Abstract
Two-component regulatory systems drive signal transduction in bacteria. The simplest of these employs a membrane sensor kinase and a cytoplasmic response regulator. Environmental sensing is typically coupled to gene regulation. The histidine kinase EnvZ and its cognate response regulator OmpR regulate expression of outer membrane proteins (porins) in response to osmotic stress. We used hydrogen:deuterium exchange mass spectrometry to identify conformational changes in the cytoplasmic domain of EnvZ (EnvZc) that were associated with osmosensing. The osmosensor localized to a seventeen amino acid region of the four-helix bundle of the cytoplasmic domain and flanked the His243 autophosphorylation site. High osmolality increased autophosphorylation of His243, suggesting that these two events were linked. The transmembrane domains were not required for osmosensing, but mutants in the transmembrane domains altered EnvZ activity. A photoactivatable fusion protein composed of EnvZc fused to the fluorophore mEos2 (EnvZc-mEos2) was as capable as EnvZc in supporting OmpR-dependent ompF and ompC transcription. Over-expression of EnvZc reduced activity, indicating that the EnvZ/OmpR system is not robust. Our results support a model in which osmolytes stabilize helix one in the four-helix bundle of EnvZ by increased hydrogen bonding of the peptide backbone, increasing autophosphorylation and downstream signaling. The likelihood that additional histidine kinases use similar cytoplasmic sensing mechanisms is discussed.
Keywords: hydrogen:deuterium exchange mass, spectrometry, Two-component regulatory system, EnvZ/OmpR, histidine kinase, Osmoregulation, Robustness, Super-resolution microscopy
1. Introduction
In bacteria, signal transduction is mainly achieved via two-component regulatory systems (TCRS). Bacterial species such as Escherichia coli that have numerous TCRS (~30) are able to adapt to and survive many environmental stresses, whereas H. pylori, for example, has only four TCRS and is severely niche-restricted. The first component of the TCRS is a sensor kinase (HK), usually an inner membrane protein, which undergoes autocatalytic phosphorylation on a conserved histidine residue. The HK transfers the phosphoryl group onto the second component called a response regulator (RR) at a conserved aspartic acid. Most RRs are DNA binding proteins and phosphorylation enhances their affinity for DNA. Much information exists as to how RRs bind DNA and how phosphorylation alters their activity. In contrast, relatively little is known as to the signals that activate HKs and how signaling occurs in response to environmental stress. This is in part because HKs are most often membrane proteins and they are not abundant compared to RRs.
The EnvZ/OmpR system is an archetype TCRS and its activation pathway has been intensely studied. In response to increasing external osmolality, EnvZ is phosphorylated, resulting in elevated OmpR ~ P levels. OmpR ~ P binds to the upstream regulatory regions of the porin genes encoding the outer membrane proteins OmpF and OmpC. OmpR ~ P binds to and activates transcription of ompC and activates and represses transcription of ompF (see Fig. 1). Both porins enable nutrient exchange, but OmpC has a slower flow rate (Nikaido and Vaara, 1985). Activation of ompC is one indicator that bacteria are inside a host and in the case of Salmonella enterica, EnvZ/OmpR activation leads to expression of virulence genes located on pathogenicity island 2 (SPI-2), which promote intracellular survival and replication (Chakraborty et al., 2015; Feng et al., 2003; Lee et al., 2000).
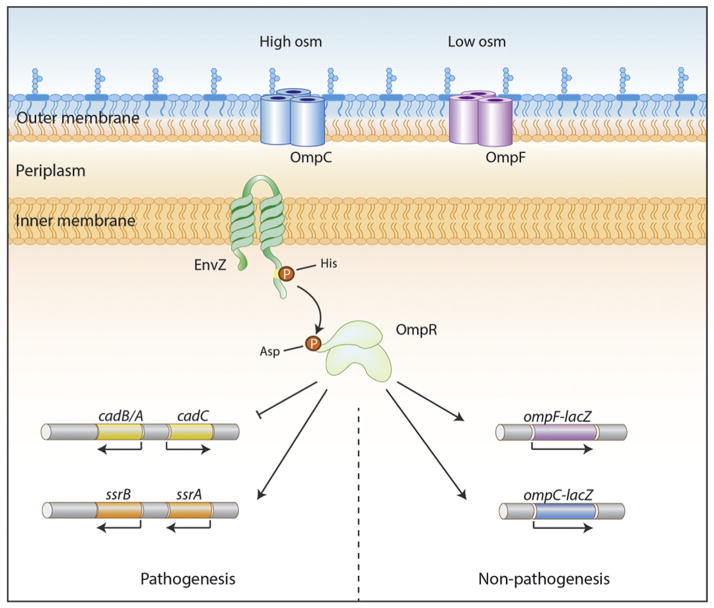
Fig. 1.
Schematic view of EnvZ/OmpR regulation. The HK EnvZ exists with N- and C-termini in the cytoplasm, contains two TMs in the inner membrane and a periplasmic loop in between TM1 and TM2. In response to intracellular osmolytes, autophosphorylation increases 10-fold at the conserved histidine residue (H243). The phosphoryl group is transferred to an aspartic acid residue on OmpR (D55) and OmpR ~ P binds to the regulatory regions of ompF and ompC porin genes to activate and/or repress transcription (“Non-pathogenesis”). In Salmonella, OmpR ~ P activates the SsrA/B TCRS that is located on pathogenicity island 2 (SPI-2) and is essential for virulence (“Pathogenesis”).
We recently examined the conformational changes that EnvZ undergoes in response to osmolytes. We were surprised to discover that the core of the osmosensing apparatus was in the cytoplasm. In the present manuscript, we summarize these findings and extrapolate to other HKs that may employ similar activation mechanisms. Questions of interest to the field of transmembrane signaling and two-component signaling are raised herein.
2. Material and methods
Bacterial culture conditions, β-galactosidase assays preparation of outer membranes and separation on urea SDS PAGE and phos-tag labeling were as previously described (Adediran et al., 2014; Mattison et al., 2002).
2.1. Plasmid construction
The cytoplasmic region of EnvZ (envZc, encoding amino acid residues 180–450), was amplified from E. coli MG1655 genomic DNA with primer pairs 5′-CCGGAATTCACCATGCGTATCCAGAACCGACCGTTGG-3′ and 5′-GGAAGATCTCCCTTCTTTTGTCGTGCCCTG-3′. This fragment was flanked with N-terminal EcoRI and C-terminal BglII restriction sites, and cloned into pMPMA6Ω that was digested with the same pair of enzymes, generating pMPMA6Ω-envZc plasmid. The gene encoding the fluorescent protein mEos2 was amplified with primers 5′GGAAGATCTATGAGTGCGATTAAGCCAGAC-3′ and 5′-CCGCTCGAGTTATCGTCTGGCATTGTCAGG-3′ then inserted downstream of envZc via BglII and XhoI sites of pMPMA6Ω-envZc, producing pMPMA6Ω-envZc-mEos2.
