ABSTRACT
Quorum sensing (QS) is a widespread process in bacteria used to coordinate gene expression with cell density, diffusion dynamics, and spatial distribution through the production of diffusible chemical signals. To date, most studies on QS have focused on model bacteria that are amenable to genetic manipulation and capable of high growth rates, but many environmentally important bacteria have been overlooked. For example, representatives of proteobacteria that participate in nitrification, the aerobic oxidation of ammonia to nitrate via nitrite, produce QS signals called acyl-homoserine lactones (AHLs). Nitrification emits nitrogen oxide gases (NO, NO2, and N2O), which are potentially hazardous compounds that contribute to global warming. Despite considerable interest in nitrification, the purpose of QS in the physiology/ecology of nitrifying bacteria is poorly understood. Through a quorum quenching approach, we investigated the role of QS in a well-studied AHL-producing nitrite oxidizer, Nitrobacter winogradskyi. We added a recombinant AiiA lactonase to N. winogradskyi cultures to degrade AHLs to prevent their accumulation and to induce a QS-negative phenotype and then used mRNA sequencing (mRNA-Seq) to identify putative QS-controlled genes. Our transcriptome analysis showed that expression of nirK and nirK cluster genes (ncgABC) increased up to 19.9-fold under QS-proficient conditions (minus active lactonase). These data led to us to query if QS influenced nitrogen oxide gas fluxes in N. winogradskyi. Production and consumption of NOx increased and production of N2O decreased under QS-proficient conditions. Quorum quenching transcriptome approaches have broad potential to identify QS-controlled genes and phenotypes in organisms that are not genetically tractable.
IMPORTANCE
Bacterial cell-cell signaling, or quorum sensing (QS), is a method of bacterial communication and gene regulation that is well studied in bacteria. However, little is known about the purpose of QS in many environmentally important bacteria. Here, we demonstrate quorum quenching coupled with mRNA-Seq to identify QS-controlled genes and phenotypes in Nitrobacter winogradskyi, a nitrite-oxidizing bacterium. Nitrite oxidizers play an important role in the nitrogen cycle though their participation in nitrification, the aerobic oxidation of ammonia to nitrate via nitrite. Our quorum quenching approach revealed that QS influences production and consumption of environmentally important nitrogen oxide gases (NO, NO2, and N2O) in N. winogradskyi. This study demonstrated a novel technique for studying QS in difficult-to-work-with microorganisms and showed that nitrite oxidizers might also contribute to nitrification-dependent production of nitrogen oxide gases that contribute to global warming.
INTRODUCTION
Quorum sensing (QS) is a widespread process that bacteria use to coordinate gene expression with cell density, diffusion dynamics, and spatial distribution through the production of diffusible chemical signals (1, 2). Generally, as the density of bacterial cells increases, so does the concentration of QS signal, leading to coordinated expression of various genes in the entire bacterial population (1). The phenotypes associated with QS-controlled genes are often cooperative and stress-associated behaviors that benefit a population of bacteria, for example, biofilm formation, nutrient acquisition, luminescence, conjugation, and adaptation to stationary phase (2–4). There has been considerable interest in the study of bacterial QS, both in model systems for studying social evolution and to better understand important microbial behaviors, such as pathogenesis (1).
To date, most studies on QS have focused on model bacteria that are amenable to genetic manipulation (1). Many well-designed studies have introduced null mutations in the QS signal synthase and/or signal receptor-transcriptional regulator genes to determine what genes and phenotypes are controlled by QS (1). While this approach has significantly increased our understanding, many other QS-proficient organisms that are not easily genetically tractable have not been thoroughly studied. Through the use of bioassays, mass spectrometry, genome sequencing, and metagenomics, putative chemical signals and QS genes have been identified in representatives of proteobacteria that participate in the process of nitrification (5–10).
During nitrification, diverse genera of chemolithotrophic bacteria and/or archaea oxidize ammonia (NH3) to nitrite (NO2−) and then to nitrate (NO3−) (11–14). Generally, NH3 is oxidized to NO2− by ammonia oxidizers, including bacteria (AOB) and archaea (AOA), while NO2− is oxidized to NO3− by nitrite-oxidizing bacteria (NOB). Recently, complete oxidation of NH3 to NO3− (comammox) was identified in representatives of the genus Nitrospira, a group previously characterized as NOB (15, 16). Nitrification is a key part of the nitrogen cycle in natural, agricultural, and industrial systems and is a contributor to gas emissions of nitric oxide (NO), nitrogen oxides (NOx), and nitrous oxide (N2O), which are hazardous gases that contribute to global warming (11, 17).
In many proteobacteria, QS is accomplished through the production of acyl-homoserine lactone (AHL) signaling compounds or autoinducers (1). AHLs represent the best-studied class of autoinducer, and they are generally produced by a LuxI homolog autoinducer synthase and detected by a LuxR homolog signal receptor-transcriptional regulator (1). The luxI and luxR genes are commonly located adjacent to each other in the genome and are generally positively autoregulated (1).
One method to study AHL QS is by specifically inactivating all AHL autoinducers through the use of recombinant lactonase to promote a QS-deficient phenotype (18, 19). AiiA, an AHL lactonase identified from Bacillus spp., is a well-characterized enzyme that specifically hydrolyzes the homoserine lactone (HSL) ring of AHLs, regardless of the chain length of the acyl group or other moiety (20, 21). So-called “quorum quenching” approaches have been implemented through both heterologous expression in a host of interest and addition of purified AHL lactonase (18, 20, 22–24). In this study, we used purified AiiA lactonase to identify QS-controlled gene expression and phenotypes in Nitrobacter winogradskyi, a well-characterized NOB that is currently not genetically tractable.
The genus Nitrobacter consists of a ubiquitous group of NOB in the family Bradyrhizobiaceae isolated from soil, water, and wastewater treatment systems (13, 14). N. winogradskyi is a well-studied example of NOB due to its superior growth rate and growth yield compared to other NOB, and it was the first NOB shown to produce AHLs (10, 13, 14). In addition, the genome of N. winogradskyi has been sequenced and this bacterium has been the subject of recent global transcriptome studies (6, 25, 26). Expression of N. winogradskyi genes nwiI, encoding an autoinducer synthase, and nwiR, encoding a receptor-transcriptional regulator, was shown to be cell density dependent and to correlate with the AHL concentration in culture (10). The structure of the predominant AHL was identified as that of an unsaturated AHL with a 10 carbon acyl chain, C10:1-HSL (10). Nuclear magnetic resonance (NMR) spectroscopy analysis of AHL extracts produced via heterologous expression in Escherichia coli identified the isomeric form of C10:1-HSL (27) but suggested a location for the double bond that is different from that previously described (10). However, heterologous expression of autoinducer synthases in E. coli often produces AHLs that are different from those in the native strain (27, 28).
Previous attempts have been made to identify QS-controlled phenotypes in N. winogradskyi (10, 27). Mellbye et al. showed that the growth rate decreased as transcription of nwiI and nwiR increased and AHLs began to accumulate (10). Shen et al. observed up to a 2-fold increase or 5-fold decrease in the expression of select genes of the nitrite oxidoreductase (NXR) gene cluster after the addition of purified C10:1-HSL to cultures at saturating concentrations but did not observe any statistically significant phenotypic changes (27).
