Abstract
Virus-like particles (VLPs) constitute a promising platform in vaccine development and targeted drug delivery. To date, most applications use simple nonenveloped VLPs as human papillomavirus or hepatitis B vaccines, even though the envelope is known to be critical to retain the native protein folding and biological function. Here, we present tagged enveloped VLPs (TagE-VLPs) as a valuable strategy for the downstream processing and monitoring of the in vivo production of specific-site-functionalized enveloped influenza VLPs. This two-step procedure allows bioorthogonal functionalization of azide-tagged nascent influenza type A hemagglutinin proteins in the envelope of VLPs through a strain-promoted [3 + 2] alkyne–azide cycloaddition reaction. Importantly, labeling does not influence VLP production and allows for construction of functionalized VLPs without deleterious effects on their biological function. Refined discrimination and separation between VLP and baculovirus, the major impurity of the process, is achieved when this technique is combined with flow cytometry analysis, as demonstrated by atomic force microscopy. TagE-VLPs is a versatile tool broadly applicable to the production, monitoring, and purification of functionalized enveloped VLPs for vaccine design trial runs, targeted drug delivery, and molecular imaging.
Virus-like particles (VLPs) hold great promise as a platform for the development of long-lasting vaccine candidates, i.e., more-effective vaccines that do not require constant updates.1 Vaccines with improved clinical activities that use recombinant VLPs as their antigens have been developed,2−4 namely against hepatitis B as well as human papillomavirus viruses.1,5 Moreover, engineered VLPs carry additional promise for the generation of a wide range of nanoscale carriers in targeted drug delivery and molecular imaging.6 Previous studies have also shown VLPs to be a safe and efficient platform to deliver active proteins to cells.7 Additionally, genetically engineered VLPs have been exploited as drug-delivery systems for the targeted delivery of cytotoxic agents to tumors.8,9
The versatility of VLP platforms has prompted development of strategies to functionalize them. In contrast with genetic methods, chemical-based approaches for the production of modified VLPs are experimentally simpler, more efficient, less time-consuming, and more cost-effective. In one report, surface modification of adenovirus vectors was achieved by metabolic incorporation of azidohomoalanine (Aha) followed by a copper(I)-catalyzed alkyne–azide cycloaddition reaction.10 Alternatively, Francis and co-workers produced synthetic MS2 viral capsids functionalized with antibodies by using a oxidative coupling strategy.11 However, to date, the production of synthetically modified VLPs has been limited to simple nonenveloped VLPs. Complex enveloped VLPs show potential as platforms for the presentation of membrane proteins. The envelope is thus essential to maintaining the proteins in their folded and biologically functional state, which is critical to vaccine efficacy.12 There are clinical trials that report efficacy and safety improvements only after the incorporation of membrane proteins on the VLP surface, which induces a more-specific antibody response.13 Enveloped VLPs have the potential to generate antibodies of high diagnostic and therapeutic relevance to target transporters, ion channels, and membrane proteins present in the human genome that lack inhibitory antibodies because of current technical limitations.14 The potential of enveloped VLP platforms as vaccine candidates and drug carriers, together with the strict constraints of regulatory agencies for higher quality and safety control of biopharmaceuticals, highlights the need for new downstream processing methods for the production of functionalized enveloped VLPs.
Herein, we present a bioorthogonal labeling strategy that enables us to successfully functionalize complex enveloped influenza VLPs within live cells. The approach reported here, termed as tagged enveloped VLPs (TagE-VLPs) is composed of four key components: (i) residue-specific replacement of methionine (Met) by Aha15,16 to access azide-tagged precursor enveloped influenza VLPs, (ii) Aha-specific modification by strain-promoted alkyne–azide [3 + 2] cycloaddition (SPAAC) reaction,17 (iii) downstream processes monitoring and optimization, and (iv) discrimination between VLPs and baculovirus. The TagE-VLP strategy uses the baculovirus expression vector system that results in a considerable increase in downstream processing complexity because routine purification procedures and analytical methods are not able to strictly discriminate between VLPs and baculovirus.14,18 The main goal is to introduce a minimal size tag that does not disrupt particle size, charge, and biological function. This is a key step toward the development and improvement of the purification process, also enabling the site-specific labeling of the VLPs with synthetic molecules of interest, not just fluorescence tags, to address unmet medical needs. The versatility and flexibility of TagE-VLP offers the potential to develop functionalized enveloped VLPs for applications in vaccine design and targeted drug-delivery systems.
Results and Discussion
Design Criteria and Implementation of the TagE-VLP Platform
The ability to achieve fully functional chemically modified enveloped VLPs relies on the efficient incorporation of a tagged noncanonical amino acid at specific residues followed by bioorthogonal modification. This strategy, if performed in live cells, enables the monitoring, characterization, and VLP quantification from the beginning of the production process. This is a valuable tool which to improve upstream and downstream processes. To achieve site-specific in vivo VLP modification and labeling, we designed a two-step approach (Figure 1). The first step involves metabolic incorporation of noncanonical amino acid Aha, a Met analogue that contains an azide tag, into the hemagglutinin (HA) protein of influenza VLPs. We chose a triplet codon for Met to code our chemical tag because of the low incidence of Met in the gene of HA. In addition, Met replacement by Aha has been shown to be a powerful tool to introduce azide tags at specific residues on recombinant proteins or newly synthesized proteins on a cell without affecting the physicochemical properties or biological functions.15,16,19,20 The second step consists of bioorthogonal labeling with a cyclooctyne derivative (in this case, Click-iT Alexa Fluor 488 DIBO alkyne) through strain-promoted alkyne–azide [3 + 2] cycloaddition that enables precise placement of a modification into the nascent enveloped VLPs in vivo. Labeling may be performed at the desired purification step to achieve optimal yields and purified VLPs. Briefly, to produce in vivo labeled enveloped VLPs, cells were grown in culture and fed with Aha during protein synthesis. Met was added to a parallel cell culture, which was used as a control. During protein synthesis, Aha is incorporated as a surrogate for Met throughout the gene sequence of HA.
Figure 1.
Site-specific in vivo labeling of enveloped influenza VLPs. (a) Schematic representation of the procedure to metabolically introduce an azide-tagged noncanonical amino acid Aha for subsequent strain-promoted alkyne–azide [3 + 2] cycloaddition (SPAAC) labeling. During cellular protein synthesis, the Aha added to the culture medium is incorporated into nascent HA proteins. Addition of the Alexa 488-cyclooctyne reagent allows site-specific modification of HA (fluorescent tag in our case), which is reflected in VLP production. The modified HAs are incorporated into the vesicles’ envelope that is secreted from the cells that carry the chemical modification with it. (b) Confocal microscopy analysis of chemically modified VLP with the fluorescent probe Alexa 488. Dilutions (100-fold) of bulk VLPs (107 particles mL–1) were deposited onto IbiTreat 8 μ well slides. Multicolor fluorescent beads (500 nm) were used as size and green signal references (converted to grayscale). Red signal was also acquired (converted to grayscale), and from green–red merge images 500 nm beads can be discriminated from VLPs (yellow and green dots, respectively). In addition to color discrimination, for each particle detected, the full width at half-maximum was determined to evaluate the approximately 500 nm size of the control beads (red) and the subdiffraction limit VLP size (yellow) (see the Experimental section for more details). The control VLP sample shows no green signal (no labeling with Alexa 488), specific to SPAAC ligation in the experiment samples. Scale bars (white) indicate 2 μm in all images. Additional information regarding particle detection and RAW confocal images can be found in Figure S1.