2.2. Coverslip cleaning
Coverslips (22 × 22 mm; Deckgläser) were first washed under running ultrapure H2O, then soaked in 1 M HCl at RT overnight. The coverslips were subsequently rinsed with ultrapure H2O and sonicated in ultrapure H2O for 30 min. They were then washed again under running ultrapure H2O followed by another round of 30 min sonication in ultrapure H2O. Next, they were washed again under running ultrapure H2O followed by dry blowing with N2 and lastly, they were placed in a 60 °C oven to completely remove the water on the coverslips before storing in a clean covered container.
2.3. Sample preparation for microscopy
Plasmid pMPMA6Ω-envZc-mEos2 was transformed into E. coli MG1655, E. coli MG1655ΔenvZ, E. coli MG1655ompR101and E. coli MG1655ΔompB by heat shock. These strains were cultured in 2 ml LB plus 100 μg/ml ampicillin overnight to prepare the seed culture. The next day, the culture was diluted 1/100 into LB medium supplemented with 0.02% L-arabinose to induce protein expression. The bacteria were harvested at 4000 × g for 5 min. The recovered cell pellet was washed once with PBS, fixed by incubation with 4% PFA in PBS for 15 min at room temperature, and washed with PBS twice. 1.5% low melting agarose was deposited onto a microscopy slide with a small indent (Toshin Riko). The agarose in the indent was covered with a cleaned coverslip for 1 h until the agarose hardened. The coverslip was then removed and the sample deposited onto the agarose pad. Using a new clean coverslip, the sample was sandwiched and immobilized between the coverslip and the agarose pad.
2.4. Microscope imaging
Super-resolution imaging was conducted with a Nikon Ti-E motorised inverted microscope equipped with a perfect focus system and total internal-reflection excitation (TIRF) scheme. Samples were activated with a 405 nm Coherent CUBE laser and excited with a 561 nm Coherent Sapphire laser by highly inclined laminated optical sheet (HILO) illumination. Fluorescence emission was collected with a CFI plane apochromat TIRF N.A. 1.49 100× oil immersion objective, and imaged onto an electron multiplying CCD camera (Andor DU897). Images were recorded at 50 ms per frame for around 10 000 frames. Super-resolution images were constructed using Quick PALM plug-in in ImageJ.
3. Results
3.1. The core of EnvZ environmental sensing is in the cytoplasm
In order to identify the region of EnvZ that was essential for osmotic signaling, we compared the hydrogen:deuterium exchange mass spectrometry profiles at low and high osmolality. For these experiments, we employed only the cytoplasmic domain of EnvZ, EnvZc, which is soluble, easy to over-express and purify and has all of the biochemical activities of the wildtype protein (Mattison and Kenney, 2002; Park et al., 1998). We compared hydrogen:deuterium exchange at low and high osmolality and then subjected EnvZc to pepsin digestion and collected the peptides for mass spectrometry. Using this approach, we identified a 17-amino acid peptide that was the core of the osmosensing apparatus (Wang et al., 2012). This region was significant, because it flanked the phosphorylated histidine and it overlapped with a region where amino acid substitutions had been isolated that dramatically influenced the kinase activity (see Fig. 2). Our results indicated that at low osmolality, residues in the vicinity of His243 were very dynamic and the helix was somewhat disordered. This region was also identified in the NMR structure as showing high rates of exchange at low osmolality (Tomomori et al., 1999). At high osmolality, the helix was stabilized by two hydrogen bonds in the peptide backbone. The same 17-amino acid peptide showed reduced exchange in the presence of sucrose and NaCl, i.e., it was independent of the osmolyte present. The increase in hydrogen bonding positions the histidine so that it is more readily phosphorylated, increasing the autokinase activity by ten-fold (Fig. 3). It was surprising that the core of the osmosensing apparatus was in the cytoplasm, but studies have shown that E. coli, unlike eukaryotes, was capable of concentrating its cytoplasm in response to osmotic stress (Cayley et al., 2000). Furthermore, EnvZc alone was capable of osmosensing, i.e., EnvZ did not need to be in the membrane in order to sense osmotic stress and activate porin gene expression (Fig. 4). With an enhanced understanding of the EnvZc signaling process, it is now evident that other kinases likely behave similarly (see below).
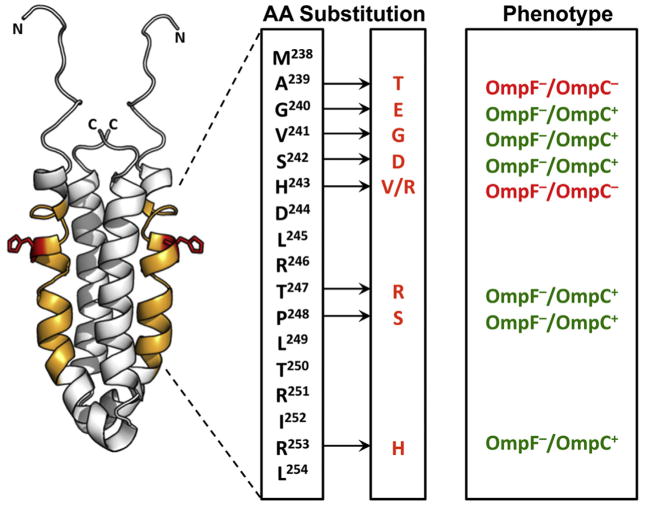
Fig. 2.
The EnvZ osmosensing core is in the cytoplasm. Structure of the EnvZc dimer (amino acids 223–289; PDB ID: 1JOY) (Tomomori et al., 1999) is shown in white. Orange residues (238–254) showed decreased deuterium exchange at high osmolality, the sequence is indicated in the box to the right. The red sidechain indicates His243, the site of autophosphorylation and phosphotransfer in EnvZ. Amino acid substitutions that alter EnvZ activity are highlighted with an arrow and the substitution is indicated, along with the porin phenotype observed. This region was also highly dynamic in the NMR structure that was solved at low osmolality (Tomomori et al., 1999). Used with permission from (Wang et al., 2012).
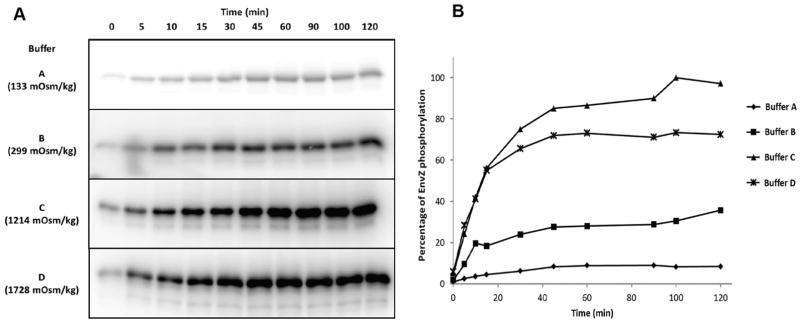
Fig. 3.
Osmolytes increase EnvZc autophosphorylation. (A) EnvZc was phosphorylated in the presence of γ-32P-ATP and after the times indicated, the reaction was stopped by the addition of sample buffer and the products were separated on SDS PAGE. Afterward, the gel was dried and exposed to film. The osmolality of the buffer is indicated to the left of the figure. (B) Densitometry from (A). Used with permission from Wang et al., 2012.