Here, we utilized a quorum quenching approach to identify both primary and secondary regulatory effects of AHL QS in N. winogradskyi. Using purified AiiA lactonase, AHLs were depleted from N. winogradskyi cultures and QS-controlled genes were identified through comprehensive mRNA sequencing (mRNA-Seq) analysis. Our transcriptome analysis showed that depletion of AHLs affected the expression of a significant percentage (52%) of the genetic inventory in N. winogradskyi and also suggested a link between QS and nitrogen oxide fluxes in this bacterium. Our experiments confirm a previous report that N. winogradskyi can produce N2O (29) and present new evidence that QS affects NOx fluxes. Our work demonstrates that AiiA-mediated quorum quenching coupled with mRNA-Seq is a useful technique to identify QS-controlled genes and phenotypes in difficult-to-study organisms.
RESULTS
AiiA lactonase treatment of N. winogradskyi cultures depletes AHLs.
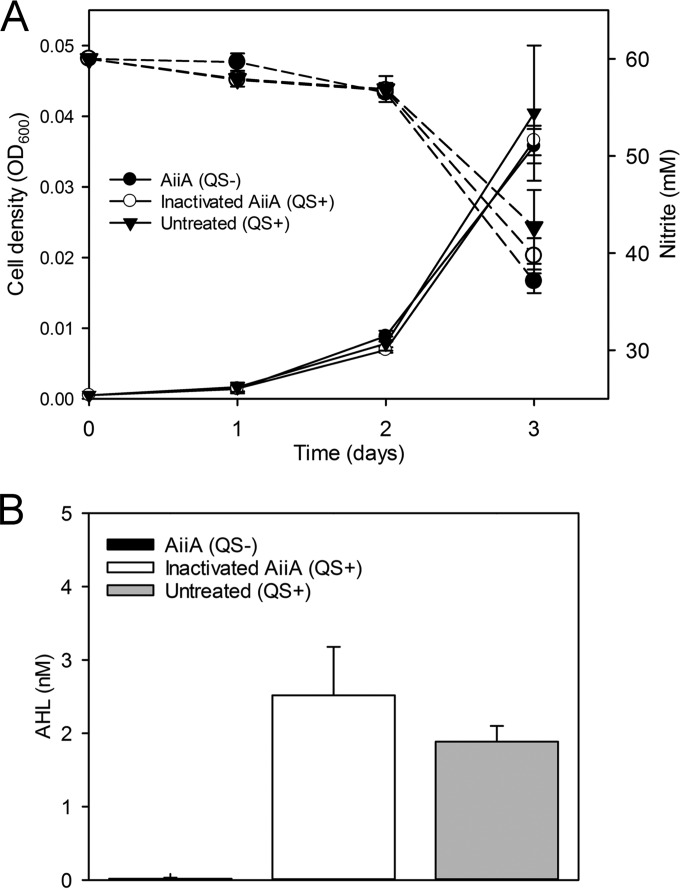
To determine the effect of QS inhibition in N. winogradskyi, we initiated and monitored three batch culture treatments: (i) AiiA lactonase treatment (QS-deficient), (ii) heat-denatured AiiA lactonase treatment (QS-proficient) (to determine if protein addition had an effect), and (iii) no-added-lactonase treatment (QS-proficient). Depending on cell density, approximately 0.28 or 0.71 µg protein ml−1 was added to both lactonase and heat-denatured lactonase treatments daily (see Text S1 in the supplemental material). Although the treatments showed no significant differences in nitrite oxidation rate, growth rate, or growth yield (Fig. 1A), the addition of AiiA lactonase prevented the accumulation of bioassay-detectable AHL (Fig. 1B). Lactonase-treated and heat-denatured lactonase-treated cultures were harvested on day 3 during peak signal production as observed in our previous work (10) to collect RNA for mRNA-Seq (Fig. 1).
FIG 1 .
Batch culturing comparison of N. winogradskyi results determined under QS-proficient and -deficient conditions. (A) Closed circles represent AiiA lactonase-treated (QS-deficient) cultures, open circles represent heat-inactivated AiiA lactonase (QS-proficient) cultures, and triangles represent untreated (QS-proficient) cultures. Solid lines correspond to cell density measured as the optical density at 600 nm (OD600; left y axis), and dashed lines correspond to the NO2− concentration in millimolar (right y axis) measured over time (days; x axis). (B) Bars indicate AHL concentrations in nanomolar (y axis) of AiiA lactonase-treated (QS-deficient) cultures, heat-inactivated AiiA lactonase-treated (QS-proficient) cultures, and untreated cultures (QS-proficient) when cultures were harvested on day 3. Depending on cell density, approximately 0.28 or 0.71 µg protein ml−1 was added to both lactonase and heat-denatured lactonase treatments daily (see Text S1 in the supplemental material). Values are the means of the results of four independent biological replicates. Error bars indicate the standard deviations of the means.
Transcriptome responses to QS inhibition.
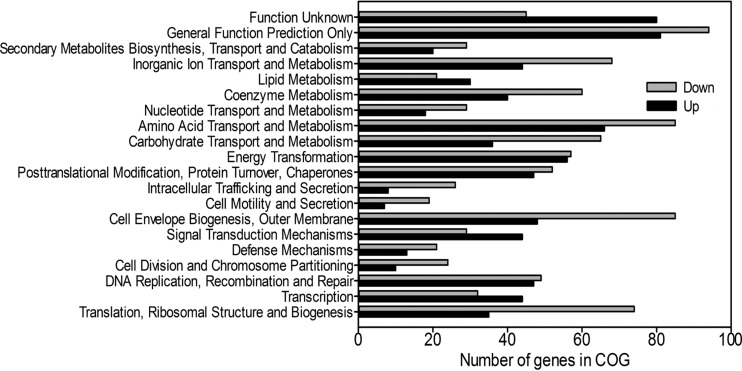
The transcriptome of N. winogradskyi under QS-deficient (AiiA-treated) conditions was compared to that present under QS-proficient (heat-denatured AiiA-treated) conditions. All changes in gene expression are expressed as the ratio of the number of transcripts seen under the QS-proficient treatment conditions to the number seen under QS-deficient treatment conditions. First, we validated our quorum quenching approach by noting an increase in the transcript abundance of the signal synthase nwiI gene and the signal receptor nwiR gene under QS-proficient conditions (Table 1). As previously noted, many bacterial QS genes, particularly the signal synthase gene, are autoregulated, creating a positive-feedback loop (1, 10). In addition, levels of methionine biosynthesis transcripts increased up to 7.7-fold, possibly due to increased use of S-adenosyl methionine for AHL biosynthesis (Table 1). The transcriptome analysis revealed 1,631 genes showing statistically significant changes in expression in QS-proficient cells, but many changes were <3-fold (see Table S1 and Dataset S1 in the supplemental material). In total, expression of 1,346 genes changed marginally and expression of 237 genes changed >3-fold between QS-deficient and QS-proficient conditions (see Dataset S1). Grouping the expression changes into clusters of orthologous groups (COG) functional categories, COGs associated with the process of translation as well as nucleotide, carbohydrate, and amino acid metabolism and transport substantially (>60% of category COGs) changed in expression (Fig. 2). We observed a similar trend whether an expression cutoff (e.g., 2-fold cutoff) was applied or not; thus, all of the data were included in the COG analysis.
TABLE 1 .
