Abstract
Acidobacteria is one of the most abundant phyla in soils and has been detected in rhizosphere mainly based on cultivation-independent approaches such as 16S rRNA gene survey. Although putative interaction of Acidobacteria with plants was suggested, so far no plant–bacterial interactions were shown. Therefore, we performed several in vitro tests to evaluate Acidobacteria–plant interactions and the possible mechanisms involved in such interaction. We observed that Arabidopsis thaliana inoculated with three strains belonging to Acidobacteria subdivision 1 showed increase in biomass of roots and shoots as well as morphological changes in root system. Our results indicate that the plant hormone indole-3-acetic acid production and iron acquisition are plausibly involved in the plant and Acidobacteria interactions. Here, we confirm for the first time that Acidobacteria can actively interact with plants and act as plant growth-promoting bacteria. In addition, we show that Acidobacteria strains produce exopolysaccharide which supports the adhesion of bacteria to the root surfaces.
Keywords: Acidobacteria, PGPB, IAA, Root biomass, Arabidopsis thaliana
Introduction
Acidobacteria is a very diverse and ubiquitous bacterial phylum. Furthermore, those bacteria seem to be especially well adapted to soil environment, often representing one the most abundant bacterial phylum (Janssen 2006; Lee et al. 2008). Although there are inconsistencies in reports regarding the preference of Acidobacteria in inhabiting bulk versus rhizosphere soils (Fierer et al. 2007; Singh et al. 2007; Kielak et al. 2008), there are clear evidences for the association of some Acidobacteria with plants (da Rocha et al. 2010, 2013). The enormous phylogenetic diversity within the phylum also suggests that Acidobacteria are genetically and, most likely, metabolically dissimilar; thus, the results of single studies cannot be generalized and easily extrapolated to the whole phylum.
Due to the still low number of sequenced genomes and difficulties associated with cultivation, the ecological role of this phylum remains rather unknown (Kielak et al. 2016). Nevertheless, a number of studies have compared distribution and diversity of Acidobacteria in relation to plant root proximity (Chow et al. 2002; Filion et al. 2004; da Rocha et al. 2010; Chaparro et al. 2014) and/or plant exudates (Shi et al. 2011; Mao et al. 2014). For example, acidobacterial strains have been obtained from internal plant tissues hinting to an endophytic lifestyle (Idris et al. 2004; Nissinen et al. 2012; Poosakkannu et al. 2015). Mendes et al. (2014) using culture-independent approach technique have shown that Acidobacteria are overrepresented in soybean rhizosphere, and da Rocha et al. (2010) have reported by means of qPCR, the Holophagae (Acidobacteria subdivision 8) being more abundant in leek rhizosphere. However, in the second case bacterial cell number was lower in spheres very proximate to roots or on the root surface itself.
Acidobacteria were also shown to be dominant in the rhizosphere of Arabidopsis thaliana, and moreover, change in terms of phylum composition and abundance during plant development, possibly due to changes in plant exudation (Chaparro et al. 2014). However, most of the studies were based on culture-independent method based on 16S ribosomal gene marker sequencing due to difficulty to culture Acidobacteria and perform experiments under laboratory conditions. Thus, those type of studies do not investigate the nature of plant–bacteria interactions. Concerning few studies on Acidobacteria physiology, non-traditional sources of carbon such as complex polysaccharides were suggested to improve cultivability of Acidobacteria (Koch et al. 2008; Pankratov and Dedysh 2010; Eichorst et al. 2011). Also some of the characterized strains were shown to be able to utilize plant-derived polymers (Pankratov et al. 2008, 2012; Eichorst et al. 2011) further suggesting close relation between plants and specific Acidobacteria subdivisions. Nevertheless, available strains can be used for attempts of studying interactions with plants under experimental conditions. In order to test the hypothesis that Acidobacteria strains effect plant growth, we assayed the interactions of three class Acidobacteria strains with A. thaliana ecotype Columbia 0 (Col 0) under in vitro conditions.