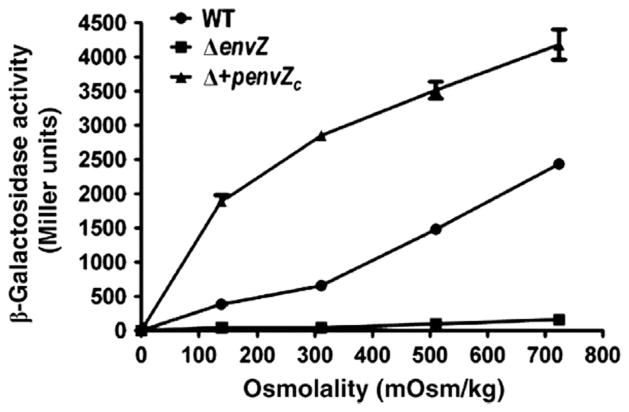
Fig. 4.
EnvZc can sense osmolality without being in the inner membrane. β-galactosidase assay of an ompC-lacZ transcriptional fusion as a function of osmolality. The activity of the wildtype strain is shown in circles, envZ null is shown in squares and the envZ null strain complemented with envZc is shown in the triangles. Measurements were made in triplicate and the experiment was repeated twice. The symbols represent the mean ± standard deviation (error bars). The absence of error bars indicates that the error was less than the symbol. Used with permission from Wang et al., 2012.
3.2. EnvZc activation is a consequence of reduced dynamics
Helix-stabilization of the His243-containing 17-amino acid peptide required the histidine, further highlighting its importance in conformational switching. An alanine substitution at His243, which was incapable of autophosphorylation and hence activation, failed to undergo this conformational switch and remained dynamic, i.e., it was stuck in the low-osmolality signaling state. Furthermore, the intact protein was required for the switch to a less dynamic conformational state, i.e., pre-digestion with pepsin and subsequent subjection of the peptides to high concentrations of osmolytes did not lead to reduced deuterium exchange. Thus, the histidine is essential for EnvZ to switch from a dynamic, low-activation state to a rigid, high-activation state. Our observations of EnvZc conformational dynamics explains why replacement of one region of EnvZc with the identical region from a thermophile enabled crystallization of EnvZc (Ferris et al., 2014), i.e., it was replaced with a less dynamic form, which enhanced crystallization. The high osmotic conditions used for crystallography also favored a stable conformation.
3.3. The OmpR binding region of EnvZc is flexible and exists in two conformations
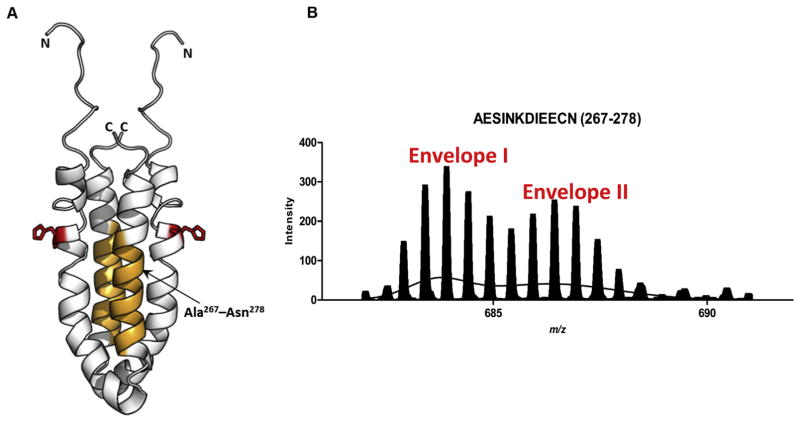
Most of the peptides of EnvZc that were analyzed by HDXMS exhibited a Gaussian distribution on m/z plots (Wang et al., 2012). This was evidence of Ex2 kinetics, in which the conformational change was faster than hydrogen:deuterium exchange. However, one peptide displayed a characteristic bimodal distribution (see Fig. 5), indicative of Ex1 kinetics in which the conformational change was slower than hydrogen:deuterium exchange. This bimodal distribution also suggests that this region (peptide AESINKDIEECN), was capable of existing in two different states (Fig. 5B). That peptide was on the second helix of the four-helix bundle (Fig. 5A) and was reported to be a site of interaction with the RR OmpR (Skerker et al., 2008; Szurmant and Hoch, 2010; Tomomori et al., 1999). The bimodal distribution of this peptide shifted to the less dynamic state with increasing osmolality. Thus, the OmpR interaction region of EnvZ is also flexible, existing in two conformations and activation reduces the flexibility of this region. Further evidence that this region was involved in OmpR binding comes from labeling of the cysteine (Cys-277) with the fluorophore Alexa 568, which prevented interaction with OmpR, as evidenced by a lack of a shift on native PAGE (Foo et al., 2015). This region also shows a high divergence among HKs (Fig. 6).
Fig. 5.
The OmpR binding site of EnvZc displays bimodal Ex1 kinetics (A) Structure of an EnvZc dimer (amino acids 223–289; PDB ID: 1JOY) (Tomomori et al., 1999) is shown in white. Orange residues (267–278) are part of the second helix proximal to the helix containing His-243 (red) and constitute an OmpR-binding site, based on cysteine labeling experiments (Foo et al., 2015), and (Skerker et al., 2008; Szurmant and Hoch, 2010). (B) The spectral envelope of residues 267–278 after 10 min deuterium exchange (charge state: +2) was fit to the sum of two Gaussian distributions. Envelope II corresponded to a significantly higher exchange than Envelope I. Used with permission from Wang et al., 2012.
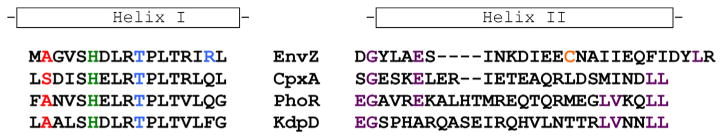
Fig. 6.
Sequence alignment of the His-containing osmosensor region of EnvZ and the OmpR interaction region. Helix I was identified by HDXMS as being highly dynamic at low osmolyte concentrations (Fig. 2). The sequence of EnvZ and some of its close homologues in E. coli is compared. The phosphorylated histidine is shown in green, in red is an alanine, where substitution with a threonine alters kinase activity (Russo and Silhavy, 1991), reducing affinity for ATP by >3- fold (Kenney, 1997). The blue threonine is a position where substitution with arginine or histidine produces a super kinase (Matsuyama et al., 1986). The second helix is predicted to be a site of interaction with RRs and modification of the cysteine in EnvZ (Cys-277, in orange) with a fluorophore prevents OmpR binding (Foo et al., 2015). It is evident that this region is highly divergent among HKs. Some conserved residues are highlighted in magenta.
