Abstract
Background
Myxozoa is a well-known economically and ecologically important group of metazoan parasites, phylogenetically related to Cnidaria. High diversity of myxosporeans has been recorded in Russia and China; however, most of the species were solely morphologically characterised. Here, we identified a new gibel carp-infecting Myxobolus species and morphologically and molecularly compared the Russian and Chinese isolates of this new myxosporean.
Results
Myxobolus pronini n. sp. was found free in the abdominal cavity of Carassius auratus gibelio (Bloch, 1782) in Lake Baikal watershed, Russia, and embedded in the visceral serous membranes of the same fish species in Lake Taibai, Hubei province, China. The morphometric data of the plasmodia and mature spores exhibited some differences between the Russian and Chinese isolates, but SSU rDNA sequences indicated that these two geographical isolates are conspecific. The mature spores from the two locations are obovate in frontal view, with wider anterior than posterior end and lemon-shaped in sutural view. Spores of the Russian isolate were 14.3–16.2 (mean 15.1 ± 0.2) μm long, 9.6–10.8 (10.1 ± 0.1) μm wide and 6.4–7.4 (6.7 ± 0.15) μm thick; those of the Chinese isolate were 13.8–15.6 (14.7 ± 0.24) μm long, 9.6–13.3 (9.6 ± 0.65) μm wide and 6.2–7.2 (6.6 ± 0.16) μm thick. The newly-generated rDNA sequences (including SSU rDNA, ITS and LSU rDNA) from the two isolates represented some variations within the intraspecific range. Homology search by BLAST showed that the newly obtained rDNA sequences do not match any sequences available on GenBank. Phylogenetic analysis based on the aligned partial SSU rDNA sequences indicated that this novel species clustered with several gibel carp-infecting Myxobolus spp. with round anterior end of spores. Additionally, phylogenetic analysis based on all obtained ITS sequences showed that distinct genetic geographical differentiation occurred for this new parasite.
Conclusions
Myxobolus pronini n. sp. is described by integrating morphological, ecological and molecular evidence. Two geographical isolates of this species showed some morphological and genetic differences but within the intraspecific range of variation.
Keywords: Myxobolus pronini n. sp, Abdominal cavity, Serous membranes, Geographical variation, Gibel carp, Carassius auratus gibelio, Russia, China
Background
Myxozoa Grassé, 1970 is an economically and ecologically important group of metazoan parasites. Although more than 2,200 species have been reported from aquatic invertebrates, fishes, reptiles, birds and mammals, including humans worldwide [1], high hidden diversity of this group of parasites is widely acknowledged [2, 3]. For instance, no malacosporean has been reported in China for no studies concerned this group of myxozoans. Among reported myxozoans, some were described and identified solely upon spore morphology [4–6]. These described species with insufficient data should be validated or revised by supplementing with detailed taxonomic characteristics. Currently, an integrated approach combining spore morphology, exact location of sporulation, tissue- and host-specificity and molecular characteristics has been widely recommended and accepted for identification of novel or cryptic myxosprean species and for discrimination of species with morphological similarity [7]. The genus Myxobolus, comprising more than 900 species is the most speciose within the phylum Myxozoa [8, 9]. Among them, about 500 Myxobolus species have been described or recorded from Russia and China [10, 11], although many species were possibly misidentified [6]. As a part of an ongoing joint project applying modern taxonomic features to approve the validity of the incompletely described existing taxa and uncover the actual species diversity of fish myxosporeans in Russia and China, a new species, designated here as Myxobolus pronini n. sp. from the abdominal cavity and the visceral serous membranes of the gibel carp Carassius auratus gibelio (Bloch, 1782) inhabiting in the watershed of Lake Baikal in Russia and Yangtze River in China, is morphologically and molecularly characterised in this paper.
Methods
Sample collection and morphological identification
Twelve gibel carp, 9.5–19.7 cm in body length, were captured from a pond near Barguzin River (53°69'N, 109°80'E), a tributary of Lake Baikal, by gill net in June of 2015. Twenty gibel carp, 16.5 to 31.3 cm in body length, were obtained from Lake Taibai (29°58'18"N, 115°23'9"E), located at the middle and lower reaches of Yangtze River, by trawl net in November of 2015. Russian and Chinese fish samples were frozen and transported in iceboxes to the laboratory in Ulan-Ude and Wuhan, respectively. Complete parasitological examination was performed. After necropsy, visual examination of the gills, muscle, liver, intestine, spleen, heart and gallbladder was performed to find the suspected plasmodia of myxosporean parasites by naked eye and further examined by preparing wet mounts of all inspected organs with stereomicroscopy and phase contrast microscopy. Frozen isolated plasmodia of the Russian isolate were transported to the laboratory of Institute of Hydrobiology, Chinese Academy of Sciences for further morphological observation and molecular characterisation.
Morphological and morphometric data of randomly selected 50 normal mature spores were obtained and measured from smear preparations of ruptured by needle plasmodia according to Lom & Arthur [12] using an Olympus BX53 microscope equipped with an ocular micrometer. Thawed spores of the two geographical isolates were photographed with the Zeiss Axiplan 2 Image and Axiophot 2. Line drawings were made based on the photographs with the aid of Adobe Photoshop CS (Adobe Systems, San Jose, CA, USA). The presence of iodinophilous vacuole and mucous envelope of the spores was checked by mixing a small drop of Lugol’s solution and black Indian ink with water suspension of fresh spores. All measurements are given in micrometres and are presented as the range followed by the mean ± standard deviation (SD) in parentheses, unless otherwise indicated.
