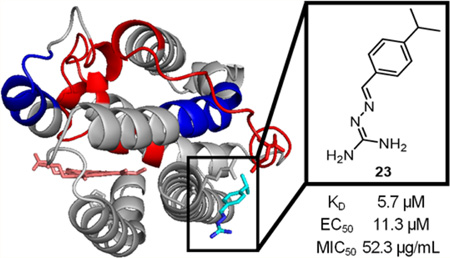
Abstract
New therapeutic targets are required to combat multidrug resistant infections, such as the iron-regulated heme oxygenase (HemO) of Pseudomonas aeruginosa, due to links between iron and virulence and dependence on heme as an iron source during infection. Herein we report the synthesis and activity of a series of iminoguanidine-based inhibitors of HemO. Compound 23 showed a binding affinity of 5.7 µM and an MIC50 of 52.3 µg/mL against P. aeruginosa PAO1. An in cellulo activity assay was developed by coupling HemO activity to a biliverdin-IXα-dependent infrared fluorescent protein, in which compound 23 showed an EC50 of 11.3 µM. The compounds showed increased activity against clinical isolates of P. aeruginosa, further confirming the target pathway. This class of inhibitors acts by binding to an allosteric site; the novel binding site is proposed in silico and supported by saturation transfer difference (STD) NMR as well as by hydrogen exchange mass spectrometry (HXMS).
Graphical Abstract
Since the introduction of antimicrobials into the clinic, resistant strains of pathogenic microbes have developed rapidly regardless of the mechanism of action of the drug class.1 The major approved antimicrobial drug classes target essential bacterial cell functions including cell wall formation,2 protein synthesis,3 and DNA synthesis.4 Targeting such essential cellular processes places the bacterial cell under significant selective pressure to acquire mutations, accelerating the development of drug resistance.5,6 To combat the decline in viable antibiotics and growing antimicrobial resistance, an alternative hypothesis suggests targeting virulence factors that are not required for survival but are essential for pathogenesis within the host. Potential antivirulence targets include quorum sensing,7 secretion systems,8 and production of exotoxins and biofilms.9 Inhibition of these virulence factors would render pathogens less infectious while preventing the stress for survival that leads to resistance.10–13
Iron is an essential micronutrient for survival and patho-genesis of almost all bacterial pathogens and is tightly regulated by the iron-regulatory protein Fur.14–16 Gram-negative bacteria acquire iron via secretion and uptake of high-affinity iron chelators called siderophores. Previous research in the area of antivirulence development focuses on the inhibition of siderophore-mediated iron-uptake;17–19 while initial therapies proved ineffective at inhibiting iron availability, current “Trojan horse” strategies using siderophore–β-lactam conjugates show improved uptake and antimicrobial activity in a wide range of Gram-negative pathogens.20–22 Although these compounds achieve cellular uptake, they rely on chelated ferric iron to promote active transport. As a result, these compounds would prove ineffective against pathogens that may have adopted to utilize ferrous iron and/or heme as a primary iron source within the host.
P. aeruginosa, a common nosocomial and drug-resistant pathogen, encodes a well-defined heme uptake and utilization system including the outer-membrane receptor PhuR, the periplasmic transfer protein PhuT, the inner membrane ABC transporter complex PhuUV, and the cytoplasmic chaperone PhuS.23,24 Heme uptake concludes with the oxidative cleavage of heme by the iron-regulated heme oxygenase (HemO), releasing Fe3+, CO(g), and β- and δ-biliverdin.25 The importance of heme utilization by chronic infections of P. aeruginosa has been validated by recent studies showing that during chronic infection of the lung longitudinal clonal isolates adapt to become more efficient at heme uptake.26 Furthermore, genetic analyses of isolates from patients with chronic lung infection have identified mutations within the promoter of the PhuR transporter that upregulate its expression.27 Additionally, we have recently shown in a PAO1 ΔhemO deletion strain that a catalytically active HemO is required to drive heme flux into the cell.28 Taken together, the experimental evidence suggests that HemO represents a viable target for the development of antivirulants targeting P. aeruginosa.