QS-dependent changes in gene expression in N. winogradskyi
| Gene category or number(s) | Gene name(s) | Description or role | Fold changea |
|---|---|---|---|
| Quorum sensing | |||
| Nwi0283, Nwi0284, Nwi0403, Nwi0586, Nwi2890 | metH, metW | Methionine biosynthesis | 1.5 to 7.7 |
| Nwi0626 | nwiI | Autoinducer synthase | 2.5 |
| Nwi0627 | nwiR | AHL-binding LuxR | 1.3 |
| Biosynthetic metabolism | |||
| Nwi0719, Nwi0720 | nirBD | Assimilatory nitrite reductase | −2.5 to −9.3 |
| Nitrogen metabolism | |||
| Nwi2648 | nirK | Putative nitrite reductase, NO production/consumption |
2.2 |
| Nwi2653–Nwi2649 | ncgABC |
nirK cluster genes, NO production/consumption |
2.7 to 19.9 |
| NO and/or guanine nucleotide signaling | |||
| Nwi0500 | Diguanylate cyclase/phosphodiesterase | 3.7 | |
| Nwi0529, Nwi0597–Nwi0599, Nwi1111, Nwi1121–Nwi1124, Nwi1130, Nwi1132–Nwi1134 |
flhA, fliH, fliG fliF, flgI, flgG, flgF, fliL, fliM, fliP, flgB, flgC, fliE |
Flagellum biosynthesis/assembly | −1.3 to −2.6 |
| Nwi0557 | nnrS | NO-related gene product | 8.1 |
| Nwi1922 | RelA/SpoT homolog | 2.3 | |
| Nwi2061 | Crp domain regulator | 8.9 | |
| Nwi2151 | Ppx/GppA phosphatase | 3.2 |
Fold change data correspond to the difference in mRNA transcript levels between AiiA-treated QS-deficient cells and QS-proficient cells (P ≤ 0.05, n = 4).
FIG 2 .
Clusters of orthologous group (COG) assignments of gene expression under QS-proficient conditions. Bars indicate the number of genes with increased expression (black) or the number of genes with decreased expression (gray) under QS-proficient conditions for each functional group. A quantity of 100 genes corresponds to 3.2% of COG assignments in the genome. In total, 56.3% of the COG assignments changed in expression level. Expression changes correspond to the difference in mRNA transcript levels between AiiA-treated QS-deficient cells and QS-proficient cells (P ≤ 0.05).
Based on previous work that suggested that QS is autoregulated in N. winogradskyi (10), we used SCOPE to search for lux-box-like inverted repeat motifs upstream of nwiI and nwiR (see Text S1 and Fig. S2 in the supplemental material) (30). Two different motifs (motif A and motif B) were identified that suggested that QS directly activates nirK cluster genes, as well as the stringent response secondary messenger system mediated through GppA phosphatase, and expression of several other genes (see Fig. S1 and Table S2).
Nitrite metabolism and signal transduction genes are induced under QS-proficient conditions.
An in-depth scan of the QS transcriptome of N. winogradskyi showed that the largest changes in expression involved genes encoding proteins associated with biosynthetic metabolism, nitrogen metabolism, and signal transduction, particularly those associated with nitrite metabolism (Table 1). Under QS-proficient conditions, assimilatory nitrite reductase gene nirBD decreased in expression up to 9.3-fold, suggesting imminent growth arrest and induction of the stringent response, as nitrite was the sole nitrogen source in the medium (Table 1). In addition, expression of nitrite reductase gene nirK increased 2.2-fold whereas expression of nirK cluster genes, including ncgABC, increased up to 19.9-fold under QS-proficient conditions (Table 1). Furthermore, expression of Nwi0557, a homolog of nnrS, a putative NO-responsive membrane protein gene, increased 8.1-fold (Table 1). These changes suggest a possible link between QS effects on N biosynthesis metabolism and NOx metabolism.
QS-proficient conditions also changed the expression levels of several genes involved in signal transduction and flagellum biosynthesis. Guanine nucleotide secondary messenger (e.g., ci-di-GMP, ppGpp) biosynthesis and response genes, including genes encoding a diguanylate cyclase/phosphodiesterase homolog (Nwi0500), a Crp domain regulator (Nwi2061), a RelA/SpoT homolog (Nwi1922), and Ppx/GppA phosphatase (Nwi2151), increased in expression by 2.3- to 8.9-fold (Table 1). Fourteen genes associated with flagellum biosynthesis and assembly decreased in expression up to 2.6-fold (Table 1). That said, no obvious phenotype differences, such as changes in motility, biofilm, or aggregate formation, were observed under QS-proficient or -deficient conditions.
Quorum sensing in N. winogradskyi influences NOx fluxes.
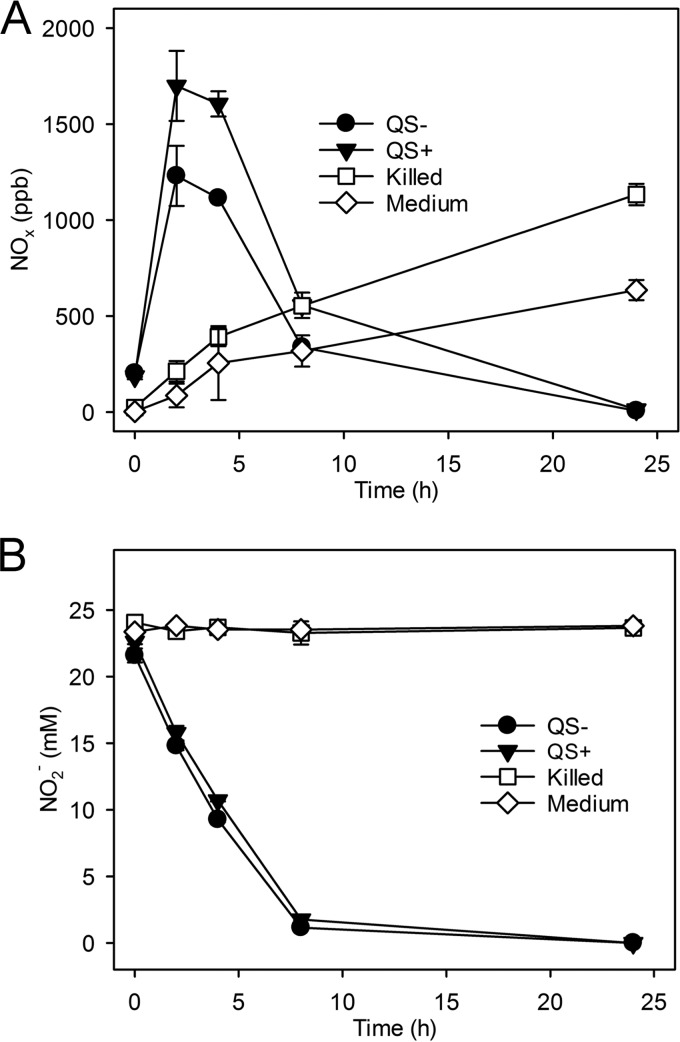
Following the transcriptome analysis prediction that QS affects nitrite metabolism through expression of nirK cluster genes, production and consumption of NOx gases by N. winogradskyi were measured. In order to observe the biggest difference in gas fluxes, cells were incubated at high cell density in sealed vials conducive to AHL accumulation. Preliminary tests detected abiotic NOx accumulation in the headspace above sterile nitrite-containing growth medium, most likely due to the aqueous chemical decomposition of protonated NO2− (nitrous acid [HNO2; also known as HONO]) to NO and NO2, collectively referred to as NOx gases (31). Therefore, we included both heat-killed cells and sterile medium controls along with our QS-proficient and -deficient treatments in the experiments. NOx gas measurements were made from such suspensions of concentrated QS-proficient or -deficient N. winogradskyi cells during 24 h of NO2− oxidation. We predicted that an increase in expression of the nirK cluster genes under QS-proficient conditions would either increase NOx production as earlier studies have suggested (32–34) or decrease NOx production by consuming NO as previously reported (34–36).