HA is a protein from the envelope; thus, the VLPs carry the azide-tagged amino acid after budding from the host cells. At this stage, a complex particle is obtained that displays Aha-tagged HA in the envelope, and that is ideally suited for post-expression bioorthogonal labeling with a cyclooctyne fluorescent probe (Figure 1a). The time of addition of amino acid was optimized through small-scale (50 mL) batch production. The incorporation of Aha into HA protein was performed 12, 24, 36, and 48 h post-infection (hpi) of cells with baculovirus and assessed by confocal microscopy and flow cytometry analysis (Figures 1b, S1, and S2). The time of addition that resulted in a higher concentration of fluorescent VLPs was found to be 24 hpi (Figure 1b). Further scale-up (500 mL) of influenza VLP production was performed with this time reference. The versatility of our TagE-VLP strategy allows for bioorthogonal labeling at different stages of the production process of influenza enveloped VLPs. We chose to perform the SPAAC labeling after the VLPs were harvested because downstream processing (DSP) is the major bottleneck of bioprocess design. Fluorescent beads (500 nm) were used in confocal microscopy analysis as fiducial markers of size and the green fluorescence signal as a visual reference of successful VLP labeling and detection. As a result of its multicolor fluorescence profile, red signal was also acquired. Green–red merged images allowed discrimination between VLPs (green) and beads (yellow) dots not only by particle size but also by color. With this methodology, one can perform quantitative analysis on the detected VLPs. The number per μm2 and mean fluorescence intensity (If) of labeled VLPs determined by imaging processing of confocal images indicates the optimal conditions to perform the bioorthogonal functionalization step.
Particles with sizes below the diffraction limit of the microscope will appear as the point-spread function (PSF) of the instrument. VLPs are subdiffraction limit particles; therefore, their signal is the PSF of the microscope (approximately 240 nm). Particle size analysis was performed, and the full width at half-maximum (fwhm) was determined, a parameter that is a better approximation of particle size. Control fluorescent beads (size of approximately 500 nm) alone present a fwhm value of approximately 540 nm. The mixture between VLPs and beads showed a bimodal size distribution, which indicates the presence of both particles (Figures 1b and S1). As mentioned, with sizes below the resolution limit of the microscope, the signal is limited by the PSF, and the value observed for the VLPs has an average size distribution of 240 nm, which is the microscope’s PSF. The control VLP sample with added Met showed no green fluorescence signal, even after incubation with Alexa probe. This means that incorporation of Aha is necessary to observe fluorescence and that azide ligation between the noncanonical amino acid and the fluorophore is site-specific. The best time to incorporate Aha into HA protein was 24 hpi (time-dependent baculovirus infection was performed and is described in Figure S2a,b) (one reached approximately 3722 labeled VLP per cm2 with an If of 670.5 ± 167 au (arbitrary units) (mean ± SD), 40% and 100% higher than 12 hpi or 36 or 48 hpi, respectively).
TagE-VLP Platform Improvement of Downstream Processing of Influenza VLPs
By the optimization of the amino acid incorporation, the system becomes suitable for scale-up production and purification of labeled VLPs, detailed in Figure S3a. During production, to label mainly the HA protein, the addition of Aha amino acid to the cell culture was only performed after the late onset of gene expression (hpi = 24).
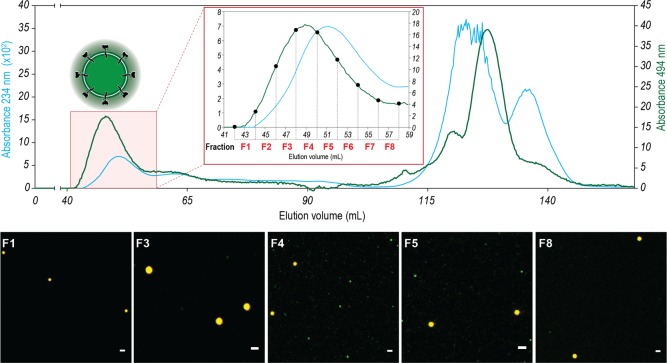
DSP proceeded with a standard protocol for influenza VLP purification already established in iBET’s laboratory. Analysis of all DSP steps to monitor the presence and concentration of modified VLP across the process was performed by confocal microscopy (Figures S3 and S4) and by flow cytometry (Figures S6 and S7) for both labeled and control VLPs. Alexa probe was added to the samples before analysis. Clarification of the supernatant, to remove any remaining cells and cell debris, was performed by means of depth filter technology.21 Intermediate purification involved an anionic exchange chromatography (AEX) and a concentration and diafiltration step by using ultrafiltration technology. AEX was operated in negative (flow-through (FT)) mode, which means that the working volume is still high. To make the process cost-effective, labeling with Alexa during purification was only performed after concentration of the flow through bulk. Size-exclusion chromatography (SEC) was used in this case as a model of a polishing step to remove a significant part of the remaining impurities such as baculovirus, DNA, or host cell proteins (Figures 2, S4, and S8). Elution of influenza VLPs was monitored by detecting the absorption of the eluted solution at 234 and 494 nm (the maximum absorption wavelength of Alexa Fluor 488). Absorption at 234 nm reports roughly all biomolecules that pass through the detector while absorption at 494 is specific for the fluorescent VLPs that incorporated the Alexa-488 probe. This dual detection allows better discrimination between the particles of interest VLP and all other contaminants such as baculovirus. Although SEC removed some baculovirus, the product still contained impurities. A fluorescence-activated cell-sorting (FACS) step was added at the end to overcome this issue and separate VLPs from baculovirus by taking advantage of their distinct particle sizes (150–200 and 300–400 nm, respectively) (Figures S3 and S5). Additional information regarding particle detection, individual green and red channels and RAW confocal images can be found in Figure S4a,b for the control and Aha addition experiments. To validate the approach, each sample of the DSP process was also studied by flow cytometry. SSC–green fluorescence and red–green fluorescence 2D correlograms are depicted in panels a and b of Figure S8, respectively. Detailed procedures for confocal microscopy and flow cytometry acquisition and apparatus are available in the Experimental section.
Figure 2.
Detailed interpretation of VLP polishing step by means of size-exclusion chromatography for the Alexa-488 labeled VLP. A pair of detection signals were used to monitor SEC. The elution profile was monitored by detecting the absorption of the eluted solution at both 234 nm (blue curve) and 494 nm (green curve) (emission wavelength of Alexa probe). The absorption at 234 is where roughly all biomolecules that pass through the detector contribute to the signal obtained either by absorption or by light scattering (DNA, proteins, and lipids). The detection of the absorption at 494 is specific for the fluorescent VLPs that incorporated the Alexa-488 probe. This dual detection allows better discrimination between the particles of interest (VLP) and all other contaminants such as baculoviruses. VLPs are contained in the column void volume. For each SEC fraction, confocal microscopy images were taken to monitor the presence of modified VLP (green fluorescent VLP). Scale bars (white) indicate 2 μm in all images. Images are ROI from larger independent images to better visualize the subdiffraction green dots. Merge (green–red) images are shown for clarity. According to the scheme highlighted in Figure 1, a red signal was also acquired, and from green–red merge images, 500 nm beads can be discriminated from VLPs (yellow and green dots, respectively). At the end of the SEC, between 115 and 130 mL of elution volume, concerning the elution of small molecules, there is evidence of detector signal saturation due to the elution of a high concentration of free Alexa-488 in the solution used in the labeling of VLPs.
Recent reports have described the ability to detect and sort lipid-based particles, exosomes, and enveloped viruses, with flow cytometry.22,23 HA concentration and number of particles measurement was performed to assess VLP production yields using the TagE-VLPs strategy. HA concentration at harvest time was 1.4 μg mL–1 for both Met control and Aha experiments. Nanoparticle tracking analysis revealed that both the control and experiment cultures produced VLPs in the same order of magnitude (1.56 × 109 and 1.39 × 109 particles mL–1), respectively, meaning that the VLP production yields were not affected.