3.4. EnvZc also responds to cytoplasmic pH
The response to osmolytes stabilized the region around the phosphorylated histidine through localized hydrogen bonding of the peptide backbone. That led us to hypothesize that EnvZ might actually be functioning as a pH sensor and that osmolytes might exert their effect by cytoplasmic acidification. To examine this possibility, we incorporated a DNA-based FRET biosensor that was pH-sensitive inside E. coli and Salmonella cells and exposed them to osmotic stress. The cytoplasm of both E. coli and Salmonella acidified, by approximately one pH unit (Chakraborty and Kenney, 2015). Furthermore, when Salmonella was present inside macrophage vacuoles, its cytoplasm acidified to ~pH 5.6, effectively becoming the pH of the vacuole. This acidification was dependent on OmpR repression of the cadC/BA operon, which normally serves to restore intracellular pH via lysine decarboxylation and cadavarine export, as well as OmpR-dependent up-regulation of the FoF1 ATP synthase (Chakraborty et al., 2015). Thus, OmpR appears to play a dual role in acid stress at least in Salmonella, by activating proton translocation via increased F1Fo ATP synthase coupled with elimination of proton consumption via repression of the cad system. Taken together, these combined effects enable wild-type Salmonella to reduce intracellular pH upon acid stress. Both of these effects require EnvZc (Chakraborty et al., 2015).
3.5. Accumulating evidence for cytoplasmic sensing by other HKs
Because amino acids in the four-helix bundle of HKs are highly conserved, as are the mechanisms of activation among TCRS, we wondered whether other HKs were capable of signaling without being in the inner membrane, i.e., how universal is this mechanism? It turns out that there are reports of cytoplasmic sensing as far back as the 1980s, but an interpretation of the results was lacking (Makino et al., 1989). An intrinsic prejudice existed that because HKs were in the membrane, the membrane was important for signaling. This was in part because the HKs are topologically similar to the chemoreceptors, which bind and respond to external (periplasmic) ligands (Ronson et al., 1987). It was therefore assumed that the periplasmic domains were essential for signaling. And clearly, in some HKs, such as those that do bind external ligands, it is. For example, CpxA binds periplasmic CpxP (Danese and Silhavy, 1998). DcuS (Etzkorn et al., 2008), CitA (Gerharz et al., 2003; Reinelt et al., 2003) and PhoQ (Bader et al., 2005) bind fumarate, citrate, and cationic antimicrobial peptides, respectively, which are also present in the periplasm. However, structural studies of DcuS also emphasize the role of protein plasticity in the cytoplasmic domain of DcuS in activation (Etzkorn et al., 2008). In other HKs, localization to the membrane is not essential for signaling. For example, in the E. coli phosphate sensor PhoR, a PhoR1084 construct lacking the TMs constitutively activated phoA to levels that were higher than the wildtype. Additional studies suggested a role for the TMs in turning off PhoR once phosphate was no longer limiting (Yamada et al., 1989). Subsequent studies that examined the PhoR topology remarked that the periplasmic region of PhoR was too small to serve as a signal-sensing area (Scholten and Tommassen, 1993). The authors proposed that a likely possibility was that the cytoplasmic domain between the membrane region and the kinase domain, i.e. between amino acids 52 and 220 (located in the cytoplasm) served this purpose, since no other function could be attributed to this extended region.
The tumor-inducing (Ti) plasmid of the plant pathogen Agrobacterium tumefaciens encodes the VirG/A TCRS. VirG senses and responds to phenolic compounds and is potentiated by acid pH and monosaccharides. In 1992, Chang and Winans reported that transmembrane domains TM1 and TM2 of VirA were not required for sensing phenolic compounds, and VirAc was also capable of potentiation by acid pH (Chang and Winans, 1992). Furthermore, virAc was able to complement tumorigenesis on K. diagremontiana leaves. However, the periplasmic domain was required for interaction with ChvE to sense monosaccharides, indicating that separate sensing domains can exist. The authors proposed that entry of acetosyringone into the cytoplasm was plausible and that both acetosyringone and acidic pH must interact with a cytoplasmic portion of VirA.
In Bacillus subtilis, the PhoP/PhoR system is homologous to the PhoB/PhoR system in E. coli and regulates the PHO regulon, including the alkaline phosphate gene under phosphate-limited conditions. Hulett and co-workers discovered that PhoRc could induce phoA under phosphate limiting conditions and that, in contrast to E. coli, repression of phoA under phosphate-replete conditions remained intact in the presence of PhoRc (Shi and Hulett, 1999). Although the authors considered it unlikely at the time, they suggested that the real sensor for the low phosphate signal triggering the Pho response may reside in the PhoR kinase domain (i.e., in the cytoplasm).
In Corynebacterium glutamicum, the cytoplasmic domain of the histidine kinase MtrB was purified and reconstituted into liposomes (Moker et al., 2007). The kinase was activated by sugars, amino acids, and polyethylene glycols. Because of the variable chemical nature of the stimulus, the authors argued that it was unlikely that activation was via a ligand binding to a specific binding site, rather they argued that osmolytes were changing the hydration state of the protein, leading to activation. Activation occurred only when solutes were added to the cytoplasmic side, indicating MtrB was sensing the cytoplasm. In support of this view, deletion of the periplasmic or HAMP domains did not alter the ability of MtrB to sense osmolytes.
A truncated derivative of KdpD, possessing both the large N- and C-terminal domain, but lacking all four TMs was still capable of sensing limiting K+ concentrations (Rothenbucher et al., 2006). The sensing locus was later discovered to be located in the C-terminal cytoplasmic domain (Heermann et al., 2003), again indicating that KdpD could sense low K+ without being in the membrane. Taken together, these results indicate that at least a subset of HKs respond to cytoplasmic signals rather than extracellular signals.
3.6. How do environmental signals lead to activation?
In some two-component signaling systems, dephosphorylation of the phosphorylated RR via the HK (the so-called “phosphatase activity”) limits the level of the activated RR and serves to reset the system. Although it was proposed that EnvZ phosphatase activity was reduced in the presence of high osmolality (Jin and Inouye, 1993), Fig. 3 clearly shows that high osmolality leads to a direct effect on EnvZ activation via autophosphorylation (Wang et al., 2012). These results are consistent with ATPase measurements of EnvZ-stimulated OmpR ~ P dephosphorylation at in-vivo ratios of EnvZ to OmpR (Kenney, 2010). It was concluded that EnvZ-dependent, OmpR ~ P turnover was too low for the phosphatase activity to be significant in signaling (Mattison and Kenney, 2002; Kenney, 2010). Furthermore, the affinity of EnvZ for OmpR ~ P is lower than its affinity for OmpR, this would drive the reaction in the wrong direction (King and Kenney, 2007; Mattison and Kenney, 2002). What then determines whether an HK will respond to environmental stimuli by altering autophosphorylation or RR ~ P dephosphorylation? In the case of the HK EnvZ, it is poised to increase autophosphorylation in response to signaling (i.e., OmpR ~ P levels are low in the absence of envZ (Slauch et al., 1988) and Fig. 3), whereas its close homologue HK CpxA is poised to regulate CpxR ~ P dephosphorylation (CpxR ~ P basal levels are high) (De Wulf and Lin, 2000). It is evident from the alignment of the EnvZ osmosensing peptide, that this region is highly conserved in HKs (Fig. 6), one notable difference between EnvZ and CpxA is the alanine at position 239. When the EnvZ alanine is replaced with a threonine, the affinity of EnvZ for ATP is reduced by >3-fold, and phosphotransfer to OmpR is substantially impaired (Kenney, 1997). Thus, this region seems unlikely to account for these differences in activity. Alternately, the second helix of the four-helix bundle (e.g. the RR binding site, Fig. 5) may contribute significantly to specificity, as there is much more divergence among HKs in this region (Fig. 6).