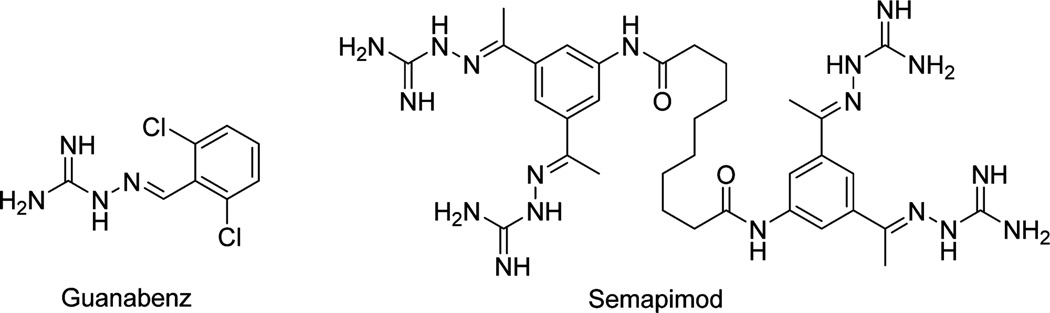
Previous work in the Wilks lab has explored HemO as a target for inhibiting heme utilization. Multiple scaffolds have been identified via a virtual screen that produced compounds with inhibitory activity both in vitro and in vivo,29 including the iminoguanidine compound 1 (Table 1).30 In addition to a modest binding affinity for HemO (31.2 µM) and antimicrobial activity against P. aeruginosa (MIC50 = 250 µg/mL), compound 1 also exhibited curing activity in a Caenorhabditis elegans infection model.30 Iminoguanidines have found application in a number of therapeutic agents. For example, iminoguanidine-based α-2 agonist guanabenz (Figure 1) is approved by the FDA for the treatment of hypertension;31 another iminoguanidine-containing compound, semapimod, has been developed as an anti-inflammatory and antiparasitic agent.32–34 Iminoguanidines have also been evaluated for their biological activities in different areas including anticoagulation,35 pain management,36 cancer therapy,37 and management of alcohol dependence.38 Current work explores inhibitory and antimicrobial activities of iminoguanidines (analogues of 1) and their activities in clinical isolates of P. aeruginosa. Further, binding modes of analogues of 1 are suggested by in silico methodologies and experimentally validated structure–activity relationships are confirmed through saturation transfer difference (STD) NMR and hydrogen– deuterium exchange mass spectrometry (HXMS).
Table 1.
Dissociation Constants, KD, and Effective Concentrations, EC50, for Compounds 1–25
| compd | R1 | R2 | R3 | KD (µM) | EC50 (µM) |
|---|---|---|---|---|---|
| 1 | H | H | N(CH3)2 | 8.8 ± 0.8 | 123 |
| 2 | F | H | H | 11 ± 1.4 | >200 |
| 3 | H | F | H | 12 ± 1.5 | 45.1 |
| 4 | H | H | F | 14 ± 2.0 | 77.5 |
| 5 | Cl | H | H | 11 ± 1.6 | 11.9 |
| 6 | H | Cl | H | 8.4 ± 0.6 | 19.4 |
| 7 | H | H | Cl | 7.0 ± 0.7 | 122 |
| 8 | H | Cl | Cl | 8.4 ± 0.7 | 20.3 |
| 9 | Br | H | H | 6.7 ± 0.4 | 35.3 |
| 10 | H | Br | H | 12 ± 1.2 | 11.2 |
| 11 | H | H | Br | 15 ± 3.1 | 35.6 |
| 12 | OH | H | H | 9.2 ± 0.8 | 102 |
| 13 | H | OH | H | 21 ± 2.2 | >200 |
| 14 | H | H | OH | 4.8 ± 0.5 | >200 |
| 15 | H | OH, OHa | H | 8.0 ± 1.3 | >200 |
| 16 | OH | Br | H | 8.0 ± 0.7 | 60.3 |
| 17 | OCH3 | H | H | NDb | 72.7 |
| 18 | H | OCH3 | H | NDb | NDb |
| 19 | H | H | OCH3 | 7.8 ± 0.9 | 77.4 |
| 20 | Naphc | Naphc | H | 12 ± 1.8 | 52.4 |
| 21 | H | Naphc | Naphc | NDb | 43.1 |
| 22 | H | H | Ph | 6.7 ± 0.4 | 21.9 |
| 23 | H | H | i-Pr | 5.7 ± 1.0 | 11.3 |
| 24 | H | OBn | H | NDb | 34.3 |
| 25 | H | H | NO2 | 6.2 ± 0.6 | 64.4 |
3,5-Disubstituted.
