Abstract
Background
The East African riverine cichlid species Astatotilapia burtoni serves as an important laboratory model for sexually dimorphic physiology and behavior, and also serves as an outgroup species for the explosive adaptive radiations of cichlid species in Lake Malawi and Lake Victoria. An astounding diversity of genetic sex determination systems have been revealed within the adaptive radiation of East African cichlids thus far, including polygenic sex determination systems involving the epistatic interaction of multiple, independently segregating sex determination alleles. However, sex determination has remained unmapped in A. burtoni. Here we present mapping results supporting the presence of multiple, novel sex determination alleles, and thus the presence of polygenic sex determination in A. burtoni.
Results
Using mapping in small families in conjunction with restriction-site associated DNA sequencing strategies, we identify associations with sex at loci on linkage group 13 and linkage group 5–14. Inheritance patterns support an XY sex determination system on linkage group 5–14 (a chromosome fusion relative to other cichlids studied), and an XYW system on linkage group 13, and these associations are replicated in multiple families. Additionally, combining our genetic data with comparative genomic analysis identifies another fusion that is unassociated with sex, with linkage group 8–24 and linkage group 16–21 fused in A. burtoni relative to other East African cichlid species.
Conclusions
We identify genetic signals supporting the presence of three previously unidentified sex determination alleles at two loci in the species A. burtoni, strongly supporting the presence of polygenic sex determination system in the species. These results provide a foundation for future mapping of multiple sex determination genes and their interactions. A better understanding of sex determination in A. burtoni provides important context for their use in behavioral studies, as well as studies of the evolution of genetic sex determination and sexual conflicts in East African cichlids.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-3177-1) contains supplementary material, which is available to authorized users.
Keywords: Sex determination, Polygenic sex determination, Cichlid, Fish, Evolution, Astatotilapia burtoni
Background
Sex determination is absolutely fundamental to the fitness of sexually reproducing organisms, yet the “switches” modulating female versus male development are incredibly diverse [1]. This diversity arises from surprisingly numerous transitions in sex determination mechanisms across taxa, yet little is know about the evolutionary and developmental forces underlying these transitions. Given the current catalog of identified sex determination mechanisms, fish are arguably the taxon with the highest diversity of sex determination mechanisms [2]. Broadly, sex determination mechanisms are categorized as environmental (e.g., temperature during development), behavioral (e.g., social status), or genetic (e.g., the Sry gene in mammals). Historically, the architecture of genetic sex determination was considered relatively simple, with a single inherited cue driving both primary gonadal sexual development, and producing resounding secondary sexual effects throughout the organism [3]. In vertebrates with genetic sex determination, sex determination alleles act as this cue. In species with XY sex determination systems, inheritance of the Y allele is associated with male development (XX females, XY males); with ZW systems the W allele is associated with female development (ZW females, ZZ males). When a sex determination allele arises on an autosome, the evolutionary trajectory of that chromosome can change, with new selection pressures potentially leading to local recombination suppression and heteromorphy over time [4]. Thus sex determination alleles can exist on highly degenerated sex chromosomes such as the Y chromosome in mammals [5], within defined sex loci with local recombination suppression [6], or as a simple polymorphism with no associated chromosomal changes [7].
As sex determination alleles are cataloged, surprising overlap suggests repeated modification of a few pathways across broad taxa to modulate sexual development. Specifically, members of the Sry-related HMG box (sox) and doublesex and mab-3 related (dmrt) families of transcription factors, and anti-Müllerian hormone (amh) pathway members, have repeatedly evolved as sex determination factors (reviewed in [8]; see also [9, 10]). Studies in fish have been key to identifying the diversity of vertebrate genetic sex determiners, with a role for dmrt first identified in medaka, and later demonstrated in chicken and frog (Xenopus) [6, 11, 12]; similarly the role of the amh pathway was first identified in pejerrey and fugu, and later in platypus [7, 13, 14]. Moreover, recent identification of sex determiners such as gsdf and sexually dimorphic on the Y-chromosome (sdY) within fish make it clear that sex determination mechanisms are not limited to the above pathways [15, 16].
It was also within fish that polygenic sex determination was discovered [17, 18], demonstrating that genetic sex determination need not be monogenic in nature. In polygenic sex determination systems, multiple, independently segregating sex determination alleles interact to determine sex within a species [3]. Historically, examples of polygenic sex determination were considered anomalous or evolutionarily transitory; however, with broader surveys and readily available genomic tools, polygenic sex determination has been identified in multiple animal taxa, including fish, mammals, insects, and copepods [3, 19], suggesting that polygenic sex determination is a common and stable evolutionary strategy.
In fish, polygenic sex determination systems exist in both single- and multi-locus configurations. In platyfish (Xiphophorus maculatus), three alleles (X, Y, and W) segregate at the sex determination locus, interacting to determine male versus female development in what is sometimes referred to as an XYW system [18, 20]. In some East African cichlids (e.g., Metriaclima spp. from Lake Malawi), XY and ZW sex determination systems reside on distinct chromosomes, and epistasis between genotypes at the two loci determines male versus female development [21, 22]. Polygenic sex determination in fish can involve more than two sex determination alleles. For example, mapping sex as a quantitative trait in a hybrid cross of two cichlid species identified at least five different loci interacting in an epistatic hierarchy to determine sex [23]. In sea bass, the genetic architecture of sex determination may be equally polygenic, with recent mapping efforts identifying at least four loci in the genome associated with sex determination [24].
In zebrafish (Danio rerio), several independent studies indicated a highly polygenic basis of sex determination in standard laboratory strains [25–27]; however, a more recent study revealed a monogenic ZW sex determination system in natural populations, with the W allele absent from laboratory strains, presumably lost during domestication [28]. The zebrafish findings serve as a cautionary tale for interpreting laboratory results without natural context, but also speak to the lability of sex determination and development in fishes. Such lability has been known for some time, with the loss of a natural sex chromosome and arisal of a novel autosomal sex determiner during selective breeding in guppies (Poecilia reticulata) described nearly a century ago [29]. In natural context, the forces underlying such transitions between sex determination systems are poorly understood.
The adaptive radiation of African cichlid fish is a powerful model system for the study of evolution and development, and an astounding diversity of genetic sex determination loci have been revealed from just a handful of studied species [22, 23, 30–35]. These sex determination loci exist in various contexts, including polygenic sex determination [22, 23, 30, 31], on supernumerary chromosomes [32], and in regions of potential recombination suppression indicative of early sex chromosome evolution [21, 36, 37]. The riverine cichlid Astatotilapia burtoni has served as a longstanding model of sexual physiology, behavior, and related gene expression [38], yet until recently sex determination in the species was largely unexamined. A recent study described sex-specific transcriptional differences in the gonad of A. burtoni [39], and another inferred the presence of an XY system in the species using crossing experiments with hormonally sex-reversed individuals [40]. However, direct genetic mapping of sex determination was not previously performed for A. burtoni.
Here, we describe use of double digest restriction associated DNA sequencing (ddRADseq) to perform genome scans for sex-associated loci in small A. burtoni families, followed by replication in additional, independent families. We order the A. burtoni genome scaffolds to two other, higher quality cichlid genomes to orient mapping. Mapping results indicate the presence of an XYW locus on linkage group 13 (LG13), and an XY locus on LG5-14 (a chromosome fusion relative to other cichlids). The evidence for at least three sex determination alleles strongly supports the presence of polygenic sex determination in A. burtoni, though we discuss caveats related to sex reversal and lability of sex determination during fish domestication. Additionally, haplotype data define an additional pair of linkage groups involved in the other A. burtoni-specific chromosome fusion.
Results and discussion
Comparative scaffolding of the A. burtoni genome
A. burtoni was one of five cichlid species sequenced as part of the cichlid genome project, but was also the species with the most fragmented genome assembly (scaffold N50 of 1.2 Mb, versus 2.5–4.4 Mb for the other four species; [41]). In order to provide chromosome-level organization of A. burtoni scaffolds for mapping, we ordered and oriented them to anchored genomes of two of other cichlid species (Additional files 1 and 2). The Nile tilapia (Oreochromis niloticus) genome assembly was anchored to a high-resolution radiation hybrid map, providing a golden path [41, 42]. Genome scaffolds of Metriaclima zebra, a Lake Malawi species, were also anchored to a RADseq-based genetic map [43]. A. burtoni scaffold assignment was consistent by linkage group for both species, but regular inconsistencies in scaffold order indicate a moderate level of intrachromosomal rearrangement between O. niloticus and M. zebra. We use the O. niloticus scaffolding for the following analysis because it provided the most parsimonious ordering based on our marker data, including producing continuous, uninterrupted mapping intervals. We number A. burtoni linkage groups based on synteny with established O. niloticus linkage group numbering, per convention in the cichlid genetics community [42].