3.7. TM domains modulate EnvZ activity, but are not essential
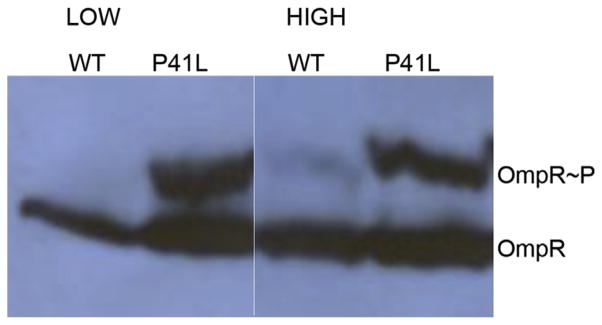
Substitutions have been isolated in the transmembrane domains of EnvZ that alter signaling. For example, envZ mutations were created by hydroxylamine (Tokishita and Mizuno, 1994) or nitrosoguanadine mutagenesis (Russo et al., 1993) or by UV irradiation (Hsing et al., 1998). Another study isolated missense mutations in envZ that were better able than wildtype E. coli MG1655 to colonize the streptomycin-treated mouse (Adediran et al., 2014; Leatham-Jensen et al., 2012). These envZ missense mutations were located in TM1 and TM2, and were more resistant to bile salts and colicin V compared to E. coli MG1655. The phenotype of the EnvZ mutants was consistent with the presence of higher OmpR ~ P levels (Adediran et al., 2014). OmpR ~ P was isolated from these strains and separated on SDS-PAGE after treatment with the Phos-tag reagent (see Fig. 7). At low osmolality, the wildtype strain had no detectible OmpR ~ P and at high osmolality OmpR ~ P levels were barely detectible (compare lanes 1 and 3, Fig. 7). This was surprising, because high osmolality increased the autokinase of EnvZ by 10-fold (see Fig. 3) (Wang et al., 2012), but produced barely detectible levels of OmpR ~ P. In contrast, the TM1 mutant P41L exhibited extremely high levels of OmpR ~ P (lane 2) that did not increase further with osmolality (lane 4). This concentration of OmpR ~ P was much higher than what was observed at high osmolality, which is considered to be a highly activating state. Mutants P148S (TM2) and V33E (TM1) behaved similarly (Adediran et al., 2014). At this high OmpR ~ P level, OmpR would be expected to exert pleiotropic effects, i.e., regulating genes normally outside of its repertoire. This situation is similar to a previously isolated envZ mutation (envZ473) that exhibited a pleiotropic phenotype and resulted in the OmpR-dependent regulation of genes that were considered to be outside of its normal regulon, including phoA, phoE, lamB and malE (Slauch et al., 1988; Verhoef et al., 1979; Wandersman et al., 1980; Wanner et al., 1979). Since the cytoplasmic domain of EnvZ (EnvZc) is capable of osmosensing (Wang et al., 2012), and pH sensing (Chakraborty et al., 2015) the trans-membrane domains likely serve to dampen the activity of the cytoplasmic domain. TM mutants such as P41L might then act to relieve the dampening effect of the transmembrane domains, activating EnvZc to phosphorylate OmpR to high levels. HDXMS experiments with intact, full-length EnvZ incorporated into nanodiscs (Ritchie et al., 2009) are in progress and will hopefully provide these insights.
Fig. 7.
A TM1 mutant of EnvZ (P41L) has significantly higher OmpR ~ P compared to wildtype. OmpR ~ P and OmpR levels in wildtype E. coli MG1655 (WT) and EnvZP41L (P41L), grown in M63 media in the absence (LOW) or presence (HIGH) of 15% sucrose. OmpR ~ P runs higher than OmpR due to the phos-tag label. Used with permission from (Adediran et al., 2014).
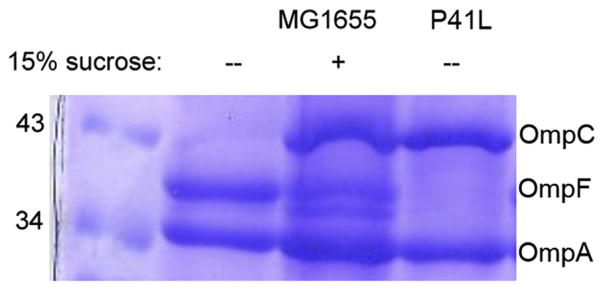
The elevated levels of OmpR ~ P observed in the TM mutant strains even at low osmolality (Fig. 7) should lead to elevated levels of OmpC. Indeed, outer membranes were isolated and the porins separated by SDS PAGE. Fig. 8 confirms this prediction. The wild-type expresses OmpF at low osmolality and OmpC at high osmolality (lanes 2, 3). The EnvZ TM mutants all expressed OmpC at low osmolality (lane 4), consistent with the elevated OmpR ~ P levels shown in Fig. 7. The question that is raised by this work is how does TM1 mutant P41L produce such high OmpR ~ P?
Fig. 8.
A TM1 mutant of EnvZ is locked in the high-osmotic signaling state and expresses OmpC at low osmolality. Outer membranes from MG1655 and the EnvZP41L mutant were isolated after growth in M63 in the absence (−) or presence of 15% sucrose (+). Molecular size markers are indicated on the left and outer membrane porins OmpC, OmpF and OmpA locations are indicated on the right. OmpA is a monomeric porin and a good loading control, since it is constitutively expressed. Used with permission from (Adediran et al., 2014).
3.8. New insights into the EnvZ/OmpR system
The TM mutants such as EnvZP41L provide new insight into the EnvZ/OmpR two-component signaling system. Previous studies determined the level of OmpR in the cell to be approximately 2 μM and OmpR concentration increased to 3.5 μM at high osmolality (Cai and Inouye, 2002). Yet, the level of OmpR ~ P in the wildtype strain at high osmolality was barely detectible (Fig. 7) (Adediran et al., 2014), a condition that is considered to be highly activating for OmpR. If there is 3.5 μM OmpR in the cell, why is there so little of it phosphorylated? This result raises the question of whether there is another yet unknown role for OmpR. Recent studies with GalR discovered that GalR was bound to many more sites on the chromosome than previously recognized and it was proposed to play a role in organizing the nucleoid (Qian et al., 2012). Perhaps OmpR behaves similarly. However, new measurements of OmpR levels made by counting fluorescent proteins or by single particle tracking suggest that the numbers obtained by quantitative western blotting may be substantially over-estimated (Durisic et al., 2014a,b; Foo et al., 2015; Wang et al., 2014). If true, then the low-level of OmpR ~ P would be more realistic physiologically. Additional super-resolution imaging experiments with OmpR will help to clarify this issue.
