Abstract
Genetically encoded FRET-based sensor proteins have significantly contributed to our current understanding of the intracellular functions of Zn2+. However, the external excitation required for these fluorescent sensors can give rise to photobleaching and phototoxicity during long-term imaging, limits applications that suffer from autofluorescence and light scattering, and is not compatible with light-sensitive cells. For these applications, sensor proteins based on Bioluminescence Resonance Energy Transfer (BRET) would provide an attractive alternative. In this work, we used the bright and stable luciferase NanoLuc to create the first genetically encoded BRET sensors for measuring intracellular Zn2+. Using a new sensor approach, the NanoLuc domain was fused to the Cerulean donor domain of two previously developed FRET sensors, eCALWY and eZinCh-2. In addition to preserving the excellent Zn2+ affinity and specificity of their predecessors, these newly developed sensors enable both BRET- and FRET-based detection. While the dynamic range of the BRET signal for the eCALWY-based BLCALWY-1 sensor was limited by the presence of two competing BRET pathways, BRET/FRET sensors based on the eZinCh-2 scaffold (BLZinCh-1 and -2) yielded robust 25–30% changes in BRET ratio. In addition, introduction of a chromophore-silencing mutation resulted in a BRET-only sensor (BLZinCh-3) with increased BRET response (50%) and an unexpected 10-fold increase in Zn2+ affinity. The combination of robust ratiometric response, physiologically relevant Zn2+ affinities, and stable and bright luminescence signal offered by the BLZinCh sensors allowed monitoring of intracellular Zn2+ in plate-based assays as well as intracellular BRET-based imaging in single living cells in real time.
Zn2+ serves important catalytic and structural roles in numerous cellular processes, acting as a Lewis acid in enzyme catalysis and enhancing protein stability in transcription factors.1,2 Since free Zn2+ ions are potent inhibitors of enzyme activity and avid protein binders, intracellular Zn2+ homeostasis is tightly controlled by a variety of transporters, channels, metallothioneins, and metal-responsive transcription factors.1,3 Zn2+ deficiency and dysregulation of intracellular Zn2+ levels have been related to various diseases including neurodegeneration, growth retardation, immunodeficiency, cancer, and diabetes.4−7 Zn2+ also plays a regulatory role by modulating intercellular signaling between neuronal cells,6,8 is involved in insulin secretion in pancreatic β-cells, and has been implicated as an intracellular second messenger that controls the activity of phosphatases, caspases, and ion channels.1,4,5,7,9−11 Both small-molecule fluorescent probes and fluorescent sensor proteins have significantly contributed to our current understanding of the intracellular role of Zn2+.12−19 Whereas small-molecule-based probes are easily applied, genetically encoded sensors offer superior control over intracellular localization and concentration. Moreover, most protein-based sensors are based on Förster Resonance Energy Transfer (FRET) and are therefore ratiometric, which allows for more reliable quantification of Zn2+ concentrations. A variety of FRET sensor proteins have been developed for intracellular Zn2+ imaging both by our group and by others, yielding cytosolic levels of free Zn2+ between 0.1 and 1 nM in (cancer) cell lines, pancreatic β cells, primary cells, and Arabidopsis thaliana root tips.15−21 Subcellular targeting of these sensors has also allowed determination of the free Zn2+ concentrations in the endoplasmic reticulum (ER), Golgi system, mitochondria, and pancreatic β cell granules, although conflicting concentrations have been reported with different sensors.5,15,17,22,23 In addition, red-shifted variants have been developed that allow multiparameter imaging together with the original CFP-YFP-based sensors, allowing simultaneous Zn2+ imaging in different cellular compartments in the same cell.5,15,24,25
Whereas fluorescent sensors have become essential tools to study Zn2+ homeostasis and signaling in real time in a single cell, they come with some inherent limitations. Laser excitation of the donor fluorophore can result in photobleaching and phototoxicity, precluding measurements over extended periods of time.26,27 External illumination gives rise to background autofluorescence and light scattering, hampering in vivo imaging and other applications in strongly absorbing or scattering media such as cell suspensions, plant cells, and blood. Fluorescence is also not compatible with applications that involve light-sensitive cells, containing either endogenous photoreceptors or engineered photosensitive proteins in optogenetic experiments. For all of these applications, bioluminescent sensor proteins based on the modulation of energy transfer between a donor luciferase and an acceptor fluorescent domain (BRET; Bioluminescence Resonance Energy Transfer) would be highly desirable. BRET has mainly been used to study dynamic protein–protein interactions or enzymatic activity in living cells.28−30 In addition, BRET sensor proteins targeting intracellular messengers such as Ca2+,31 cAMP,32 and cGMP33 have been developed. These sensors consist of a specific ligand binding domain (CaM-M13, Epac, GAF) fused in between Renilla luciferase (RLuc or RLuc8) and GFP or YFP (or its improved variants Venus or Citrine). The limited brightness of Renilla and other luciferases typically restricts their application to populations of cells and precludes the use of bioluminescence in single-cell experiments. Nagai and co-workers recently showed that the brightness of luciferases can be enhanced by making tight chimeras with fluorescent acceptor domains.34 The improved brightness of these so-called Nanolanterns was shown to improve the sensitivity and even allow single-cell bioluminescence imaging. Sensors for Ca2+, cAMP, and ATP were developed based on Nanolanterns, but these sensors operate by modulating the activity of the luciferase and are therefore not ratiometric.34,35
The recent introduction of the stable and bright luciferase NanoLuc (NLuc) has substantially decreased the sensitivity gap between fluorescence and bioluminescence, allowing applications for BRET sensors that are inconceivable with other luciferases. NLuc is a small (19 kDa) engineered luciferase derived from the deep sea shrimp Oplophorus gracilirostris.36 Its glow-type blue luminescence is stable for hours and ∼150-fold brighter than that of RLuc or firefly luciferase (Photinus pyralis). Its emission peak at 450 nm makes it an ideal donor luciferase for cyan fluorescent proteins and synthetic dyes such as Cy3, but the tail in the emission peak above 500 nm also allows it to be used as a BRET donor for green and orange fluorescent proteins and red-shifted dyes.29,37−39 As a result, BRET assays using NLuc have recently been reported for diagnostic applications such as therapeutic drug monitoring37 and antibody detection in blood plasma (LUMABS),40 as well as screening for ligand–protein interactions.38,41 In this work, we used NLuc to create the first BRET sensors for measuring intracellular Zn2+. A novel approach was used in which the NLuc domain was fused to the CFP donor domain of two FRET sensor protein families, eCALWY and eZinCh-2. In addition to preserving the excellent Zn2+ affinity and specificity of their predecessors, these newly developed sensor proteins enable both BRET- and FRET-based imaging. BRET/FRET- and BRET-only variants of eZinCh-2 allowed robust monitoring of intracellular Zn2+ in plate-based assays as well as intracellular BRET-based imaging in single living cells in real time.