Materials and methods
Bacterial strains
Three Acidobacteria strains belonging to the class Acidobacteria from the NIOO-KNAW microorganisms’ collection were used in this study. Two of the strains are affiliated with genus Granulicella, namely Granulicella sp. WH15 (Valášková et al. 2009) and 5B5 (KM979383), and one is a type strain of the genus Acidicapsa, A. ligni WH120T (Valášková et al. 2009; Kulichevskaya et al. 2012). Pseudomonas putida IAC-RBal4 (KJ590499) and Escherichia coli WA321 (DSM no. 4509) strains were used as positive and negative controls of plant growth-promoting bacteria, respectively.
Plant–bacteria interaction experiment
Arabidopsis thaliana ecotype Columbia 0 seeds were surface sterilized by washing in 70 % ethanol for 5 min followed by submerging for 10 min in 50 % bleach and rinsing four times with sterile distilled water. Sterile seeds were placed on half-strength Murashige and Skoog (MS) medium pH 5.7 (Murashige and Skoog 1962) supplemented with 12 g L−1 plant agar (Duchefa Biochemie bv) and 5 g L−1 sucrose. Six plants were grown per plate. Seedlings were incubated at 21 °C with the light cycle of photoperiod 16 h/8 h day/night. The root tips of 5-day-old seedlings were inoculated with 2.5 µL of bacterial suspension (phosphate saline buffer, pH 5.5) of OD600 = 1 corresponding to 1.7 × 106, 1.5 × 107 and 1.3 × 107 CFU for A. ligni WH120T, Granulicella sp. 5B5 and WH15, respectively, or by direct transfer from the solid medium (0.1 × TSA, pH 5.0 see below) as an alternative method. The effect of bacteria on plant growth was evaluated 3 weeks post-inoculation.
Indole acetic acid (IAA) production
IAA production was determined based on the method described by Bric et al. (1991). Acidobacteria strains and P. putida IAC-RBal4 (positive control) were inoculated on 0.1 × tryptone soy agar (TSA) plates (pH 5.0 and 5.7) supplemented with 5 mM L−1 tryptophan and covered with a cellulose nitrate filter (0.45 µm pore size, Sartorius). TSA contained 1 g L−1 NaCl, 3.0 g L−1 TSB (Oxoid), 1.95 g L−1 MES, 20 g L−1 agar (Boom, Netherlands). Plates were incubated at 20 °C until colonies reached approximately 4–5 mm diameter (5 days), and then the membranes were washed in the Salkowski reagent (1.2 % FeCl2 in 37 % sulfuric acid). The reaction was allowed to proceed for 30 min at RT until purple color appeared. All strains were tested in triplicates on separate plates.
Phosphate solubilization assay
Acidobacteria strains were tested for their ability to solubilize a mineral form of phosphate. P. putida IAC-RBal4 was used as a positive control. Tests were performed on the National Botanical Research Institute’s phosphate growth medium (NBRIPM) containing per liter 15 g agar–agar ultrapure (Merck KGaA), 10 g glucose, 5 g Ca3(PO4)2, 5 g MgCl2·6H2O, 0.25 g MgSO4·7H2O, 0.2 g KCl and 0.1 g (NH4)2SO4 (Nautiyal 1999). All strains were inoculated by transfer from the 0.1 × TBA pH 5.0 media using inoculation loop and incubated for 6 weeks at 20 °C. The clearing zones around the colonies indicated phosphate solubilization by the isolates. The experiment was carried out in triplicate on separate plates.
Siderophore production
Detection of siderophore production was carried out in chrome azurol S (CAS) agar plates. Removal of iron from the CAS dye by iron-chelating compounds results in a color change from blue to yellow/orange. The CAS medium was prepared according to the method described by Schwyn and Neilands (1987). Bacteria were collected from 0.1 × TSA plates, resuspended and washed twice with phosphate-buffered saline pH 6.5. An aliquot of 10 µL of bacterial suspension was spotted on CAS agar plates. Plates were checked daily for color change around each colony. E. coli WA321 was used as a positive control.
nifH targeting PCR
The nifH gene targeting PCR was performed according to the modified protocol by Brankatschk et al. (2012). PCR amplification was performed in a 25-μL reaction mixture including DNA template, 0.6 μM of primers (nifHF/nifHR), 200 μM dNTPs, 1× of Taq buffer and 0.04 U FastStart High Fidelity Taq Enzyme Blend (Roche). The PCR were performed under the following conditions: initial denaturation step 5 min at 95 °C, followed by touchdown cycles of denaturation for 15 s at 95 °C, annealing starting at 63 °C with temperature decreases of 2 °C per two cycles and elongation at 72 °C for 45 s followed by 30 cycles with annealing at 53 °C. The final extension was extended to 10 min at 72 °C.