Further analysis of total protein, DNA concentration, and baculovirus content were also performed during upstream and downstream processing. These analytical methods are essential to characterize the bioprocess and to make sure that control and labeled VLP data are comparable (data not shown). The polishing step (SEC; Figures 2 and S4) and FACS (Figures 3, S6, S7, and S8) are discussed later. It is consistently observed (Figure 1b) that the control VLPs, with Met, does not exhibit significant green fluorescence signal, as seen in the flow cytometry 2D correlograms from Figures S6, S7, and S8. (The VLP gate for every DSP step of the control reveals no increase in positive Alexa 488 population, whether or not a positive Alexa 488 signal is observed for the Aha experiment). Moreover, the concentrated Aha-labeled VLP sample showed an If of 670.5 ± 167 au in the confocal images, whereas only background fluorescence intensity levels were detected in the control VLP samples. The residual green signal detected at the control concentration step is mainly a result of unspecific binding of the probe (incorporation onto hydrophobic moieties of lipid membranes) to process impurities that are more easily observed at higher concentrations. Furthermore, the concentration detected was residual relative to the labeled VLPs (Figures S6, S7, and S8). The data confirmed the presence of labeled VLPs across DSP, with levels of concentration and purity consistent with the evaluated step. Ultrafiltration permeates and column-wash fractions from both AEX and SEC were analyzed, and no loss of labeled VLP was detected. These results demonstrate that this methodology is a powerful tool to monitor, online or at-line, each of the steps during manufacture of the product of interest, which can play an important role in DSP optimization.24 Online and at-line process analyzers are inserted in one of the major categories of process analytical technology (PAT) tools, having important applications in the biopharmaceutical industry.
Figure 3.

Discrimination between VLPs and baculovirus by FACS analysis. (a) Flow cytometry of a baculovirus sample (used for infection and VLP production). A 2D correlogram of side scatter and green fluorescence signals are shown with 5% contour plots of each population. A size-scatter size ruler was made with 100, 200, and 500 nm size fluorescent beads (grayscale). Gate thresholds for negative and positive populations were performed using 100 nm bead signals. The top-right quadrant indicates green fluorescent positive >100 nm particles (VLP). In each chart, the [100–200] nm per Alexa 488 positive population gate (VLP) was built to quantify and sort the presence of labeled VLP. This analysis monitors the scatter profile of the 200–400 nm rods (red) of baculoviruses that have no green fluorescence. (b) Flow cytometry of a VLP sample before the DSP steps (blue) shows that there are clearly two particle populations: one green positive population at ≈200 nm and one with lower and nonexistent green fluorescence that has a wider size distribution. (c) Flow cytometry of F4 from the VLP SEC purification step. Analysis of the green fluorescent signal shows that the >200 nm fraction is reduced relative to A as a result of the VLP-specific green fluorescence signal. This sample was sorted with populations P1 (<200 nm population, VLP-rich) and P2 (>200 nm population, baculovirus-rich). (d) 2D correlogram of red and green fluorescence signals are shown for each population depicted in I (baculovirus), II (before DSP), and III (SEC F4). Gate thresholds for negative and positive populations were performed using 100 nm bead signals: the bottom-right quadrant is the VLP-positive quadrant (green, positive particles and red, negative particles). A significant green signal and no red signal correlates with modified VLP samples. Figures S6a,b; S7a,b; and S8a,b depict additional flow cytometry performed in the study for all steps of the DSP process.
The purification process flowchart was chosen as a proof of concept for the applied methodology, which means that other schemes and types of chromatography can also be exploited. As already discussed, labeling was performed prior to the SEC step to decrease the process cost. Because the previous chromatographic step was performed in a negative mode, the SEC step allowed the optimization process to be fine-tuned. Each fraction of this polishing step was interpreted in detail by confocal microscopy (Figures 2 and S4) and confirmed by flow cytometry analysis (Figure S8). One of the drawbacks of the baculovirus expression system is that it is difficult to remove baculovirus from the purified complex enveloped product.25,26 The rod-shape form of baculovirus makes it difficult to differentiate them from VLPs because, even with different detection methods, there are angles at which their sizes appear similar. Like VLPs, baculovirus also bud out of the cell to give an envelope content that is similar in the two species.26 Control VLPs were injected onto a SEC column, and the elution profile was followed at 234 nm (280 and 260 nm were also tested but showed lower absorbance intensity and signal-to-noise ratio) (data not shown). The elution profiles of both the labeled VLP and control samples are very similar, with the VLP sample eluting at the void volume of the SEC column as expected (Figure 2). For modified VLP bulk, the absorbance intensity at 494 nm was also evaluated (emission wavelength of Alexa probe). Usually, VLP samples are contained in the void volume peak of the chromatogram as a result of their high Stoke radius. However, analysis of the two wavelengths shows that the two peaks are not superimposable. Fraction F4 presented a higher fluorescence (494 nm) value, which does not correspond to the peak maximum at 234 nm. Confocal microscopy images also revealed that F4 contains a higher concentration of labeled VLPs, which is in agreement with results from SEC chromatogram and flow cytometry analyses (Figures 4a,b and S8). This means that a mixture of VLPs and other components elute in the void volume. Due to its rod shape, baculovirus elutes in different volumes of the chromatogram.25 Notably, this labeling methodology enables better discrimination between VLPs and other process impurities, in particular baculovirus, which is the major contaminant in this process. The online detection of VLPs leads to a more-informed decision as to which fractions should be selected to continue in the purification process, an important step to obtain a higher recovery yield with improved VLP purity. The peak at the end of the chromatogram corresponds to free probe (494 nm) or DNA and low-molecular-weight contaminants (234 nm).
Figure 4.
Integrity and functionality of modified VLPs. (a) Quantification of the number of fluorescent particles detected in each DSP step in the control, unlabeled VLP, and in the labeled VLP (steps from Figure S3a). (b) Quantification of the number of fluorescent particles detected in each SEC fraction in the labeled VLP purification (SEC from Figure 2). (c) Hemagglutination assay for each step of the modified VLP purification process to assess preservation of HA biological function. (d) Hemagglutination assay for each fraction of the SEC step. (e) TEM analysis of control VLPs from the concentration step of the purification process. Scale bar indicates 100 nm. (f) TEM analysis of modified VLPs from the concentration step of the purification process. Scale bar indicates 100 nm. Uncropped and additional TEM images are available in Figure S10. The determination of the concentration of the labeled VLP solution based on the particle detection in panels c and d was also performed using eq 2 from the Experimental section and is available in Figure S4b.
TagE-VLP Maintenance of Integrity and Functionality
Modified VLP integrity and HA biological function were assessed by means of a hemagglutination assay (Figure 4c,d). The correlation of their biological integrity with the number of fluorescent particles is also demonstrated by the quantification of the acquired images and particle counting (Figure 4a,b). Control and modified VLP HA concentration values are comparable for each step of the production and purification processes. This assay evaluates the biological interaction between sialic acid receptors present in erythrocytes and HA protein.27−29 The same interactions happen under our conditions, which proves that the HA biological function is preserved even after chemical functionalization and labeling. The ability of these enveloped VLPs to maintain their characteristics may indicate that this methodology could be used to functionalize these particles with distinct targets. HA content increases as fluorescence intensity increases (Figures 2 and 4d), which means that the labeling is specific for Aha-containing influenza VLPs. SEC fractions F4 and F5 give a higher percentage of HA recovery, which is in agreement with confocal microscopy results and number of fluorescent VLP detected (Figures 2 and 4a,b) and flow cytometry data (Figure S8).