Results
Design Considerations
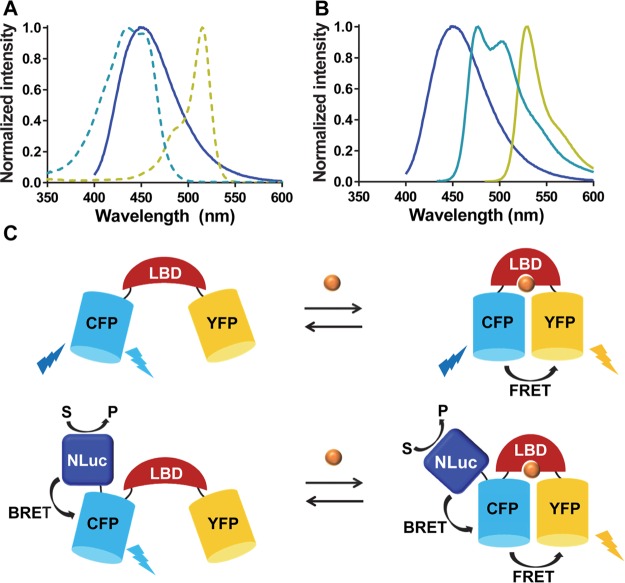
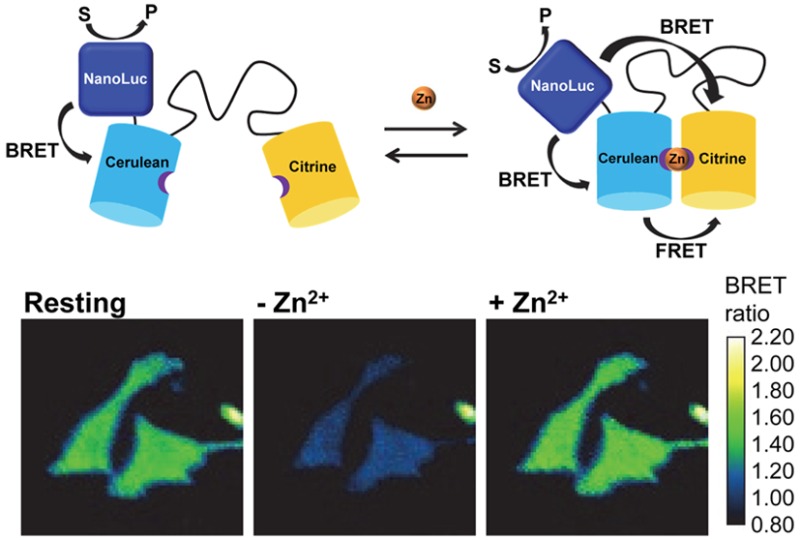
The strong overlap between the emission of NLuc and the excitation spectrum of Cerulean (cyan fluorescent protein (CFP) variant used in our sensors) should enable efficient energy transfer (BRET) from NLuc to Cerulean, which can subsequently be transferred to the yellow acceptor fluorescent protein (YFP) Citrine in a second energy transfer step (FRET; Figure 1). In this mechanism, ligand binding would primarily affect FRET between the fluorescent domains. BRET between NLuc and Cerulean is used to allow excitation of Cerulean without the need of an external light source. Figure 1A shows that the emission spectrum of NLuc also overlaps with the excitation spectrum of Citrine, allowing direct energy transfer between NLuc and Citrine. Unlike BRET between NLuc and Cerulean, this second BRET pathway is likely to be modulated by ligand binding as well and can contribute to the overall properties of the BRET sensor. The emission peak of Citrine is well separated from both Cerulean and NLuc, allowing it to be easily distinguished from the two others (Figure 1B).
Figure 1.
(A) Emission spectrum of NLuc (dark blue) displayed together with the excitation spectra of Cerulean (dashed light blue) and Citrine (dashed yellow). (B) Overlap of emission spectrum of NLuc (dark blue) with the emission spectra of Cerulean (light blue) and Citrine (yellow). (C) Sensor mechanisms of FRET sensor (top) and BRET/FRET sensor (bottom). The binding of the ligand to its binding domain (LBD) induces a change of FRET between CFP and YFP. Upon fusion of NLuc to the FRET sensor, bioluminescence readout is introduced while the ligand reporting properties of the sensor remain unchanged.
The approach outlined in Figure 1 assumes that introduction of the luciferase domain will not affect the properties of the FRET sensor part. This assumption may not apply to all FRET sensors, as some FRET sensors are known to rely on subtle conformational changes. However, the two FRET sensors used in this study, eCALWY and eZinCh-2, both rely on the modulation of intramolecular domain interactions. The eCALWY FRET sensor consists of the two metal binding domains Atox1 and WD4, connected by a long and flexible linker and flanked by self-associating variants of Cerulean and Citrine.19 The Zn2+ binding domains each contain two cysteines that together can coordinate one Zn2+ ion in a tetrahedral fashion. Formation of the Zn2+ complex disrupts the interaction between the fluorescent domains, resulting in a substantial decrease of FRET. The eZinCh-2 FRET sensor does not contain distinct Zn2+ binding domains but displays a Zn2+ coordinating histidine and cysteine residue on the surface of both Cerulean and Citrine.15 Together, these four residues allow the formation of a stable tetrahedral Zn2+ complex, bringing both fluorescent domains closer together and inducing an increase in FRET. To minimize the perturbation of the FRET sensor properties as much as possible, we introduced the NLuc domain at the N-terminus of the Cerulean domain in both sensors.
BLCALWY-1
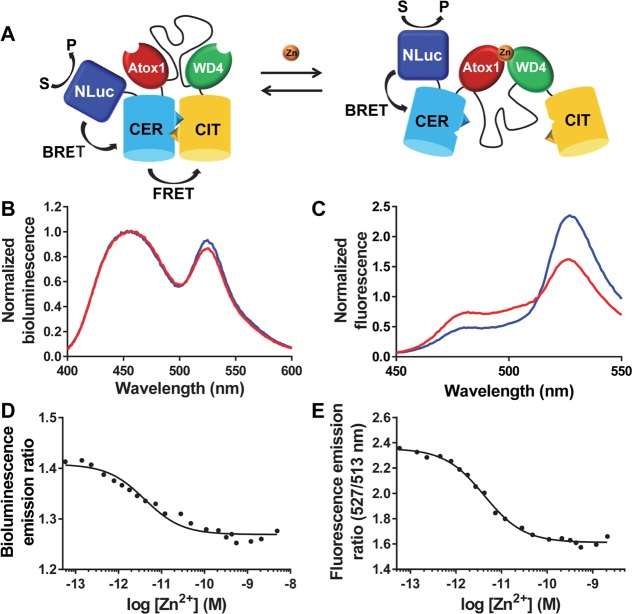
BLCALWY-1 was created by introducing NLuc between the N-terminal His-tag and the Cerulean domain in eCALWY-1 using a short linker consisting of flexible glycine and serine residues (GSGSG-linker) between NLuc and Cerulean (Figure 2A). To allow efficient purification of the full size protein, a Strep-tag was added to the C-terminus of the Citrine domain. Recombinant expression of BLCALWY-1 in E. coli BL21(DE3) and subsequent purification using Ni2+ affinity chromatography and Strep-Tactin affinity chromatography yielded pure BLCALWY-1 in low but sufficient yield (∼0.1 mg/L) for characterization using bioluminescence and fluorescence spectroscopy (Figure S1). The bioluminescence spectrum of BLCALWY-1 in the absence of Zn2+ shows a broad peak centered around 460 nm with contributions from both NLuc and Cerulean, and a second peak at 530 nm originating from Citrine (Figure 2B). The addition of Zn2+ resulted in a small (7%) but significant decrease in emission ratio, consistent with the expected decrease in FRET. Monitoring of the BRET ratio as a function of free Zn2+ concentration yielded a Kd of 4.1 ± 0.9 pM (Figure 2D), which is similar to the Kd of 2 pM that was previously determined for the parental eCALWY-1 sensor.19 To assess the origin of the small dynamic range, we also monitored Zn2+ binding using fluorescence spectroscopy. Unlike the BRET experiments, FRET detection revealed a robust 2-fold decrease in Citrine/Cerulean emission ratio upon Zn2+ binding (Figure 2C). Fitting the emission ratio as a function of free Zn2+ concentration yielded a Kd of 4.2 ± 0.3 pM (Figure 2E). Since these sensor properties are almost indistinguishable from the eCALWY-1-sensor, we conclude that both the affinity and conformational properties of the sensor part were not significantly affected in BLCALWY-1. The relatively modest decrease in emission ratio could be due to direct energy transfer between NLuc and Citrine. This second BRET pathway may be much less affected by Zn2+ binding and could even be increased, attenuating the spectral changes due to FRET between Cerulean and Citrine. A second form of attenuation would be inefficient BRET between NLuc and Cerulean, which would mean that the spectral change needs to be detected on top of a background of NLuc emission that is Zn2+-insensitive.
Figure 2.
(A) Sensor mechanism of the BLCALWY-1 sensor (CER = Cerulean, CIT = Citrine). (B,C) Bioluminescence (B; normalized to emission at 455 nm) and fluorescence (C; normalized to emission at 513 nm) emission spectra of BLCALWY-1 in Zn2+-depleted (blue) and Zn2+-saturated states (red). Measurements were performed using 0.5 nM BLCALWY-1 and 3000-fold diluted furimazine (B) or 50 nM BLCALWY-1 (C) in 150 mM HEPES (pH 7.1); 100 mM NaCl, 10% (v/v) glycerol, 5 μM DTT, 1 mM TCEP, and 1 mg mL–1 BSA, at 20 °C (B) or 25 °C (C). (D,E) Bioluminescence emission ratio (emission 500–545 nm/emission 400–455 nm; D) and fluorescence emission ratio (527/513 nm; E) of BLCALWY-1, in the presence of a range of free Zn2+ concentrations buffered using 1 mM EDTA, 1 mM HEDTA, 1 mM DHPTA, 5 mM EGTA, or 1 mM EGTA. Measurements were performed using 0.2 nM BLCALWY-1 and 3200-fold diluted furimazine (D) or 50 nM BLCALWY-1 (E) in 150 mM HEPES (pH 7.1), 100 mM NaCl, 10% (v/v) glycerol, 5 μM DTT, 1 mM TCEP, and 1 mg mL–1 BSA, at 23–25 °C. Data points represent the average of two measurements, and the solid lines are fits using eq 1, from which Kd’s of 4.1 ± 0.9 pM (D) and 4.2 ± 0.3 pM (E) were determined, respectively.