Identification of two sex determination loci
Two A. burtoni families with near-equal sex ratios were genotyped using a ddRADseq strategy, and resulting SNP marker genotypes were tested for association with gonadal sex (Table 1). Markers with significant p values (p < 0.01) were further sorted by allele frequency in each sex, to search for minor alleles at intermediate frequency in one sex, and absent in the other, following a dominant model of sex determination (Additional file 3). Both families had numerous markers with significant associations with phenotypic sex, and alignment of marker sequences to the tilapia genome revealed clustering of the associated markers by linkage group. In one family, marker genotypes in a region of LG13 show near perfect association with sex, as do markers on unanchored portions of the tilapia assembly including UNK25 and UNK107 (Fig. 1, Additional file 3). Based on association and haplotype patterns we predict that sex-associated scaffolds aligning to UNK25 and UNK107 should be placed on LG13, within or adjacent to the sex associated scaffolds aligning there. Because maternal alleles at LG13 co-segregate with sex, we designate this family as having sex determined by a ZW system.
Table 1.
Summary of ddRADseq mapping families
| Family | M | F | Readsa | Stacksa | Polymorphic stacksb | Lowest p-valuec | Corresponding Tilapia linkage groupsd |
|---|---|---|---|---|---|---|---|
| RAD 1 | 12 | 12 | 9.7 × 106 | 435,006 | 4,777 | 9.6 × 10−6 | LG13, UNK25, UNK107 |
| RAD 2 | 9 | 8 | 5.9 × 106 | 341,384 | 4,268 | 1.4 × 10−3 | LG5, LG14 |
Families with number of male (M) and female (F) offspring. aAverage reads and stacks per offspring. bTotal number of polymorphic stacks used for association mapping, requiring a minimum stack depth of 10 to call homozygotes, and genotypes called for 20 of 24 offspring for RAD 1, and 15 of 17 offspring for RAD 2. cThe lowest p-value obtained for Fisher’s exact test of allele association with sex, shared by multiple markers (Additional file 3). dLinkage groups with markers with lowest p-value association
Fig. 1.
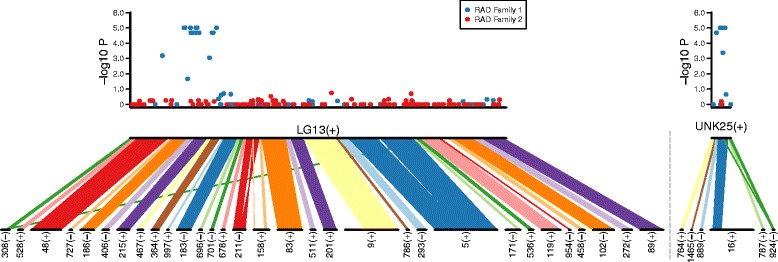
Association of sex with SNP markers on LG13 in family 1. Association plotted as the negative log of Fisher’s exact test p value of marker alleles with sex (top), aligned to A. burtoni genome scaffolds ordered and oriented to LG13 and UNK25 of the anchored O. niloticus genome assembly (bottom). Associated scaffolds aligning to O. niloticus UNK25 (right) should likely be placed on LG13, in or adjacent to the peak of association with sex
In a second family, associated RAD markers consistently aligned to tilapia LG5 and LG14 (Fig. 2a, Additional file 3). Previous karyotype analysis demonstrated that A. burtoni has two fused chromosome pairs relative to all other haplochromine cichlids surveyed [44]. Examination of our associated marker genotypes at LG5 and LG14 revealed clear haplotypes with no recombinants spanning the two linkage groups (Additional file 4), demonstrating that LG5 and LG14 constitute one of the chromosome fusions in the A. burtoni karyotype relative to tilapia; we refer to this fusion as LG5-14 in A. burtoni (Fig. 2). Paternal alleles co-segregate with sex at LG5-14, and thus we designate it as carrying an XY sex determination system.
Fig. 2.
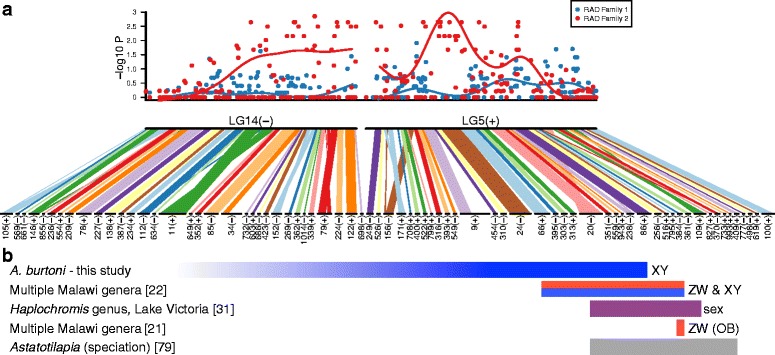
Association of sex with SNP markers on LG5-14 in family 2, with comparisons to other mapping studies. a Association plotted as the negative log of Fisher’s exact test p value of marker alleles with sex (top), aligned to A. burtoni genome scaffolds ordered and oriented to LG5 and LG14 of the O. niloticus genome assembly (bottom). In A. burtoni, LG5 and LG14 are fused relative to other cichlids. Lines on association plot indicate sliding window analysis of association data (window, 10 SNPs; step, 1 SNP). b Comparison of sex-associated markers with previous mapping studies in cichlids, with numbers linking to references. Colored bars indicate range of sex-associated markers relative to A. burtoni scaffolds above in a. Kudo et al. [31] described association with sex without indicating an XY or ZW system. Roberts et al. [21] mapped the orange blotch (OB) pigmentation allele, tightly linked to a W sex determination allele
Because the two discovery families were relatively small and provided relatively low power to detect associations, we confirmed association of these loci with phenotypic sex by testing 25 additional families with simple sequence repeat (SSR) markers on LG13 and LG5-14 (Table 2, Additional files 5 and 6). Segregation of parental alleles at either LG13 or LG5-14 is associated with sex determination in the majority of families tested. In families where genotypes at LG5-14 are associated with sex, the sire’s alleles are consistently associated with sex of offspring (Table 2), replicating findings from the discovery family, and providing strong evidence for an XY sex determination system on LG5-14. Where multiple families from the same LG5-14 XY sire were tested, the same paternal haplotype is consistently associated with male sex in offspring (Additional file 6). We infer that this paternal haplotype is tightly linked to a dominant Y male sex determination allele.
Table 2.
Association of genetic markers with sex in families
| Family | Sire | Dam | M | F | M:F | LG5-14 Y p | LG13 Y p | LG13 W p | System |
|---|---|---|---|---|---|---|---|---|---|
| RAD 1 | 1 | 1 | 12 | 12 | 0.50 | 0.6843 | NI | 0.0001 | LG13 ZW |
| RAD 2 | 2 | 2 | 9 | 8 | 0.53 | 0.0023 | 0.6199 | 1 | LG5-14 XY |
| 1 | 1 | 1 | 10 | 36 | 0.22 | 0.1639 | NI | 0.0012 | LG13 ZW |
| 2 | 3 | 3 | 8 | 8 | 0.50 | 0.0087 | NI | 0.0001 | LG13 ZW |
| 3 | 3 | 3 | 9 | 8 | 0.53 | 0.3469 | NI | 0.0152 | LG13 ZW |
| 4 | 4 | 4 | 6 | 26 | 0.19 | NI | NI | 0.0192 | LG13 ZW |
| 5 | 5 | 5 | 4 | 6 | 0.40 | 1 | 0.5714 | 0.0048 | LG13 ZW |
| 6 | 6 | 6 | 4 | 10 | 0.29 | 0.2208 | 1 | 0.015 | LG13 ZW |
| 7 | 6 | 7 | 10 | 9 | 0.53 | 0.1534 | NI | 0.009 | LG13 ZW |
| 8 | 7 | 8 | 2 | 13 | 0.13 | 1 | 0.0128 | NI | LG13 XY |
| 9 | 7 | 9 | 16 | 22 | 0.42 | 0.72 | 0.0001 | 0.752 | LG13 XY |
| 10 | 8 | 10 | 12 | 14 | 0.46 | 0.0001 | 0.2177 | 1 | LG5-14 XY |
| 11 | 8 | 11 | 12 | 10 | 0.55 | 0.003 | 0.0092 | 0.6914 | LG5-14 XY |
| 12 | 8 | 12 | 15 | 11 | 0.58 | 0.0001 | 0.4283 | 0.4279 | LG5-14 XY |
| 13 | 8 | 13 | 12 | 7 | 0.63 | 0.0002 | 0.3261 | 0.3498 | LG5-14 XY |
| 14 | 8 | 14 | 4 | 2 | 0.67 | 0.0667 | 0.4 | 1 | LG5-14 XY |
| 15 | 9 | 15 | 12 | 10 | 0.55 | 0.0015 | 0.3246 | 0.1312 | LG5-14 XY |
| 16 | 9 | 16 | 13 | 8 | 0.62 | 0.0237 | NI | 0.2031 | LG5-14 XY |
| 17 | 9 | 17 | 15 | 8 | 0.65 | 0.0083 | 0.2213 | NI | LG5-14 XY |
| 18 | 10 | 18 | 11 | 5 | 0.69 | 0.0256 | 0.5962 | 1 | LG5-14 XY |
| 19 | 11 | 19 | 5 | 4 | 0.56 | 0.444 | 0.0079 | NI | LG13 XY + LG5-14 XY |
| 20 | 12 | 20 | 9 | 11 | 0.45 | 1 | 1 | 0.1789 | Unknown |
| 21 | 6 | 21 | 12 | 26 | 0.32 | 0.6792 | 0.2305 | NI | Unknown |
| 22 | 13 | 22 | 7 | 9 | 0.44 | 0.3024 | 0.1262 | 0.3147 | Unknown |
| 23 | 13 | 23 | 14 | 17 | 0.45 | 1 | 1 | 0.4809 | Unknown |
| 24 | 13 | 24 | 20 | 23 | 0.47 | 0.3652 | 0.7619 | 0.0755 | Unknown |
| 25 | 13 | 25 | 22 | 20 | 0.52 | 0.3783 | 0.0414 | 1 | Unknown |
Families including identification number of sire and dam, number of male (M) and female (F) offspring, and sex ratio (M:F). p values are from Fisher’s exact test of association of SSR marker data with phenotypic sex, testing the sire’s alleles for Y associations, and dam’s alleles for W association, with bold values indicating p < 0.05, and NI indicating markers tested were not informative. System indicates inferred sex determination system in family from marker data within and among families. An expanded version of this table with markers used, and a table with marker genotype data is available in Additional files 5 and 6.