Genomic DNA isolation and sequencing
Preserved isolated plasmodia or myxospores in 95 % ethanol were centrifuged at 4,000× g for 10 min and washed two times with distilled water to remove the ethanol remnants. Genomic DNA was isolated from the obtained pellet using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Düsseldorf, Germany), following the manufacturer’s recommended protocol for animal tissue. gDNA concentration was determined using a NanoDrop2000 spectrophotometer (Thermo Scientific) at 260 nm. Ribosomal DNA fragments, including small subunit ribosomal DNA (SSU rDNA), internal transcribed spacer (ITS) and large subunit ribosomal DNA (LSU rDNA) were amplified using the primers listed in Table 1. PCR was carried out in a 25 μl reaction mixture which comprised 30 ng template DNA, 1× PCR mixture (CWbiotech, Beijing, China) and 10 pmol of each primer. The partial SSU rDNA was amplified using the primers MyxospecF-18R [13, 14] and the PCR cycle consisted of an initial denaturation step at 95 °C for 4 min, followed by 35 cycles at 95 °C for 1 min, 48 °C for 1 min, 72 °C for 2 min, and a final extension at 72 °C for 10 min.
Table 1.
Primers used for PCR amplification or sequencing of SSU rDNA, ITS and LSU rDNA of Myxobolus pronini n. sp
The complete ITS was amplified using the primer pair 18R-V/Myxo28S1F-V [13, 15] and the PCR cycle comprised by an initial denaturation step at 94 °C for 5 min, followed by 35 cycles at 94 °C for 50 s, 54 °C for 50 s, 72 °C for 2 min and a final extension at 72 °C for 10 min. The complete LSU rDNA sequence was obtained by assembling two overlapping parts. The first part of LSU was obtained using the NLF160/NLR1694 primer pair [16] and the second part using the NLF1050-NLR3284 primer pair [17]. PCR cycling parameters for LSU rDNA amplification were 95 °C for 3 min, then 30 cycles of 95 °C for 1 min, 48 °C for 1 min, 72 °C for 2 min, followed by 10 min incubation at 72 °C. PCR products were separated by agarose gel electrophoresis, purified with a PCR purification kit (CWBiotech, Beijing, China) and then cloned into PMD-18 T vector system (Takara, Tokyo, Japan). Then positive clones were selected and sequenced with the ABI BigDye Terminator v3.1 Cycle Sequencing Kit with an ABI 3100 Genetic Analyzer. Potential intraspecific geographical variations of the ITS were tested by sequencing of three clones per isolate.
Phylogenetic analysis
All sequences were assembled in BioEdit [18] and the consensus sequences obtained were determined as myxozoan by a GenBank BLAST search. To explore the phylogenetic relationships of the present species with the existing myxobolids, 37 SSU rDNA sequences of selected myxosporean species were aligned with Clustal X version 1.8 [19] by default setting. All newly-generated ITS sequences of Myxobolus pronini n. sp. and several cyprinid-infecting myxosporean species were also aligned to explore the possible geographical variation of this species. The alignment was corrected manually using the alignment editor of the software MEGA 6.0 [20]. Pairwise sequence distances and similarity of clones of the two geographical isolates based on ITS were calculated in MEGA 6.0 [20]. Phylogenetic analysis was performed using maximum likelihood (ML) analysis in PhyML 3.0 [21] and Bayesian analysis in MrBayes [22]. Optimal evolutionary model for ML and Bayesian analysis was determined using Modeltest 3.7 [23] which identified the best evolutionary model for SSU rDNA and ITS dataset were the general time reversible model (GTR + I + G) and HKY + I, respectively, judging by the Akaike information criterion. Two independent runs were conducted with four chains for a million generations for Bayesian analysis. Phylogenetic trees were sampled every 100 generations. The first 25 % of the samples were discarded from the cold chain (burninfrac = 0.25). Bootstrap confidence values were calculated with 100 pseudoreplicates for ML. Ceratonova shasta Atkinson, Foott & Bartholomew, 2014 (AF001579) and Myxobolus cerebralis Hofer, 1903 (AY479922) were used as the outgroup taxa for SSU rDNA and ITS dataset, respectively. Trees were initially examined in TreeView X [24] and then edited and annotated in Adobe Illustrator (Adobe Systems Inc.).
Results
No clinical symptoms were observed for the infected fish from China, but slightly swelling abdomen was observed in infected fish from Russia. After dissection, a single big oval and yellowish plasmodium, measuring 1.4 × 0.93 cm was found free in the abdominal cavity of each infected host from Russia, while several small oval and whitish plasmodia, measuring 0.85 × 0.65 mm was found to protrude out of the surface of visceral serous membranes of each infected host from China (Fig. 1). The prevalence of this parasite was 25 % (3/12) and 5 % (1/20) for the Russian and Chinese isolates, respectively. Plasmodium wall consisted of a layer of collagen fibres in both isolates and no host inflammatory response was observed.
Fig. 1.
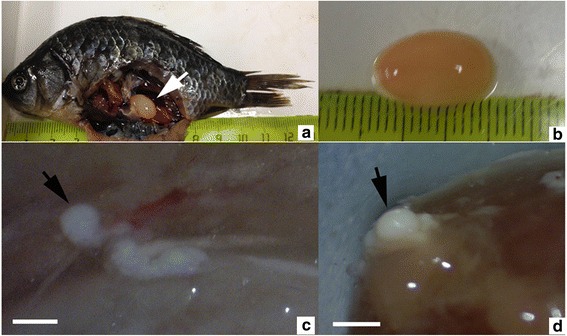
The plasmodium of Myxobolus pronini n. sp. a, b Large yellowish plasmodium (arrow) free in the abdominal cavity of Carassius auratus gibelio sampled from a pond near Lake Baikal, Russia; c, d A single small whitish plasmodium (arrow) in the serous membrane of the liver of C. auratus gibelio sampled from Lake Taibai of Yangtze River watershed, China. Scale-bars: 1 mm
Family Myxobolidae Thélohan, 1892
Genus Myxobolus Bütschli, 1882
Myxobolus pronini n. sp.