BLZinCh Sensors
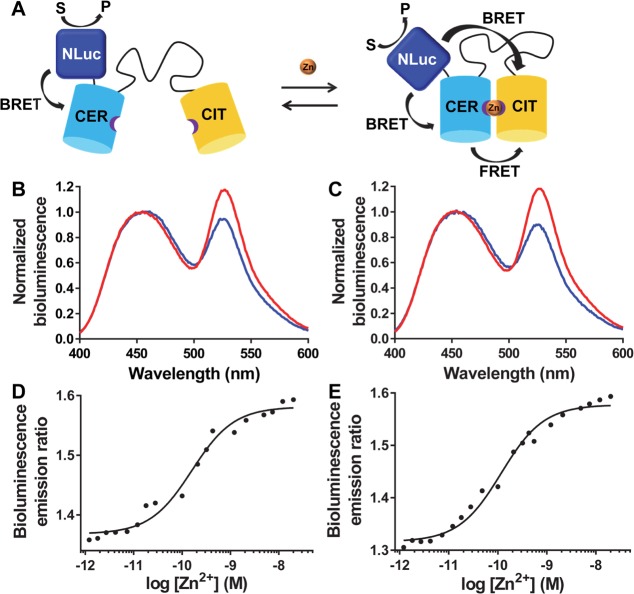
The eZinCh-2 sensor can be considered a large chelating ligand in which Zn2+ binding switches the sensor from an open state with a distribution of conformations that is governed by the length and the flexibility of the linker, to a compact Zn2+-bound state. Based on this mechanism, both FRET between the fluorescent domains and BRET between NLuc and Citrine would be expected to increase upon Zn2+ binding in BLZinch, which should result in a larger BRET response (Figure 3A). To ensure high BRET efficiency between NLuc and Cerulean without perturbing the stability of both protein domains, two different linkers were tested. In BLZinCh-1, NLuc and Cerulean were fused together directly, leaving out the three most C-terminal residues (ILA) of NLuc. These C-terminal residues were predicted to be flexible based on a homology model of NLuc, which was confirmed very recently by the crystal structure of NLuc that shows that these residues are located outside the twisted β-sheet of NLuc (PDB: 5IBO). In BLZinCh-2, the full-sized NLuc domain was used, and a short flexible GGSGGS linker was inserted between NLuc and Cerulean to minimize the potential for steric hindrance between the two domains and provide more conformational freedom for the NLuc domain. BLZinCh-1 and -2 were recombinantly expressed in E. coli and purified following the same two-step affinity chromatography purification procedure as used for BLCALWY-1, yielding pure sensor proteins in reasonable yields (∼1–2 mg/L; Figure S1). Bioluminescence spectra showed a substantial increase of the emission ratio upon Zn2+ binding for both sensor variants, with the BLZinCh-2 sensor showing a slightly larger BRET response (30% vs 25%) upon Zn2+ binding (Figure 3B,C). Monitoring the BRET ratio as a function of free Zn2+ concentration yielded Kd’s of 160 ± 29 pM and 117 ± 16 pM for BLZinCh-1 and BLZinCh-2, respectively (Figure 3D,E), which are similar to the Zn2+ affinity previously reported for the parental eZinCh-2 FRET sensor (Kd = 270 ± 50 pM).15,42 As expected, Zn2+ binding could also be monitored by measuring FRET between the two fluorescent domains, showing a dynamic range and Zn2+ affinity that are comparable to those of the parental eZinCh-2 FRET sensor (Figure S2A–D).15,42
Figure 3.
(A) Sensor mechanism of BLZinCh-1 and BLZinCh-2 (CER = Cerulean, CIT = Citrine). (B,C) Bioluminescence emission spectra (normalized to emission at 455 nm) of BLZinCh-1 (B) and BLZinCh-2 (C) in Zn2+-depleted (blue) and Zn2+-saturated state (red). Measurements were performed using 0.5 nM protein and 3000-fold diluted furimazine in 150 mM HEPES (pH 7.1), 100 mM NaCl, 10% (v/v) glycerol, 5 μM DTT, 1 mM TCEP, and 1 mg mL–1 BSA, at 20 °C. (D,E) Bioluminescence emission ratio (emission 500–545 nm/emission 400–455 nm) of BLZinCh-1 (D) and BLZinCh-2 (E), in the presence of a range of free Zn2+ concentrations buffered using 1 mM HEDTA, 1 mM DHPTA, 5 mM EGTA, or 1 mM EGTA. Measurements were performed using 0.2 nM protein and 3200-fold diluted furimazine in 150 mM HEPES (pH 7.1), 100 mM NaCl, 10% (v/v) glycerol, 5 μM DTT, 1 mM TCEP, and 1 mg mL–1 BSA, at 23–25 °C. Data points represent the average of two measurements, and the solid lines are fits assuming single binding events using eq 1, from which Kd’s of 160 ± 29 pM (D) and 117 ± 16 pM (E) were determined, respectively.
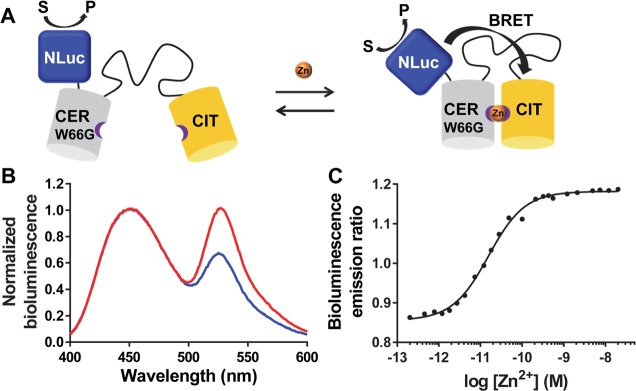
As the emission spectrum of NLuc overlaps with the excitation spectra of both Cerulean and Citrine, energy transfer may take place directly to Citrine, or via Cerulean. To investigate the contribution of each pathway, we mutated tryptophan-66 in the Cerulean chromophore of BLZinCh-1 to glycine (BLZinCh-3; Figure 4A). While the Cerulean domain in this sensor variant is no longer fluorescent, it should still participate in Zn2+ binding, modulating the efficiency of BRET between NLuc and Citrine. BLZinCh-3 showed a significantly larger change in emission ratio upon binding Zn2+ (50%), which is mainly due to the substantially lower emission ratio in the absence of Zn2+ (Figure 4B). These results show that direct BRET between NLuc and Citrine is an important BRET pathway and sufficient to yield a robust change in emission ratio. The increased dynamic range can be explained by the better spectral separation of the NLuc and Citrine emission peaks compared to Cerulean and Citrine (see also Figure 1A). Remarkably, monitoring of the BRET ratio as a function of free Zn2+ concentration yielded a Kd of 15.6 ± 1.0 pM for BLZinCh-3, which represents a 10-fold increase in Zn2+ affinity compared to BLZinCh-1 (Figure 4C). Close inspection of the crystal structure of Cerulean (PDB: 2WSO)43 reveals the presence of a hydrogen bond between the tryptophan NH in the chromophore and the side chain OH of Ser-205, which is located on the same β-strand as the metal coordinating His-206 and Cys-208 (Figure S3). The loss of this hydrogen bond in the W66G mutant and/or the loss of packing in the core of the protein may have resulted in increased flexibility of this β-strand, possibly allowing His-206 or Cys-208 to adopt a more favorable coordination geometry around the Zn2+ ion. To explore whether the increased affinity is due to the lack of a large hydrophobic residue, we also mutated Cerulean-Trp-66 to Leu and His. Both mutants displayed a similar Zn2+ affinity (W66L mutant Kd = 11.1 ± 1.1 pM; W66H mutant Kd = 15.0 ± 1.6 pM) and dynamic range as the W66G mutant (Figure S4A,B), suggesting that the increased Zn2+ affinity may be primarily due to a loss of the specific hydrogen bond between Trp-66 and Ser-205 in Cerulean. The importance of such subtle differences in geometry and/or second coordination sphere was noticed previously during the development of eZinCh-2, where the relative positions of the Cys and His residues on the two fluorescent domains were also found to have a large effect not only on FRET efficiency but also on Zn2+ affinity.15
Figure 4.