A statistically significant (P < 2 × 10−6) accumulation of NOx (measured as parts per billion [ppb] by volume) was registered for both QS-proficient and -deficient treatments (approximately 1,699 and 1,240 ppb, respectively) after 2 h of incubation, compared to the accumulations seen with medium alone and with heat-killed cell controls (approximately 87 and 213 ppb, respectively) (Fig. 3A). The peak NOx accumulation in both the QS-proficient and -deficient treatments was transient and was followed by the disappearance of NOx as NO2− was consumed by N. winogradskyi (Fig. 3). In contrast, heat-killed cells and medium-alone controls slowly accumulated NOx in the headspace over time and NO2− concentrations did not change significantly (Fig. 3). The pH of N. winogradskyi cultures and controls did not change significantly during the experiments (data not shown). Although these data suggest that some abiotic NOx accumulation occurred in the controls without N. winogradskyi cells, transient NOx production occurring during active NO2− oxidation by N. winogradskyi was significantly greater than that seen with the controls. QS-proficient cells both produced and consumed NOx at significantly greater rates (P < 0.003) than AiiA-treated, QS-deficient cells (Fig. 3A). Addition of heat-denatured AiiA to either growth medium or heat-killed cell controls did not affect the rate of NOx accumulation or consumption (data not shown). QS-proficient cells produced approximately 756 ppb NOx h−1 during the initial 2 h of the experiment, while QS-deficient cells produced 514 ppb NOx h−1 (Fig. 3A). Between h 4 and h 8, the net levels of consumption of NOx by QS-proficient and QS-deficient cells were 262 ppb h−1 and 191 ppb h−1, respectively (Fig. 3A).
FIG 3 .
QS-dependent production and consumption of NOx gases in N. winogradskyi. Closed circles (QS−) indicate AiiA lactonase-treated (QS-deficient) cells, closed triangles (QS+) indicate untreated cells (QS-proficient), open squares (killed) indicate heat-inactivated cell controls, and open diamonds (medium) indicate sterile medium controls. All measurements were made over 24 h (x axis). (A) Values correspond to NOx gases accumulated in the headspace measured as parts per billion (ppb; y axis). (B) Values show NO2− concentrations (millimolar; y axis) in solution. Values are the means of the results of four independent biological replicates. Error bars indicate the standard deviations of the means.
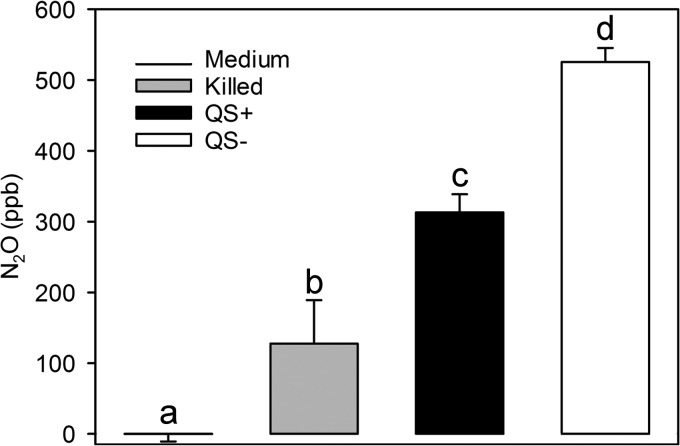
QS inhibition increases N2O production by N. winogradskyi.
Following a previous unsubstantiated report of N2O production by Nitrobacter species (29), we measured N2O accumulation by both QS-proficient and -deficient concentrated N. winogradskyi cells after 24 h of NO2− oxidation. Despite the absence of a known nitric oxide reductase gene (nor) in the N. winogradskyi genome, N. winogradskyi cells accumulated significantly (P < 0.002) more N2O than either growth medium alone or heat-killed cell controls (Fig. 4). AiiA-treated, QS-deficient cells accumulated approximately 525 ppb N2O, 1.68-fold more N2O than QS-proficient cells (313 ppb N2O) (P < 0.0001) (Fig. 4). In addition, the molar ratio of NOx-N at 2 h to normalized N2O-N accumulated at 24 h (after all NOx was consumed) was considerably higher (6.90 NO-N/N2O-N ratio) than that seen under QS-deficient conditions (2.78 NO-N/N2O-N ratio). These data suggest that QS also affects N2O production by N. winogradskyi, likely through changes in NOx fluxes.
FIG 4 .
QS-dependent production of N2O by N. winogradskyi. N2O accumulation in headspace is shown as parts per billion above atmospheric N2O (ppb; y axis). The dark line (medium) indicates medium controls, the light gray bar (killed) indicates heat-inactivated cell controls, the black bar (QS+) indicates untreated QS-proficient cells, and the white bar (QS−) indicates AiiA lactonase-treated QS-deficient cells. Values are the means of the results of four independent biological replicates. Error bars indicate the standard deviations of the means. Different letters represent significant differences between treatments determined by a one-way analysis of variance (P < 0.0001, n = 4).
DISCUSSION
Quorum quenching transcriptomics is a novel technique to identify QS-controlled genes and phenotypes.
We used a quorum quenching transcriptomic technique to investigate the role of QS in N. winogradskyi, a nitrite-oxidizing bacterium that participates in the nitrogen cycle. Our experiments identified a QS-controlled phenotype and revealed a link between QS and NOx metabolism in a nitrifying microorganism. While mutational analysis is the preferred method to study QS, there are many bacteria, including those of environmental importance, with no established genetic system and new techniques are needed to determine the purpose of QS in these organisms. Quorum quenching also represents a different methodology to study animal pathogens and social evolution (20, 22–24). Mutant construction in most bacteria takes several generations of selection that may introduce unintended changes, particularly during social-evolution experiments. As previously suggested, quorum quenching through addition of a lactonase or chemical inhibitor is a quicker method to induce a QS-deficient phenotype without the need for several generations of selection and the possibility of pleiotropic effects (23, 24, 37).
NOx metabolism in N. winogradskyi.
NO production and consumption in N. winogradskyi have generated considerable interest and confusion for almost 30 years. Earlier work by Freitag et al. and Freitag and Bock suggested that Nitrobacter strains consume NO to generate NADH and produce N2O under various conditions (29, 35). Starkenburg et al. published the sequenced genome of N. winogradskyi and noted that it lacked the gene(s) known to produce N2O but possessed a putative NO-producing gene, nirK, and ncgABC, closely related to homologs in N. europaea (6). However, later work by Starkenburg et al. could not confirm production of NO by N. winogradskyi when nirK was expressed but did show that N. winogradskyi was able to consume NO (36). Coculture studies of the AOB Nitrosomonas europaea and N. winogradskyi suggest that N. winogradskyi may consume NO, as less NO accumulated during coculture but expression of nirK in N. winogradskyi increased (25, 38). Finally, the studies described above and work on other NOB and AOB were incorporated into a multispecies metabolic model to assess sources and sinks of NO in relation to N2O production during wastewater treatment (34). The model predicts that NOB likely oxidize NO to NO2− but do not substantially contribute to N2O production (34).