Type-host : Gibel carp Carassius auratus gibelio Bloch, 1782 (Cypriniformes: Cyprinidae).
Type-locality : Pond near Barguzin River (53°69'N, 109°80'E), Lake Baikal watershed in Russia.
Other localities : Lake Taibai in Hubei Province, China.
Site in host : Abdominal cavity and visceral serous membranes.
Prevalence : 25 % (3/12) in Russia and 5 % (1/20) in China.
Type-material : Hapantotype specimens of spores in glycerine gelly, phototypes, fixed in 10 % formalin and 95 % ethanol were deposited in the Laboratory of Fish Diseases, Institute of Hydrobiology, Chinese Academy of Science [collection numbers MP20150719 (Russian isolate) and MP20151110 (Chinese isolate)].
Representative DNA sequences : Representative rDNA sequences were deposited in the GenBank database under the accession numbers KU524889–KU524890 (SSU); KU524891–KU524892 (LSU); and KU524893–KU524898 (ITS).
ZooBank registration : To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN), details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:A7B0E5DD-69BE-4C62-B1F6-354FB84689BA. The LSID for the new name Myxobolus pronini is urn:lsid:zoobank.org:act:6774338 F-6897-410A-8CB1-EB6211CF647E.
Etymology : The species is named after Professor Nikolai Martemyanovich Pronin (Institute of General and Experimental Biology of Siberian Branch, Russian Academy of Sciences) in recognition of his significant contribution to the knowledge of fish parasites in Siberia, Russia.
Description (Figs. 1, 2, 3, 4 and 5)
Fig. 2.
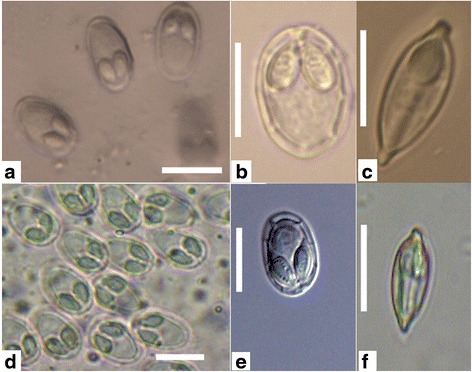
Myxospores of Myxobolus pronini n. sp. under light microscopy. a-c The Russian isolate. d-f The Chinese isolate. a, b, d, e, frontal view; c, f, sutural view; Scale-bars: 10 μm
Fig. 3.
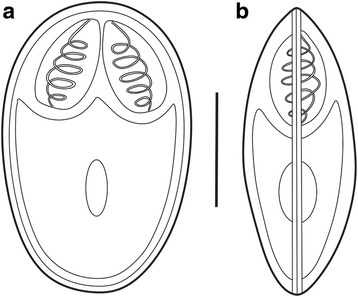
Schematic drawings of myxospores of Myxobolus pronini n. sp. a Frontal view. b Sutural view. Scale-bar: 5 μm
Fig. 4.
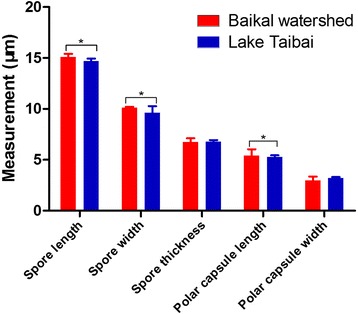
Morphometric differences between the Russian and Chinese isolates of Myxobolus pronini n. sp. Asterisks indicate significant differences
Fig. 5.
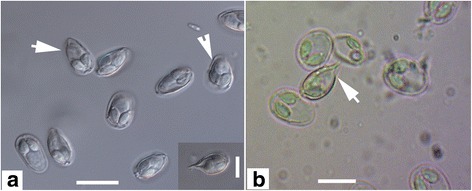
Abnormal myxospores of Myxobolus pronini n. sp. under light microscopy. a Incompletely developed abnormal spores (arrows). b Henneguya-like spore (arrow). Scale-bars: 10 μm
Myxospores elongated obovate in frontal view, lemon-shaped in sutural view, with blunt anterior end wider than posterior end (Figs. 2 and 3). Spore measurements as follows: Russian isolate [14.3–16.2 (15.1 ± 0.3) long, 9.6–10.8 (10.1 ± 0.1) wide, 6.4–7.4 (6.7 ± 0.15) thick]; Chinese isolate [13.8–15.6 (14.7 ± 0.24) long, 9.0–13.3 (9.6 ± 0.65) wide, 6.2–7.2 (6.6 ± 0.16) thick]. Spore valves relatively thin, symmetrical, smooth. Polar capsules 2, pyriform, equal in size, converging anteriorly, measuring as follows: Russian isolate [4.3–6.7 (5.4 ± 0.63) long, 4.8–5.6 (3.0 ± 0.16) wide]; Chinese isolate [4.8–5.6 (5.3 ± 0.16) long, 2.9–3.4 (3.0 ± 0.12) wide] (Table 2). Polar filaments coiled with 5–6 turns, situated perpendicularly to the longitudinal axis of polar capsules; 3–5 folds observed at posterior end of some spores. Intercapsular appendix and mucous envelope not observed, but a round iodinophilous vacuole remarkable.
Table 2.