Results and discussion
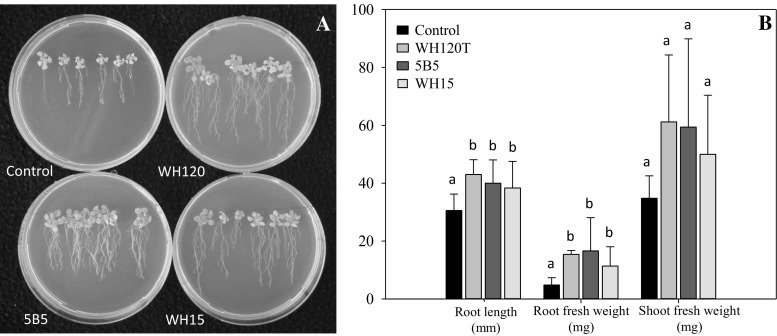
In this study, we tested three acidobacterial strains for possible interactions with A. thaliana (Col 0) roots. The growth of the plantlets was clearly positively affected by the presence of bacteria (Fig. 1). The presented results are shown with bacteria transferred directly from the solid media since this method of inoculation resulted in a stronger plant response. Root length, lateral root formation and root hair number were increased in plants exposed to Acidobacteria strains used in this study (Fig. 1A). Moreover, the root biomass increased significantly for plantlets inoculated with all three strains (Fig. 1B). The improved root architecture, more lateral branches and/or higher number of root hairs assist in more efficient water and nutrients uptake (Herder et al. 2010). Increased shoot biomass was also observed; however, the differences were not significant. We hypothesize that the stronger effect observed on plant growth with bacteria from the growth media in comparison with the bacterial suspension is not only related to higher bacterial biomass in the inoculum but also to the stress and longer adaptation time experienced by bacteria under the unfavorable culture conditions.
Fig. 1.
Effects of inoculation with Acidobacteria strains on Arabidopsis thaliana seedlings. Root tips of 5-day-old seedlings were inoculated by direct transfer with a loop of bacteria grown on 0.1 × TSA medium, pH 5.0. A Changes in morphology. Image was taken 3 weeks post-inoculation. B Changes in root length and fresh biomass. Different letters (a, b) indicate statistically significant differences (P < 0.05) between inoculated and control plants according to t test. Error bars represent SD (n = 6 plates each with six plants)
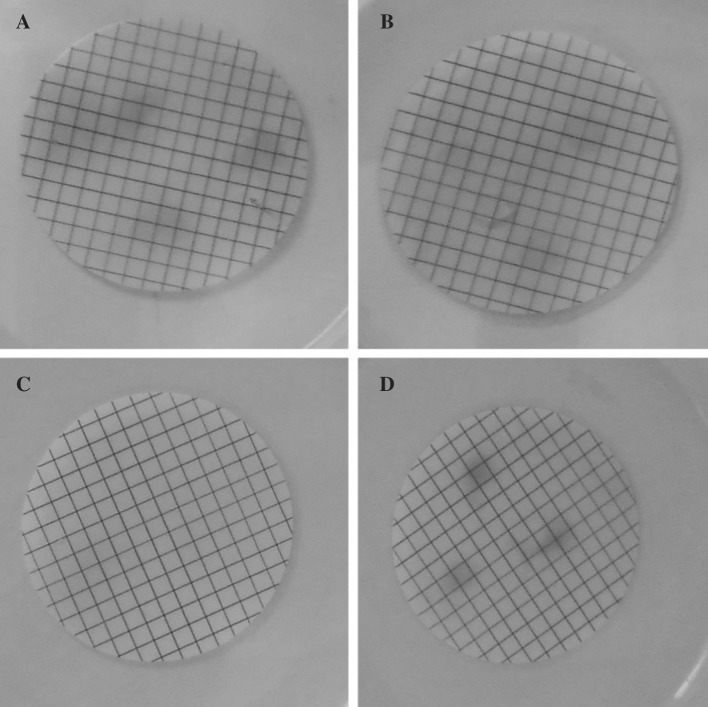
Bacterial adhesion, biofilm formation and growth along the root surfaces were observed for all three strains (Fig. 2). Acidobacteria strains Granulicella paludicola, G. pectinivorans, G. aggregans and G. rosea (Pankratov and Dedysh 2010), Acidicapsa borealis and A. ligni (Kulichevskaya et al. 2012) and Terriglobus tenax (Whang et al. 2014) were proven to produce extracellular polysaccharide. By genome mining, Ward et al. (2009) have suggested Acidobacteria being involved in soil matrix formation, water and nutrition trapping, or bacterial adhesion that lead to soil aggregate formation. However, here, for the first time we show that Acidobacteria strains produce exopolysaccharide (EPS) in the adhesion of bacteria to the root surfaces.