Our quorum quenching transcriptomics approach led us to investigate production and consumption of NOx gases in N. winogradskyi and showed that N. winogradskyi may function as a source and/or a sink of NOx and N2O. We hypothesized that if increased expression of nirK and associated genes under QS-proficient conditions increased the concentration of NOx gases, then more NO would be available for abiological or nonspecific mechanism-based reduction to N2O. Contrary to our prediction, QS-deficient cells produced significantly more N2O than QS-proficient cells. A closer inspection of the molar ratio of peak NOx-N and N2O-N produced shows that QS-proficient cells not only produced more NOx than QS-deficient cells but also directed considerably less of the NOx to the N2O pool than QS-deficient cells. These data possibly suggest consumption of NOx via oxidation back to nitrite and, subsequently, nitrate, as previously suggested (36), or that there are alternate fates for NO such as unspecified signaling roles.
Note that N. winogradskyi produced considerably less N2O than NO, as previously predicted (34). N. winogradskyi does not contain a known nitric oxide reductase (nor), and, while our data show that some of the N2O formation is dependent on live cells, there may be abiotic reactions of NO with cellular components as has been recently suggested during ammonia oxidation by Thaumarchaeota (39). This hypothesis may partially explain why QS-deficient cells produce more N2O, since their consumption of NOx is slower than that by QS-proficient cells.
Considering the close homologies between nirK and ncgABC of N. winogradskyi and N. europaea, the two gene clusters may serve similar purposes. In N. europaea, both nirK and ncgABC were shown to confer tolerance to NO2−, but nirK had a negative fitness effect in ncgABC mutants (40, 41). Our observation of an up to 19.9-fold increase in ncgABC transcripts (Table 1; see also Table S1 and Dataset S1 in the supplemental material) under QS-proficient conditions suggests that these genes may play a role in NOx consumption or detoxification in N. winogradskyi, but future work is needed to investigate this prediction.
One interesting observation arising from our work on NOx flux is the abiotic generation of NOx from NO2−, likely via chemical decomposition of aqueous HNO2 or of its gaseous analog, HONO. Although this phenomenon was studied extensively in the past (31), reviewed in the nitrification and engineering fields (33), and recently appreciated in soils (42, 43), most studies in the nitrification and environmental engineering fields have still largely ignored it. Many studies on nitrifying microorganisms, particularly NH3 oxidizers, routinely measure NO but do not account for NOx generated abiotically from the NO2− end product of NH3 oxidation. In addition, many metabolic modeling studies, including an important study cited here modeling NO and N2O turnover (34), have not included abiotic formation of NO as a significant source. Clearly, future models and studies need to consider the contribution of abiotic reactions to NOx production.
Why does QS regulate NOx metabolism?
The immediate rapid generation of NO by concentrated N. winogradskyi cells is an initially puzzling response for a NO2− oxidizer, since reductant would be required to reduce NO2− to NO. However, metabolic modeling of electron flow in Nitrobacter has suggested that generation and consumption of NO would help explain previous experimental data (32). Some support for QS regulation of this metabolic response recently emerged, as Nitrobacter accumulated fewer AHLs under mixotrophic than under autotrophic growth conditions, perhaps also suggesting less generation of NO when organic carbon is available (27). Another possible explanation is the model proposed by Starkenburg et al. that suggests that NO production is a strategy to reversibly block the terminal cytochrome oxidase and redirect electrons toward generation of reductant (36). This hypothesis would make sense if one role for QS is to promote redirection of electrons away from reductive cellular biosynthesis and toward generation of electron-rich storage compounds such as poly-β-hydroxybutyrate as previously reported (6).
In other bacteria, QS generally controls production of a public good that can be used by the entire population (1). According to previous reports that Nitrobacter generates NADH from NO oxidation (35), an increase in NO generation at higher cell densities (QS-proficient conditions) may function as a public good for energy generation. NO generated in a large population of Nitrobacter is more likely to be utilized by nearby Nitrobacter cells and may benefit the population.
Another possible function for QS regulation of the nirK cluster and other genes could be preparation for stationary phase, partially through NO signaling. Signal integration of QS and stress responses has been previously demonstrated in Pseudomonas aeruginosa and other bacterial species (3). Transcriptome data suggest that QS-proficient conditions both prepare for growth arrest via repression of assimilatory nitrite reductase nirBD and induction of the stringent response and promote transition from a motile to sessile state through inhibition of flagellar expression and possible guanine secondary messenger signaling (Table 1). Many of these and other changes in transcription could be indirect effects of NO signaling. According to transcriptome data, QS activation triggers transcription of a putative nnrS homolog (Table 1). While the role of nnrS in N. winogradskyi is unknown, this gene was previously shown to be transcribed during exposure to NO and to regulate chemotaxis in Rhodobacter sphaeroides (44, 45). NnrS has recently been proposed to function as an NO sensor (46), and we speculate that it may serve a similar role in N. winogradskyi. NO signaling would also be a convenient way to detect nearby AOB, since some NO is produced via NH3 oxidation.
The identification of QS regulation of NOx metabolism in N. winogradskyi raises questions about NOx metabolism in other NOB. A cursory search of genomic databases shows that all NOB and comammox bacteria contain nirK homologs and that all except Nitrococcus and Nitrolancea contain nnrS homologs but that only Nitrobacter species contain both ncgABC and clearly annotated autoinducer synthase and receptor genes associated with QS. Since Nitrobacter species are r-strategists with the ability to exploit higher substrate concentrations and sporadically grow to higher densities, they may make better use of cell-density-dependent QS genetic regulation (10, 47). In addition, as r-strategists, Nitrobacter species might use QS-controlled preparation for starvation as an important strategy to recognize transitions to an unfavorable energy-limited situation (48). Future research into nirK function in NOB is needed to confirm the role of nirK and nnrS in NOx fluxes and to determine if NO signaling occurs in these microorganisms.
MATERIALS AND METHODS
Chemicals.
N-Decanoyl-dl-homoserine lactone (C10-HSL) was purchased from Sigma-Aldrich (St. Louis, MO). Acetic acid and high-performance-liquid-chromatography (HPLC)-grade ethyl acetate were purchased from VWR International (Radnor, PA) and EMD Chemicals (Darmstadt, Germany), respectively.
Bacterial strains, plasmids, and growth medium.
Bacterial strains and plasmids used in this study are outlined in Table S3 in the supplemental material. N. winogradskyi was routinely cultivated in 60 mM NaNO2-supplemented mineral salts medium as described previously (26), with minor modifications for NOx and N2O measurements (see Text S1 in the supplemental material and the descriptions of AiiA QS inhibition and culturing experiments below). N. winogradskyi cultures were routinely screened for heterotrophic contamination by plating 200-µl aliquots of culture on Luria-Bertani (LB) agar plates. Agrobacterium tumefaciens was prepared and cultivated as described elsewhere (49, 50). Escherichia coli strains were grown in LB medium on a rotatory shaker at 200 rpm and 37°C.