Comparative data for spore measurements (range ± standard deviation in micrometres) of Myxobolus pronini n. sp. and morphologically similar species
| Species | Host | Site in host | SL | SW | ST | PCL | PCW | PFC | Reference |
|---|---|---|---|---|---|---|---|---|---|
| M. pronini n. sp. Russian isolate | Carassius auratus gibelio | Abdominal cavity | 14.3–16.2 (15.1 ± 0.3) | 9.6–10.8 (10.1 ± 0.1) | 6.4–7.4 (6.7 ± 0.15) | 4.3–6.7 (5.4 ± 0.63) | 2.2–3.6 (3.1 ± 0.05) | 5–6 | This study |
| M. pronini n. sp. Chinese isolate | C. auratus gibelio | Mesentery | 13.8–15.6 (14.7 ± 0.24) | 9.0–13.3 (9.6 ± 0.65) | 6.2–7.2 (6.6 ± 0.16) | 4.8–5.6 (5.3 ± 0.16) | 2.9–3.4 (3.0 ± 0.12) | 5–6 | This study |
| M. ellipsoides | Tinca tinca | Gills, liver, intestine | 12–15 | 9–11 | 4 | [9] | |||
| M.qiankiangensis | C. auratus auratus | Spleen, body cavity | 15.6–18.0 (17.3) | 10.6–12.0 (11.4) | 8.0–8.4 (8.3) | 7.8–9.6 (8.6) | 3.6–4.2 (3.8) | 7–9 | [8] |
| M. sigini | Hypophthalmichthys molitrix | Body cavity | 9.8–11.3 (10.6) | 7.2–7.8 (7.4) | 4.8–6.0 (5.3) | 4.8–5.0 (4.9) | 2.4–2.6 (2.5) | 6–7 | [11] |
| M. pfrille | Chrosomus neogaeus | Body cavity | 12.0–14.4 (12.9) | 8.4–10.2 (9.5) | (7.0) | 5.0–6.0 (5.6) | 3.6 | 5 | [11] |
| M. hypseleotris | Micropercops cinctus | Skin, muscle, intestine | 12.6–15.6 (14.6) | 8.4–10.8 (9.7) | 6.0–7.2 (6.6) | 4.8–6.0 (5.4) | 2.8–3.6 (3.3) | 5–6 | [11] |
| M. lentisuturalis | C. gibelio | Muscle | 11.2–12.4 (11.8) | 7.2–8.4 (7.6) | (5.2) | 4.0–4.4 (4.2) | 2.0–2.8 (2.5) | 4 | [29] |
Abbreviations: SL spore length, SW spore width, ST spore thickness, PCL polar capsule length, PCW polar capsule width, PFC polar filament coils
In addition to the above described normal spores, there were less than 5 % abnormal spores on average in all individual plasmodia of the two geographical isolates, among which some possessed Henneguya-like caudal appendages (Fig. 5).
Morphometric variation
The mature spores from the fish host in the two localities showed typical characteristics of the genus of Myxobolus and were very similar in shape. There was an overlap in the morphometric data although some features exhibited significant differences (Table 2, Fig. 4). The length and width of spores and the length of the polar capsules of the Russian isolates was significantly greater than those of the Chinese isolates (spore length: t (98) = 15.25, P < 0.001; spore width: t (78) = 6.462, P < 0.001; polar capsule length: t (98) = -3.833, P < 0.001). However, the thickness of the spores and the width of the polar capsules of the two isolates were almost identical (spore thickness: t (98) = -0.889, P = 0.377; polar capsule width: t (98) = 1.460, P = 0.150).
Molecular characterisation
Ribosomal RNA gene sequences of the two geographical isolates (6,068 and 6,056 bp for the Russian and Chinese isolate, respectively) were obtained after trimming the ambiguous positions in the two sequencing ends and the applied amplified primers, both with about 45.7 % G + C content. The SSU rDNA sequences of the two isolates (1,734 bp and 1,739 bp for the Russian and Chinese isolate, respectively; accession numbers KU524890 and KU524889, respectively) exhibited a similarity of 99.5 %, whereas the LSU rDNA sequences of the two isolates (3,619 bp and 3,611 bp for the Russian and Chinese isolate, respectively; accession numbers KU524892 and KU524891, respectively) showed 99.3 % similarity. All six ITS sequences (accession numbers KU524893–KU524898) were 715 bp in length, but the intra-isolate genetic distances were generally lower than inter-isolate distances (Table 3).
Table 3.
Genetic distance (number of different positions and p-distance; below diagonal) and percent sequence similarity (%) (above diagonal) obtained from the distance matrix based on a 715 bp ITS fragment of the Russian and Chinese isolates of Myxobolus pronini n. sp
| Isolatea | 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| 1 | KU524896 | – | 99.7 | 99.6 | 99.6 | 99.6 | 99.6 |
| 2 | KU524897 | 2 (0.0029) | – | 99.6 | 99.6 | 99.6 | 99.1 |
| 3 | KU524898 | 3 (0.0043) | 2 (0.0043) | – | 99.4 | 99.4 | 99.0 |
| 4 | KU524895 | 3 (0.0043) | 3 (0.0043) | 3 (0.0058) | – | 100 | 99.6 |
| 5 | KU524894 | 3 (0.0043) | 3 (0.0043) | 4 (0.0058) | 0 (0.0000) | – | 99.6 |
| 6 | KU524893 | 6 (0.0043) | 6 (0.0086) | 7 (0.0101) | 3 (0.0043) | 3 (0.0043) | – |
aKU524893–KU524895 isolated from Lake Taibai, China; KU524896–KU524898 isolated from the coast of Lake Baikal watershed, Russia
Bayesian analysis of aligned full length ITS sequences revealed that M. pronini n. sp. forms an independent branch, clustering within the gibel carp-infecting Henneguya-Myxobolus clade and remarkable genetic geographical differentiation occurred for this new parasite (Fig. 6). A BLAST search using SSU rDNA of this species as queries indicated that these sequences were distinguished from all myxosporean sequences available in the GenBank database but most similar to those of M. nielii (Nie & Li, 1973) Landsberg & Lom, 1991 (JQ690358; 98 %), H. doneci Schulman, 1962 (LC011456; 98 %), M. hearti Chen, 1998 (GU574808; 96 %), and H. globulata Ye & Wang, 2012 (JQ690355; 93 %). ML and BI analyses produced similar tree topologies, although with somewhat different support values at some evolutionary nodes. Therefore, only the Bayesian tree is present here, along with the bootstrap values of ML tree. Phylogenetic analysis clearly placed M. pronini n. sp. in the clade of gibel carp-infecting Myxobolus species with round anterior end of spores under the support of high bootstrap and posterior probability value (Fig. 7).