As mentioned above, modified VLPs can be fluorescently labeled using Alexa Fluor488 probe. When this labeling is taken advantage of, both control and TagE-VLPs were incubated with Alexa, separated in a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel, and revealed using a fluorescent image analyzer (Figures 5c and S11). No fluorescent bands were detected for control VLPs, meaning that no labeling occurs without the noncanonical amino acid incorporation, as previously described. However, in VLP samples with Aha modification, it was possible to detect three fluorescent bands. These bands were excised from the gel and identified by nano liquid chromatography–mass spectrometry (nanoLC–MS). Bands identified with (2) and (3) were confirmed by mass spectrometry as Hemagglutinin of influenza A virus. Band (3) is probably a result of protein degradation during time. Band (4) was identified by mass spectrometry as a Telokin-like protein of Autographa californica nuclear polyhedrosis virus, i.e., a protein from the baculovirus. Because this virus replicates during infection and VLP production, it is possible to obtain some residual baculovirus Aha incorporation. However, Aha addition to the cell culture was only performed after the late onset of gene expression to minimize this possibility. Gel fluorescent data supports the specificity of Aha incorporation into HA. The fluorescence intensity of baculovirus (band (4)) is clearly lower than HA corresponding ones and only appears when samples were precipitated resulting in VLP degradation. Mass spectrometry also detected Met–Aha modification in several peptides of the fluorescent bands, observed by a shift in the spectra (Supporting Table 1). Therefore, Aha incorporation is preferentially made into HA proteins, a result of amino acid time of addition optimization. TEM analysis was performed to assess the presence, integrity, and morphology of both control (Figure 4e) and modified VLP samples (Figure 4f). The morphology is maintained; their size (∼170 nm) and spherical shape are similar (Figure S5a,b). Furthermore, ultrastructural details of both VLP envelopes revealed characteristic influenza HA spikes.30,31 Moreover, Western blot analysis for HA- and M1-specific detection was performed (Figures 5a,b and S11), revealing that both control and modified VLPs have the two influenza proteins. Protein identity was confirmed by mass spectrometry (Band ID 1 and 2 from Supporting Table 1). This result further confirms the intact composition of modified VLPs.
Figure 5.

Identification of HA and M1 proteins by Western blot analysis and fluorescent band detection of labeled influenza VLPs’ proteins. (a) M1 influenza protein detection on control and labeled VLPs by Western blot analysis. M1 protein from influenza A H1N1 strain was used as positive control (M1 standard). Band (1) was excised and identified as M1 by mass spectrometry. (b) HA influenza protein detection on control and labeled VLPs by Western blot analysis. H3 VLP from influenza A H3 strain was used as the positive control (H3 standard). Band (2) was excised and identified as HA by mass spectrometry. (c) SDS-PAGE gel fluorescence detection of control and labeled VLPs incubated with Alexa 488 probe. Bands (2) and (3) were excised and detected as HA by mass spectrometry. Band (4) was detected as a Telokin-like protein of baculoviruses. The term “pp” means precipitated sample.
FACS Analysis Enabling of VLP and Baculovirus Discrimination
Flow cytometry analysis allowed the detection and characterization of labeled VLPs and size discrimination between these particles (100–200 nm spheres) and baculovirus (200–400 nm rods). Fluorescent beads of 100, 200, and 500 nm were used as a particle size ruler in flow cytometry with the side-scatter signal,22,23 which was then used to evaluate the VLP sample size distribution. It is possible to do a direct correlation between bead size and VLP samples because their refractive indexes are similar.
A 2D correlogram of side scatter and green fluorescence signals was acquired for each bead (100, 200, and 500 nm) for the VLP and control samples to detect the presence of baculovirus and evaluate further particle separation by cell sorting. A baculovirus sample (used for cell infection and VLP production) was analyzed to monitor the scatter profile of these 200–400 nm length rod-shape particles. Sizes that ranged from 200 to 500 nm were observed, which indicates that these rods are polydisperse (Figure 3a). A sample from the harvest step showed that at this stage there are clearly two particle populations: one green positive population at ∼200 nm and one with lower green fluorescence that has a wider size distribution (Figures 3b and S7). The size heterogeneity comes from a VLP–baculovirus mixture because the sample is from an early purification step still rich in baculovirus contaminants. A fraction from the SEC step (Figures 3c and S8) shows that the green fluorescent signal of the >200 nm fraction is reduced relative to baculovirus and harvest panels (Figure 3a,b). The presence of baculovirus is reduced relative to VLP in the SEC sample that is from a final purification step. However, the SEC fraction still contains some baculovirus because the baculovirus has a broad elution profile as a result of their rod-like shape. VLP sorting of the SEC F4 fraction sample was performed to separate the VLPs from baculovirus. Fluorescent beads (200 nm) were used to define two sorting populations: P2 (>200 nm particles) is a baculovirus-rich population, and P1 (<200 nm particles) is VLP-rich (Figure 3c). This strategy increased the yield on VLP production and minimized the presence of baculovirus in the final DSP product.
TEM analysis of baculovirus control (used to infect cells) and modified VLP (after concentration) was used to evaluate the size and heterogeneity of samples (Figure 6a). Baculovirus samples are characteristically rod-shaped with an average size of approximately 250 nm. As expected, the concentrated VLP sample contained both small and large particles, which corresponds to VLPs or baculovirus and process impurities, respectively. The size of the VLPs is different from the one presented previously (Figure 4e,f), which confirms the heterogeneity of the system.32 It is clear that there are unwanted larger particles at this stage of the process that are not VLPs, or at least not complete ones, because of the lack of HA spikes. This result provides an indication of what to expect from both sorting populations. The presence of VLP and baculovirus was performed for the baculovirus control and P1 and P2 sorting populations by atomic force microscopy (AFM; Figure 6b). AFM images of the baculovirus control samples revealed the well-known rod-like structure. Images of the P1 sorting population (<200 nm, VLP-rich fraction) revealed only the presence of spherical particles. However, the P2 sorting population (>200 nm, baculovirus-rich fraction) presented only large particles, which mainly consisted of baculovirus rod-shaped particles and other large process impurities. The baculovirus size in the P2 sorting population is similar to that calculated from the control sample. This data confirms that our system is suitable as a FACS purification step, and represents important progress to meet the increasing demand for VLP–baculovirus separation and DSP quality control requirements.
Figure 6.
Modified VLP detailed analysis. (a) TEM images of the major impurity in VLP production, a baculovirus, and a VLP sample before sorting revealing optimal VLP and large undesirable particles. Scale bars indicate 100 nm in all images. Uncropped and additional TEM images are available in Figure S10. (b) AFM images (error and 3D images) of a baculovirus control sample, showing rod-like morphology of this virus. Samples from each DSP step were sorted into P1 and P2, as described in Figure 3c. AFM images of P2 and P1 samples clearly show <200 nm spherical particles, consistent with VLP, and on the opposite side, the >200 nm show rod-shaped, nonspherical particles more akin to baculovirus morphology, as shown in the left AFM panels. The longitudinal (fill line) and transversal (dashed lines) cross-sections were performed to better illustrate the spherical and rod shapes of each particle visualized in each sample.
Conclusions
Herein, we report a straightforward two-step strategy to chemically functionalize and label complex enveloped VLPs in vivo by using SPAAC. Unlike previous reports, this methodology is designed for VLPs that are able to display membrane proteins in their lipid bilayer and potentiate the plethora of antigens that can be presented to cells in vaccine design. The functionalization of these particles, particularly those with membrane proteins, is not straightforward with current methods. The technical challenges and antigen choice limitations can be overcome by using the TagE-VLPs strategy described here. This flexible and site-specific system does not have an impact on biological function of the VLPs studied and can be potentially used in several virus or VLPs. The reported strategy can be used to functionalize these particles and expand their utility in exciting applications, such as vaccine design, drug delivery or molecular-imaging agents for diagnostics.
Importantly, the use of flow cytometry to analyze polydisperse lipid suspensions that contain VLPs has greatly enhanced our knowledge of their heterogeneity. Techniques that characterize, discriminate, and accurately separate each individual population with particle counting and concentration determination are scarce for nanosized particles. The extended use of FACS to characterize VLPs provides a better description and understanding of purified VLPs produced under different methods and systems and enables proper identification and separation of contaminants and desired particles. This easy-to-use and fast methodology only requires fluorescent beads for size calibration, which lends this technique to be used as an at-line, high-throughput, nondisruptive method with which to monitor all stages of VLP production in addition to current techniques, which are time-consuming and typically do not allow analysis of the VLPs in their native form. Both fluorescent labeling and FACS methods described in this manuscript are powerful tools for DSP monitoring and optimization that allow the improvement of product recovery yields and increase VLP purity levels.
It is worth to note that this method is not exclusively dependent on two-dimension particle discrimination. The new bioorthogonal labeling method here reported allows to engineer biologically functional VLPs, for instance, by conjugation of synthetic epitopes that are nonfluorescent, as the size signal enables one-dimension functionalized VLP–baculovirus separation. We have sorted VLPs from baculovirus using their distinctive size and green color discrimination (see Figure S9). Having both differentiating parameters only increased the accuracy of the method, but restricting to one variable does not obviates its application and multifunctionalize the Aha moiety with other bioactive molecules.