3.9. Where is EnvZc located?
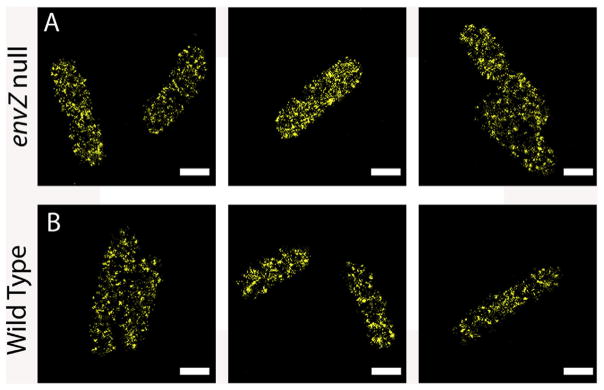
Because EnvZc was sufficient for osmosensing, we wondered whether it was weakly associated with the inner membrane or randomly distributed throughout the cytoplasm. To address this question, we constructed a fluorescent-protein fusion of EnvZc in which envZc was fused to mEos2. The PALM super-resolution images are shown in Fig. 9. When envZc-mEos2 was expressed in an envZ null background (Fig. 9A), the protein was distributed throughout the cytoplasm. It can clearly been seen in the figure that EnvZc was not localized to a discrete region of the cell. Similarly, when envZc-mEos2 was expressed in the wildtype strain containing EnvZ in the membrane, EnvZc-mEos2 was also distributed throughout the cytoplasm (Fig. 9B). It is worthy to note that there are “dark” regions where EnvZc is absent. This is most likely due to the presence of the nucleoid occupying these regions (Foo et al., 2015; Endesfelder et al., 2013). Although the presence of wildtype EnvZ did not alter the localization of EnvZc (Fig. 9B), over-expression of envZc in the presence of wildtype EnvZ increased ompC-lacZ activity by ~20%. This increase in activity by over-expression of EnvZc suggested that it might be worth reexamining whether the EnvZ/OmpR system is robust (Kenney, 2010); see below).
Fig. 9.
Localization of EnvZc by super-resolution microscopy. EnvZc-mEos2 was plasmid expressed as described in Methods in an envZ null strain (A) or the wildtype strain (B). The absence or presence of wildtype EnvZ did not alter the distribution of EnvZc-mEos2.
4. Is EnvZ/OmpR robust?
Robustness models predict that the steady state level of OmpR ~ P is insensitive to variations in [EnvZ] and [OmpR] (Shinar et al., 2007). The ratio of EnvZ to OmpR is approximately 1:35, based on quantitative western blotting, although single molecule estimates of fluorescent protein fusions suggest that this value may be severely over-estimated (Foo et al., 2015). A previous study examined whether altering EnvZ levels by plasmid over-expression altered the level of OmpR ~ P. OmpR ~ P levels were not measured, but rather inferred by ratiometric imaging of OmpF-YFP and OmpC-CFP fluorescence (Batchelor and Goulian, 2003). Additional copies of EnvZ were plasmid-expressed. By varying the IPTG inducer concentration, it was claimed that changing EnvZ 10-fold below and above EnvZ wildtype levels did not affect the ratio of OmpF/OmpC, implying that OmpR ~ P levels were unaffected. The authors concluded that the EnvZ/OmpR system was robust. However, other in vitro studies have measured OmpR ~ P turnover as a function of [EnvZc] and found that turnover was highly sensitive to changing EnvZc concentration (Kenney, 2010).
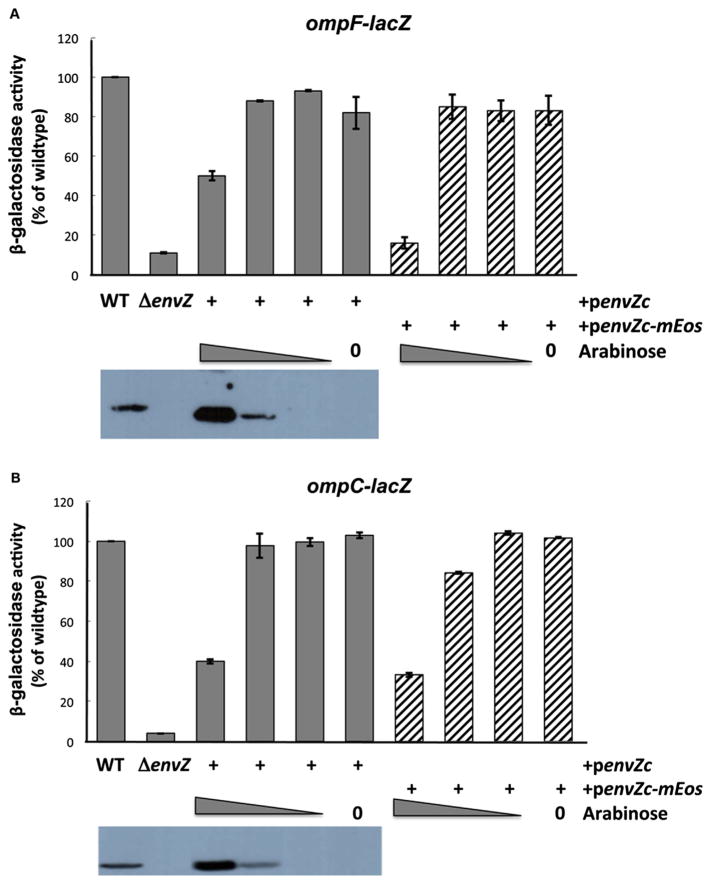
We re-examined this issue as we altered the concentration of EnvZc in order to produce enough EnvZc-mEos to generate the super-resolution images shown in Fig. 9. For these experiments, EnvZc-mEos was cloned into a pBAD plasmid and expression was induced by the addition of arabinose (Fig. 10). At the lowest concentration of arabinose, EnvZc was not visible on the western blot (Fig.10, shown below the graphs of β-galactosidase activity), yet the activities of transcriptional fusions to ompF- or ompC-lacZ were equivalent to the wildtype protein (compare columns 1 with columns 5–6). This concentration of arabinose was also used to produce the super-resolution image in Fig. 9. At the highest concentration of arabinose (0.2%, column 3), EnvZ protein was abundant and readily detectible (see blot), but at this concentration, the β-galactosidase activity was reduced to 50–40% of the wildtype activity (ompF vs ompC, respectively). Thus, the EnvZ/OmpR system is clearly not robust under these conditions of EnvZc over-expression. This experiment also demonstrates that the EnvZc-mEos2 fusion protein constructed for super-resolution microscopy has similar levels of activity compared to EnvZc (compare striped bars with shaded bars), indicating that the fusion does not affect EnvZc function.
Fig. 10.