(A) Sensor mechanism of BLZinCh-3 (CER = Cerulean, CIT = Citrine). (B) Bioluminescence emission spectra (normalized to emission at 455 nm) of BLZinCh-3 in Zn2+-depleted (blue) and Zn2+-saturated state (red). The measurement was performed using 0.1 nM protein and 3000-fold diluted furimazine in 150 mM HEPES (pH 7.1), 100 mM NaCl, 10% (v/v) glycerol, 5 μM DTT, 1 mM TCEP, and 1 mg mL–1 BSA, at 20 °C. (C) Bioluminescence emission ratio (emission 500–545 nm/emission 400–455 nm) of BLZinCh-3 in the presence of a range of free Zn2+ concentrations buffered using 1 mM HEDTA, 1 mM DHPTA, 5 mM EGTA, or 1 mM EGTA. Measurements were performed using 0.2 nM protein and 3200-fold diluted furimazine in 150 mM HEPES (pH 7.1), 100 mM NaCl, 10% (v/v) glycerol, 5 μM DTT, 1 mM TCEP, and 1 mg mL–1 BSA, at 23 °C. Data points represent the average of two measurements, and the solid line depicts a fit of the data assuming a single binding event using eq 1, from which the Kd of 15.6 ± 1.0 pM was determined.
BLZinCh Sensors Allow Intracellular [Zn2+] Measurements Using a Plate Reader
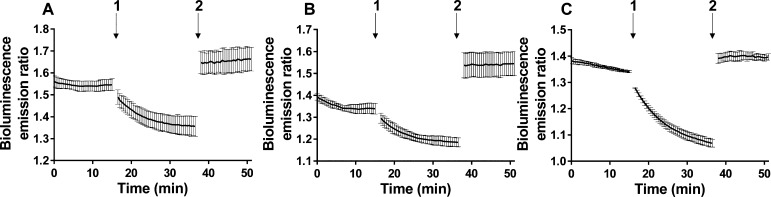
One of the potential benefits of using BRET compared to FRET is the ability to perform measurements on a population of cells using a standard plate reader. Similar measurements using FRET sensors would suffer from autofluorescence and light scattering, effectively prohibiting their application in high-throughput plate-reader-based assays. To test the performance of the BLZinCh sensors for these applications, we transiently transfected HeLa cells with sensors BLZinCh-1, -2, and -3 and measured the bioluminescence emission ratio of cells in suspension. The emission ratios measured in situ were very reproducible and similar to those obtained using purified sensor proteins measured on the same plate reader in a buffer. Figure 5A,B shows that the sensors responded to changing Zn2+ levels displaying a decrease of the ratio upon addition of the chelator TPEN, followed by an increase when adding a saturating amount of Zn2+ in the presence of the Zn2+ ionophore pyrithione. A comparison of the emission ratio in the resting state to the ratios in empty and fully occupied states yielded sensor occupancies of 61 ± 10% for BLZinCh-1 and 43 ± 6% for BLZinCh-2. These binding levels correspond to free cytosolic Zn2+ concentrations of 281 ± 108 pM (BLZinCh-1) and 92 ± 22 pM (BLZinCh-2), which are in agreement with values obtained using FRET sensors in single-cell imaging experiments.15,19 As expected based on its Zn2+ affinity of 15 pM, BLZinCh-3 was found to be almost completely saturated with Zn2+ under normal conditions (occupancy = 85 ± 4% corresponding to free [Zn2+] = 91 ± 36 pM; Figure 5C). The emission ratios shown in Figure 5 were not normalized and thus show good reproducibility and high signal-to-noise ratios. The ability to measure both BRET and FRET using the same sensor protein allowed us to directly compare the performance of bioluminescence and fluorescence detection in plate reader assays. A head-to-head comparison confirmed the superior performance of bioluminescence detection, which showed very similar bioluminescence spectra and reproducible performance at a range of cell dilutions, whereas fluorescence was dominated by light scattering, precluding the determination of meaningful FRET data (Figures S5–S7). Genetically encoded BRET sensors thus provide an attractive and easily accessible alternative to live-cell microscopy imaging or FACS for monitoring intracellular Zn2+ concentrations.
Figure 5.
Bioluminescence emission ratio (emission 500–545 nm/emission 400–455 nm) of HeLa cells expressing BLZinCh-1 (A), BLZinCh-2 (B), and BLZinCh-3 (C) in a resting state and after subsequent addition of 50 μM TPEN (1) and 100 μM Zn2+/5 μM pyrithione (2). Bioluminescence of a suspension of HeLa cells was measured on a plate reader using 3000-fold diluted furimazine in 20 mM HEPES (pH 7.4), 140 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, and 1.0 mM MgCl2, at 37 °C. Measurements were stopped for ∼1 min during the additions at time points 1 and 2. All traces represent the average of four measurements. Error bars represent standard deviation (SD).
Intracellular Zn2+ Imaging in Single Living Cells Using BRET
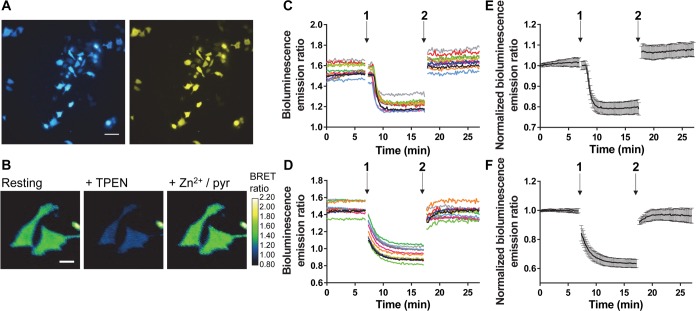
Having established the robust performance of the BLZinCh sensors to monitor intracellular Zn2+ on cell populations, we next tested whether the bright luminescence of the BLZinCh sensors would also allow live-cell imaging on single cells. Figure 6A shows images of HeLa cells expressing the BLZinCh-1 sensor obtained using an Olympus LV200 microscope equipped with filters specific for NLuc/Cerulean and Citrine emission. Measurements with this microscope, which is optimized for bioluminescence imaging, required exposure times of less than 1 s. Figure 6B–D show the BRET ratio of single cells expressing BLZinCh-1 or -3 in their resting state, followed by the subsequent addition of TPEN and excess Zn2+/pyrithione. Compared to similar FRET traces obtained using BLZinCh sensor precursor eZinCh-2 (Figure S8), the BRET traces are very stable and show less cell-to-cell variation in the absolute emission ratio. Average ratio analysis results in 78 ± 5% sensor occupancy for BLZinCh-1 and full occupancy for BLZinCh-3 (Figure 6E,F), which is in agreement with the cell suspension data. These results show that intracellular bioluminescence imaging of free Zn2+ using the BLZinCh sensors rivals intracellular fluorescence imaging using FRET sensors in terms of resolution and signal-to-noise, without the drawbacks associated with external illumination.
Figure 6.
(A) Bioluminescence images of NLuc-Cerulean emission (left; 420–460 nm) and Citrine emission (right; 510–550 nm) of HeLa cells expressing the BLZinCh-1 sensor. Scale bar = 80 μm. (B) False-colored ratiometric images of two HeLa cells expressing BLZinCh-1, in resting state and after additions of 50 μM TPEN and 100 μM Zn2+/5 μM pyrithione. Scale bar = 20 μm. (C,D) Bioluminescence emission ratio traces of 12 cells expressing BLZinCh-1 (C) or BLZinCh-3 (D), in a resting state and after additions of 50 μM TPEN (1) and 100 μM Zn2+/5 μM pyrithione (2). (E,F) Average bioluminescence emission ratio of the data shown in C and D, respectively, after normalization of the emission ratio at t = 0 min. Experiments were done using 500-fold diluted furimazine in live-cell imaging buffer (20 mM HEPES (pH 7.4), 140 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, and 1.0 mM MgCl2), at 37 °C and 5% CO2. Imaging was stopped for ∼1 min during the additions at time points 1 and 2. Error bars represent SD.