Fig. 2.
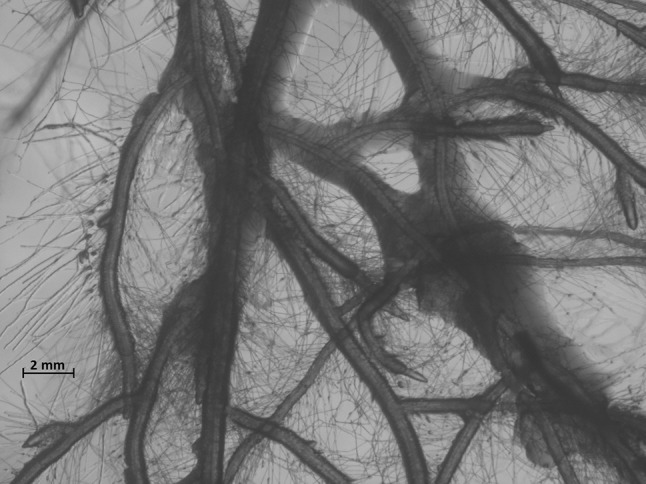
Root colonization and biofilm formation around Arabidopsis thaliana roots by strain 5B5
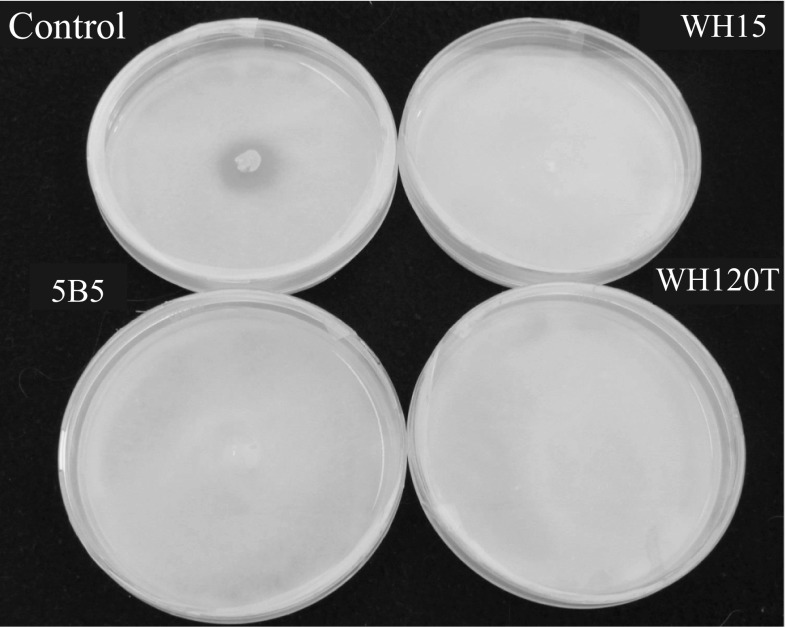
Bacteria can have positive effect on plant growth indirectly by acting as a biocontrol agent or directly by modulating plant hormone levels (Hayat et al. 2010) or/and by facilitating resource acquisition (mostly nitrogen, phosphorus and iron). Among the plant hormones, the auxin indole-3-acetic acid (IAA) has received most of the attention. As auxin production could best explain the observed changes in the plant phenotypes, we tested our strains for production of this phytohormone. A color change due to IAA production was observed for all three strains indicating all of them as positive for indolic substances production (Fig. 3).