AiiA lactonase production and activity measurement.
Plasmid pDSK519 carrying aiiA was kindly provided by Max Teplitski and Mengsheng Gao of the University of Florida. The aiiA gene was cloned, and AiiA was expressed and purified as outlined in Text S1 in the supplemental material. AiiA-specific activity units were determined by measuring reductions of AHL concentrations after 4 hours as outlined in Text S1 in the supplemental material.
AHL bioassay.
AHLs were extracted from N. winogradskyi cultures and quantified in Miller units by broad-range Agrobacterium tumefaciens bioassay as described previously (10, 49, 50). AHL concentrations (nanomolar) were estimated using standard concentrations of C10-HSL (see Fig. S2 in the supplemental material). For determining AiiA activity, 200 µl of the assay solution was directly added to A. tumefaciens culture as described previously (49, 50).
AiiA QS inhibition and culturing experiments.
Batch cultures of N. winogradskyi were prepared in 100 ml 60 mM NO2−-supplemented medium at pH 7.5 as outlined above, inoculated to an optical density at 600 nm (OD600) of 0.001 from mid-exponential-phase cultures, and grown in Erlenmeyer flasks on a rotatory shaker at 100 rpm and 30°C. All experiments, including mRNA-Seq experiments, included 4 biological replicates. For NOx and N2O measurements, batch cultures were either treated with AiiA lactonase as outlined in Text S1 in the supplemental material or left untreated and were harvested by centrifugation. Harvested cells were suspended to an OD600 of 0.2 in 25 mM (NOx measurement) or 60 mM (N2O measurement) NO2−-supplemented medium and treated with AiiA lactonase or left untreated or subjected to heat killing by incubation at 110°C for 20 min. Cultures were placed in 41-ml serum vials. The serum vial cultures were then capped with gray-butyl stoppers, crimp sealed, and incubated for 24 h as outlined. NOx and N2O levels were routinely measured in the headspace as outlined below. Experimental cultures were routinely monitored every 24 h to check cell density (OD600), NO2− concentration by the Griess assay (51), and AHL concentration as described above.
For QS inhibition transcriptome experiments and other experiments, a specific number of activity units of AiiA lactonase (filtered using a 0.2-µm-pore-size filter) was added into N. winogradskyi batch cultures every 24 h (QS-deficient conditions) as outlined in Text S1 in the supplemental material.
RNA preparation and sequencing.
RNA was extracted using an RNeasy minikit (Qiagen, Germantown, MD) (10), and mRNA was enriched and prepared for Illumina mRNA-Seq as described previously (52). The libraries were sequenced (150mer paired-end sequencing) on a HiSeq 3000 Sequencer (Illumina, San Diego, CA) at the Center for Genome Research and Biocomputing Core Laboratories at Oregon State University.
Transcriptome data analysis.
The mRNA-Seq data were analyzed using the CLC Genomics Workbench (CLC bio, Prismet, Denmark) as previously described (52). Briefly, mRNA-Seq reads were normalized to reads per kilobase of transcript per million mapped reads (RPKM) and the module for empirical analysis of differential gene expression (DGE) was used as described previously (53, 54). Quantitative reverse transcription-PCR (qRT-PCR) was used to corroborate gene expression of selected genes with total RNA from biological replicates and primers as described in Text S1 in the supplemental material and outlined in Table S4 and Table S5.
Analytical methods.
NO2− levels were determined by chemical assay as described previously (51). NOx levels were measured using a portable NOx detector (LMA-3D and LNC-3D; Unisearch Associates Ltd., Concord, Ontario, Canada) that passes air through a CrO3 filter to convert NO to NO2 and then measures parts per billion by volume of NO2 by chemiluminescence as described previously (55), with modifications. Briefly, 1 ml or 5 ml of samples was injected into the intake line of the instrument and the peak NOx level recorded. NOx peaks were quantified by comparison to both NO and acidified NO2− standards. N2O was measured by gas chromatography as described previously (52). Cell density was measured as OD600, and the protein concentration was measured with a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockford, IL).
Accession number(s).
Raw datasets and processed datasets are available at Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) under accession no. GSE84969.
SUPPLEMENTAL MATERIAL
Supplemental Materials and Methods: detailed methods, including medium formulation, promoter element prediction, corroboration of mRNA-Seq results by qRT-PCR, AiiA purification, activity measurement, and QS inhibition. Download
mRNA-Seq expression data generated by Illumina HiSeq 3000 Sequencer (Illumina, San Diego, CA) and analyzed using the CLC Main Workbench (CLC bio, Prismet, Denmark). Fold change data indicate changes in expression between AiiA-treated, QS-deficient cells and QS-proficient cells treated with heat-inactivated AiiA. Download
Putative lux-box-like promoter elements in N. winogradskyi. Sequence logos shown are graphic representations of aligned sets of putative promoter DNA sequences displaying frequencies of bases at each position as the relative heights of letters. The sequence logo also shows the degree of sequence conservation as the total height of a stack of letters, measured in bits. (a) Promoter motif A found upstream of nwiRI, the nirK cluster, and other genes. (b) Promoter motif B found upstream of nwiRI, the GppA phosphatase gene, and other genes. Table S2 shows all genes with these promoter motifs. Download
N-Decanoyl-l-homoserine lactone (C10-HSL) acyl-homoserine lactone (AHL) bioassay standard curve. Known concentrations of C10-HSL were added to bioassay cultures (see Materials and Methods in the main text) for estimation of the concentration of AHLs in N. winogradskyi cultures. The black line corresponds to the nonlinear regression of C10-HSL concentration (nanomolar, y axis) compared to AHL bioassay Miller units (x axis), and dotted lines indicate the 95% confidence band. The regression was calculated as follows: y = 0.0047x + 5 × 10− 5x2, R2 = 0.99. Download
Statistically significant changes in gene expression under QS-proficient conditions in N. winogradskyi
Putative QS-controlled genes with upstream lux-box-like promoter elements.
Bacterial strains and plasmids.
Comparison of fold changes in expression between QS-deficient and proficient treatments analyzed by quantitative PCR (qPCR) and mRNA-Seq.
Genes and primers used to corroborate gene expression.
ACKNOWLEDGMENTS
Special thanks to Ajai Dandekar for helpful advice on the use of AiiA lactonase to inhibit AHL QS, Max Teplitski and Mengsheng Gao for providing the pDSKaiiA and pDSK constructs, and Jun Zhu for providing the bioassay strain A. tumefaciens KYC55(pJZ372) (pJZ384) (pJZ410) for this and previous projects. We also thank David Myrold for gas chromatography instrument use, Steve Perakis for use of a NOx detector, and Chris Catricala, Rebecca Ferrell, Anne Taylor, Michael Dobie, Neeraja Vajrala, and Aiden Maxwell for helpful discussions and laboratory assistance.
This work was supported by Department of Energy (DOE) award ER65192 (co-principal investigators, L.A.S-S. and P.J.B.), USDA-NIFA award no. 2012-67019-3028 (P.J.B.), and USDA-NIFA postdoctoral fellowship award no. 2016-67012-24691 (B.L.M.).
The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Mellbye BL, Giguere AT, Bottomley PJ, Sayavedra-Soto LA. 2016. Quorum quenching of Nitrobacter winogradskyi suggests that quorum sensing regulates fluxes of nitrogen oxide(s) during nitrification. mBio 7(5):e01753-16. doi:10.1128/mBio.01753-16.