Discussion
In this work, we successfully modified two previously developed FRET-based Zn2+ sensor proteins to allow BRET-based detection of intracellular Zn2+, in addition to their fluorescence readout. Fusion of the bright and stable blue-light emitting luciferase NLuc to the Cerulean domain provided an efficient mechanism to allow bioluminescent excitation of these FRET sensors. While the dynamic range of the BRET signal for the eCALWY-based BLCALWY-1 sensor was limited by the presence of two competing BRET pathways, BRET/FRET sensors based on the eZinCh-2 scaffold yielded robust BRET sensors with 25–30% changes in BRET ratio. In addition, introduction of a chromophore-silencing mutation, yielded a BRET-only sensor with increased BRET response (50%) and an unexpected 10-fold increase in Zn2+ affinity. The combination of robust ratiometric response, physiologically relevant Zn2+ affinities, and stable and bright luminescence signal offered by the BLZinCh sensors, provides an attractive bioluminescent alternative to fluorescence-based detection for measuring intracellular Zn2+ concentrations in live cells. Whereas background fluorescence and scattering of excitation light precludes FRET-based plate reader analysis, such assays were found to be easily performed with the BLZinCh sensors using bioluminescence detection. The bioluminescence brightness of the BLZinCh sensors was also shown to be sufficient for intracellular imaging, providing one of the first examples of single-cell imaging using a genetically encoded BRET sensor.
BRET sensors are often developed by replacing the donor fluorescent domain of a FRET sensor by a luciferase with sufficient spectral overlap with the acceptor fluorophore. The alternative approach introduced here was to add the NLuc luciferase by fusing it to the donor fluorescent domain. In addition to offering both FRET- and BRET-based detection, this approach does not require extensive sensor development, as it builds upon the already optimized reporting properties of existing sensors. In principle, this approach should be more broadly applicable, as the majority of previously developed FRET sensors are based on the CFP-YFP FRET pair. Two important aspects need to be taken into account, however. First, our approach is best suited for FRET sensors that do not rely on a subtle allosteric mechanism, where introduction of an additional 20 kDa protein domain would most likely perturb the sensor mechanism. Examples include FRET sensors that are based on mutually exclusive domain interactions and self-associating fluorescent domains,44 FRET sensors based on helper domain interaction,45 FRET sensors based on affinity clamps,46 and the semisynthetic SNIFIT protein switches based on SNAP- and CLIP-tag technology.47−50 While the original FRET sensor properties in BLCALWY-1, BLZinCh-1, and BLZinCh-2 remained unperturbed, the changes in BRET emission ratio were found to be attenuated compared to the changes in FRET ratio. This is partly due to the fact that BRET between NLuc and Cerulean is not 100% efficient, which introduces a background emission that is not modulated by Zn2+ binding. This explains why the change in bioluminescence emission ratio in the BLZinCh sensors is smaller than the change in FRET ratio. The small change in bioluminescence emission ratio observed for BLCALWY-1 is probably also related to its sensor mechanism. BLCALWY-1 switches between two alternative conformations in which the average distance between Cerulean and Citrine increases upon Zn2+ binding. However, BRET also occurs directly between NLuc and Citrine, and this pathway is apparently not modulated by Zn2+ to the same extent. In the BLZinCh sensors Zn2+ binding decreases the distance between both Cerulean and Citrine and NLuc and Citrine, such that both FRET between Cerulean and Citrine and BRET between NLuc and Citrine contribute to the increase in emission ratio. The importance of direct BRET between NLuc and Citrine was further corroborated by the BLZinCh-3 sensor variant, which lacks the donor fluorophore. This sensor showed an increased BRET response, which can be explained by the better spectral separation of the emission of NLuc and Citrine. The 50% dynamic range observed for this BRET sensor rivals that of previously reported ratiometric BRET sensors, which following extensive optimization reported a maximal dynamic range of 60–70%.31,32 Guided by the recently determined crystal structure of NLuc, similar optimization of the linker length between NLuc and Cerulean or their relative position (C- vs N-terminal) can be envisioned to further improve the dynamic response of the BLZinCh sensors. Recent work on other BRET applications such as in vitro diagnostics and cell-based screening for protein–protein interactions showed that NLuc can also be effectively used in combination with more red-shifted FRET pairs, either red fluorescent proteins or synthetic fluorophores such as Cy3 or NCT.29,37−39 Although the light output of these systems would be lower due to less efficient BRET, this disadvantage may be compensated by less spectral overlap in the emission spectrum and the better penetration length of red light in biological tissues, in particular for in vivo applications.
The sensor proteins reported here represent the first bioluminescent sensors for intracellular measurement of free Zn2+ concentrations. These sensors should be useful in all those applications where external illumination is either unwanted or gives rise to high background signals. Although the genetic encoding of FRET-based sensor proteins allows easy distribution, their widespread application has been hampered by the requirement for live-cell fluorescence microscopy imaging facilities. The ability to now carry out these measurements in a plate reader assay using bioluminescence detection should make such measurements accessible to a broader group of scientists. In addition, BRET-based detection is ideally suited for high-throughput screening,51 providing the opportunity to identify novel pathways involved in the regulation of Zn2+ homeostasis and signaling. The affinities of BLZinCh-1 and BLZinCh-2 are similar to that of the parental eZinCh-2 sensor and should therefore also be applicable to measure organelle-specific free Zn2+ concentrations in, e.g., the mitochondria and the ER. The combined BRET/FRET response provides the added value that fluorescence could be used to verify the correct intracellular locations and allow FACS-based sorting of specific cell populations. The BRET-only sensor BLZinCh-3 provides a better BRET response and has the potential to be used in conjunction with other CFP-YFP-based sensors for multiparameter imaging. The unexpected 10-fold higher Zn2+ affinity of BLZinCh-3 means that the sensor is fully saturated when expressed in the cytosol of a mammalian cell line such as HeLa cells, making it unsuitable to detect perturbations that would result in an increase in the free Zn2+ concentration. However, free Zn2+ concentrations in other cell types such as bacteria have been postulated to be lower and in a concentration regime (10–40 pM)52 that is similar to this sensor’s Kd. This sensor could also be useful to further investigate the discrepancies reported for the free Zn2+ concentrations in specific organelles such as the ER.
While single-cell imaging using FRET may remain the preferred imaging method for standard cell imaging, our demonstration that live single-cell imaging is also feasible using BRET detection is important for all those applications where laser excitation is unwanted. These applications include avoiding photobleaching and phototoxicity during prolonged imaging experiments, imaging in optogenetic measurements, and imaging in cell systems that are naturally light-sensitive, such as retinal cells, chloroplasts, and systems triggered by light such as circadian rhythms. A practical advantage of BRET vs FRET detection is the lower cell-to-cell variability in emission ratio for BRET imaging, which is caused by the absence of background fluorescence. The latter effect also results in similar absolute BRET ratios observed in vitro and in situ, which is advantageous for in vivo applications.
Materials and Methods
Construction of Expression Plasmids
Plasmids pET28a-eZinCh-2 and pET28a-eCALWY-4 were available in our lab.15,19 In these plasmids, an NheI restriction site was introduced in the thrombin cleavage site, and the original stop codon was “removed” using the QuikChange Lightning Multi-Site-Directed Mutagenesis Kit (Stratagene) and primers S1 and S2 (Figures S9 and S10, Table S1, Eurofins Genomics). In addition, a Strep-Tag was inserted at the C-terminus of the gene constructs using a previously published cloning technique53 employing primers S3 and S4 (Figures S9 and S10, Table S1). The NLuc gene was PCR amplified from an existing plasmid40 employing primers S5–S9 (Figures S9 and S10, Table S1) introducing NheI and NdeI restriction sites at the 5′- and 3′-ends, respectively, and the chosen NLuc-Cerulean linker upstream of the NdeI site at the 3′-end. Ligation of NheI and NdeI digested FRET sensor vectors and NLuc PCR product was carried out using T4 DNA ligase (New England Biolabs). pET28a-BLCALWY-1 was cloned from pET28a-BLCALWY-4 by reversing the Cys-to-Ser mutation of the second Zn2+ binding cysteine in WD4 using the QuikChange Lightning Multi-Site-Directed Mutagenesis Kit and primer S10 (Figure S9, Table S1). The same procedure and primers S11–S13 (Figure S10, Table S1) were used to create the tryptophan mutants of the Cerulean chromophore in pET28a-BLZinCh-1. BLZinCh-1, -2, and -3 were cloned into a pCMV vector for mammalian cell expression. The plasmid sequence outside the encoded gene of pCMV-VVDZn-754 was PCR amplified introducing restriction sites NheI and XhoI at the distal ends using primers S14 and S15 (Figure S11, Table S1). The resulting PCR product and gene constructs pET28a-BLZinCh-1, -2, and -3 were digested using NheI and XhoI, and ligation was carried out using T4 DNA ligase. The correct open reading frames of all gene constructs in the plasmids were confirmed by Sanger sequencing (StarSEQ GmbH, Mainz, Germany).