In families where LG13 is associated with sex determination, either a maternal or a paternal allele can be found associated with sex, depending on the family tested (Table 2). In several families, a maternal haplotype segregates with sex in the offspring, and the same maternal haplotype is associated with female sex in multiple families, supporting the presence of a dominant W female sex determination allele (Additional file 6). Surprisingly, paternal alleles at LG13 co-segregate with sex of offspring in families from two sires, suggesting that a dominant Y male sex determination allele may also be present on LG13 (Table 2, Additional file 6). Differences in parental inheritance of sex determination alleles among families with LG13 sex determination suggest that an XYW sex determination system akin to that found in platyfish may be present at LG13; however, occasional sex reversal could also account for observed patterns in the absence of a LG13 Y allele (see below).
Several families showed no association of sex with markers at LG13 or LG5-14 (Table 2). These families allow the possibility of additional, unidentified sex determination loci in A. burtoni. Alternatively, interactions among identified sex determination alleles, or between sex determination alleles and the environment, could confound interpretation.
Interactions between sex loci
Because we find multiple loci and alleles that appear to impact sex, we would expect some families to be segregating multiple alleles that would interact to influence sex determination. Studies in both tilapias and Lake Malawi cichlids with polygenic sex determination have revealed clear patterns of epistasis relating to phenotypic sex [21–23, 30]. Unfortunately, we find no clear examples of such interactions in our families. One small family (Family 19) has perfect segregation of sex with paternal alleles at LG13, supporting the sire having an LG13 XY genotype. Two sons but no daughters in the family also inherit a multi-marker haplotype at LG5-14 associated with the LG5-14 Y in other families (Additional file 7). We hypothesize that the genotype of the sire and two sons in this family is LG5-14 XY, LG13 XY, and thus that individuals with a Y allele at each locus are viable and develop as males; however, this interpretation is based on a single, small family and is thus very tentative.
In families tested where sex is not clearly associated with alleles at either LG13 or LG5-14 alone (Table 2), inter- and intra-locus interactions may be present that do not conform to our expected patterns of allelic dominance. Additionally, some families were not included in this study because they had extreme sex ratios (sometimes single-sex), and/or few offspring, preventing meaningful interpretation of inheritance patterns; however, interaction of multiple sex alleles may produce skewed sex ratios or non-viable genotypes. In cichlids and other fish species, co-segregation of both Y and W alleles in families produces female-biased sex ratios [18, 22, 30], and in pygmy mice with an XYW system, mating of some sex genotypes results in inviability of a portion of their offspring [45]. Parnell and Streelman described five distinct sex determination loci (three XY and two ZW systems) in a Lake Malawi cichlid mapping cross, demonstrating that sex ratios represent the outcome of complex interactions between relatively strong and weak sex determination alleles [23]. Thus, epistasis may be difficult to characterize, particularly in small families, and further work is needed to understand the phenotypic outcomes of interactions between the sex determination alleles identified.
Sex ratios and penetrance
Previous studies have demonstrated that polygenic sex determination systems in cichlids produce skewed sex ratios. In a survey of species from Lake Malawi, families with sex determined by an XY system had equal sex ratios, while those with a ZW system generally had a female-biased sex ratio [22]. In families where both XY and ZW systems segregated, a 1:3 (male:female) sex ratio is expected [22]. In some Lake Victoria cichlids, sex determination factors on supernumerary chromosomes also produce female-skewed sex ratios [32]. Comparing the sex ratios of our LG5-14 XY families and LG13 ZW A. burtoni families reveals an intriguing and significant pattern: XY families have a male-biased sex ratio, and ZW families have a female-biased sex ratio (Fig. 3a). Because we only identified a few putative LG13 XY families, we do not test for significant divergence in sex ratio or penetrance.
Fig. 3.
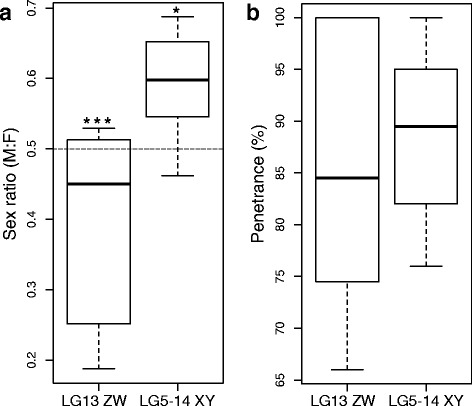
Sex ratios and penetrance by sex determination system. a Sex ratio by sex determination system. Dashed line indicates a 1:1 sex ratio, with values lower than the line indicating a female-biased sex ratio, and those above the line a male-biased sex ratio. (LG13 ZW, n = 8 families, 63 males and 115 females total; LG5-14 XY, n = 10 families, 115 males and 83 females total; χ 2 test for divergence from expected 1:1 sex ratio: *, p = 0.023, χ 2 = 5.2; ***, p < 0.0001, χ 2 = 15.2; LG13 ZW and LG5-14 XY family sex ratio differ from each other, p = 0.0053, Welch’s unequal variances t-test). b Penetrance by sex determination system, not significant
Comparing penetrance of each sex determination allele, we find no significant differences (Fig. 3b), and a mean penetrance of 88 % for all sex determination alleles. Incomplete penetrance could be explained by a number of factors, including incorrect phenotype data, recombination between our sex-linked markers and the sex determination polymorphism, or sex reversal. If sex reversal is the case, it presents an alternative hypothesis regarding the interpretation that both W and Y alleles are present at LG13. The presence of a W allele has greater support based on replication with a number of dams, but only two sires were found to have their LG13 alleles segregate with sex of their offspring. Phenotypic males in some families with LG13 ZW dams occasionally carry a W haplotype at tested markers. For example, see Family 3, where two males exhibit the W-associated haplotype at LG13 (Additional file 6). Uncommon males that are genetically female (ZW at LG13) would have their LG13 alleles co-segregate with the sex of their offspring, lending the appearance of an XY system even though a distinct, male-determining Y allele might not exist at the locus.
Genetic intervals for sex loci
Incomplete penetrance makes it difficult to determine mapping intervals via recombination breakpoint mapping, because it is unclear if genotype-phenotype mismatch at a particular marker is due to recombination or sex reversal. Though our ddRADseq families included relatively few individuals, we estimate genetic intervals based on strongly associated genome scaffolds. For the LG13 ZW locus, scaffolds spanning approximately 4.6 Mb show the highest association with sex (Fig. 1, Additional file 3). At LG5-14, a broad plateau of association spans 43 Mb of the linkage group based on data from the initial discovery family (Fig. 2). We further interrogated the association data with a sliding window analysis on LG5-14, to look for regions with the highest density of associated SNPs, which may correspond to the region of highest sequence divergence. Sliding window analysis reveals a broad peak of the highest density of associated SNPs on several scaffolds that align to LG5 of tilapia. In one family with LG5-14 XY sex determination, recombination breakpoints in one individual support that the region under this peak contains the sex determination locus (Individual 8, Family 18, Additional file 6; spanning scaffold 156 to scaffold 66).