Over-expression of EnvZc and EnvZc-mEos inhibits wildtype EnvZ activity. (A) β-galactosidase activity of ompF-lacZ stimulated by EnvZc (shaded bars) and EnvZc-mEos (striped bars). Cells were grown in low osmolality A media for 4 h (157 mOsm/Kg). Measurements were performed in triplicate and the mean and standard deviation was determined. Data were plotted as a percentage of the wildtype activity, where wildtype is 100%. Below the graph is a western blot. EnvZ wildtype protein was detectible (left lane), but was absent in the envZ null strain (blot lane 2) and was visible only at 0.2% and 0.002% arabinose (blot lanes 3 and 4, respectively). At lower arabinose concentrations, EnvZc protein was not detected, but the EnvZc activity was similar to wildtype (82–93%). The EnvZc-mEos activity was similar to wildtype (83–85%). (B) β-galactosidase activity of ompC-lacZ stimulated by EnvZc (shaded bars) and EnvZc-mEos (striped bars). Cells were grown in A media +15% sucrose (736 mOsm/Kg) for 4 h and measurements were performed as described in (A). As was observed with ompF-lacZ, EnvZc-mEos was essentially identical to wildtype in supporting OmpR-dependent transcription (84–104%).
4.1. Is EnvZc a mechanosensor?
When E. coli encounters a high osmotic environment, it responds by taking up potassium (if available) or by stimulating water efflux that leads to a concentrated cytoplasm (Csonka and Hanson, 1991). Previous studies have measured the concentration of the cytoplasm up to 1.8 osmoles (Cayley et al., 2000) in response to osmolytes. This enormous change in osmolality is also accompanied by large changes in volume; estimates of changes in spheroplast volumes suggest that this can be up to 3-fold (Sun et al., 2014). Perhaps when E. coli undergoes this large volume change, it transmits mechanical stress to the TM domains of EnvZ, altering its signaling properties. Stretching experiments using magnetic tweezers and various EnvZ constructs are underway in our laboratory that will test this hypothesis. Recent studies of bacterial cytoplasmic membranes indicate that their basic properties are significantly different from red cell membranes and lipid vesicles. For example, the spheroplast membranes are tensionless and the surface area appears to be controlled by a membrane reservoir equivalent to membrane folds (Sun et al., 2014). This finding may challenge our existing notion that mechanosensitive channels play an important role during volume changes in E. coli (Martinac et al., 1987; Perozo and Rees, 2003).
5. Summary
In the present manuscript, we have reviewed the evidence that EnvZ senses the cytoplasm, rather than responding to an external stimulus. We also present evidence that other HKs likely function similarly. Thus, not all HKs need to be present in the inner membrane in order to carry out their signal transducing functions. Super-resolution images establish that EnvZc was not localized to the inner membrane whether or not the wildtype membrane-embedded protein was present. Thus, cytoplasmic sensing by EnvZc is not a consequence of being associated with the inner membrane. Lastly, we demonstrate that the EnvZ/OmpR system is not robust when EnvZc is expressed to high levels. This raises the intriguing question as to why are these HK proteins present in the inner membrane?
Acknowledgments
We are grateful to Scott Stibitz for pointing out the previous work of Steve Winans on VirA. This work was supported by VA 5IO1BX-000372 to LJK and a Research Centre of Excellence in Mechanobiology from the Ministry of Education, Singapore.
abbreviations
- HDXMS
hydrogen:deuterium exchange mass spectrometry
- PAGE
polyacrylamide gel electrophoresis
- h
hours
- RT
room temperature
- HK
histidine kinase
- RR
response regulator
References
- Adediran J, Leatham-Jensen MP, Mokszycki ME, Frimodt-Moller J, Krogfelt KA, Kazmierczak K, Kenney LJ, Conway T, Cohen PS. An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic. Infect Immun. 2014;82:670–682. doi: 10.1128/IAI.01149-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Goulian M. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc Natl Acad Sci U S A. 2003;100:691–696. doi: 10.1073/pnas.0234782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SJ, Inouye M. EnvZ-OmpR interaction and osmoregulation in E coli. J Biol Chem. 2002;277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- Cayley DS, Guttman HJ, Record MT., Jr Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys J. 2000;78:1748–1764. doi: 10.1016/s0006-3495(00)76726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Kenney LJ. pH Regulation in E. coli and Salmonella. 2015 (In Preparation) [Google Scholar]
- Chakraborty S, Mizusaki H, Kenney LJ. A FRET-based biosensor tracks OmpR acidification of Salmonella in the macrophage vacuole. PLOS Biol. 2015 doi: 10.1371/journal.pbio.1002116. http://dx.doi.org/10.1371/journal.pbio.1002116. [DOI] [PMC free article] [PubMed]
- Chang CH, Winans SC. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J Bacteriol. 1992;174:7033–7039. doi: 10.1128/jb.174.21.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka LN, Hanson AD. Prokaryotic osmoregulation: genetics and physiology. Ann Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol. 1998;180:831–839. doi: 10.1128/jb.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P, Lin EC. Cpx two-component signal transduction in Escherichia coli: excessive CpxR-P levels underlie CpxA* phenotypes. J Bacteriol. 2000;182:1423–1426. doi: 10.1128/jb.182.5.1423-1426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durisic N, Cuervo LL, Lakadamyali M. Quantitative super-resolution microscopy: pitfalls and strategies for image analysis. Curr Opin Chem Biol. 2014a;20:22–28. doi: 10.1016/j.cbpa.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Durisic N, Laparra-Cuervo L, Sandoval-Alvarez A, Borbely JS, Lakadamyali M. Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate. Nat Methods. 2014b;11:156–162. doi: 10.1038/nmeth.2784. [DOI] [PubMed] [Google Scholar]
- Endesfelder U, Finan K, Holden SJ, Cook PR, Kapanidis AN, Heilemann M. Nanoscale organization of RNA polymerase in E. coli. Biophys J. 2013;105:172–181. doi: 10.1016/j.bpj.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzkorn M, Kneuper H, Dunnwald P, Vijayan V, Kramer J, Griesinger C, Becker S, Unden G, Baldus M. Plasticity of the PAS domain and a potential role for signal transduction in the histidine kinase DcuS. Nat Struct Mol Biol. 2008;15:1031–1039. doi: 10.1038/nsmb.1493. [DOI] [PubMed] [Google Scholar]
- Feng X, Oropeza R, Kenney LJ. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol Microbiol. 2003;48:1131–1143. doi: 10.1046/j.1365-2958.2003.03502.x. [DOI] [PubMed] [Google Scholar]
- Ferris HU, Coles M, Lupas AN, Hartmann MD. Crystallographic snapshot of the Escherichia coli EnvZ histidine kinase in an active conformation. J Struct Biol. 2014;186:376–379. doi: 10.1016/j.jsb.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Foo YH, Spahn C, Heilemann M, Kenney LJ. Single cell super-resolution imaging of E. coli OmpR during environmental stress. Integr Biol. 2015 doi: 10.1039/c5ib00077g. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerharz T, Reinelt S, Kaspar S, Scapozza L, Bott M. Identification of basic amino acid residues important for citrate binding by the periplasmic receptor domain of the sensor kinase CitA. Biochemistry. 2003;42:5917–5924. doi: 10.1021/bi0340595. [DOI] [PubMed] [Google Scholar]