Expression and Purification of Sensor Proteins
The pET28a plasmids encoding for the BLZinCh sensors and BLCALWY-1 were transformed into E. coli BL21(DE3) competent bacteria (Novagen). Single colonies were picked and used to inoculate 8 mL of Luria–Bertani (LB) medium cultures supplemented with 30 μg/mL kanamycin (Calbiochem). The bacteria were grown overnight at 37 °C and 250 rpm, followed by transfer of the cells to 0.5–2 L of LB medium cultures containing 30 μg/mL kanamycin. After growing the bacteria at 37 °C and 150 rpm until OD600 ≈ 0.6, expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG, Sigma-Aldrich), and bacteria were further grown for 20–24 h at 18 °C and 150 rpm. Bacteria were harvested by centrifugation for 10 min at 10 000g. The pellets were resuspended in 10–16 mL (BLZinCh sensors) or 40 mL (BLCALWY-1) of BugBuster protein extraction agent (Novagen) supplemented with 10–16 or 40 μL of Benzonase (Novagen), respectively, and 1 mM tris(2-carboxyethyl)phosphine (TCEP, Sigma-Aldrich) and incubated for ∼1 h at RT. Next, the Bugbuster suspension was centrifuged for 20 min at 16 000g. Further purification was performed via Ni2+-affinity chromatography using the N-terminal 6xHis-tag employing Ni-NTA HisBind resin (Novagen) and the C-terminal Strep-tag using Strep-Tactin Superflow high capacity resin (IBA Life Sciences). All wash and elution buffers were supplemented with 1 mM TCEP, and 50 μM DL-dithiothreitol (DTT, Sigma-Aldrich) was additionally added to the Strep-Tactin elution buffer. Using Amicon Ultra-4 Centrifugal Filter Units (molecular weight cutoff 10 kDa; Millipore), the elution fractions were concentrated and dialyzed to 150 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.1), 100 mM NaCl, 10% (v/v) glycerol, 50 μM DTT, and 1 mM TCEP. Subsequently, the proteins were frozen in liquid nitrogen for storage at −80 °C. Storage was done in small aliquots (∼50 uL) to prevent protein aggregation as a result of repetitive freeze–thaw cycles. Protein concentrations were determined using the absorption of Citrine at 516 nm (extinction coefficient = 77 000 M–1 cm–1).55
Bioluminescence Measurements and Zn2+ Titrations
Bioluminescence emission spectra were recorded in a Varian Cary Eclipse spectrophotometer in a 200 μL volume. Bioluminescence readout of the Zn2+ titration experiments was performed on Tecan Infinite F500 microplate reader using filters for detection of NLuc-Cerulean (400–455 nm) and Citrine (500–545 nm) emission and integration times of 1 s. Zn2+ titrations were carried out with 0.2 nM sensor protein in a buffer consisting of 150 mM HEPES (pH 7.1), 100 mM NaCl, 10% (v/v) glycerol, 5 μM DTT, 1 mM TCEP, and 1 mg mL–1 bovine serum albumin (BSA), together with different Zn2+ chelators (1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM N-(2-hydroxyethyl)ethylenediamine-N,N′,N′-triacetic acid (HEDTA), 1 mM 1,3-diamino-2-hydroxypropane-N,N,N′,N′-tetraacetic acid (DHPTA), 1 or 5 mM ethylene glycol-bis(2-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA, all from Sigma-Aldrich)) and different concentrations of ZnCl2 (Sigma-Aldrich), as previously described.19 Titrations were performed in a white flat-bottom 96-well plate (Nunc, Thermo Scientific) in a 150 μL volume. The mixed titration samples were incubated for 10–15 min, after which 3200-fold diluted furimazine (Nano-Glo substrate) was added followed by another 10 min incubation. The dissociation constants of the sensor proteins for binding Zn2+ were determined by fitting the ratio of the emission of Citrine to the emission of NLuc-Cerulean using eq 1. [Zn2+] is the calculated free Zn2+ concentration; P1, the difference between the ratio in the Zn2+-saturated and Zn2+-depleted states; and P2, the emission ratio in the Zn2+-depleted state.
| 1 |
Zn2+ titrations using fluorescence readout were performed using 50–100 nM sensor protein in the same buffer as the bioluminescence measurements, without the addition of furimazine, in a black flat-bottom 96-well plate (Greiner Bio-One). Fluorescence emission was measured from 450 to 550 nm (5 nm bandwidth) on a Tecan Safire2 fluorescence microplate reader when exciting at 430 nm (10 nm bandwidth).
Mammalian Cell Culture and in Situ [Zn2+] Measurements in Cell Suspension
HeLa cells were grown at 37 °C and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 4.5 g/L d-glucose, 0.58 g/L l-glutamine, 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Life Technologies). One day before transfection, 150 000 cells were seeded per well of a six-well plate (Corning). Cells were transfected with 2 μg of pCMV-BLZinCh-1, -2, or -3 and 10 μL of Lipofectamine 2000 (Life Technologies) in Opti-MEM Reduced Serum Media (Life Technologies). After 4 h, the media were changed to DMEM. Two days after transfection, cells were released from the wells using trypsin (Thermo Fisher Scientific) and resuspended in 1 mL of preheated (37 °C) live-cell imaging buffer (20 mM HEPES (pH 7.4), 140 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, and 1.0 mM MgCl2). A total of 15 μL of cell suspension was transferred to a 96-well plate (Nunc, Thermo Scientific) to which buffer was added to a final volume of 150 μL. After the addition of 1 μL of 20-fold diluted furimazine (Nano-Glo substrate), bioluminescence was monitored on a Tecan Infinite F500 microplate reader using filters for the detection of NLuc-Cerulean (400–455 nm) and Citrine (500–545 nm) emission and integration times of 1 s, at 37 °C. The measurement was performed for 50 min, during which 50 μM N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN, Sigma-Aldrich) was loaded onto the cells after 15 min and 100 μM ZnCl2 and 5 μM 2-mercaptopyridine N-oxide (pyrithione, Acros Organics) after 35 min. During the additions of reagents, the measurement was stopped for ∼1 min.
Sensor occupancies were calculated using eq 2, and corresponding Zn2+ levels were calculated using eq 3, in which Rmin and Rmax are the steady-state emission ratios after the addition of TPEN and Zn2+/pyrithione, respectively, and R is the steady-state emission ratio of the cells in the resting state.
| 2 |
| 3 |
Live-Cell Bioluminescence Imaging
One or 2 days before transfection, HeLa cells were plated on glass bottom dishes (Greiner Bio-One). Cells were transfected with pCMV-BLZinCh-1 or pCMV-BLZinCh-3 as described above. Imaging was performed 2 days after transfection. A total of 1 mL of live-cell imaging buffer was loaded onto the cells, and 2 μL of furimazine (Nano-Glo substrate) was added. Bioluminescence imaging was done on a LV200 bioluminescence microscope (Olympus) at 60× magnification (UPlanSApo 60x/1.35 Oil ∞/0.17/FN26.5 UIS2 BFP1 objective microscope lens), using BFP (420–460 nm) and GFP (510–550 nm) emission filters, in a fully closed chamber at 37 °C and 5% CO2. For both recording channels, an exposure time of 0.5–3 s was used. The emission was measured every 10 s for 27 min, during which 50 μM TPEN was loaded onto the cells after 7 min, and 100 μM ZnCl2 and 5 μM pyrithione after 17 min. During the additions of reagents, the measurement was stopped for ∼1 min. Image analysis was performed using ImageJ software.56 Sensor occupancies were calculated using eq 2. False-colored ratiometric images were obtained by subtracting the background signal from the Citrine emission channel, dividing the resulting Citrine and NLuc-Cerulean emission channel, and thresholding the resulting image at 0.8 (lower threshold) and 2.2 (upper threshold).
Acknowledgments
The authors thank Appie Kalaç, Jan-Willem Muller, Laura van Smeden, Kwankwan Zhu, Hans C. de Ferrante, Esther M. van Leeuwen, Sjoerd A. S. Nooijens, Femke L. A. Vaassen, Yeshi A. de Bruin, Elles P. Elschot, Laura L. E. Jacobs, Simone F. A. Wouters, and Dr. Anne M. Hessels for assistance with initial cloning and characterization of the sensors. The authors thank Prof. Dr. Takeharu Nagai (Osaka University) for sharing a homology model of the NLuc structure. This work was supported by an ECHO grant from The Netherlands Organization of Scientific Research (700.59.013) and an ERC starting grant (ERC-2011-StG 280255).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.6b00453.