Candidate genes
Though precise genetic intervals are not clear, we examined gene content on the A. burtoni scaffolds with the highest association in our RAD family data (Additional file 8). Notably, genes with established roles in sex determination in multiple vertebrate species (e.g., sox3, dmrt, amh, and gsdf) are not found on these scaffolds, suggesting the use of novel sex determination genes by A. burtoni. Here we highlight intriguing candidate genes on associated scaffolds based on known roles in sexual development or fertility.
LG13
The total length of sex-associated scaffolds at LG13 ZW is relatively small compared to those at LG5-14. One strong candidate for sex determination in the region is cytochrome P450 17alpha-hydroxylase (cyp17a1, scaffold 16), a key steroidogenic enzyme in the production of sex hormones in vertebrates [46]. Mutations of CYP17A1 in humans produce low androgen levels and ambiguous or female external genitalia in XY individuals [47]. In amphibians (Rana frogs), CYP17 has been repeatedly implicated as one of the key players in gonadal differentiation [48]. In the cichlid Nile tilapia, strong sexually dimorphic expression of cyp17a1 is found during sexual differentiation [49]. Moreover, strong changes in cyp17a1 expression accompany both natural sex reversal in field rice eel, a sequential hermaphrodite [50], and induced sex reversal in zebrafish [51]. Across species, activity of cyp17a1 is associated with male sexual development, suggesting that a deleterious mutation to the gene in A. burtoni could produce a W allele inducing feminization of the gonad.
Another compelling candidate on LG13 is type I bone morphogenetic protein receptor (bmpr1a, scaffold 467), required for Amh-induced regression of female gonadal tissues in mammals; targeted disruption of Bmpr1a in Mullerian ducts in mice leads to pseudohermaphrodite males retaining oviducts and uteri [52]. Amh pathway members have been identified as sex determination genes in multiple fish species [7, 13, 34], demonstrating the importance of the pathway in sex determination. An evolved bmpr1a loss-of-function mutation in A. burtoni could reasonably shift gonadal development towards ovarian development.
A few genes found in the sex-associated region of LG13 are involved in production or response to gonadotropins. Luteinizing hormone beta (lhb, scaffold 186) is a subunit of luteinizing hormone, a gonadotropin secreted from the pituitary to direct gonadal development in fishes [53]. Notably, knock-out of lhb in zebrafish (Danio) produces female infertility via oocyte maturation defects, while testes and male fertility are normal, providing a sex-specific role for the gene in reproductive biology in fish [54]. Ventral anterior homeobox 1 (vax1, scaffold 16) is essential for expression of gonadotropin-releasing hormone, with loss of Vax1 in mice leading to hypogonadism and infertility in both sexes [55]. Glial cell line derived neurotrophic factor family receptor alpha 1 (gfra1, scaffold 186) is expressed in spermatogonial stem cells in fishes in response to gonadotropins [56].
LG5-14
The sex-associated region at LG5-14 XY is broad, and thus we focus consideration of candidate genes on scaffolds on the region of LG5-14 with the highest density of sex-associated SNPs (Fig. 2, Additional file 8). Perhaps the most interesting candidate in the region is wingless-type MMTV integration site family member 4 (wnt4, scaffold 156), required for female development in vertebrates, and considered one of the central female-promoting factors in sex differentiation networks [57–59]. While loss of Wnt4 in mice causes partial female-to-male sex reversal [57], and there has been at least one case of sex reversal in an XY human carrying a WNT4 duplication [60], to date wnt4 has not been implicated as the primary sex determination gene in any species. wnt7a (scaffold 526) is also found in the XY region on LG5-14 in A. burtoni, and has been implicated in sexually dimorphic gonad development in mammals [61].
R-spondin 4 (rspo4, scaffold 351) is a member of the R-spondin gene family, where rspo1 is also a key member of vertebrate sex determination networks, with mutation in humans producing XX sex reversal [59, 62]. While a direct role for rspo4 in sex differentiation has not yet been demonstrated, it exhibits sexual dimorphic expression in mammalian gonad, with higher levels in the ovary [63].
Multiple genes in the LG5 XY region modulate fundamental aspects of gonadal development. Bone morphogenetic protein 7 (Bmp7, scaffold 9) regulates germ cell proliferation in mouse gonad [64], and has been implicated in gonadal divergence in chickens [65]. Further, bmp7 is involved in androgen-dependent development of secondary sexual characteristics in medaka [66]. GATA binding protein 2 (gata2, scaffold 9) is a zinc finger transcription factor that is expressed in a sexually dimorphic fashion during mouse gonadogenesis [67], and is known to regulate the expression of gonadotropins including lhb [68, 69], found in the LG13 ZW interval. Cyp27b1 (scaffold 66) encodes 25-Hydroxyvitamin D3 1-alpha-hydroxylase, the rate-limiting enzyme in synthesis of the active form of Vitamin D3; female Cyp27b1 knock out mice have extensive reproductive defects, including delayed puberty, uterine hypoplasia, altered estrogen and gonadotropin levels, and infertility [70–72].
A second chromosome fusion
Because our ddRADseq haplotype data was able to resolve the linkage groups involved in one of two the A. burtoni chromosome fusions relative to other cichlids [44], we attempted to identify the other known fusion by exploring linkage between markers in our RAD genotype data. Building an outcross linkage map with only 24 offspring from RAD family 1 produced a poorly resolved linkage map, with 30 linkage groups containing greater than five markers. Though the map was very rough, it identified candidates for linkage group fusions that were then examined by manual comparison of haplotypes. Markers aligning to two tilapia linkage groups, LG8-24 and LG16-21, were linked in our map output. (The nomenclature for both LG8-24 and LG16-21 in tilapia does not indicate fused chromosomes, but is instead a historical artifact: the standard O. niloticus linkage map originally had 24 linkage groups [73], but later improvement of the genetic map identified 22 linkage groups [42], collapsing the former LG8 and LG24 into a single linkage group, as well as LG16 and LG21.) We find continuous haplotypes spanning LG8-24 and LG16-21, with significant linkage between markers aligning to the two linkage groups (LOD = 10.3, Additional file 4), supporting the fusion of the two associated chromosomes in A. burtoni relative to other cichlids. Maintaining the community naming convention based on the original O. niloticus linkage map, this linkage group would be given the rather cumbersome designation LG8-24-16-21. A previous report suggested that one of the fusions in A. burtoni consisted of LG15 and LG19 relative to other cichlids [31], but we find no evidence of linkage between markers mapping to those linkage groups (LOD = 0.8, Additional file 4). This discrepancy may stem from interchromosomal rearrangements more complex than simple fusions, where different portions of a particular tilapia LG map to different A. burtoni LGs. Thus, use of different genetic markers could provide patterns suggesting fusion of different LGs.
Comparisons among cichlid sex determination systems
A previous study inferred the mode of sex determination system in A. burtoni using hormonal sex reversal [40], in a line derived from the same lab strain we use here. Estradiol sex reversal of males and subsequent testcrosses strongly indicated an XY sex determination system, and ruled out a ZW system [40]. These results are consistent with our mapping data, because it is quite possible that only the XY sex determination system was present in the four families used for sex reversal experiments. Indeed, bottlenecks during transfer of the A. burtoni lab strain between labs may inadvertently lead to complete loss of some sex determination alleles. Concurrent mapping experiments by another group using the same lab line as the sex reversal experiments above identified co-segregation of sex with markers on LG5 indicating an XY system [74], completely consistent with the mapping results we describe here. While the concurrent study found no association of sex with LG13, it did identify an association of sex with LG18 in the offspring of a pair of wild-caught individuals from a Lake Tanganyika estuary [74]. Together, our differing but overlapping results support the presence of complex, polygenic, and potentially rapidly evolving sex determination in A. burtoni.
A number of sex determination loci have been identified in cichlids. In tilapia, sex determination has been associated with LG3, LG20, LG23, and two distinct loci on LG1, depending on the species and population [30, 35, 36, 75]. In Lake Malawi haplochromine cichlid species, LG3, LG5, LG20, and two loci on LG7 have been associated with sex determination [21, 23]. In haplochromine cichlids from Lake Victoria, LG2, LG5, and supernumerary B chromosomes have been associated with sex [31, 32]. Here we identify two loci impacting sex determination in the riverine haplochromine A. burtoni, LG5-14 and LG13. While this is the first time LG13 and LG14 have been associated with sex determination, it is intriguing that LG5 has now been associated with haplochromine sex determination in A. burtoni, Malawi, and Victoria cichlids. To provide finer scale comparison of sex association with LG5, we identified A. burtoni scaffolds containing sex-associated markers from other studies (Fig. 2b), revealing overlap of sex loci among multiple species. As on-going mapping studies identify specific polymorphism underlying sex determination in each context, we should be able to distinguish reuse of ancestral polymorphism versus repeated de novo evolution of sex determination on LG5. However, both W [21, 22] and Y (this study; also suggestive patterns in two families in [22]) acting alleles have been associated with LG5, suggesting at least two evolutionary events. LG5 has been dubbed a “speciation chromosome” because of the number of adaptive and species-delineating phenotypes mapped there, including pigmentation, vision, jaw shape, and tooth shape [76], and it may exhibit dynamic evolution of sex determination with both W and Y alleles now clearly identified. The identification of the LG5-14 fusion in A. burtoni should provide additional context to consider the role of this chromosome in adaptation and speciation, particularly in comparing the evolutionary trajectory of LG14 among lineages with and without the fusion. Indeed, the LG5-14 fusion in A. burtoni may represent karyotype evolution driven in part by the presence of a sex determination locus on LG5. For example, if novel alleles under strong sexually antagonistic selection arose on LG14, the resulting sexual conflict could have been resolved by fusion to the LG5 sex chromosomes, ensuring that sex-specific fitness benefits were only expressed in the appropriate sex.