Fig. 3.
Isolates were tested for indole acetic acid (IAA) production by bacteria immobilization on a nitrocellulose membrane followed by washing the membrane with Salkowski reagent. P. putida IAC-RBal4 was used as a positive control. A P. putida IAC-RBal4, B strain WH120T, C strain 5B5 and D strain WH15
Moreover, we tested our strains for nutrient acquisition abilities. Phosphorus (P) is an important macronutrient for plant growth and development. However, in general, the concentration of soluble P in soil is quite low. It is postulated that bacteria can enhance the P acquisition of plants (Richardson and Simpson 2011). All three strains were proven to be not able to solubilize mineral phosphate (Fig. 4). Nevertheless, the enhanced P uptake by plants can be also achieved via hormonal stimulation of root growth, branching or root hair development mediated by IAA among others (Richardson and Simpson 2011).
Fig. 4.
Isolates were tested for phosphate solubilization on agar plate using National Botanical Research Institute’s phosphate medium. P. putida IAC-RBal4 was used as a positive control
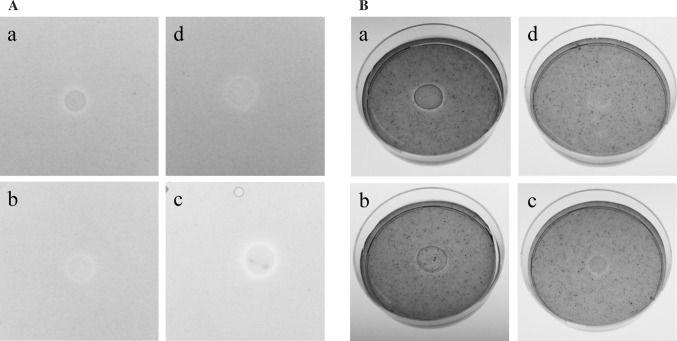
On the other hand, the assay for siderophore production showed that all three strains react with the media (Fig. 5). Detection of siderophore production was carried out on chrome azurol S (CAS) agar plates. Removal of iron from the CAS dye by iron-chelating compounds results in a color change. The discoloration was observed for all strains with stronger discoloration zones observed around 5B5 and WH120T colonies than around colonies of strain WH15. Plants have high iron requirement but similarly to P, most of the iron in soil is in ferric form, which is unavailable for plant uptake (Hayat et al. 2010). The strategies of iron uptake by plants are similar to those from bacteria. Those include acidification of the rhizosphere resulting in reduction of Fe3–Fe2 or synthesis of Fe3 chelators (Morrissey and Guerinot 2009; Saha et al. 2013). Additional advantage of bacterial siderophore production is competition with pathogens by removing iron from the environment (Saha et al. 2013).
Fig. 5.
Isolates were tested for siderophore production on CAS medium. E. coli WA321 was used as a positive control. A CAS plates prepared with the MM9, pH 6.8. B CAS plates prepared with the MM9, pH 6.0 and supplemented with casamino acid. a E. coli WA321, b strain WH120T, c strain 5B5 and d strain WH15
In order to test the ability of the strains to fix N2, we have carried out PCR targeting nitrogenase (nifH) gene. However, the results showed no evidence of such ability of tested strains. The absence of nifH is in agreement with acidobacterial genome mining studies (Ward et al. 2009). Up to now, there is no experimental evidence for the ability of Acidobacteria type strains to fix nitrogen.