Fig. 6.
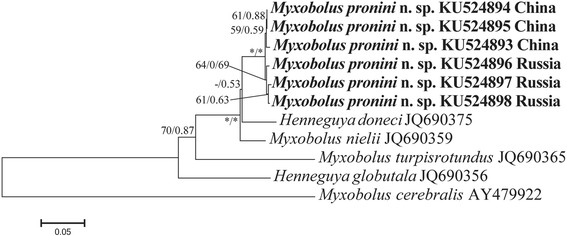
Bayesian analysis of genetic geographical differentiation of Myxobolus pronini n. sp. based on ITS sequences rooted with Myxobolus cerebralis (AY479922). Numbers at nodes indicate Bayesian posterior probability and ML bootstrap values, respectively. Asterisks indicate support values > 95 % and dashes indicate values < 50 %
Fig. 7.
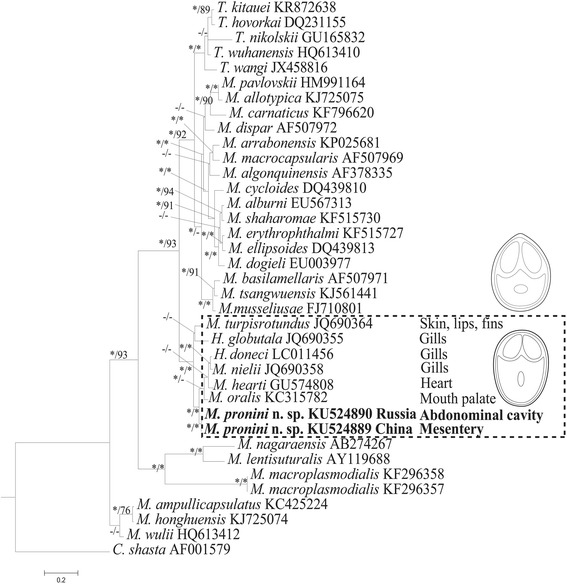
Phylogenetic tree generated by Bayesian analysis of aligned partial SSU rRNA gene sequences of Myxobolus pronini n. sp. and related species, rooted with Ceranova shasta (AF001579). Numbers at nodes indicate Bayesian posterior probability and ML bootstrap values, respectively. Asterisks indicate support values > 95 % and dashes indicate values < 50 %. The new species (indicated in bold) clustered within a cyprinid-infecting Henneguya-Myxobolus clade of parasites with round or blunt anterior ends of spores (dashed box)
Discussion
Within the phylum Myxozoa, Myxobolus Bütschli, 1882 is the largest genus [8, 9] and the diversity keeps increasing with more attention paid by fish parasitologists over the world, especially recently from South America, Africa and India [25–27]. Out of more than 900 Myxobolus species reported to date, M. pronini n. sp. closely resembles in spore shape M. ellipsoids Thélohan, 1892; M. qiankiangensis (Chen, 1998) Eiras, Molnár & Lu, 2005; M. sigini (Chen, 1988) Landsberg & Lom, 1991; M. pfrille (Fantham, 1939) Landsberg & Lom, 1991; M. hypseleotris Chen & Ma, 1998; and M. lentisuturalis Dyková, Fiala & Nie, 2002 (Table 2). However, spores of M. ellipsoides are remarkably smaller than those of M. pronini n. sp. Additionally, M. ellipsoides was originally reported from gills of Tinca tinca in France and then recorded in most of organs of 58 distantly related fish hosts [11, 28]. So, it can be suspected that M. ellipsoides actually represents a cryptic species complex. Narrower anterior end of spores, longer spore length, a greater number of polar filament coils and irregular bigger plasmodium can significantly discriminate M. qiankiangensis from M. pronini n. sp., although both species were found in the abdominal cavity of C. auratus. Myxobolus sigini infects silver carp, Hypophthalmichthys molitrix Valenciennes, 1884 rather than gibel carp and can be further clearly differentiated from M. pronini n. sp. by the smaller size of the spores and the smaller ratio of polar capsule length to polar capsule width. Myxobolus pfrille and M. hypseletris are very similar to M. pronini n. sp. by having elliptical spores, but the host and spore size of these two species are different. Furthermore, a distinct triangular intercapsule appendix occurs in M. pfrille, but is lacking in M. pronini n. sp. The spore shape of M. lentisuturalis is very similar to that of M. pronini n. sp., but the two species differ in the spore size, infection site and the number of coils of the polar filament [29].