Evolutionary context
Evolutionarily, the genus Astatotilapia represents an immediate outgroup to the adaptive radiations of cichlid species in Lake Malawi and Lake Victoria [41, 77], providing important context for studies of sex determination evolution in those flocks. Ongoing studies may reveal if the sex determination locus on LG5/LG5-14 is the first found both within and without the lake adaptive radiations, and/or shared among radiations. Additionally, a recent report implicated the same region of LG5 as one of a handful of genomic islands of speciation in Astatotilapia species, based on sequence divergence between ecomorphs ([78]; Fig. 2b). One hypothesis is that the signal of divergence at LG5 in that study is due to divergence related to the sex determination locus, with the LG5 XY system potentially present in one ecomorph and absent in the other.
Ultimately, our mapping results must be confirmed in natural populations. The recent discovery that the natural sex determination chromosome in zebrafish (Danio) was lost in lab strains urges caution in interpreting genetic mapping of sex determination in domesticated fish [28]. More recently, large-scale changes in gene expression were identified after only a single generation of domestication in trout [79], suggesting that domestication can act rapidly on the phenotypic output of the fish genome. The standard lab strain of A. burtoni we use here has been maintained in the lab for nearly four decades [80], representing over 100 generations in captivity in which loss or gain of sex determination alleles could have taken place. Indeed, recently collected stocks of A. burtoni exhibit a number of phenotypic differences from the classic lab strain [81], suggesting underlying genetic differences between wild and lab strain A. burtoni.
In general, the evolutionary pressures that maintain polygenic sex determination systems remain unknown. We favor the hypothesis that the multiple sex genotypes produced by polygenic sex determination may provide alternative fitness strategies, for example, alternate sex genotypes may be associated with differences in color (yellow vs. blue male morphs) or behavior (potential sneaker vs. territorial mating strategies) in A. burtoni [82, 83]. In any case, A. burtoni should serve as an excellent model for understanding the outcomes of polygenic sex determination for primary and secondary sexual characteristics. The availability of genome editing tools in the species permits mechanistic dissection of genetic variation [84, 85], while decades of research in the species provides a wealth of information regarding sexually dimorphic behavior, physiology, and related gene expression [38, 81, 86–89].
Conclusions
Examples of polygenic sex determination are accumulating across taxa [3], and particularly among cichlid fish species. Here we present evidence for polygenic sex determination in the cichlid A. burtoni, a longstanding model of sex-related physiology and behavior, and a close relative of the explosive adaptive radiations of species in Lake Malawi and Lake Victoria. Our results strongly indicate an XY system on LG5-14 and a ZW system on LG13. Additionally, inheritance in some families indicates a Y allele on LG13, which would support the presence of an XYW system; however, occasional apparently sex reversed individuals make the conclusion that both W and Y alleles are present at LG13 tentative. The locus on LG13 represents a novel association with sex, while LG5 has been implicated in sex determination, adaptation, and speciation in a number of contexts. Our results add to the evidence that LG5 may have played an unusual role in cichlid diversification, including fusion of LG5 and LG14 in A. burtoni. Ongoing investigation of sex determination in A. burtoni aims to identify the sex determination genes and understand their action and interaction on primary and secondary sexual characteristics. Understanding the complex genetics underlying sex determination in A. burtoni should provide additional insight and context to an already fundamental model of sexual dimorphism and behavioral evolution.
Methods
Husbandry and phenotyping
Fish husbandry and procedures were performed under Institutional Animal Care and Use Committee approved protocols at North Carolina State University (Roberts Lab) and Stanford University (Fernald Lab), with all fish originating from Fernald Lab lines. Breeding tanks were set up with a single male and multiple females. The species is a maternal mouth brooder, allowing parentage to be determined. Fertilized eggs were collected in the first week post-fertilization and tumbled in flasks until fry were free swimming. At approximately 6 weeks post-fertilization, families were transferred to 50 gal aquaria until sexual maturity, between 4 and 5 months of age. Fish were euthanized in a bath of 250 mg/L tricaine methanesulfonate (Tricaine-S, Western Chemical) and dissected for gonad collection. Gonads clearly containing eggs were scored as female, while other gonads were squashed between slides and visualized under a microscope to determine the presence of sperm (male) or immature ovarian follicles (female). Sex ratio was calculated as the number of male offspring divided by the total number of offspring in the family.
Genotyping
DNA was extracted from finclips taken from parents and offspring using standard methods (GeneJET Genomic DNA Purification Kit, Thermofisher). ddRADseq libraries were built as previously described [90] using SphI and MluCI enzymes, and sequenced on a single Illumina HiSeq lane with 100 bp paired-end reads (North Carolina State University Genomic Sciences Laboratory). Reads were de-multiplexed, allowing zero mismatches with the variable length barcode sequence [90]. Barcodes were trimmed from each read, and 10-N bases were trimmed from the end of each read, where N is the number of bases in the barcode. Thus, all reads were exactly 90 bases long for alignment. Reads were aligned to reference (A. burtoni v1; [41]) using m = 1 and genotypes called in Stacks [91]. For further analysis we used markers with genotypes called in at least 20 of 24 offspring for RAD family 1, and 15 of 17 offspring for RAD family 2, using a minimum stack depth of 10 to call homozygous genotypes. Association of ddRADseq markers with phenotypic sex was performed in PLINK [92], using Fisher’s exact test to determine p-values of association. Genotyping of SSR markers used PCR with fluorophore-labeled primers (Additional file 9) and separation on an Applied Biosystems 3730 capillary sequencer as previously described [21]. Fisher’s exact test was used to determine significant co-segregation of parental alleles with offspring sex. Where a significant association was found in a family, penetrance was calculated as the proportion of offspring with the expected genotype-phenotype association at the marker with the lowest Fisher’s exact test p value. We note this is an imperfect measure of penetrance because linked markers are used rather than causative sex determination polymorphisms. Onemap was used to order RAD genotypes into linkage groups using outcross mapping, linking markers at LOD >4 and a maximum recombination fraction of 0.5 [93].
Comparative genomics
The A. burtoni genome (v1; [41]) was partitioned into non-overlapping 5 kb sequence windows and mapped to reference genomes (Tilapia_v1.1 and Mzebra_v0; [41]) using the MEM algorithm implemented in bwa (v. 0.7.5a; [94]). The resulting bam file was filtered to include only sequences mapping to a single coordinate in the reference genome and with fewer than 5 suboptimal hits. Aligned sequences with less than 90 % sequence identity to the reference reference were also removed. This strategy allowed us to place 637 scaffolds from the unanchored A. burtoni genome onto the 22 assembled Tilapia linkage groups. An additional 99 A. burtoni scaffolds were confidently placed onto unmapped contigs in the Tilapia assembly. The resulting alignments and association data were visualized using custom R scripts. For sliding window analysis, windows of 10 SNPs with 1 SNP step size were used, and the resulting line smoothed for visualization purposes.
Acknowledgements
We thank Martha Burford-Reiskind, Jennifer Baltzegar, Megan Fritz, Emily Moore, Ashley Elias, Erin Peterson, and members of our labs for valuable insight. We thank Danielle Blakkan and Jodie White for aquaculture support.
Funding
The work was supported by National Science Foundation Grant IOS-1456765 to RBR, and by the National Institutes of Health grants NIH F32HD071755 to SAJ, and NIH NS034950 and NIH NS093277 to RDF.
Availability of data and material
Coordinates of all genetic markers reference publicly available genomes and are provided in additional files (Aburtoni v1; Tilapia_v1.1; and Mzebra_v0; [41]). Raw ddRADseq sequence and scripts are available on request.
Authors’ contributions
NBR, SAJ, RDF, and RBR designed the study. NBR, SAJ, BLD, RDF, and RBR wrote the manuscript. NBR, SAJ, and MKS performed animal husbandry and dissections. NBR, MKS, and AQR performed molecular biology and genetic mapping experiments. NBR produced ddRADseq libraries. KPC analyzed ddRADseq data. BLD performed comparative genomics. NBR, BLD, KPC, and RBR interpreted mapping data. All authors approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All vertebrate animal work was performed under the approval and oversight of the Institutional Animal Care and Use Committee at North Carolina State University (protocol 15-043-O) and the Administrative Panel on Laboratory Animal Care at Stanford University (protocol 9882).