Conclusion
Based on our findings, we provide for the first time a direct evidence of active Acidobacteria–plant interaction and data indicating growth-promoting effects by Acidobacteria. We verified that a possible auxin production is involved in plant growth promotion. Although commonly used, the test conducted to unravel possible mechanisms of this phenomenon, for the first time was applied for Acidobacteria. Further studies are needed to better understanding the beneficial Acidobacteria–plant interaction as well as the mechanisms involved in such interaction. In addition, we conclude that EPS production during root colonization by Acidobacteria might be helpful in root adhering to soil particles and in root protection. Taking into account the dominance in abundance of this phylum in soil environment, the overall impact of Acidobacteria on plant growth may be significant and the results shown here indicate for the first time that Acidobacteria can act as plant growth-promoting bacteria.
Acknowledgments
We thank Manuela Di Lorenzo and Tanja R. Scheublin for technical advices. This work was financially supported by the Dutch Ministry of Economic Affairs, Agriculture and Innovation (729.004.016) and the BE-Basic partner organization (www.be-basic.org). Publication 6101 of the Netherlands Institute of Ecology (NIOO-KNAW).
References
- Brankatschk R, Bodenhausen N, Zeyer J, Bürgmann H. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl Environ Microbiol. 2012;78(12):4481–4489. doi: 10.1128/AEM.07878-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bric JM, Bostock RM, Silverstone SE. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol. 1991;57(2):535–538. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8(4):790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow ML, Radomski CC, McDermott JM, Davies J, Axelrood PE. Molecular characterization of bacterial diversity in Lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol Ecol. 2002;42(3):347–357. doi: 10.1111/j.1574-6941.2002.tb01024.x. [DOI] [PubMed] [Google Scholar]
- da Rocha UN, van Elsas JD, van Overbeek LS. Real-time PCR detection of Holophagae (Acidobacteria) and Verrucomicrobia subdivision 1 groups in bulk and leek (Allium porrum) rhizosphere soils. J Microbiol Methods. 2010;83(2):141–148. doi: 10.1016/j.mimet.2010.08.003. [DOI] [PubMed] [Google Scholar]
- da Rocha UN, Plugge CM, George I, van Elsas JD, van Overbeek LS. The rhizosphere selects for particular groups of Acidobacteria and Verrucomicrobia. PloS One. 2013;8(12):e82443. doi: 10.1371/journal.pone.0082443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichorst SA, Kuske CR, Schmidt TM. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl Environ Microbiol. 2011;77(2):586–596. doi: 10.1128/AEM.01080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88(6):1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Filion M, Hamelin R, Bernier L, St-Arnaud M. Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl Environ Microbiol. 2004;70(6):3541–3551. doi: 10.1128/AEM.70.6.3541-3551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol. 2010;60(4):579–598. [Google Scholar]
- Herder GD, Van Isterdael G, Beeckman T, De Smet I. The roots of a new green revolution. Trends Plant Sci. 2010;15(11):600–607. doi: 10.1016/j.tplants.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Idris R, Trifonova R, Puschenreiter M, Wenzel WW, Sessitsch A. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl Environ Microbiol. 2004;70(5):2667–2677. doi: 10.1128/AEM.70.5.2667-2677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72(3):1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielak AM, Pijl AS, van Veen JA, Kowalchuk GA. Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol Ecol. 2008;63(3):372–382. doi: 10.1111/j.1574-6941.2007.00428.x. [DOI] [PubMed] [Google Scholar]
- Kielak AM, Barreto CC, Kowalchuk GA, van Veen JA, Kuramae EE. The ecology of Acidobacteria: moving beyond genes and genomes. Front Microbiol. 2016 doi: 10.3389/fmicb.2016.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch IH, Gich F, Dunfield PF, Overmann J. Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., Acidobacteria isolated from alpine and forest soils. Int J Syst Evolut Microbiol. 2008;58(5):1114–1122. doi: 10.1099/ijs.0.65303-0. [DOI] [PubMed] [Google Scholar]