Experimental Section
Cell Culture
High Five cell line (Trichoplusia ni derived BTI-Tn-5B1-4) was obtained from Invitrogen (B855-02, Invitrogen Corporation, Paisley, UK). Cells were routinely cultured in ESF921 protein-free medium (960-001-01, Expression Systems) in 125 mL Erlenmeyer flasks (430421, Corning) with a working volume of 10 mL. High Five cells were kept in a humidified incubator at 27 °C and 110 rpm. Every 3–4 days, after reaching a cell concentration of 2–3 × 106 cells mL–1, they were reinoculated at 3 × 105 cells mL–1. Cell concentration and viability were determined by hemocytometer cell counts (Brandt, Wertheinmain, Germany) and trypan blue exclusion dye method (Merck, Darmstadt, Germany). High Five cells were chosen as producer cell line due to the higher productivity achieved for the proof-of-concept of the method described. However, when considering late-stage biopharmaceutical production, it is crucial to evaluate the safety of this host cell line because it was reported the presence of latent alphanodavirus in the High Five genome.33,34 By itself, the virus may not constitute a burden; however, if it assumes the replicative form, it may contaminate final product preparations, requiring extra downstream processing efforts.35 Nevertheless, it has been described as there being no contamination present in the Invitrogen master High Five cells bank,36 the cell source used, and additionally, there are not any reports indicating the infection of human hosts by the alphanodavirus. In fact, both High Five and Sf9 cell lines have regulatory acceptance for manufacturing of biologicals, such as Cervarix, the GSK HPV vaccine, or Flublock, the Protein Sciences influenza vaccine.33
VLP Production and Metabolic Labeling Optimization
For production studies, cells were cultured in 500 mL Erlenmeyer flasks (431145, Corning) with a working volume of 50 mL or in 2000 mL Erlenmeyer flasks (431255, Corning) with a working volume of 250 mL. High Five cells infection with recombinant baculovirus (kindly provided by Redbiotec AG) encoding the H3 subtype influenza A–Johannesburg/33/94 and M1 A–California/06/2009 influenza virus strains was performed at a cell concentration at infection (CCI) of 2 × 106 cells mL–1,with a multiplicity of infection (MOI) of 15 IP cell–1. After 12 h post-infection, the culture medium was removed by centrifugation at 200g for 10 min, and the cells were washed with D-PBS (14190–169, Gibco). ESF921 methionine deficient and protein-free medium (96-200-01, Expression Systems) was then added to the infected cells. Noncanonical amino acid incorporation was tested at several hpi values (18, 24, 36, and 42 hpi) to identify the best condition for VLP production. The culture medium was supplemented with 4 mM Aha (AS-63669, AnaSpec). To generate appropriate controls, this study was also carried out with 4 mM l-Methionine (M2893, Sigma-Aldrich) at the same conditions.
Harvest and Clarification
High Five infected cells were harvested at 48 hpi by centrifugation at 200g for 10 min (JA10 rotor, Avanti J25I centrifuge, Beckman Coulter). Harvest was set to 48 hpi, at which time productivity was highest, a parameter that was screened and optimized previously (data not shown). The pellet was discarded and Benzonase (101654, Merck Millipore) was added to the supernatant at a final concentration of 50 U mL–1 and incubated at room temperature (22 °C) for 15 min. The clarification of supernatant was performed by dead-end filtration using a Sartopore filter with 0.45 + 0.2 μm pore size (SART5445307H7-SS-A, Sartorius, Germany). The clarification of VLP-containing bulk was performed at a constant flow rate of 100 mL min–1 using a Tandem 1081 Pump (Sartorius Stedim Biotech). The pressure was monitored by an in-line pressure transducer (080-699PSX-5, SciLog) to control possible overpressure. The filtration module was previously conditioned with three capsule volumes of buffer (50 mM HEPES, pH 7.4, and 300 mM NaCl as working buffer).
Anion-Exchange Chromatography
Sartobind Q MA 75 (93IEXQ42DB-12 V, Sartorius) membrane adsorber was used as the first purification step, operated in negative mode (FT). The membrane adsorber was equilibrated with 50 mM HEPES, pH 7.4, and 400 mM of NaCl equilibration buffer. The VLP clarified suspension was diluted with concentrated NaCl buffer to match the conductivity of equilibration buffer. The flow rate was set to 4.76 MV min–1, and the VLPs were collected in the FT pool. A final elution step was performed with 50 mM HEPES, pH 7.4, and 1.0 M NaCl elution buffer to guarantee that all particles were removed from the membrane adsorber. VLP concentration along these fractions was determined by hemagglutination assay and nanoparticle tracking analysis. All chromatographic steps were performed at room temperature (RT) (22 °C).
Ultrafiltration and Diafiltration
Sartobind Q FT pool containing VLPs were concentrated using tangential flow filtration (TFF). Ultrafiltration experiments were conducted using flat sheet Pellicon XL Ultrafiltration Module Biomax 300 kDa 0.005 m2 (PXB300C50, Merck Millipore).The membrane module was set up accordingly with the manufacturer’s instructions. The ultrafiltration module was preconditioned with deionized water to eliminate trace preservatives and equilibrated with working buffer before the concentration step. To ensure sterility, the TFF system was sanitized with 0.1 M NaOH and incubated with this solution for 60 min. A Tandem 1081 Pump (Sartorius Stedim Biotech,Germany) was used on the feed side set up to a fixed flow rate of 40 mL min–1. Transmembrane pressure (TMP) was controlled by adjusting the retentate flow rate using a flow-restriction valve. The pressure was monitored at feed inlet, retentate outlet, and permeate outlet by in-line pressure transducers (080-699PSX-5, SciLog) The feed–retentate and the permeate volumes were monitored using a technical scale (TE4101, Sartorius Stedim Biotech). At a proper feed volume, three diafiltration volumes with working buffer were performed. After achieving the desired concentration factor, the TFF loop was completely drained, and the VLP retentate was recovered. Samples of the final retentate and permeate were taken to assess process yield.
Size-Exclusion Chromatography
Concentrated VLPs were labeled with 20 μM of Alexa Fluor 488 (C-10405, Life Technologies) for 60 min, according to manufacturers’ instructions and prior to the polishing step. Size-exclusion chromatography was performed using a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) coupled to an ÄKTA Explorer 10 liquid chromatography system (GE Healthcare) equipped with UV and conductivity–pH monitors. System operation and data gathering and analysis was done using the UNICORN 5.0 software (GE Healthcare).
The column was loaded with 5 mL of concentrated VLPs, using a 5 mL capillary loop, at a constant flow rate of 0.5 mL min–1. Working buffer was used as eluent, and the eluted fractions were collected for further analyses. Elution of influenza VLPs was monitored by detecting the absorption of the eluted solution at 234 and 494 nm (maximum absorption wavelength of Alexa Fluor 488). Absorption at 234 reports roughly, all biomolecules that pass through the detector either by absorption or by light scattering (DNA, proteins, and lipids). The detection of the absorption at 494 is specific for the fluorescent VLP that incorporated the Alexa-488 probe. This dual detection allows better discrimination between the particles of interest VLP and all other contaminants, such as baculovirus.
Hemagglutination Assay
Hemagglutinin protein was quantified using a hemagglutination assay. The assay was carried out based on the protocol described elsewhere37 with some modifications. Briefly, 25 μL of D-PBS (14190-169, Gibco) were added in each well of a clear, V-bottom 96 well microtiter plate (611 V96, Sterilin). In the first well (upper left), 25 μL of each sample was added, and then 2-fold serial dilutions (25 μL of sample in an equal volume of PBS) were performed. The excess 25 μL from the final dilution was discarded. After this step, 25 μL of 1% chicken erythrocytes (Lohmann Tierzucht GmbH, Germany) was added to each well of each serial dilution series. The plate was incubated at 4 °C for 30 min without disturbance. As a positive control, influenza vaccine (Influvac, Abbott) was used. The level of hemagglutination was inspected visually for all of the wells, and the highest dilution capable of agglutinating chicken erythrocytes was determined.