Abbreviations
- ddRADseq
Double-digest restriction site associated DNA sequencing
- LG
Linkage group
- LOD
Logarithm of odds
- Mb
Megabases
- SNP
Single nucleotide polymorphism
- SSR
Simple sequence repeat
- UNK
Unknown linkage group; portion of genome assembly not included on a linkage group
Additional files
Alignment of A. burtoni scaffolds to anchored O. niloticus (Nile tilapia) genome. (PDF 725 kb)
Alignment of A. burtoni scaffolds to anchored M. zebra genome. (PDF 619 kb)
Association results of sex with RAD markers in two families. (XLSX 653 kb)
Linkage tests and RAD marker haplotypes spanning two chromosome fusions in two families. (XLSX 110 kb)
Expanded summary table of association of genetic markers with sex in families, including genetic markers used. (XLSX 53 kb)
SSR marker data at LG13 and LG5-14 for 25 families. (XLSX 145 kb)
SSR marker data at LG13 and LG5-14 for Family 19, with inferred Y haplotype and offspring sex genotypes. (XLSX 51 kb)
Gene annotations for scaffolds on LG13 and LG5-14 associated with sex. (XLSX 440 kb)
Primer sequence and scaffold location for SSR markers used. (XLSX 35 kb)
Contributor Information
Natalie B. Roberts, Email: nbhodges@ncsu.edu
Scott A. Juntti, Email: sjuntti@stanford.edu
Kaitlin P. Coyle, Email: kate.coyle@ncsu.edu
Bethany L. Dumont, Email: bldumont@ncsu.edu
M. Kaitlyn Stanley, Email: mkstanle@ncsu.edu.
Allyson Q. Ryan, Email: allyson.ryan@embl.de
Russell D. Fernald, Email: rfernald@stanford.edu
Reade B. Roberts, Email: rbrober2@ncsu.edu
References
- 1.Beukeboom LW, Perrin N. The Evolution of Sex Determination. 1. Oxford: Oxford Press; 2014. [Google Scholar]
- 2.Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R, Perrin N, Ross L, Valenzuela N, Vamosi JC. Tree of Sex Consortium. Sex determination: why so many ways of doing it? PLoS Biol. 2014;12(7):e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore EC, Roberts RB. Polygenic sex determination. Curr Biol. 2013;23(12):R510–R512. doi: 10.1016/j.cub.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24(2):94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355(1403):1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondo M, Hornung U, Nanda I, Imai S, Sasaki T, Shimizu A, Asakawa S, Hori H, Schmid M, Shimizu N, Schartl M. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 2006;16:815–826. doi: 10.1101/gr.5016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, Mizuno N, Fujita M, Suetake H, Suzuki S, Hosoya S, Tohari S, Brenner S, Miyadai T, Venkatesh B, Suzuki Y, Kikuchi K. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu) PLoS Genet. 2012;8(7):e1002798. doi: 10.1371/journal.pgen.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi K, Hamaguchi S. Novel sex-determining genes in fish and sex chromosome evolution. Dev Dyn. 2013;242(4):339–353. doi: 10.1002/dvdy.23927. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff JN, Hong Y, Li Q, Sha Z, Zhou H, Xie M, Yu Q, Liu Y, Xiang H, Wang N, Wu K, Yang C, Zhou Q, Liao X, Yang L, Hu Q, Zhang J, Meng L, Jin L, Tian Y, Lian J, Yang J, Miao G, Liu S, Liang Z, Yan F, Li Y, Sun B, Zhang H, Zhang J, Zhu Y, Du M, Zhao Y, Schartl M, Tang Q, Wang J. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 2014;46(3):253–260. doi: 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- 10.Takehana Y, Matsuda M, Myosho T, Suster ML, Kawakami K, Shin-I T, Kohara Y, Kuroki Y, Toyoda A, Fujiyama A, Hamaguchi S, Sakaizumi M, Naruse K. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat Commun. 2014;5:4157. doi: 10.1038/ncomms5157. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, Nishida-Umehara C, Matsuda Y, Takamatsu N, Shiba T, Ito M. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A. 2008;105(7):2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461(7261):267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 13.Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, Sakamoto T, Fernandino JI, Somoza GM, Yokota M, Strüssmann CA. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci U S A. 2012;109(8):2955–2959. doi: 10.1073/pnas.1018392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508(7497):488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 15.Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, Fujiyama A, Naruse K, Hamaguchi S, Sakaizumi M. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics. 2012;191(1):163–170. doi: 10.1534/genetics.111.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, Klopp C, Cabau C, Bouchez O, Fostier A, Guiguen Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr Biol. 2012;22(15):1423–1428. doi: 10.1016/j.cub.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 17.Kosswig C. Polygenic sex determination. Experientia. 1964;20(4):190–199. doi: 10.1007/BF02135395. [DOI] [PubMed] [Google Scholar]
- 18.Volff JN, Schartl M. Variability of genetic sex determination in poeciliid fishes. Genetica. 2001;111:101–110. doi: 10.1023/A:1013795415808. [DOI] [PubMed] [Google Scholar]
- 19.Alexander HJ, Richardson JM, Edmands S, Anholt BR. Sex without sex chromosomes: genetic architecture of multiple loci independently segregating to determine sex ratios in the copepod Tigriopus californicus. J Evol Biol. 2015;28(12):2196–2207. doi: 10.1111/jeb.12743. [DOI] [PubMed] [Google Scholar]
- 20.Schultheis C, Böhne A, Schartl M, Volff JN, Galiana-Arnoux D. Sex determination diversity and sex chromosome evolution in poeciliid fish. Sex Dev. 2009;3(2–3):68–77. doi: 10.1159/000223072. [DOI] [PubMed] [Google Scholar]
- 21.Roberts RB, Ser J, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlids. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ser JR, Roberts RB, Kocher TD. Multiple interacting loci control sex determination in lake Malawi cichlid fish. Evolution. 2010;64:486–501. doi: 10.1111/j.1558-5646.2009.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parnell NF, Streelman JT. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity. 2013;110:239–246. doi: 10.1038/hdy.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palaiokostas C, Bekaert M, Taggart JB, Gharbi K, McAndrew BJ, Chatain B, Penman DJ, Vandeputte M. A new SNP-based vision of the genetics of sex determination in European sea bass (Dicentrarchus labrax) Genet Sel Evol. 2015;47:68. doi: 10.1186/s12711-015-0148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley KM, Breyer JP, Melville DB, Broman KW, Knapik EW, Smith JR. An SNP-Based Linkage Map for Zebrafish Reveals Sex Determination Loci. G3 (Bethesda) 2011;1(1):3–9. doi: 10.1534/g3.111.000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liew WC, Bartfai R, Lim Z, Sreenivasan R, Siegfried KR, Orban L. Polygenic sex determination system in zebrafish. PLoS One. 2012;7(4):e34397. doi: 10.1371/journal.pone.0034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson JL, Rodríguez Marí A, Braasch I, Amores A, Hohenlohe P, Batzel P, Postlethwait JH. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One. 2012;7:e40701. doi: 10.1371/journal.pone.0040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson CA, High SK, McCluskey BM, Amores A, Yan YL, Titus TA, Anderson JL, Batzel P, Carvan MJ, 3rd, Schartl M, Postlethwait JH. Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics. 2014;198(3):1291–1308. doi: 10.1534/genetics.114.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winge O. The nature of sex chromosomes. Proc Sixth Int Congr Genet. 1932;1:343–355. [Google Scholar]
- 30.Lee BY, Hulata G, Kocher TD. Two unlinked loci controlling the sex of blue tilapia (Oreochromis aureus) Heredity. 2004;92:543–549. doi: 10.1038/sj.hdy.6800453. [DOI] [PubMed] [Google Scholar]