- Heermann R, Fohrmann A, Altendorf K, Jung K. The transmembrane domains of the sensor kinase KdpD of Escherichia coli are not essential for sensing K+ limitation. Mol Microbiol. 2003;47:839–848. doi: 10.1046/j.1365-2958.2003.03348.x. [DOI] [PubMed] [Google Scholar]
- Hsing W, Russo FD, Bernd KK, Silhavy TJ. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Inouye M. Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. J Mol Biol. 1993;232:484–492. doi: 10.1006/jmbi.1993.1404. [DOI] [PubMed] [Google Scholar]
- Kenney LJ. Kinase activity of EnvZ, an osmoregulatory signal transducing protein of Escherichia coli. Arch Biochem Biophys. 1997;346:303–311. doi: 10.1006/abbi.1997.0315. [DOI] [PubMed] [Google Scholar]
- Kenney LJ. How important is the phosphatase activity of sensor kinases? Curr Opin Microbiol. 2010;13:168–176. doi: 10.1016/j.mib.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ST, Kenney LJ. Application of fluorescence resonance energy transfer to examine EnvZ/OmpR interactions. Methods Enzymol. 2007;422:352–360. doi: 10.1016/S0076-6879(06)22017-2. [DOI] [PubMed] [Google Scholar]
- Leatham-Jensen MP, Frimodt-Moller J, Adediran J, Mokszycki ME, Banner ME, Caughron JE, Krogfelt KA, Conway T, Cohen PS. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect Immun. 2012;80:1716–1727. doi: 10.1128/IAI.06193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AK, Detweiler CS, Falkow S. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J Bacteriol. 2000;182:771–781. doi: 10.1128/jb.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- Martinac B, Buechner M, Delcour AH, Adler J, Kung C. Pressure-sensitive ion channel in Escherichia coli. Proc Natl Acad Sci U S A. 1987;84:2297–2301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Mizuno T, Mizushima S. Interaction between two regulatory proteins in osmoregulatory expression of ompF and ompC genes in Escherichia coli: a novel ompR mutation suppresses pleiotropic defects caused by an envZ mutation. J Bacteriol. 1986;168:1309–1314. doi: 10.1128/jb.168.3.1309-1314.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Kenney LJ. Phosphorylation alters the interaction of the response regulator OmpR with its sensor kinase EnvZ. J Biol Chem. 2002;277:11143–11148. doi: 10.1074/jbc.M111128200. [DOI] [PubMed] [Google Scholar]
- Mattison K, Oropeza R, Byers N, Kenney LJ. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J Mol Biol. 2002;315:497–511. doi: 10.1006/jmbi.2001.5222. [DOI] [PubMed] [Google Scholar]
- Moker N, Reihlen P, Kramer R, Morbach S. Osmosensing properties of the histidine protein kinase MtrB from corynebacterium glutamicum. J Biol Chem. 2007;282:27666–27677. doi: 10.1074/jbc.M701749200. [DOI] [PubMed] [Google Scholar]
- Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Saha SK, Inouye M. Two-domain reconstitution of a functional protein histidine kinase. Proc Natl Acad Sci U S A. 1998;95:6728–6732. doi: 10.1073/pnas.95.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E, Rees DC. Structure and mechanism in prokaryotic mechanosensitive channels. Curr Opin Struct Biol. 2003;13:432–442. doi: 10.1016/s0959-440x(03)00106-4. [DOI] [PubMed] [Google Scholar]
- Qian Z, Dimitriadis EK, Edgar R, Eswaramoorthy P, Adhya S. Galactose repressor mediated intersegmental chromosomal connections in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:11336–11341. doi: 10.1073/pnas.1208595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Chapter 11-Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson CW, Nixon BT, Ausubel FM. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987;49:579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Rothenbucher MC, Facey SJ, Kiefer D, Kossmann M, Kuhn A. The cytoplasmic C-terminal domain of the Escherichia coli KdpD protein functions as a K+ sensor. J Bacteriol. 2006;188:1950–1958. doi: 10.1128/JB.188.5.1950-1958.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo FD, Silhavy TJ. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- Russo FD, Slauch JM, Silhavy TJ. Mutations that affect separate functions of OmpR the phosphorylated regulator of porin transcription in Escherichia coli. J Mol Biol. 1993;231:261–273. doi: 10.1006/jmbi.1993.1281. [DOI] [PubMed] [Google Scholar]
- Scholten M, Tommassen J. Topology of the PhoR protein of Escherichia coli and functional analysis of internal deletion mutants. Mol Microbiol. 1993;8:269–275. doi: 10.1111/j.1365-2958.1993.tb01571.x. [DOI] [PubMed] [Google Scholar]
- Shi L, Hulett FM. The cytoplasmic kinase domain of PhoR is sufficient for the low phosphate-inducible expression of pho regulon genes in Bacillus subtilis. Mol Microbiol. 1999;31:211–222. doi: 10.1046/j.1365-2958.1999.01163.x. [DOI] [PubMed] [Google Scholar]
- Shinar G, Milo R, Martinez MR, Alon U. Input output robustness in simple bacterial signaling systems. Proc Natl Acad Sci U S A. 2007;104:19931–19935. doi: 10.1073/pnas.0706792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch JM, Garrett S, Jackson DE, Silhavy TJ. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J Bacteriol. 1988;170:439–441. doi: 10.1128/jb.170.1.439-441.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Sun TL, Huang HW. Physical properties of Escherichia coli spheroplast membranes. Biophys J. 2014;107:2082–2090. doi: 10.1016/j.bpj.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Hoch JA. Interaction fidelity in two-component signaling. Curr Opin Microbiol. 2010;13:190–197. doi: 10.1016/j.mib.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokishita S, Mizuno T. Transmembrane signal transduction by the Escherichia coli osmotic sensor, EnvZ: intermolecular complementation of trans-membrane signalling. Mol Microbiol. 1994;13:435–444. doi: 10.1111/j.1365-2958.1994.tb00438.x. [DOI] [PubMed] [Google Scholar]
- Tomomori C, Tanaka T, Dutta R, Park H, Saha SK, Zhu Y, Ishima R, Liu D, Tong KI, Kurokawa H, Qian H, Inouye M, Ikura M. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- Verhoef C, Lugtenberg B, van Boxtel R, de Graaff P, Verheij H. Genetics and biochemistry of the peptidoglycan-associated proteins b and c of Escherichia coli K12. Mol Gen Genet. 1979;169:137–146. doi: 10.1007/BF00271664. [DOI] [PubMed] [Google Scholar]
- Wandersman C, Moreno F, Schwartz M. Pleiotropic mutations rendering Escherichia coli K-12 resistant to bacteriophage TP1. J Bacteriol. 1980;143:1374–1383. doi: 10.1128/jb.143.3.1374-1383.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based super resolution imaging. Proc Natl Acad Sci U S A. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner BL, Sarthy A, Beckwith J. Escherichia coli pleiotropic mutant that reduces amounts of several periplasmic and outer membrane proteins. J Bacteriol. 1979;140:229–239. doi: 10.1128/jb.140.1.229-239.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Makino K, Amemura M, Shinagawa H, Nakata A. Regulation of the phosphate regulon of Escherichia coli: analysis of mutant phoB and phoR genes causing different phenotypes. J Bacteriol. 1989;171:5601–5606. doi: 10.1128/jb.171.10.5601-5606.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]