SDS-PAGE analysis of the purified sensor proteins; fluorescence analysis of Zn2+ binding to BLZinCh-1 and BLZinCh-2; Cerulean crystal structure with hydrogen bond between Ser-205 and chromophore-tryptophan highlighted; bioluminescence analysis of Zn2+ binding to Cerulean-W66L and -W66H mutants of BLZinCh-1; plate-based fluorescence and bioluminescence spectral analysis of HeLa cell suspensions expressing BLZinCh-1 exposed to changing Zn2+ levels; fluorescence emission ratio traces of 12 Hela cells expressing eZinCh-2 exposed to changing Zn2+ levels; nucleotide and amino acid sequences of BLCALWY-1, BLZinCh-1, and BLZinCh-2 in pET28a and/or pCMV vectors; primers used during the different cloning steps (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Fukada T.; Yamasaki S.; Nishida K.; Murakami M.; Hirano T. (2011) Zinc homeostasis and signaling in health and diseases: Zinc signaling. JBIC, J. Biol. Inorg. Chem. 16, 1123–1134. 10.1007/s00775-011-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L.; Falchuk K. H. (1993) The biochemical basis of zinc physiology. Physiol Rev. 73, 79–118. [DOI] [PubMed] [Google Scholar]
- Maret W. (2009) Molecular aspects of human cellular zinc homeostasis: redox control of zinc potentials and zinc signals. BioMetals 22, 149–157. 10.1007/s10534-008-9186-z. [DOI] [PubMed] [Google Scholar]
- Fukada T.; Hojyo S.; Furuichi T. (2013) Zinc signal: a new player in osteobiology. J. Bone Miner. Metab. 31, 129–135. 10.1007/s00774-012-0409-6. [DOI] [PubMed] [Google Scholar]
- Hessels A. M.; Taylor K. M.; Merkx M. (2016) Monitoring cytosolic and ER Zn2+ in stimulated breast cancer cells using genetically encoded FRET sensors. Metallomics 8, 211–217. 10.1039/C5MT00257E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sensi S. L.; Paoletti P.; Koh J. Y.; Aizenman E.; Bush A. I.; Hershfinkel M. (2011) The neurophysiology and pathology of brain zinc. J. Neurosci. 31, 16076–16085. 10.1523/JNEUROSCI.3454-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. M.; Hiscox S.; Nicholson R. I.; Hogstrand C.; Kille P. (2012) Protein kinase CK2 triggers cytosolic zinc signaling pathways by phosphorylation of zinc channel ZIP7. Sci. Signaling 5, ra11. 10.1126/scisignal.2002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A.; Nakamura M.; Fujii H.; Tamano H. (2013) Synaptic Zn2+ homeostasis and its significance. Metallomics 5, 417–423. 10.1039/c3mt20269k. [DOI] [PubMed] [Google Scholar]
- Bellomo E.; Massarotti A.; Hogstrand C.; Maret W. (2014) Zinc ions modulate protein tyrosine phosphatase 1B activity. Metallomics 6, 1229–1239. 10.1039/C4MT00086B. [DOI] [PubMed] [Google Scholar]
- Kaltenberg J.; Plum L. M.; Ober-Blobaum J. L.; Honscheid A.; Rink L.; Haase H. (2010) Zinc signals promote IL-2-dependent proliferation of T cells. Eur. J. Immunol. 40, 1496–1503. 10.1002/eji.200939574. [DOI] [PubMed] [Google Scholar]
- Yamasaki S.; Hasegawa A.; Hojyo S.; Ohashi W.; Fukada T.; Nishida K.; Hirano T. (2012) A novel role of the L-type calcium channel alpha1D subunit as a gatekeeper for intracellular zinc signaling: zinc wave. PLoS One 7, e39654. 10.1371/journal.pone.0039654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozym R. A.; Thompson R. B.; Stoddard A. K.; Fierke C. A. (2006) Measuring picomolar intracellular exchangeable zinc in PC-12 cells using a ratiometric fluorescence biosensor. ACS Chem. Biol. 1, 103–111. 10.1021/cb500043a. [DOI] [PubMed] [Google Scholar]
- Carter K. P.; Young A. M.; Palmer A. E. (2014) Fluorescent sensors for measuring metal ions in living systems. Chem. Rev. 114, 4564–4601. 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer P. J.; Miranda J. G.; Gorski J. A.; Palmer A. E. (2009) Genetically encoded sensors to elucidate spatial distribution of cellular zinc. J. Biol. Chem. 284, 16289–16297. 10.1074/jbc.M900501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessels A. M.; Chabosseau P.; Bakker M. H.; Engelen W.; Rutter G. A.; Taylor K. M.; Merkx M. (2015) eZinCh-2: A versatile, genetically encoded FRET sensor for cytosolic and intraorganelle Zn imaging. ACS Chem. Biol. 10, 2126–2134. 10.1021/acschembio.5b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessels A. M.; Merkx M. (2015) Genetically-encoded FRET-based sensors for monitoring Zn2+ in living cells. Metallomics 7, 258–266. 10.1039/C4MT00179F. [DOI] [PubMed] [Google Scholar]
- Qin Y.; Dittmer P. J.; Park J. G.; Jansen K. B.; Palmer A. E. (2011) Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc. Natl. Acad. Sci. U. S. A. 108, 7351–7356. 10.1073/pnas.1015686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y.; Miranda J. G.; Stoddard C. I.; Dean K. M.; Galati D. F.; Palmer A. E. (2013) Direct comparison of a genetically encoded sensor and small molecule indicator: implications for quantification of cytosolic Zn2+. ACS Chem. Biol. 8, 2366–2371. 10.1021/cb4003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenborg J. L.; Nicolson T. J.; Bellomo E. A.; Koay M. S.; Rutter G. A.; Merkx M. (2009) Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat. Methods 6, 737–740. 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V.; Grossmann G.; Vinkenborg J. L.; Merkx M.; Thomine S.; Frommer W. B. (2014) Dynamic imaging of cytosolic zinc in Arabidopsis roots combining FRET sensors and RootChip technology. New Phytol. 202, 198–208. 10.1111/nph.12652. [DOI] [PubMed] [Google Scholar]
- Maret W. (2015) Analyzing free zinc(II) ion concentrations in cell biology with fluorescent chelating molecules. Metallomics 7, 202–211. 10.1039/C4MT00230J. [DOI] [PubMed] [Google Scholar]
- Chabosseau P.; Tuncay E.; Meur G.; Bellomo E. A.; Hessels A.; Hughes S.; Johnson P. R.; Bugliani M.; Marchetti P.; Turan B.; Lyon A. R.; Merkx M.; Rutter G. A. (2014) Mitochondrial and ER-targeted eCALWY probes reveal high levels of free Zn2+. ACS Chem. Biol. 9, 2111–2120. 10.1021/cb5004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. G.; Qin Y.; Galati D. F.; Palmer A. E. (2012) New sensors for quantitative measurement of mitochondrial Zn2+. ACS Chem. Biol. 7, 1636–1640. 10.1021/cb300171p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenburg L. H.; Hessels A. M.; Ebberink E. H.; Arts R.; Merkx M. (2013) Robust red FRET sensors using self-associating fluorescent domains. ACS Chem. Biol. 8, 2133–2139. 10.1021/cb400427b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. G.; Weaver A. L.; Qin Y.; Park J. G.; Stoddard C. I.; Lin M. Z.; Palmer A. E. (2012) New alternately colored FRET sensors for simultaneous monitoring of Zn2+ in multiple cellular locations. PLoS One 7, e49371. 10.1371/journal.pone.0049371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boute N.; Jockers R.; Issad T. (2002) The use of resonance energy transfer in high-throughput screening: BRET versus FRET. Trends Pharmacol. Sci. 23, 351–354. 10.1016/S0165-6147(02)02062-X. [DOI] [PubMed] [Google Scholar]
- Welsh D. K.; Noguchi T. (2012) Cellular bioluminescence imaging. Cold Spring Harb Protoc 2012, 852–866. 10.1101/pdb.top070607. [DOI] [PubMed] [Google Scholar]
- Bacart J.; Corbel C.; Jockers R.; Bach S.; Couturier C. (2008) The BRET technology and its application to screening assays. Biotechnol. J. 3, 311–324. 10.1002/biot.200700222. [DOI] [PubMed] [Google Scholar]