In addition to the above mentioned cypriniform-infecting species and species recorded in Russia and China, M. macroplasmodialis Molnár, Ranzani-Paiva, Eiras & Rodrigues, 1998, with a unique big free plasmodium (above 1 cm) in the abdominal cavity of Salminus brasiliensis Cuvier, 1816 (Characiformes) in Brazil [30] is morphologically similar with M. pronini n. sp., but the oval and antero-laterally diverged opening of the polar capsules and smaller spore size of the former undoubtedly differentiate it from the latter. Regarding the specificity of infection site of the Russian isolate of M. pronini n. sp., no conclusion can be made due to the lack of the early developmental stages. But, it could be presumed that plasmodia of the Russian isolate start their development in the serous membranes of the abdominal wall or visceral organs and then become detached from those sites at an advanced stage of development, similar to M. macroplasmodialis. Since the smaller plasmodia of the Chinese isolate were at an advanced developmental stage, determined by the presence of most of mature spores, we thought that the sporulation in the Russian isolate occurred in the serous membranes of the abdomen rather than in the visceral serous membranes and the different microhabitat produced differently sized plasmodia. Size and shape plasticity of plasmodia and spores have been widely reported for many myxosporean parasites [7, 31]. However, both isolates of M. pronini n. sp. represented a strict specificity to connective tissues which is consistent with most of cyprinid-infecting Myxobolus species [7, 32].
Simple and variable morphological features of myxospores were acknowledged to be the main factors responsible for the difficulty of myxosporean taxonomy and species identification [6, 33, 34]. As such, the two geographical isolates of M. pronini n. sp. showed some significant variations in the length and width of spores and the length of the polar capsules. Regional genetic differentiation of several myxozoans has been reported by phylogenetic analysis of ITS [14, 33, 35, 36]. We also demonstrated here that a distinct genetic variation occurred for two isolates of M. pronini n. sp. by this molecular marker. Gibel carp is widely distributed in Eurasia; its origin is still unclear, although some researchers suggest that the native habitat of the gibel carp is the Far East [37]. Due to the fact that the evolutionary migration route of the gibel carp remains unknown, the origin of speciation of this new parasite remains enigmatic. Also, some subspecies of gibel carp, including allogynogentic gibel carp have been introduced via anthropogenic means to wide areas for intensive culture, especially in China. Previous reports showed that pathogens from populations of cultured fish spilled-over to wild watersheds and produced genetic exchanges with wild populations and then caused genetic variations of pathogenic microorganisms [38]. However, no infection of cultured gibel carp by the new myxosporean was found despite continuing intensive investigations of the myxosporean fauna of gibel carp cultured in China during the past ten years. Therefore, factors driving this geographical genetic variation await further research.
Molecular characteristics have been widely accepted as important parameters for myxosporean taxonomy, species identification and discrimination of cryptic species [7]. Due to the lack of sufficient LSU rDNA data available on GenBank and almost congruent phylogenetic results based on LSU rDNA and SSU rDNA [17], only phylogenetic analysis of SSU rDNA sequences was performed here. The result showed that two isolates of M. pronini n. sp. had an unambiguously independent position within the freshwater Myxobolus-Henneguya clade and clustered with several gibel carp-infecting Myxobolus species to form an independent lineage. However, this lineage represented diverse sites of sporulation, i.e. M. hearti dwelling in the heart ventricle, M. oralis Liu, Whipps, Nie & Gu, 2014 in the palate of the mouth, M. nielii, H. doneci and H. globulata in the gill filaments [39] and M. turpisrotundus Zhang, Wang, Li & Gong, 2010 in the subepidermal tissues of the body surface, fins and gill arches [6]. So, the present result supported the viewpoint that phylogenetic affinities of the fish hosts provide stronger evolutionary signal for evolutionary relationships of histozotic myxobolids than tissue-specificity to some extent [27]. However, all species involved in this lineage possess spores with a blunt anterior end thus highlighting the importance of spore morphology as taxonomic criterion for myxosporeans. Additionally, Myxobolus species with caudal appendages dispersed in different groups among this lineage rather than forming an independent cluster, indicates the presence of this feature independently evolved several times from Myxobolus to Henneguya or vice versa [5].
Similar to other histozotic myxosporeans [11, 40], some proportion of morphological abnormal spores also occurred in most of the plasmodia of the two geographical isolates of M. pronini n. sp. Insufficient nutrients for normal development could not completely explain their occurrence for some abnormal spores located in the centre rather than the periphery of plasmodia. Henneguya-like caudal appendages of M. pronini n. sp. confirm once more that Myxobolus is closely genetically related to Henneguya [41], although we disagree with the suggestion of suppression of Hennguya by some researchers [5] to avoid further taxonomic confusion of these two speciose myxobolid groups.
Conclusions
The two isolates infecting the serous membranes of abdominal cavity and visceral organs of the gibel carp form Russia and China are conspecific and represent a species new to science, M. pronini n. sp., delimitated by integrating morphological, ecological and molecular evidence. Two geographical isolates of this species represented some variations in the length and width of spores and the length of polar capsules, but within the range of intraspecific variation.
Acknowledgements
The authors thank two anonymous reviewers and Prof. Aneta Kostadinova for their comments and corrections which improved much the manuscript.
Funding
The present work is financially supported by the Jiangsu Province Fund of Sciences (BK2012240), Jiangsu Fishery Project (D2015–11), Chinese Natural Sciences Fund (31411130191, 31472296), Basic project SB Russian Academy of Science (VI.51.1.3) and the Russian Fund of Basic Research (14–04–91176 GFEN_a).
Availability of data and material
The rRNA gene sequences for Myxobolus pronini n. sp. were submitted to GenBank under accession numbers KU524890–KU524898.
Authors’ contributions
XHL, MDB collected fish samples, performed parasitological examination and molecular analysis and helped write the manuscript; YLZ, QQZ, TTL and AHL performed morphological comparisons and helped write the manuscript. JYZ designed this study and drafted the manuscript. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Animal Ethics Committee of Institute of Hydrobiology, Chinese Academy of Sciences (Approval No. ISH FISH2015-0614-9).