We have plotted the hemagglutination assay according to the percentage (%) of HA recovery in each analyzed sample. This percentage is determined according to eq 1:
| 1 |
Total Protein Quantification
Total protein was quantified using the BCA Protein Assay Kit (23225, Thermo Fisher Scientific) according to the manufacture’s protocol. Bovine serum albumin (BSA) was used for the calibration curve (23209, Thermo Fisher Scientific). To avoid matrix interference, the samples were diluted between 2- and 256-fold. The assay took place in a clear 96 well microplate (260895, Nunc) and the absorbance at 562 nm was measured on Infinite 200 PRO NanoQuant (Tecan) microplate multimode reader.
Total dsDNA Quantification
Total DNA was quantified using the fluorescent-based Quant-iT Picogreen dsDNA assay kit (P7589, Invitrogen) according to the manufacturer’s instructions. To avoid matrix interference, the samples were diluted between 2- and 256-fold with the provided reaction buffer. The assay took place in a black 96 well microplate, flat transparent (3603, Corning), and the fluorescence was measured on Infinite 200 PRO NanoQuant (Tecan) microplate multimode reader.
Nanoparticle Tracking Analysis
Total virus-like particles concentration and size distribution were measured using the NanoSight NS500 (Nanosight Ltd.). The samples were diluted in D-PBS (14190–169, Gibco) so that virus-like particles concentration would be in the 108–109 particles mL–1 range (the instrument’s linear range). All measurements were performed at 22 °C. Sample videos were analyzed with the Nanoparticle Tracking Analysis (NTA) 2.3 analytical software (release version build 0025). Capture settings (shutter and gain) were adjusted manually. For each sample, 60 s videos were acquired, and particles between 70 and 130 nm were considered.
Confocal Microscopy
Using Life Technologies (Carlsbad, CA) Tetraspeck beads, one can use as visual reference of successful VLP labeling and detection. Due to its four-color fluorescence using green (which also detects labeled VLP) and red (specific for beads) fluorescence, one can perform quantitative analysis on the detected VLP. An inverted confocal point-scanning Zeiss LSM 710 microscope equipped with 405, 458, 488, 561, and 633 nm lasers was used. Due to the diffraction limit associated with microscopy techniques, no particle below that threshold can be visualized with high resolution. Thus, it would appear the point-spread function (PSF) of the instrument. VLP are subdiffraction limit particles; thus, their signal in the microscope would be the PSF of the microscope (approximately 240 nm). By using 500 nm size fluorescent beads as a control, together with their dual-fluorescence emission spectra, one can perform an accurate detection of subdiffraction limit particles minus VLP. This methodology was used to evaluate the best time for amino acid (Aha or Met) addition after baculovirus infection: 12, 24, 36, and 48 hpi were evaluated. Dilutions of each condition’s supernatant (100-fold) were deposited into IbiTreat 8 microwell slides (Ibidi, Martinsried, Germany) and allowed to attach for 1 h. Each preparation was then labeled with 20 μM of Alexa Fluor 488 (C-10405, Life Technologies) for 30 min according to manufacturers’ instructions. The sample was washed three times with PBS, and a 500-fold dilution of 500 nm fluorescent beads was added to each sample for 30 min. Medium was changed for fresh PBS. In all steps, the PBS used for dilution preparation, wash steps, and sample acquisition, was filtered with a 0.1 μM nylon filter. Control VLP (methionine amino acid added during VLP production; M2893, Sigma-Aldrich), modified VLP (with the Click-it noncanonical amino acid, Aha; AS-63669, AnaSpec), and 500 nm beads were imaged using a 63× oil objective and green and orange channels were acquired. From each independent experiment at least three images at different viewfields in the microslide were taken for all samples. ImageJ software was used to perform merge images as well as to perform particle count and detection and size analysis in each preparation from which the full width at half-maximum was determined (reflects particle size). From the number of particles detected, we estimated the concentration of fluorescent VLP [VLPfluo] in each DSP step and SEC fraction according to the relationship in eq 2:
| 2 |
where N is the number of fluorescent VLP detected in the microscopy image, acoverslip and aimage are, respectively the area of the microscope coverslip (9.4 × 10.7 mm for each microwell) and the area of the acquired image (44.5 × 44.5 μm), dil is the dilution factor of the added VLP (100-fold in our case), and V is the sample volume.
Flow Cytometry
Detection and characterization of labeled VLP and size discrimination between VLP (spheres of 100–200 nm) and baculovirus (rods of 200–400 nm) with flow cytometry were performed using a BD LSR Fortessa (BD Biosciences, San Jose, CA). It is equipped with three lasers (violet, 405 nm; blue, 488 nm; red, 640 nm), forward- and side-scatter detectors, and nine fluorescence emission detectors (530/30; the green channel was used for VLP-A488 and 100, 200, and 500 nm Tetraspeck fluorescent beads). The side-scatter detector was used to define the detection threshold. Using 100, 200, and 500 nm beads, one can build a particle size ruler in flow cytometry with the scatter signal,23 which can then be used to evaluate the VLP samples size distribution. The refractive index depends on the material of the scattered solution; thus, the direct correlation of bead size and VLP can only be achieved if each sample has approximately the same RI. The RI for PBS; UF retentate; SEC fractions (100-fold dilution); baculovirus (100-fold dilution); and 100 (2000-fold dilution), 200 (1000-fold dilution), and 500 nm beads (500-fold dilution) are, respectively, 1.334, 1.335, 1.335, 1.336, 1.334, 1.334, and 1.334, measured using a digital refractometer (13950000, AR 200 Digital Refractometer, Leica). The side-scatter–Alexa-488 correlograms were acquired for each bead and VLP sample, at the dilutions previously stated, to detect baculovirus presence and evaluate further particle separation by sorting. In all steps, the PBS used for dilution preparation, wash steps, and sample acquisition was filtered with a 0.1 μM nylon filter.
VLP Sorting
Sorting of the SEC F4 sample from the downstream processing was performed to separate VLPs from the baculovirus-rich fraction (>200 nm). Fluorescence activated sorting was performed in a BD FACS Aria III equipped with three lasers (blue, 488 nm; yellow-green, 561 nm; and red, 633 nm). The 200 nm fluorescent beads were used to define two sorting populations: P2, corresponding to the >200 particles detected, which is a baculovirus-rich population; and P1, which is the <200 nm particles that, in contrast to P2, are VLP-rich. Each population was acquired in vials filled with PBS, and the assessment for the presence of VLP and baculovirus was performed by atomic force microscopy.
Atomic Force Microscopy
AFM images of VLPs and baculovirus were acquired using a JPK Nano Wizard II (Berlin, Germany) mounted on a Zeiss Axiovert 200 inverted microscope (Göttingen, Germany). The AFM head is equipped with a 15 μm z-range linearized piezoelectric scanner and an infrared laser. All samples were prepared in freshly cleaved mica. For scanning in liquid environment, the mica was pretreated with poly-d-lysine for 20 min and rinsed with miliQ water. A 50 μL drop of each sample was added to the mica and rinsed with PBS buffer at least four times. The sample was then allowed to air-dry or maintained in buffer for subsequent imaging. Scanning was performed using uncoated silicon ACL cantilevers from Applied NanoStructure for air-dried samples and uncoated silicon cantilevers HQ:CSC38–No Al from MikroMasch for samples in liquid medium. The ACL cantilevers had typical resonance frequencies between 145 and 230 kHz and an average spring constant of 45 N/m. HQ:CSC38 cantilevers had typical resonance frequencies between 5 and 17 kHz and an average spring constant of 0.03 N/m. All measurements were carried out in contact mode. All images were obtained with the same or similar AFM parameters (set point, gain, and scan rate) values. Set point and gain were continuously adjusted during scanning to maintain the lowest possible value and reduce sample damage.