- Machleidt T.; Woodroofe C. C.; Schwinn M. K.; Mendez J.; Robers M. B.; Zimmerman K.; Otto P.; Daniels D. L.; Kirkland T. A.; Wood K. V. (2015) NanoBRET-A novel BRET platform for the analysis of protein-protein interactions. ACS Chem. Biol. 10, 1797–1804. 10.1021/acschembio.5b00143. [DOI] [PubMed] [Google Scholar]
- Paley M. A.; Prescher J. A. (2014) Bioluminescence: a versatile technique for imaging cellular and molecular features. MedChemComm 5, 255–267. 10.1039/C3MD00288H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K.; Hatsugai N.; Horikawa K.; Kobayashi K.; Matsu-Ura T.; Mikoshiba K.; Nagai T. (2010) Auto-luminescent genetically-encoded ratiometric indicator for real-time Ca2+ imaging at the single cell level. PLoS One 5, e9935. 10.1371/journal.pone.0009935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. I.; Collins J.; Davis R.; Lin K. M.; DeCamp D.; Roach T.; Hsueh R.; Rebres R. A.; Ross E. M.; Taussig R.; Fraser I.; Sternweis P. C. (2007) Use of a cAMP BRET sensor to characterize a novel regulation of cAMP by the sphingosine 1-phosphate/G13 pathway. J. Biol. Chem. 282, 10576–10584. 10.1074/jbc.M609695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K. H.; Sopory S.; Visweswariah S. S. (2008) The GAF domain of the cGMP-binding, cGMP-specific phosphodiesterase (PDE5) is a sensor and a sink for cGMP. Biochemistry 47, 3534–3543. 10.1021/bi702025w. [DOI] [PubMed] [Google Scholar]
- Saito K.; Chang Y. F.; Horikawa K.; Hatsugai N.; Higuchi Y.; Hashida M.; Yoshida Y.; Matsuda T.; Arai Y.; Nagai T. (2012) Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun. 3, 1262. 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai A.; Nakano M.; Saito K.; Haruno R.; Watanabe T. M.; Ohyanagi T.; Jin T.; Okada Y.; Nagai T. (2015) Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc. Natl. Acad. Sci. U. S. A. 112, 4352–4356. 10.1073/pnas.1418468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. P.; Unch J.; Binkowski B. F.; Valley M. P.; Butler B. L.; Wood M. G.; Otto P.; Zimmerman K.; Vidugiris G.; Machleidt T.; Robers M. B.; Benink H. A.; Eggers C. T.; Slater M. R.; Meisenheimer P. L.; Klaubert D. H.; Fan F.; Encell L. P.; Wood K. V. (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7, 1848–1857. 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss R.; Schena A.; Reymond L.; Patiny L.; Werner D.; Tinberg C. E.; Baker D.; Johnsson K. (2014) Bioluminescent sensor proteins for point-of-care therapeutic drug monitoring. Nat. Chem. Biol. 10, 598–603. 10.1038/nchembio.1554. [DOI] [PubMed] [Google Scholar]
- Robers M. B.; Dart M. L.; Woodroofe C. C.; Zimprich C. A.; Kirkland T. A.; Machleidt T.; Kupcho K. R.; Levin S.; Hartnett J. R.; Zimmerman K.; Niles A. L.; Ohana R. F.; Daniels D. L.; Slater M.; Wood M. G.; Cong M.; Cheng Y. Q.; Wood K. V. (2015) Target engagement and drug residence time can be observed in living cells with BRET. Nat. Commun. 6, 10091. 10.1038/ncomms10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena A.; Griss R.; Johnsson K. (2015) Modulating protein activity using tethered ligands with mutually exclusive binding sites. Nat. Commun. 6, 7830. 10.1038/ncomms8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.; den Hartog I.; Zijlema S. E.; Thijssen V.; van der Beelen S. H. E.; Merkx M. (2016) Detection of antibodies in blood plasma using bioluminescent sensor proteins and a smartphone. Anal. Chem. 88, 4525–4532. 10.1021/acs.analchem.6b00534. [DOI] [PubMed] [Google Scholar]
- Stoddart L. A.; Johnstone E. K.; Wheal A. J.; Goulding J.; Robers M. B.; Machleidt T.; Wood K. V.; Hill S. J.; Pfleger K. D. (2015) Application of BRET to monitor ligand binding to GPCRs. Nat. Methods 12, 661–663. 10.1038/nmeth.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessels A. M.; Merkx M. (2016) Simple method for proper analysis of FRET sensor titration data and intracellular imaging experiments based on isosbestic points. ACS Sens 1, 498–502. 10.1021/acssensors.6b00078. [DOI] [Google Scholar]
- Lelimousin M.; Noirclerc-Savoye M.; Lazareno-Saez C.; Paetzold B.; Le Vot S.; Chazal R.; Macheboeuf P.; Field M. J.; Bourgeois D.; Royant A. (2009) Intrinsic dynamics in ECFP and Cerulean control fluorescence quantum yield. Biochemistry 48, 10038–10046. 10.1021/bi901093w. [DOI] [PubMed] [Google Scholar]
- van der Velden L. M.; Golynskiy M. V.; Bijsmans I. T.; van Mil S. W.; Klomp L. W.; Merkx M.; van de Graaf S. F. (2013) Monitoring bile acid transport in single living cells using a genetically encoded Forster resonance energy transfer sensor. Hepatology 57, 740–752. 10.1002/hep.26012. [DOI] [PubMed] [Google Scholar]
- Grunberg R.; Burnier J. V.; Ferrar T.; Beltran-Sastre V.; Stricher F.; van der Sloot A. M.; Garcia-Olivas R.; Mallabiabarrena A.; Sanjuan X.; Zimmermann T.; Serrano L. (2013) Engineering of weak helper interactions for high-efficiency FRET probes. Nat. Methods 10, 1021–1027. 10.1038/nmeth.2625. [DOI] [PubMed] [Google Scholar]
- Huang J.; Koide S. (2010) Rational conversion of affinity reagents into label-free sensors for peptide motifs by designed allostery. ACS Chem. Biol. 5, 273–277. 10.1021/cb900284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun M. A.; Griss R.; Reymond L.; Tan K. T.; Piguet J.; Peters R. J.; Vogel H.; Johnsson K. (2011) Semisynthesis of fluorescent metabolite sensors on cell surfaces. J. Am. Chem. Soc. 133, 16235–16242. 10.1021/ja206915m. [DOI] [PubMed] [Google Scholar]
- Brun M. A.; Tan K. T.; Griss R.; Kielkowska A.; Reymond L.; Johnsson K. (2012) A semisynthetic fluorescent sensor protein for glutamate. J. Am. Chem. Soc. 134, 7676–7678. 10.1021/ja3002277. [DOI] [PubMed] [Google Scholar]
- Brun M. A.; Tan K. T.; Nakata E.; Hinner M. J.; Johnsson K. (2009) Semisynthetic fluorescent sensor proteins based on self-labeling protein tags. J. Am. Chem. Soc. 131, 5873–5884. 10.1021/ja900149e. [DOI] [PubMed] [Google Scholar]
- Masharina A.; Reymond L.; Maurel D.; Umezawa K.; Johnsson K. (2012) A fluorescent sensor for GABA and synthetic GABA(B) receptor ligands. J. Am. Chem. Soc. 134, 19026–19034. 10.1021/ja306320s. [DOI] [PubMed] [Google Scholar]
- Kelkar M.; De A. (2012) Bioluminescence based in vivo screening technologies. Curr. Opin. Pharmacol. 12, 592–600. 10.1016/j.coph.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Wang D.; Hurst T. K.; Thompson R. B.; Fierke C. A. (2011) Genetically encoded ratiometric biosensors to measure intracellular exchangeable zinc in Escherichia coli. J. Biomed. Opt. 16, 087011. 10.1117/1.3613926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91. 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aper S. J.; Merkx M. (2016) Rewiring multidomain protein switches: transforming a fluorescent Zn2+ sensor into a light-responsive Zn2+ binding protein. ACS Synth. Biol. 5, 698. 10.1021/acssynbio.6b00027. [DOI] [PubMed] [Google Scholar]
- Griesbeck O.; Baird G. S.; Campbell R. E.; Zacharias D. A.; Tsien R. Y. (2001) Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276, 29188–29194. 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Schneider C. A.; Rasband W. S.; Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.