- 31.Kudo Y, Nikaido M, Kondo A, Suzuki H, Yoshida K, Kikuchi K, Okada N. A microsatellite-based genetic linkage map and putative sex-determining genomic regions in Lake Victoria cichlids. Gene. 2015;560(2):156–164. doi: 10.1016/j.gene.2015.01.057. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida K, Terai Y, Mizoiri S, Aibara M, Nishihara H, Watanabe M, Kuroiwa A, Hirai H, Hirai Y, Matsuda Y, Okada N. B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 2011;7:e1002203. doi: 10.1371/journal.pgen.1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eshel O, Shirak A, Weller JI, Hulata G, Ron M. Linkage and Physical Mapping of Sex Region on LG23 of Nile Tilapia (Oreochromis niloticus) G3 (Bethesda) 2012;2:35–42. doi: 10.1534/g3.111.001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, Sun Y, Zhao J, Shi H, Zeng S, Ye K, Jiang D, Zhou L, Sun L, Tao W, Nagahama Y, Kocher TD, Wang D. A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 2015;11(11):e1005678. doi: 10.1371/journal.pgen.1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palaiokostas C, Bekaert M, Khan MG, Taggart JB, Gharbi K, McAndrew BJ, Penman DJ. A novel sex-determining QTL in Nile tilapia (Oreochromis niloticus) BMC Genomics. 2015;16:171. doi: 10.1186/s12864-015-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cnaani A, Lee BY, Zilberman N, Ozouf-Costaz C, Hulata G, Ron M, D’Hont A, Baroiller JF, D’Cotta H, Penman DJ, Tomasino E, Coutanceau JP, Pepey E, Shirak A, Kocher TD. Genetics of sex determination in tilapiine species. Sex Dev. 2008;2:43–54. doi: 10.1159/000117718. [DOI] [PubMed] [Google Scholar]
- 37.Gammerdinger WJ, Conte MA, Aquah EA, Roberts RB, Kocher TD. Structure and Decay of a Proto-Y Region in Tilapia, Oreochromis niloticus. BMC Genomics. 2014;15:975. doi: 10.1186/1471-2164-15-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernald RD, Maruska KP. Social information changes the brain. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17194–17199. doi: 10.1073/pnas.1202552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Böhne A, Sengstag T, Salzburger W. Comparative transcriptomics in East African cichlids reveals sex- and species-specific expression and new candidates for sex differentiation in fishes. Genome Biol Evol. 2014;6(9):2567–2585. doi: 10.1093/gbe/evu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heule C, Göppert C, Salzburger W, Böhne A. Genetics and timing of sex determination in the East African cichlid fish Astatotilapia burtoni. BMC Genet. 2014;15:140. doi: 10.1186/s12863-014-0140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, Turner-Maier J, Johnson J, Alcazar R, Noh HJ, Russell P, Aken B, Alföldi J, Amemiya C, Azzouzi N, Baroiller JF, Barloy-Hubler F, Berlin A, Bloomquist R, Carleton KL, Conte MA, D’Cotta H, Eshel O, Gaffney L, Galibert F, Gante HF, Gnerre S, Greuter L, Guyon R, Haddad NS, Haerty W, Harris RM, Hofmann HA, Hourlier T, Hulata G, Jaffe DB, Lara M, Lee AP, MacCallum I, Mwaiko S, Nikaido M, Nishihara H, Ozouf-Costaz C, Penman DJ, Przybylski D, Rakotomanga M, Renn SC, Ribeiro FJ, Ron M, Salzburger W, Sanchez-Pulido L, Santos ME, Searle S, Sharpe T, Swofford R, Tan FJ, Williams L, Young S, Yin S, Okada N, Kocher TD, Miska EA, Lander ES, Venkatesh B, Fernald RD, Meyer A, Ponting CP, Streelman JT, Lindblad-Toh K, Seehausen O, Di Palma F. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guyon R, Rakotomanga M, Azzouzi N, Coutanceau JP, Bonillo C, D’Cotta H, Pepey E, Soler L, Rodier-Goud M, D’Hont A, Conte MA, van Bers NE, Penman DJ, Hitte C, Crooijmans RP, Kocher TD, Ozouf-Costaz C, Baroiller JF, Galibert F. A high-resolution map of the Nile tilapia genome: a resource for studying cichlids and other percomorphs. BMC Genomics. 2012;13:222. doi: 10.1186/1471-2164-13-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Quin CT, Drilea AC, Conte MA, Kocher TD. Mapping of pigmentation QTL on an anchored genome assembly of the cichlid fish, Metriaclima zebra. BMC Genomics. 2013;14:287. doi: 10.1186/1471-2164-14-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poletto AB, Ferreira IA, Cabral-de-Mello DC, Nakajima RT, Mazzuchelli J, Ribeiro HB, Venere PC, Nirchio M, Kocher TD, Martins C. Chromosome differentiation patterns during cichlid fish evolution. BMC Genet. 2010;11. Erratum in: BMC Genet. 2012;13(1):2. [DOI] [PMC free article] [PubMed]
- 45.Veyrunes F, Chevret P, Catalan J, Castiglia R, Watson J, Dobigny G, Robinson TJ, Britton-Davidian J. A novel sex determination system in a close relative of the house mouse. Proc Biol Sci. 2010;277(1684):1049–1056. doi: 10.1098/rspb.2009.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller WL, Auchus RJ, Geller DH. The regulation of 17,20 lyase activity. Steroids. 1997;62(1):133–142. doi: 10.1016/S0039-128X(96)00172-9. [DOI] [PubMed] [Google Scholar]
- 47.Van Den Akker EL, Koper JW, Boehmer AL, Themmen AP, Verhoef-Post M, Timmerman MA, Otten BJ, Drop SL, De Jong FH. Differential inhibition of 17alpha-hydroxylase and 17,20-lyase activities by three novel missense CYP17 mutations identified in patients with P450c17 deficiency. J Clin Endocrinol Metab. 2002;87(12):5714–5721. doi: 10.1210/jc.2001-011880. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura M. Is a sex-determining gene(s) necessary for sex-determination in amphibians? Steroid hormones may be the key factor. Sex Dev. 2013;7(1–3):104–114. doi: 10.1159/000339661. [DOI] [PubMed] [Google Scholar]
- 49.Ijiri S, Kaneko H, Kobayashi T, Wang DS, Sakai F, Paul-Prasanth B, Nakamura M, Nagahama Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol Reprod. 2008;78(2):333–341. doi: 10.1095/biolreprod.107.064246. [DOI] [PubMed] [Google Scholar]
- 50.Yu H, Cheng H, Guo Y, Xia L, Zhou R. Alternative splicing and differential expression of P450c17 (CYP17) in gonads during sex transformation in the rice field eel. Biochem Biophys Res Commun. 2003;307(1):165–171. doi: 10.1016/S0006-291X(03)01137-9. [DOI] [PubMed] [Google Scholar]
- 51.Liang YQ, Huang GY, Liu SS, Zhao JL, Yang YY, Chen XW, Tian F, Jiang YX, Ying GG. Long-term exposure to environmentally relevant concentrations of progesterone and norgestrel affects sex differentiation in zebrafish (Danio rerio) Aquat Toxicol. 2015;160:172–179. doi: 10.1016/j.aquatox.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Jamin SP, Arango NA, Mishina Y, Hanks MC, Behringer RR. Requirement of Bmpr1a for Müllerian duct regression during male sexual development. Nat Genet. 2002;32(3):408–410. doi: 10.1038/ng1003. [DOI] [PubMed] [Google Scholar]
- 53.Levavi-Sivan B, Bogerd J, Mañanós EL, Gómez A, Lareyre JJ. Perspectives on fish gonadotropins and their receptors. Gen Comp Endocrinol. 2010;165(3):412–437. doi: 10.1016/j.ygcen.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 54.Chu L, Li J, Liu Y, Hu W, Cheng CH. Targeted gene disruption in zebrafish reveals noncanonical functions of LH signaling in reproduction. Mol Endocrinol. 2014;28(11):1785–1795. doi: 10.1210/me.2014-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmann HM, Trang C, Gong P, Kimura I, Pandolfi EC, Mellon PL. Deletion of Vax1 from Gonadotropin-Releasing Hormone (GnRH) Neurons Abolishes GnRH Expression and Leads to Hypogonadism and Infertility. J Neurosci. 2016;36(12):3506–3518. doi: 10.1523/JNEUROSCI.2723-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bellaïche J, Goupil AS, Sambroni E, Lareyre JJ, Le Gac F. Gdnf-Gfra1 pathway is expressed in a spermatogenetic-dependent manner and is regulated by Fsh in a fish testis. Biol Reprod. 2014;91(4):94. doi: 10.1095/biolreprod.114.119834. [DOI] [PubMed] [Google Scholar]
- 57.Vainio S, Heikkilä M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397(6718):405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 58.DiNapoli L, Capel B. SRY and the standoff in sex determination. Mol Endocrinol. 2008;22(1):1–9. doi: 10.1210/me.2007-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Munger SC, Capel B. Sex and the circuitry: progress toward a systems-level understanding of vertebrate sex determination. Wiley Interdiscip Rev Syst Biol Med. 2012;4(4):401–412. doi: 10.1002/wsbm.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wieacker P, Missbach D, Jakubiczka S, Borgmann S, Albers N. Sex reversal in a child with the karyotype 46, XY, dup (1) (p22.3p32.3) Clin Genet. 1996;49(5):271–273. doi: 10.1111/j.1399-0004.1996.tb03786.x. [DOI] [PubMed] [Google Scholar]