Transmission Electron Microscopy
To analyze the presence, integrity and morphology (shape and size) of the VLPs, electron microscopy was performed as follows: a drop (5 μL) of sample was adsorbed onto a Formvar coated 150 mesh copper grid from Veco (Science Services) for 2 min. The grid was washed five times with sterile filtered distilled H2O. Next, it was soaked in 2% uranyl acetate for 2 min and dried in air at room temperature (22 °C). The samples were examined with a Hitachi H-7650 120 Kv electron microscope (Hitachi High-Technologies Corporation).
PCR
Baculovirus viral DNA was extracted and purified using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics) using the manufacturer’s instructions. The number of genome containing particles were monitored by real time quantitative PCR (RT-qPCR) following the protocol described elsewhere.38
Western Blot Analysis
Western blot analysis was performed for control and modified influenza VLPs with both precipitated and nonprecipitated samples. As a control, M1 protein from influenza A H1N1 strain (SinoBiological) and H3 influenza VLP (produced and purified at iBET) were used. Protein precipitation was performed with 20% (v/v) ethanol overnight. Loading buffer (LDS sample buffer and reducing agent (Invitrogen)) was added, and protein samples were incubated at 70 °C for 10 min. Influenza VLPs were separated in a 4–12% (w/v) polyacrylamide NuPAGE gradient precast gel (Invitrogen). Samples were resolved for 60 min at a constant voltage of 200 V and transferred into a PVDF membrane using iBlot dry blotting system (Invitrogen). Membranes were blocked with 5% (w/v) of dry milk (Merck Millipore) in Tris-buffered saline with 0.1% (w/v) of Tween 20 (T-TBS buffer) for 1 h. After blocking, membranes were incubated overnight with the respective primary antibody: anti-influenza A virus M1 goat antibody (dilution 1:2000) (Abcam ab20910) or a 1:1 mixture of anti-A–Johannesburg/33/94 sheep serum (dilution 1:1000) and anti-A–Nanchang/933/95 (H3N2) HA sheep serum (dilution 1:1000) (both provided by NIBSC). Western blot detection was performed with the corresponding anti-goat or anti-sheep secondary antibody (dilution 1:2000, 1 h incubation) conjugated to horseradish peroxidase and developed using the ECL detection reagent protocol (GE Healthcare).
Fluorescence Imaging
Control and labeled VLP samples were incubated with 20 μM of Click-iT Alexa Fluor488 fluorescent probe for 30 min prior to SDS-PAGE gel running. A FLA-5100 fluorescent imaging system (Fujifilm Life Sciences) was used to reveal the gel and analyze the presence of fluorescent bands. The 473 nm laser was used, and images were acquired with 25 μ of resolution and at a voltage of 600 V.
Mass Spectrometry
HA and M1 protein bands, detected by Western blot, and fluorescent bands were destained, reduced, alkylated, and digested with trypsin (Promega, 6.7 ng/μL) overnight at 37 °C. The tryptic peptides were desalted using POROS R2 (Applied Biosystems) and analyzed by nanoLC–MS using TripleTOF 6600 (ABSciex). External calibration was performed using β-galactosidase digest (ABSciex). The 40 most intense precursor ions from the MS spectra were selected for tandem mass spectrometry (MS/MS) analysis. Data were acquired with the Analyst software TF 1.7 (ABSciex). The raw MS and MS/MS data were analyzed using Protein Pilot software version 5.0 (ABSciex) for protein identification. The search was performed against the HA and M1 protein sequences and against the Swissprot viruses database plus the protein sequences of influenza VLP proteins HA and M1. Protein identification was considered with an unused score greater than 1.3 (95% confidence). To detect modified peptides, data were also analyzed using the BioPharmaView software version 1.0 (ABSciex) considering a Met–Aha modification (mass shift of −4.986 Da) with a m/z tolerance of ±10 ppm.
Acknowledgments
The authors acknowledge funding from the European Union (EDUFLUVAC project FP7-HEALTH-2013-INNOVATION), the Fundação para a Ciência e Tecnologia (FCT, Portugal; project HIVERA/0002/2013 and FCT Investigator to G.J.L.B.), EPSRC (to G.J.L.B.), the European Commission, Marie Skłodowska-Curie Actions (MSCA), and RISE project grant 644167. S. B. C., J. M. F., F. M., and D. G. acknowledge FCT for fellowships SFRH/BD/52302/2013, SFRH/BD/70423/2010, SFRH/BD/70139/2010, and SFRH/BPD/73500/2010, respectively. The authors acknowledge Ricardo Silva for all his help in fluorescence analysis implementation and fruitful discussions. The authors also acknowledge Patrícia Gomes-Alves for her help for mass spectrometry analysis. Mass spectrometry data was obtained by the Mass Spectrometry Unit (UniMS), ITQB/iBET, Oeiras, Portugal. G. J. L. B. is a Royal Society University Research Fellow and the recipient of a European Research Council Starting Grant (TagIt).
Glossary
ABBREVIATIONS
- VLPs
virus-like particles
- TagE-VLPs
tagged enveloped VLPs
- Aha
azidohomoalanine
- SPAAC
strain-promoted alkyne–azide [3 + 2] cycloaddition
- HA
hemagglutinin
- fwhm
full width at half-maximum
- hpi
hours post-infection
- DSP
downstream processing
- PSF
point-spread function
- AEX
anionic-exchange chromatography
- SEC
size-exclusion chromatography
- FACS
fluorescence-activated cell sorting
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.6b00372.
Figures showing fluorescence microscopy of VLP, evaluation of the best time to add non-canonical azidohomoalanine after baculovirus infection, a flowchart showing specific-site-functionalized enveloped VLP production and purification steps, follow-up of DSP steps by confocal microscopy, detailed follow-up of SEC DSP step by confocal microscopy, extrapolation of VLP population particle size diameter from SSC signal using standard fluorescent beads, flow cytometry SSC signal and the size of polydisperse samples, supporting data for the flow cytometry experiment shown in Figures 2 and 3, TEM images, and raw data of Figure 5 for the identification of HA and M1 proteins by Western blot analysis and fluorescent band detection of labelled influenza VLP proteins. Additional detail on Western blod analysis and MS proteomics of Ha- and M1-labelled VLP. (PDF)
Author Present Address
¶ (J.M.F.) Institut Pasteur, Unité de Virologie Structurale, Département de Virologie, F-75724 Paris Cedex 15, France.