REFERENCES
- 1.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. 2013. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol 67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 2.Hense BA, Schuster M. 2015. Core principles of bacterial autoinducer systems. Microbiol Mol Biol Rev 79:153–169. doi: 10.1128/MMBR.00024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellbye B, Schuster M. 2011. More than just a quorum: integration of stress and other environmental cues in acyl-homoserine lactone signaling networks. In Storz G, Hengge R (ed), Bacterial stress responses, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 4.Mellbye B, Schuster M. 2014. Physiological framework for the regulation of quorum sensing-dependent public goods in Pseudomonas aeruginosa. J Bacteriol 196:1155–1164. doi: 10.1128/JB.01223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton EO, Read HW, Pellitteri MC, Hickey WJ. 2005. Identification of acyl-homoserine lactone signal molecules produced by Nitrosomonas europaea strain Schmidt. Appl Environ Microbiol 71:4906–4909. doi: 10.1128/AEM.71.8.4906-4909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starkenburg SR, Chain PS, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW, Malfatti SA, Klotz MG, Bottomley PJ, Arp DJ, Hickey WJ. 2006. Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol 72:2050–2063. doi: 10.1128/AEM.72.3.2050-2063.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starkenburg SR, Larimer FW, Stein LY, Klotz MG, Chain PS, Sayavedra-Soto LA, Poret-Peterson AT, Gentry ME, Arp DJ, Ward B, Bottomley PJ. 2008. Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl Environ Microbiol 74:2852–2863. doi: 10.1128/AEM.02311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasuno E, Kimura N, Fujita MJ, Nakatsu CH, Kamagata Y, Hanada S. 2012. Phylogenetically novel LuxI/LuxR-type quorum sensing systems isolated using a metagenomic approach. Appl Environ Microbiol 78:8067–8074. doi: 10.1128/AEM.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Ma A, Zhuang X, Zhuang G. 2014. An N-acyl homoserine lactone synthase in the ammonia-oxidizing bacterium Nitrosospira multiformis. Appl Environ Microbiol 80:951–958. doi: 10.1128/AEM.03361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellbye BL, Bottomley PJ, Sayavedra-Soto LA. 2015. Nitrite-oxidizing bacterium Nitrobacter winogradskyi produces N-acyl-homoserine lactone autoinducers. Appl Environ Microbiol 81:5917–5926. doi: 10.1128/AEM.01103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward BB. 2011. Nitrification: an introduction and overview of the state of the field, p 3–8. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 12.Sayavedra-Soto LA, Arp DJ. 2011. Ammonia-oxidizing Bacteria: their biochemistry and molecular biology, p 11–37. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 13.Starkenburg SR, Spieck E, Bottomley PJ. 2011. Metabolism and genomics of nitrite-oxidizing Bacteria: emphasis on studies of pure cultures and of Nitrobacter species, p 267–293. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 14.Daims H, Lucker S, Paslier DL, Wagner M. 2011. Diversity, environmental genomics, and ecophysiology of nitrite-oxidizing Bacteria, p 295–322. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 15.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B, Jetten MS, Lücker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein LY. 2011. Heterotrophic nitrification and nitrifier denitrification, p 95–114. In Ward BB, Arp D, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 18.Dong YH, Zhang LH. 2005. Quorum sensing and quorum-quenching enzymes. J Microbiol 43:101–109. [PubMed] [Google Scholar]
- 19.Grandclément C, Tannières M, Moréra S, Dessaux Y, Faure D. 2016. Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev 40:86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 20.Dong YH, Xu JL, Li XZ, Zhang LH. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci U S A 97:3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LH, Weng LX, Dong YH, Zhang LH. 2004. Specificity and enzyme kinetics of the quorum-quenching N-acyl homoserine lactone lactonase (AHL-lactonase). J Biol Chem 279:13645–13651. doi: 10.1074/jbc.M311194200. [DOI] [PubMed] [Google Scholar]
- 22.Gao M, Chen H, Eberhard A, Gronquist MR, Robinson JB, Connolly M, Teplitski M, Rolfe BG, Bauer WD. 2007. Effects of AiiA-mediated quorum quenching in Sinorhizobium meliloti on quorum-sensing signals, proteome patterns, and symbiotic interactions. Mol Plant Microbe Interact 20:843–856. doi: 10.1094/MPMI-20-7-0843. [DOI] [PubMed] [Google Scholar]
- 23.Feltner JB, Wolter DJ, Pope CE, Groleau M, Smalley NE, Greenberg EP, Mayer-Hamblett N, Burns J, Déziel E, Hoffman LR, Dandekar AA. 2016. LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. mBio 7(5):e01753-16. doi: 10.1128/mBio.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majerczyk C, Kinman L, Han T, Bunt R, Greenberg EP. 2013. Virulence of Burkholderia mallei quorum-sensing mutants. Infect Immun 81:1471–1478. doi: 10.1128/IAI.00048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez J, Buchanan A, Mellbye B, Ferrell R, Chang JH, Chaplen F, Bottomley PJ, Arp DJ, Sayavedra-Soto LA. 2015. Interactions of Nitrosomonas europaea and Nitrobacter winogradskyi grown in co-culture. Arch Microbiol 197:79–89. doi: 10.1007/s00203-014-1056-1. [DOI] [PubMed] [Google Scholar]
- 26.Sayavedra-Soto L, Ferrell R, Dobie M, Mellbye B, Chaplen F, Buchanan A, Chang J, Bottomley P, Arp D. 2015. Nitrobacter winogradskyi transcriptomic response to low and high ammonium concentrations. FEMS Microbiol Lett 362:1–7. doi: 10.1093/femsle/fnu040. [DOI] [PubMed] [Google Scholar]
- 27.Shen Q, Gao J, Liu J, Liu S, Liu Z, Wang Y, Guo B, Zhuang X, Zhuang G. 2016. A new acyl-homoserine lactone molecule generated by Nitrobacter winogradskyi. Sci Rep 6:22903. doi: 10.1038/srep22903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol 184:6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freitag A, Rudert M, Bock E. 1987. Growth of Nitrobacter by dissimilatoric nitrate reduction. FEMS Microbiol Lett 48:105–109. doi: 10.1111/j.1574-6968.1987.tb02524.x. [DOI] [Google Scholar]
- 30.Carlson JM, Chakravarty A, DeZiel CE, Gross RH. 2007. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res 35:W259–W264. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park JY, Lee YN. 1988. Solubility and decomposition kinetics of nitrous acid in aqueous solution. J Phys Chem 92:6294–6302. doi: 10.1021/j100333a025. [DOI] [Google Scholar]
- 32.Poughon L, Dussap CG, Gros JB. 2001. Energy model and metabolic flux analysis for autotrophic nitrifiers. Biotechnol Bioeng 72:416–433. [PubMed] [Google Scholar]
- 33.Schreiber F, Wunderlin P, Udert KM, Wells GF. 2012. Nitric oxide and nitrous oxide turnover in natural and engineered microbial communities: biological pathways, chemical reactions, and novel technologies. Front Microbiol 3:372. doi: 10.3389/fmicb.2012.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Garcia O, Chandran K, Villas-Boas SG, Singhal N. 2016. Assessment of nitric oxide (NO) redox reactions contribution to nitrous oxide (N2O) formation during nitrification using a multispecies metabolic network model. Biotechnol Bioeng 113:1124–1136. doi: 10.1002/bit.25880. [DOI] [PubMed] [Google Scholar]