- 61.Parr BA, McMahon AP. Sexually dimorphic development of the mammalian reproductive tract requires Wnt-7a. Nature. 1998;395(6703):707–710. doi: 10.1038/27221. [DOI] [PubMed] [Google Scholar]
- 62.de Lau WB, Snel B, Clevers HC. The R-spondin protein family. Genome Biol. 2012;13(3):242. doi: 10.1186/gb-2012-13-3-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kocer A, Pinheiro I, Pannetier M, Renault L, Parma P, Radi O, Kim KA, Camerino G, Pailhoux E. R-spondin1 and FOXL2 act into two distinct cellular types during goat ovarian differentiation. BMC Dev Biol. 2008;8:36. doi: 10.1186/1471-213X-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ross A, Munger S, Capel B. Bmp7 regulates germ cell proliferation in mouse fetal gonads. Sex Dev. 2007;1(2):127–137. doi: 10.1159/000100034. [DOI] [PubMed] [Google Scholar]
- 65.Hoshino A, Koide M, Ono T, Yasugi S. Sex-specific and left-right asymmetric expression pattern of Bmp7 in the gonad of normal and sex-reversed chicken embryos. Dev Growth Differ. 2005;47(2):65–74. doi: 10.1111/j.1440-169x.2004.00783.x. [DOI] [PubMed] [Google Scholar]
- 66.Ogino Y, Hirakawa I, Inohaya K, Sumiya E, Miyagawa S, Denslow N, Yamada G, Tatarazako N, Iguchi T. Bmp7 and Lef1 are the downstream effectors of androgen signaling in androgen-induced sex characteristics development in medaka. Endocrinology. 2014;155(2):449–462. doi: 10.1210/en.2013-1507. [DOI] [PubMed] [Google Scholar]
- 67.Siggers P, Smith L, Greenfield A. Sexually dimorphic expression of Gata-2 during mouse gonad development. Mech Dev. 2002;111(1–2):159–162. doi: 10.1016/S0925-4773(01)00602-5. [DOI] [PubMed] [Google Scholar]
- 68.Bernard DJ, Fortin J, Wang Y, Lamba P. Mechanisms of FSH synthesis: what we know, what we don’t, and why you should care. Fertil Steril. 2010;93(8):2465–2485. doi: 10.1016/j.fertnstert.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 69.Lo A, Zheng W, Gong Y, Crochet JR, Halvorson LM. GATA transcription factors regulate LHβ gene expression. J Mol Endocrinol. 2011;47(1):45–58. doi: 10.1530/JME-10-0137. [DOI] [PubMed] [Google Scholar]
- 70.Panda DK, Miao D, Tremblay ML, Sirois J, Farookhi R, Hendy GN, Goltzman D. Targeted ablation of the 25-hydroxyvitamin D 1alpha -hydroxylase enzyme: evidence for skeletal, reproductive, and immune dysfunction. Proc Natl Acad Sci U S A. 2001;98(13):7498–7503. doi: 10.1073/pnas.131029498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun W, Xie H, Ji J, Zhou X, Goltzman D, Miao D. Defective female reproductive function in 1,25(OH)2D-deficient mice results from indirect effect mediated by extracellular calcium and/or phosphorus. Am J Physiol Endocrinol Metab. 2010;299(6):E928–E935. doi: 10.1152/ajpendo.00378.2010. [DOI] [PubMed] [Google Scholar]
- 72.Dicken CL, Israel DD, Davis JB, Sun Y, Shu J, Hardin J, Neal-Perry G. Peripubertal vitamin D(3) deficiency delays puberty and disrupts the estrous cycle in adult female mice. Biol Reprod. 2012;87(2):51. doi: 10.1095/biolreprod.111.096511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee BY, Lee WJ, Streelman JT, Carleton KL, Howe AE, Hulata G, Slettan A, Stern JE, Terai Y, Kocher TD. A second-generation genetic linkage map of tilapia (Oreochromis spp.) Genetics. 2005;170:237–244. doi: 10.1534/genetics.104.035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Böhne A, Wilson CA, Postlethwait JH, Salzburger W. Variations of a theme: Genomics of sex determination in the cichlid fish Astatotilapia burtoni. BMC Genomics. doi:10.1186/s12864-016-3178-0. [DOI] [PMC free article] [PubMed]
- 75.Eshel O, Shirak A, Weller JI, Slossman T, Hulata G, Cnaani A, Ron M. Fine-mapping of a locus on linkage group 23 for sex determination in Nile tilapia (Oreochromis niloticus) Anim Genet. 2011;42(2):222–224. doi: 10.1111/j.1365-2052.2010.02128.x. [DOI] [PubMed] [Google Scholar]
- 76.Streelman JT, Albertson RC. Evolution of novelty in the cichlid dentition. J Exp Zool B Mol Dev Evol. 2006;306(3):216–226. doi: 10.1002/jez.b.21101. [DOI] [PubMed] [Google Scholar]
- 77.Joyce DA, Lunt DH, Genner MJ, Turner GF, Bills R, Seehausen O. Repeated colonization and hybridization in Lake Malawi cichlids. Curr Biol. 2011;21(3):R108–R109. doi: 10.1016/j.cub.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 78.Malinsky M, Challis RJ, Tyers AM, Schiffels S, Terai Y, Ngatunga BP, Miska EA, Durbin R, Genner MJ, Turner GF. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350(6267):1493–1498. doi: 10.1126/science.aac9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Christie MR, Marine ML, Fox SE, French RA, Blouin MS. A single generation of domestication heritably alters the expression of hundreds of genes. Nat Commun. 2016;7:10676. doi: 10.1038/ncomms10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fernald RD. Quantitative behavioural observations of Haplochromis burtoni under semi-natural conditions. Anim Behav. 1977;25(3):643–653. doi: 10.1016/0003-3472(77)90115-4. [DOI] [Google Scholar]
- 81.Renn SC, Carleton JB, Magee H, Nguyen ML, Tanner AC. Maternal care and altered social phenotype in a recently collected stock of Astatotilapia burtoni cichlid fish. Integr Comp Biol. 2009;49(6):660–673. doi: 10.1093/icb/icp085. [DOI] [PubMed] [Google Scholar]
- 82.Kustan JM, Maruska KP, Fernald RD. Subordinate male cichlids retain reproductive competence during social suppression. Proc Biol Sci. 2012;279(1728):434–443. doi: 10.1098/rspb.2011.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maruska KP. Social regulation of reproduction in male cichlid fishes. Gen Comp Endocrinol. 2014;207:2–12. doi: 10.1016/j.ygcen.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 84.Juntti SA, Hu CK, Fernald RD. Tol2-Mediated Generation of a Transgenic Haplochromine Cichlid, Astatotilapia burtoni. PLoS One. 2013;8:e77647. doi: 10.1371/journal.pone.0077647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Juntti SA, Hilliard AT, Kent KR, Kumar A, Nguyen A, Jimenez MA, Loveland JL, Mourrain P, Fernald RD. A Neural Basis for Control of Cichlid Female Reproductive Behavior by Prostaglandin F2α. Curr Biol. 2016;26(7):943–949. doi: 10.1016/j.cub.2016.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Renn SC, Aubin-Horth N, Hofmann HA. Fish and chips: functional genomics of social plasticity in an African cichlid fish. J Exp Biol. 2008;211(Pt 18):3041–3056. doi: 10.1242/jeb.018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Renn SC, Fraser EJ, Aubin-Horth N, Trainor BC, Hofmann HA. Females of an African cichlid fish display male-typical social dominance behavior and elevated androgens in the absence of males. Horm Behav. 2012;61(4):496–503. doi: 10.1016/j.yhbeh.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Desjardins JK, Klausner JQ, Fernald RD. Female genomic response to mate information. Proc Natl Acad Sci U S A. 2010;107(49):21176–21180. doi: 10.1073/pnas.1010442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burmeister SS, Kailasanath V, Fernald RD. Social dominance regulates androgen and estrogen receptor gene expression. Horm Behav. 2007;51(1):164–170. doi: 10.1016/j.yhbeh.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burford Reiskind MO, Coyle K, Daniels HV, Labadie P, Reiskind MH, Roberts NB, Roberts RB, Vargo EL, Schaff J. Development of a universal double-digest RAD sequencing approach for a group of non-model, ecologically and economically important insect and fish taxa. Mol Ecol Resour. 2016;16(6):1303–1314. [DOI] [PubMed]
- 91.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: building and genotyping Loci de novo from short-read sequences. G3 (Bethesda) 2011;1(3):171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Margarido GR, Souza AP, Garcia AA. OneMap: software for genetic mapping in outcrossing species. Hereditas. 2007;144(3):78–79. doi: 10.1111/j.2007.0018-0661.02000.x. [DOI] [PubMed] [Google Scholar]
- 94.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ARXIV: 1303.3997; 2013.(v. 0.7.5a, http://arxiv.org/abs/1303.3997).