Author Contributions
∇ All authors have given approval to the final version of the manuscript. S.B.C. and J.M.F. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Pitoiset F.; Vazquez T.; Bellier B. (2015) Enveloped virus-like particle platforms: Vaccines of the future?. Expert Rev. Vaccines 14, 913–915. 10.1586/14760584.2015.1046440. [DOI] [PubMed] [Google Scholar]
- Plummer E. M.; Manchester M. (2011) Viral nanoparticles and virus-like particles: Platforms for contemporary vaccine design. Wires Nanomed. Nanobiotech. 3, 174–196. 10.1002/wnan.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes F.; Teixeira A. P.; Carinhas N.; Carrondo M. J. T; Alves P. M. (2013) Insect cells as a production platform of complex virus-like particles. Expert Rev. Vaccines 12, 225–236. 10.1586/erv.12.153. [DOI] [PubMed] [Google Scholar]
- Lua L. H. L.; Connors N. K.; Sainsbury F.; Chuan Y. P.; Wibowo N.; Middelberg A. P. J. (2014) Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 111, 425–440. 10.1002/bit.25159. [DOI] [PubMed] [Google Scholar]
- Zhao Q. J.; Li S. W.; Yu H.; Xia N. S.; Modis Y. (2013) Virus-like particle-based human vaccines: Quality assessment based on structural and functional properties. Trends Biotechnol. 31, 654–663. 10.1016/j.tibtech.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Mateu M. G. (2011) Virus engineering: Functionalization and stabilization. Protein Eng., Des. Sel. 24, 53–63. 10.1093/protein/gzq069. [DOI] [PubMed] [Google Scholar]
- Kaczmarczyk S. J.; Sitaraman K.; Young H. A.; Hughes S. H.; Chatterjee D. K. (2011) Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. U. S. A. 108, 16998–17003. 10.1073/pnas.1101874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.; Hsiao S. C.; Carrico Z. M.; Francis M. B. (2009) Genome-free viral capsids as multivalent carriers for taxol delivery. Angew. Chem., Int. Ed. 48, 9493–9497. 10.1002/anie.200902426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov S. G.; Gaponenko V.; Howard O. M. Z.; Chen Y. H.; Oppenheim J. J.; Dyba M. A.; Subramaniam S.; Lee Y.; Michejda C.; Tarasova N. I. (2011) Structural plasticity of a transmembrane peptide allows self-assembly into biologically active nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 108, 9798–9803. 10.1073/pnas.1014598108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P. S.; Ostapchuk P.; Hearing P.; Carrico I. S. (2011) Unnatural amino acid incorporation onto adenoviral (Ad) coat proteins facilitates chemoselective modification and retargeting of Ad type 5 vectors. J. Virol. 85, 7546–7554. 10.1128/JVI.00118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElSohly A. M.; Netirojjanakul C.; Aanei I. L.; Jager A.; Bendall S. C.; Farkas M. E.; Nolan G. P.; Francis M. B. (2015) Synthetically modified viral capsids as versatile carriers for use in antibody-based cell targeting. Bioconjugate Chem. 26, 1590–1596. 10.1021/acs.bioconjchem.5b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grgacic E. V. L; Anderson D. A. (2006) Virus-like particles: Passport to immune recognition. Methods 40, 60–65. 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madalinski K.; Sylvan S. P. E.; Hellström U.; Mikolajewicz J.; Zembrzuska-Sadkowska E.; Piontek E. (2001) Antibody responses to pres components after immunization of children with low doses of biohepb. Vaccine 20, 92–97. 10.1016/S0264-410X(01)00312-7. [DOI] [PubMed] [Google Scholar]
- Ludwig C.; Wagner R. (2007) Virus-like particles–universal molecular toolboxes. Curr. Opin. Biotechnol. 18, 537–545. 10.1016/j.copbio.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hest J. C. M.; Kiick K. L.; Tirrell D. A. (2000) Efficient incorporation of unsaturated methionine analogues into proteins in vivo. J. Am. Chem. Soc. 122, 1282–1288. 10.1021/ja992749j. [DOI] [Google Scholar]
- Kiick K. L.; Saxon E.; Tirrell D. A.; Bertozzi C. R. (2002) Incorporation of azides into recombinant proteins for chemoselective modification by the staudinger ligation. Proc. Natl. Acad. Sci. U. S. A. 99, 19–24. 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard N. J.; Prescher J. A.; Bertozzi C. R. (2004) A strain-promoted [3 + 2] azide–alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 126, 15046–15047. 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- Kost T. A.; Condreay J. P.; Jarvis D. L. (2005) Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 23, 567–575. 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strable E.; Prasuhn D. E. Jr.; Udit A. K.; Brown S.; Link A. J.; Ngo J. T.; Lander G.; Quispe J.; Potter C. S.; Carragher B.; Tirrell D. A.; Finn M. G. (2008) Unnatural amino acid incorporation into virus-like particles. Bioconjugate Chem. 19, 866–875. 10.1021/bc700390r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tom Dieck S.; Kochen L.; Hanus C.; Heumuller M.; Bartnik I.; Nassim-Assir B.; Merk K.; Mosler T.; Garg S.; Bunse S.; et al. (2015) Direct visualization of newly synthesized target proteins in situ. Nat. Methods 12, 411–414. 10.1038/nmeth.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.; Pizzelli K.; Romero J. K.; Chrostowski J.; Evangelist G.; Hamzik J.; Soice N.; Cheng K. S. (2013) Clarification of recombinant proteins from high cell density mammalian cell culture systems using new improved depth filters. Biotechnol. Bioeng. 110, 1964–1972. 10.1002/bit.24848. [DOI] [PubMed] [Google Scholar]
- Nielsen M. H.; Beck-Nielsen H.; Andersen M. N.; Handberg A. (2014) A flow cytometric method for characterization of circulating cell-derived microparticles in plasma. J. Extracell. Vesicles 3, 3. 10.3402/jev.v3.20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlist E. J.; Nolte-’t Hoen E. N.; Stoorvogel W.; Arkesteijn G. J.; Wauben M. H. (2012) Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat. Protoc. 7, 1311–1326. 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- Undey C., Low D., Menezes J. C., and Koch M. (2011) On-Line and At-Line Process Analyzers Are Inserted in One of the Major Categories of Process Analytical Technology (Pat) Tools, Having Important Applications in the Biopharmaceutical Industry, CRC Press, Boca Raton, FL. [Google Scholar]
- Peixoto C.; Sousa M. F.; Silva A. C.; Carrondo M. J.; Alves P. M. (2007) Downstream processing of triple layered rotavirus like particles. J. Biotechnol. 127, 452–461. 10.1016/j.jbiotec.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Monteiro F.; Carinhas N.; Carrondo M. J.; Bernal V.; Alves P. M. (2012) Toward system-level understanding of baculovirus-host cell interactions: From molecular fundamental studies to large-scale proteomics approaches. Front. Microbiol. 3, 391. 10.3389/fmicb.2012.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter N. K.; Hanson J. E.; Glick G. D.; Brown J. H.; Crowther R. L.; Park S. J.; Skehel J. J.; Wiley D. C. (1992) Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: Analysis by proton nuclear magnetic resonance spectroscopy and x-ray crystallography. Biochemistry 31, 9609–9621. 10.1021/bi00155a013. [DOI] [PubMed] [Google Scholar]
- Eisfeld A. J.; Neumann G.; Kawaoka Y. (2014) Influenza a virus isolation, culture and identification. Nat. Protoc. 9, 2663–2681. 10.1038/nprot.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stencel-Baerenwald J. E.; Reiss K.; Reiter D. M.; Stehle T.; Dermody T. S. (2014) The sweet spot: Defining virus-sialic acid interactions. Nat. Rev. Microbiol. 12, 739–749. 10.1038/nrmicro3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T. (2012) Native morphology of influenza virions. Front. Microbiol. 2, 269. 10.3389/fmicb.2011.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana J.; Cardone G.; Heymann J. B.; Winkler D. C.; Steven A. C. (2012) Structural changes in influenza virus at low ph characterized by cryo-electron tomography. J. Virol. 86, 2919–2929. 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. M., Petiot E., Mullick A., Aucoin M. G., Henry O., and Kamen A. A. (2015) Critical assessment of influenza VLP production in Sf9 and HEK293 expression systems. BMC Biotechnol. 15. 10.1186/s12896-015-0152-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwang T. W.; Zeng X.; Wang S. (2016) Manufacturing of acmnpv baculovirus vectors to enable gene therapy trials. Mol. Ther.--Methods Clin. Dev. 3, 15050. 10.1038/mtm.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.-C.; Scotti P. D.; Miyamura T.; Takeda N. (2007) Latent infection of a new alphanodavirus in an insect cell line. J. Virol. 81, 10890–10896. 10.1128/JVI.00807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merten O.-W. (2007) Attention with virus contaminated cell lines. Cytotechnology 55, 1–2. 10.1007/s10616-007-9095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClenahan S. D.; Uhlenhaut C.; Krause P. R. (2014) Evaluation of cells and biological reagents for adventitious agents using degenerate primer pcr and massively parallel sequencing. Vaccine 32, 7115–7121. 10.1016/j.vaccine.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Hirst G. K. (1941) The agglutination of red cells by allantoic fluid of chick embryos infected with influenza virus. Science 94, 22–23. 10.1126/science.94.2427.22. [DOI] [PubMed] [Google Scholar]
- Vicente T.; Peixoto C.; Carrondo M. J. T; Alves P. M. (2009) Purification of recombinant baculoviruses for gene therapy using membrane processes. Gene Ther. 16, 766–775. 10.1038/gt.2009.33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.