Abbreviations
- BI
Bayesian Inference analysis
- ITS
Internal transcribed spacer
- LSU
rDNA, large subunit ribosomal deoxyribonucleic acid
- ML
Maximum Likelihood
- SD
Standard deviation
- SSU rDNA
Small subunit ribosomal deoxyribonucleic acid
Contributor Information
Xin-Hua Liu, Email: liuxinhua1990@163.com.
Marina-D Batueva, Email: mbadm@biol.bscnet.ru.
Yuan-Li Zhao, Email: 6263198@qq.com.
Jin-Yong Zhang, Email: zhanjy@ihb.ac.cn.
Qian-Qian Zhang, Email: zhangqiantianyu623@163.ocm.
Tong-Tong Li, Email: zhangjyong@126.com.
Ai-Hua Li, Email: liaihua@ihb.ac.cn.
References
- 1.Fiala I, Bartošová–Sojková P, Whipps CM. Classification and phylogenetics of myxozoa. In: Okamura B, Gruhl A, Bartholomew JL, editors, Myxozoan Evolution, Ecology and Development. Cham: Springer International Publishing; 2015. pp. 85–110.
- 2.Bartošová-Sojková P, Hrabcová M, Pecková H, Patra S, Kodádková A, Jurajda P, et al. Hidden diversity and evolutionary trends in malacosporean parasites (Cnidaria: Myxozoa) identified using molecular phylogenetics. Int J Parasitol. 2014;44:565–77. doi: 10.1016/j.ijpara.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Shin SP, Shirakashi S, Hamano S, Kato K, Lasso LT, Yokoyama H. Phylogenetic study of the genus Kudoa (Myxozoa: Multivalvulida) with a description of Kudoa rayformis sp. nov. from the trunk muscle of Pacific sierra Scomberomorus sierra. Mol Phylo Evol. 2016;98:337–45. doi: 10.1016/j.ympev.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Kent ML, Andree KB, Bartholomew JL, El-Matbouli M, Desser SS, Devlin RH, et al. Recent advances in our knowledge of the Myxozoa. J Eukaryot Microbiol. 2001;48:395–413. doi: 10.1111/j.1550-7408.2001.tb00173.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Whipps CM, Gu ZM, Zeng LB. Myxobolus turpisrotundus (Myxosporea: Bivalvulida) spores with caudal appendages: investigating the validity of the genus Henneguya with morphological and molecular evidence. Parasitol Res. 2010;107:699–706. doi: 10.1007/s00436-010-1924-9. [DOI] [PubMed] [Google Scholar]
- 6.Zhang JY, Wang JG, Li AH, Gong XN. Infection of Myxobolus turpisrotundus n. sp. in allogynogenetic gibel carp, Carassius auratus gibelio (Bloch), with revision of Myxobolus rotundus (s. l.) Nemeczek reported from C. auratus auratus (L.) J Fish Dis. 2010;33:625–38. doi: 10.1111/j.1365-2761.2010.01161.x. [DOI] [PubMed] [Google Scholar]
- 7.Atkinson SD, Bartošová-Sojková P, Whipps CM, Bartholomew JL. Apporaches for characterizing myxozoan species. In: Okamura B, Gruhl A, Bartholomew JL, editors, Myxozoan Evolution, Ecology and Development. Cham: Springer International Publishing; 2015. pp. 111–124.
- 8.Eiras JC, Molnár K, Lu YS. Synopsis of the species of Myxobolus Bütschli, 1882 (Myxozoa: Myxosporea: Myxobolidae) Syst Parasitol. 2005;61:1–46. doi: 10.1007/s11230-004-6343-9. [DOI] [PubMed] [Google Scholar]
- 9.Eiras JC, Zhang JY, Molnár K. Synopsis of the species of Myxobolus Butschli, 1882 (Myxozoa: Myxosporea: Myxobolidae) described between 2005 and 2014. Syst Parasitol. 2014;88:11–36. doi: 10.1007/s11230-014-9484-5. [DOI] [PubMed] [Google Scholar]
- 10.Donec ZS, Shulman SS. Key to the determination of parasites of freshwater fishes of the USSR. In: Bauer ON, editor. Knidosporidii (Cnidosporidia) Russian: Leningrad: Nauka; 1984. pp. 88–251. [Google Scholar]
- 11.Chen QL, Ma CL, Fauna S. Myxozoa: Myxosporea. Beijing: Science Press (In Chinese); 1998. [Google Scholar]
- 12.Lom J, Arthur JR. A guideline for preparation of species descriptions in Myxosporea. J Fish Dis. 1989;12:151–156. doi: 10.1111/j.1365-2761.1989.tb00287.x. [DOI] [Google Scholar]
- 13.Whipps CM, Adlard RD, Bryant MS, Lester RJG, Findlay V, Kent ML. First report of three Kudoa species from eastern Australia: Kudoa thyrsites from mahi mahi (Coryphaena hippurus), Kudoa amamiensis and Kudoa minithyrsites n. sp. from sweeper (Pempheris ypsilychnus) J Eukuryot Microbiol. 2003;50:215–19. doi: 10.1111/j.1550-7408.2003.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 14.Whipps CM, Matbouli ME, Hedrick RP, Blazer V, Kent ML. Myxobolus cerebralis internal transcribed spacer 1 (ITS-1) sequences support recent spread of the parasite to North America and within Europe. Dis Aquat Org. 2004;60:105–8. doi: 10.3354/dao060105. [DOI] [PubMed] [Google Scholar]
- 15.Fiala I. The phylogeny of Myxosporea (Myxozoa) based on small subunit ribosomal RNA gene analysis. Int J Parasitol. 2006;36:1521–34. doi: 10.1016/j.ijpara.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Auwera GVD, Chapella S, Wächter RD. Structure of the large ribosomal subunit RNA of Phytophthora megasperma, and phylogeny of the oomycetes. FEBS Lett. 1994;338:133–6. doi: 10.1016/0014-5793(94)80350-1. [DOI] [PubMed] [Google Scholar]
- 17.Bartošová P, Fiala I, Hypša V. Concatented SSU and LSU rDNA data confirm the main evolutionary trends within myxosporeans (Myxozoa: Myxosporea) and provide an effective tool for their molecular phylogenetics. Mol Phylogenet Evol. 2009;3:81–93. doi: 10.1016/j.ympev.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 19.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–29. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 22.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogeneticinference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 23.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–56. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 24.Page RDM. TREEVIEW: an application to display phylogenetictrees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 25.Fomena A, Bouix G. Myxosporea (Protozoa: Myxozoa) of freshwater fishes in Africa: keys to genera and species. Syst Parasitol. 1997;37:161–78. doi: 10.1023/A:1005839220014. [DOI] [Google Scholar]
- 26.Kalavati C, Nandi NC. Handbook on myxosporean parasites of Indian fishes. Kolkata: Zoological Survey of India; 2007. [Google Scholar]
- 27.Carriero MM, Adriano EA, Silva MRM, Ceccarelli PS, Maia AAM. Molecular phylogeny of the Myxobolus and Henneguya genera with several new South American species. PLos One. 2013;8:1–12. doi: 10.1371/journal.pone.0073713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shulman SS. Myxosporidii fauny SSSR. Leningrad: Nauka; 1966 (In Russian).