- 35.Freitag A, Bock E. 1990. Energy conservation in Nitrobacter. FEMS Microbiol Lett 66:157–162. doi: 10.1111/j.1574-6968.1990.tb03989.x. [DOI] [Google Scholar]
- 36.Starkenburg SR, Arp DJ, Bottomley PJ. 2008. Expression of a putative nitrite reductase and the reversible inhibition of nitrite-dependent respiration by nitric oxide in Nitrobacter winogradskyi Nb-255. Environ Microbiol 10:3036–3042. doi: 10.1111/j.1462-2920.2008.01763.x. [DOI] [PubMed] [Google Scholar]
- 37.Welsh MA, Blackwell HE. 2016. Chemical probes of quorum sensing: from compound development to biological discovery. FEMS Microbiol Rev 40:774–794 doi: 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kester RA, De Boer W, Laanbroek HJ. 1997. Production of NO and N2O by pure cultures of nitrifying and denitrifying bacteria during changes in aeration. Appl Environ Microbiol 63:3872–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozlowski JA, Stieglmeier M, Schleper C, Klotz MG, Stein LY. 2016. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J 10:1836–1845. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaumont HJ, Lens SI, Reijnders WN, Westerhoff HV, van Spanning RJ. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol Microbiol 54:148–158. doi: 10.1111/j.1365-2958.2004.04248.x. [DOI] [PubMed] [Google Scholar]
- 41.Beaumont HJ, Lens SI, Westerhoff HV, van Spanning RJ. 2005. Novel nirK cluster genes in Nitrosomonas europaea are required for NirK-dependent tolerance to nitrite. J Bacteriol 187:6849–6851. doi: 10.1128/JB.187.19.6849-6851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su H, Cheng Y, Oswald R, Behrendt T, Trebs I, Meixner FX, Andreae MO, Cheng P, Zhang Y, Pöschl U. 2011. Soil nitrite as a source of atmospheric HONO and OH radicals. Science 333:1616–1618. doi: 10.1126/science.1207687. [DOI] [PubMed] [Google Scholar]
- 43.Oswald R, Behrendt T, Ermel M, Wu D, Su H, Cheng Y, Breuninger C, Moravek A, Mougin E, Delon C, Loubet B, Pommerening-Röser A, Sörgel M, Pöschl U, Hoffmann T, Andreae MO, Meixner FX, Trebs I. 2013. HONO emissions from soil bacteria as a major source of atmospheric reactive nitrogen. Science 341:1233–1235. doi: 10.1126/science.1242266. [DOI] [PubMed] [Google Scholar]
- 44.Kwiatkowski AV, Laratta WP, Toffanin A, Shapleigh JP. 1997. Analysis of the role of the nnrR gene product in the response of Rhodobacter sphaeroides 2.4.1 to exogenous nitric oxide. J Bacteriol 179:5618–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartnikas TB, Wang Y, Bobo T, Veselov A, Scholes CP, Shapleigh JP. 2002. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a haem-copper protein. Microbiology 148:825–833. doi: 10.1099/00221287-148-3-825. [DOI] [PubMed] [Google Scholar]
- 46.Vaccaro BJ, Thorgersen MP, Lancaster WA, Price MN, Wetmore KM, Poole FL II, Deutschbauer A, Arkin AP, Adams MW. 2016. Determining roles of accessory genes in denitrification by mutant fitness analyses. Appl Environ Microbiol 82:51–61. doi: 10.1128/AEM.02602-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowka B, Daims H, Spieck E. 2015. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Environ Microbiol 81:745–753. doi: 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrews JH, Harris RF. 1986. r- and K-selection and microbial ecology. Adv Microb Ecol 9:99–147. doi: 10.1007/978-1-4757-0611-6_3. [DOI] [Google Scholar]
- 49.Zhu J, Chai Y, Zhong Z, Li S, Winans SC. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol 69:6949–6953. doi: 10.1128/AEM.69.11.6949-6953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joelsson AC, Zhu J. 2006. LacZ-based detection of acyl-homoserine lactone quorum-sensing signals. Curr Protoc Microbiol 1:Unit 1C 2. doi: 10.1002/9780471729259.mc01c02s3. [DOI] [PubMed] [Google Scholar]
- 51.Hood-Nowotny R, Umana NH, Inselbacher E, Oswald-Lachouani P, Wanek W. 2010. Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Sci Soc Am J 74:1018–1027. doi: 10.2136/sssaj2009.0389. [DOI] [Google Scholar]
- 52.Mellbye BL, Giguere A, Chaplen F, Bottomley PJ, Sayavedra-Soto LA. 2016. Steady-state growth under inorganic carbon limitation conditions increases energy consumption for maintenance and enhances nitrous oxide production in Nitrosomonas europaea. Appl Environ Microbiol 82:3310–3318. doi: 10.1128/AEM.00294-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, Smyth GK. 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erickson HE, Perakis SS. 2014. Soil fluxes of methane, nitrous oxide, and nitric oxide from aggrading forests in coastal Oregon. Soil Biol Biochem 76:268–277. doi: 10.1016/j.soilbio.2014.05.024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods: detailed methods, including medium formulation, promoter element prediction, corroboration of mRNA-Seq results by qRT-PCR, AiiA purification, activity measurement, and QS inhibition. Download
mRNA-Seq expression data generated by Illumina HiSeq 3000 Sequencer (Illumina, San Diego, CA) and analyzed using the CLC Main Workbench (CLC bio, Prismet, Denmark). Fold change data indicate changes in expression between AiiA-treated, QS-deficient cells and QS-proficient cells treated with heat-inactivated AiiA. Download
Putative lux-box-like promoter elements in N. winogradskyi. Sequence logos shown are graphic representations of aligned sets of putative promoter DNA sequences displaying frequencies of bases at each position as the relative heights of letters. The sequence logo also shows the degree of sequence conservation as the total height of a stack of letters, measured in bits. (a) Promoter motif A found upstream of nwiRI, the nirK cluster, and other genes. (b) Promoter motif B found upstream of nwiRI, the GppA phosphatase gene, and other genes. Table S2 shows all genes with these promoter motifs. Download
N-Decanoyl-l-homoserine lactone (C10-HSL) acyl-homoserine lactone (AHL) bioassay standard curve. Known concentrations of C10-HSL were added to bioassay cultures (see Materials and Methods in the main text) for estimation of the concentration of AHLs in N. winogradskyi cultures. The black line corresponds to the nonlinear regression of C10-HSL concentration (nanomolar, y axis) compared to AHL bioassay Miller units (x axis), and dotted lines indicate the 95% confidence band. The regression was calculated as follows: y = 0.0047x + 5 × 10− 5x2, R2 = 0.99. Download
Statistically significant changes in gene expression under QS-proficient conditions in N. winogradskyi
Putative QS-controlled genes with upstream lux-box-like promoter elements.
Bacterial strains and plasmids.
Comparison of fold changes in expression between QS-deficient and proficient treatments analyzed by quantitative PCR (qPCR) and mRNA-Seq.
Genes and primers used to corroborate gene expression.