of mesenchymal stem cells producing IL-12 in a mouse melanoma model. Exp Dermatol 2006; 15: 865–874. [DOI] [PubMed] [Google Scholar]
- Guo L, Guo H, Gao C, Mi Z, Russell WB, Kuo PC. Stat1 acetylation inhibits inducible nitric oxide synthase expression in interferon-gamma-treated RAW264.7 murine macrophages. Surgery 2007; 142: 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M, Ginter T, Brand P, Heinzel T, Kramer OH. Acetylation modulates the STAT signaling code. Cytokine Growth Factor Rev 2012; 23: 293–305. [DOI] [PubMed] [Google Scholar]
- Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev 2009; 23: 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell 2001; 104: 731–741. [DOI] [PubMed] [Google Scholar]
- Wang K, Brems JJ, Gamelli RL, Holterman AX. C/EBPalpha and C/EBPbeta binding proteins modulate hepatocyte apoptosis through iNOS signaling pathway. Biochim Biophys Acta 2011; 1813: 1395–1403. [DOI] [PubMed] [Google Scholar]
- Kleinert H, Wallerath T, Fritz G, Ihrig-Biedert I, Rodriguez-Pascual F, Geller DA et al. Cytokine induction of NO synthase II in human DLD-1 cells: roles of the JAK-STAT, AP-1 and NF-kappaB-signaling pathways. Br J Pharmacol 1998; 125: 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin S, Lekmine F, Sharma N, Majchrzak B, Mayer I, Young PR et al. The Rac1/p38 mitogen-activated protein kinase pathway is required for interferon alpha-dependent transcriptional activation but not serine phosphorylation of Stat proteins. J Biol Chem 2000; 275: 27634–27640. [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003; 3: 859–868. [DOI] [PubMed] [Google Scholar]
- Li Y, Batra S, Sassano A, Majchrzak B, Levy DE, Gaestel M et al. Activation of mitogen-activated protein kinase kinase (MKK) 3 and MKK6 by type I interferons. J Biol Chem 2005; 280: 10001–10010. [DOI] [PubMed] [Google Scholar]
- Tanos T, Marinissen MJ, Leskow FC, Hochbaum D, Martinetto H, Gutkind JS et al. Phosphorylation of c-Fos by members of the p38 MAPK family. Role in the AP-1 response to UV light. J Biol Chem 2005; 280: 18842–18852. [DOI] [PubMed] [Google Scholar]
- Yu Z, Kone BC. Targeted histone H4 acetylation via phosphoinositide 3-kinase- and p70s6-kinase-dependent pathways inhibits iNOS induction in mesangial cells. Am J Physiol Renal Physiol 2006; 290: F496–F502. [DOI] [PubMed] [Google Scholar]
- Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP et al. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol 2012; 129: 232–239 e1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrone D, Kuriakose J, Moors K, Jiang H, Niedzwiecki M, Perera F et al. Reproducibility and intraindividual variation over days in buccal cell DNA methylation of two asthma genes, interferon gamma (IFNgamma) and inducible nitric oxide synthase (iNOS). Clin Epigenet 2012; 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol 2008; 8: 726–736. [DOI] [PubMed] [Google Scholar]
- Galderisi U, Giordano A, Paggi MG. The bad and the good of mesenchymal stem cells in cancer: Boosters of tumor growth and vehicles for targeted delivery of anticancer agents. World J Stem Cells 2010; 2: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckermann BM, Kallifatidis G, Groth A, Frommhold D, Apel A, Mattern J et al. VEGF expression by mesenchymal stem cells contributes to angiogenesis in pancreatic carcinoma. Br J Cancer 2008; 99: 622–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djouad F, Plence P, Bony C, Tropel P, Apparailly F, Sany J et al. Immunosuppressive effect of mesenchymal stem cells favors tumor growth in allogeneic animals. Blood 2003; 102: 3837–3844. [DOI] [PubMed] [Google Scholar]
- Ohkouchi S, Block GJ, Katsha AM, Kanehira M, Ebina M, Kikuchi T et al. Mesenchymal stromal cells protect cancer cells from ROS-induced apoptosis and enhance the Warburg effect by secreting STC1. Mol Ther 2011; 20: 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, Xu Z, Zhao T, Zhao Z, Shi M, Zhao RC et al. Suppression of tumorigenesis by human mesenchymal stem cells in a hepatoma model. Cell Res 2008; 18: 500–507. [DOI] [PubMed] [Google Scholar]
- Khakoo AY, Pati S, Anderson SA, Reid W, Elshal MF, Rovira II et al. Human mesenchymal stem cells exert potent antitumorigenic effects in a model of Kaposi's sarcoma. J Exp Med 2006; 203: 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, Xu ZL, Zhao TJ, Ye LH, Zhang XD. Dkk-1 secreted by mesenchymal stem cells inhibits growth of breast cancer cells via depression of Wnt signalling. Cancer Lett 2008; 269: 67–77. [DOI] [PubMed] [Google Scholar]
- Du J, Zhou L, Chen X, Yan S, Ke M, Lu X et al. IFN-gamma-primed human bone marrow mesenchymal stem cells induce tumor cell apoptosis in vitro via tumor necrosis factor-related apoptosis-inducing ligand. Int J Biochem Cell Biol 2012; 44: 1305–1314. [DOI] [PubMed] [Google Scholar]
- Ren G, Su J, Zhang L, Zhao X, Ling W, L'Huillie et al. Species variation in the mechanisms of mesenchymal stem cell-mediated immunosuppression. Stem Cells 2009; 27: 1954–1962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.