- Kulichevskaya IS, Kostina LA, Valášková V, Rijpstra WIC, Damsté JSS, de Boer W, Dedysh SN. Acidicapsa borealis gen. nov., sp. nov. and Acidicapsa ligni sp. nov., subdivision 1 Acidobacteria from Sphagnum peat and decaying wood. Int J Syst Evolut Microbiol. 2012;62(7):1512–1520. doi: 10.1099/ijs.0.034819-0. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ka JO, Cho JC. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol Lett. 2008;285(2):263–269. doi: 10.1111/j.1574-6968.2008.01232.x. [DOI] [PubMed] [Google Scholar]
- Mao Y, Li X, Smyth EM, Yannarell AC, Mackie RI. Enrichment of specific bacterial and eukaryotic microbes in the rhizosphere of switchgrass (Panicum virgatum L.) through root exudates. Environ Microbiol Rep. 2014;6(3):293–306. doi: 10.1111/1758-2229.12152. [DOI] [PubMed] [Google Scholar]
- Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014;8:1577–1587. doi: 10.1038/ismej.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J, Guerinot ML. Iron uptake and transport in plants: the good, the bad, and the ionome. Chem Rev. 2009;109(10):4553–4567. doi: 10.1021/cr900112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. [Google Scholar]
- Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170(1):265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- Nissinen RM, Männistö MK, Elsas JD. Endophytic bacterial communities in three arctic plants from low arctic fell tundra are cold-adapted and host-plant specific. FEMS Microbiol Ecol. 2012;82(2):510–522. doi: 10.1111/j.1574-6941.2012.01464.x. [DOI] [PubMed] [Google Scholar]
- Pankratov TA, Dedysh SN. Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading Acidobacteria from Sphagnum peat bogs. Int J Syst Evolut Microbiol. 2010;60(12):2951–2959. doi: 10.1099/ijs.0.021824-0. [DOI] [PubMed] [Google Scholar]
- Pankratov TA, Serkebaeva YM, Kulichevskaya IS, Liesack W, Dedysh SN. Substrate-induced growth and isolation of Acidobacteria from acidic Sphagnum peat. ISME J. 2008;2(5):551–560. doi: 10.1038/ismej.2008.7. [DOI] [PubMed] [Google Scholar]
- Pankratov TA, Kirsanova LA, Kaparullina EN, Kevbrin VV, Dedysh SN. Telmatobacter bradus gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria, and emended description of Acidobacterium capsulatum Kishimoto et al. 1991. Int J Syst Evolut Microbiol. 2012;62(2):430–437. doi: 10.1099/ijs.0.029629-0. [DOI] [PubMed] [Google Scholar]
- Poosakkannu A, Nissinen R, Kytöviita MM. Culturable endophytic microbial communities in the circumpolar grass, Deschampsia flexuosa in a sub-Arctic inland primary succession are habitat and growth stage specific. Environ Microbiol Rep. 2015;7(1):111–122. doi: 10.1111/1758-2229.12195. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Simpson RJ. Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol. 2011;156(3):989–996. doi: 10.1104/pp.111.175448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Saha N, Donofrio RS, Bestervelt LL. Microbial siderophores: a mini review. J Basic Microbiol. 2013;53(4):303–317. doi: 10.1002/jobm.201100552. [DOI] [PubMed] [Google Scholar]
- Schwyn B, Neilands J. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160(1):47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shi S, Richardson AE, O’Callaghan M, DeAngelis KM, Jones EE, Stewart A, Firestone MK, Condron LM. Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol. 2011;77(3):600–610. doi: 10.1111/j.1574-6941.2011.01150.x. [DOI] [PubMed] [Google Scholar]
- Singh BK, Munro S, Potts JM, Millard P. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl Soil Ecol. 2007;36(2–3):147–155. [Google Scholar]
- Valášková V, de Boer W, Gunnewiek PJK, Pospíšek M, Baldrian P. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 2009;3(10):1218–1221. doi: 10.1038/ismej.2009.64. [DOI] [PubMed] [Google Scholar]
- Ward NL, Challacombe JF, Janssen PH, Henrissa B, Coutinho PM, Wu M, Xie G, Haft DH, Sait M, Badger J, et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol. 2009;75(7):2046–2056. doi: 10.1128/AEM.02294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang K-S, Lee J-C, Lee H-R, Han S-I, Chung S-H. Terriglobus tenax sp. nov., a novel exopolysaccharide producing acidobacterium isolated from rhizosphere soil of a medicinal plant. Int J Syst Evolut Microbiol. 2014;64(2):431–437. doi: 10.1099/ijs.0.053769-0. [DOI] [PubMed] [Google Scholar]