- 29.Dyková I, Fiala I, Nie P. Myxobolus lentisuturalis n. sp. (Myxozoa: Myxobolidae), a new muscle-infecting species from the Prussian carp, Carassius gibelio from China. Folia Parasitol. 2002;49:253–8. doi: 10.14411/fp.2002.048. [DOI] [PubMed] [Google Scholar]
- 30.Molnár K, Ranzani–Paiva MJ, Eiras JC, Rodrigues EL. Myxobolus macroplasmodialis n. sp. (Myxozoa: Myxosporea), a parasite of the abdominal cavity of the Characid Teleost, Salminus maxillosus in Brazil. Acta Protozool. 1998;37:241–5. [Google Scholar]
- 31.Fiala I, Bartošová–Sojková P, Okamura B, Hartikainen H. Adaptive radiation and evolution within the Myxozoa. In: Okamura B, Gruhl A, Bartholomew JL, editors, Myxozoan Evolution, Ecology and Development. Cham: Springer International Publishing; 2015. pp. 69–84.
- 32.Eszterbauer E. Genetic relationships among gill-infecting Myxobolus species (Myxosporea) of cyprinids: molecular evidence of importance of tissue-specificity. Dis Aquat Org. 2004;58:35–40. [DOI] [PubMed]
- 33.Urawa S, Freeman MA, Johnson SC, Jones SRM, Yokoyama H. Geographical variation in spore morphology, gene sequences, and host specificity of Myxobolus arcticus (Myxozoa) infecting salmonid nerve tissues. Dis Aquat Org. 2011;96:229–37. doi: 10.3354/dao02398. [DOI] [PubMed] [Google Scholar]
- 34.Zhao YJ, Li NN, Tang FH, Dong JL. Remarks on the validity of Myxobolus ampullicapsulatus and Myxobolus honghuensis (Myxozoa: Myxosporea) based on SSU rDNA sequences. Parasitol Res. 2013;112:3817–23. doi: 10.1007/s00436-013-3569-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whipps CM, Kent ML. Phylogeography of the cosmopolitan marine parasite Kudoa thyrsites (Myxozoa: Myxosporea) J Eukaryot. Microbiol. 2006;53:364–73. doi: 10.1111/j.1550-7408.2006.00114.x. [DOI] [PubMed] [Google Scholar]
- 36.Henderson M, Okamura B. The phylogeography of salmonid proliferative kidney disease Europe and North America. Proc Biol Sci. 2004;271:1729–36. doi: 10.1098/rspb.2004.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vetešník L, Papoušek I, Halačka K, Lusková V, Mendel J. Morphometric and genetic analysis of Carassius auratus complex from an artificial wetland in Morava River floodplain, Czech Republic. Fisheries Sci. 2007;73:817–22. doi: 10.1111/j.1444-2906.2007.01401.x. [DOI] [Google Scholar]
- 38.Jones SRM, Bartholomew JL, Zhang JY. Mitigating myxozoan disease impacts on wild fish populations. In: Okamura B, Gruhl A, Bartholomew JL, editors, Myxozoan Evolution, Ecology and Development. Cham: Springer International Publishing; 2015 pp. 397–414.
- 39.Ye LT. Morphology, molecular phylogenetics and seasonality of several myxosporean species from the common carp (Cyprnius carpio) and Crucian carp (Carassius auratus). Beijing: Doctoral dissertation, Chinese Academy of Sciences; 2012 (In Chinese with English abstract).
- 40.Bahri S. Abnormal forms of Myxobolus bizerti and Myxobolus mülleri (Myxosporea: Bivalvulida) spores with caudal appendages. Bull Eur Assoc Fish Pathol. 2008;28:252–55. [Google Scholar]
- 41.Liu XH, Zhang JY, Batueva MD, Voronin VN. Supplemental description and molecular characterisation of Myxobolus miyarii Kudo, 1919 (Myxosporea: Myxobolidae) infecting intestine of Amur catfish (Silurus asotus) Parasitol Res. 2016;115:1297–1306. doi: 10.1007/s00436-015-4866-4. [DOI] [PubMed] [Google Scholar]