Not determined.
Indicates bridging carbons.
Figure 1.
Molecular structures of guanabenz, an FDA-approved compound for the treatment of hypertension, and semapimod, an investigational compound with anti-inflammatory and antiparasitic properties. Both compounds contain the iminoguanidine moiety found in 1 and its analogues.
RESULTS AND DISCUSSION
Chemical Synthesis
Target compounds 1–25 (Table 1) were synthesized using a one-step condensation reaction (Scheme 1) of aminoguanidine hydrochloride and selected benzaldehyde derivatives. The products were isolated as hydrochloride salts. Although synthesis of compound 1 was reported as early as 1933,39 more recent methodologies perform the reaction in refluxing methanol followed by filtration, resulting in a monosalt and reducing the appearance of the reported biguanide side product.40 This reaction provides good yields for most aromatic substitutions, but product may be lost upon filtration for more lipophilic compounds such as compounds 22–25.
Scheme 1.
Synthesis of Inhibitors 1–25
Computational Analysis by Site Identification by Ligand Competitive Saturation (SILCS) and Docking Studies
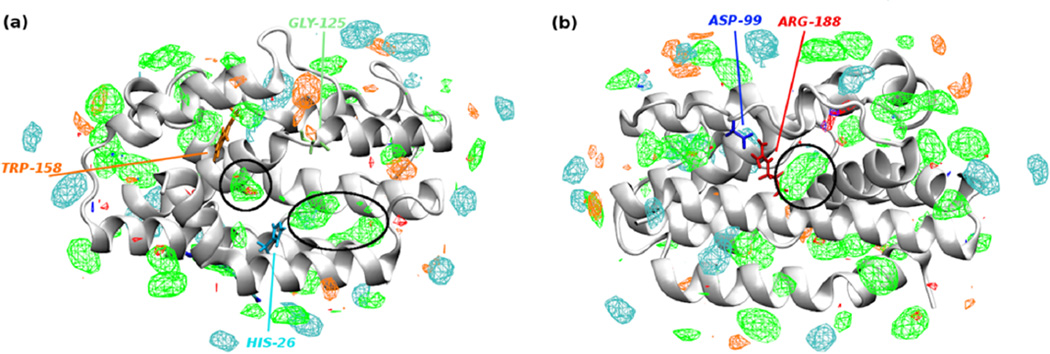
In order to instruct future inhibitor design, HemO was modeled using SILCS.41 Fragment maps, or FragMaps, which represent the binding affinity pattern of functional groups with the entire protein surface, were generated based on MD simulations of HemO in aqueous solution containing a variety of small organic solutes, such as benzene, propane, and methanol. As shown in Figure 2, when contoured at grid free energy (GFE) cutoffs of −1.2 kcal/mol, regions of significant affinity for functional group binding can be seen both in the heme-binding pocket and on the back side of the HemO. Probes of high hydrophilicity, as indicated by hydrogen-bond donor and acceptor maps, show fewer areas of favorable binding compared to their hydrophobic counterparts. The heme-binding pocket shows two distinct binding areas for hydrophobic compounds, one of which overlaps with the binding areas previously suggested for small molecules by NMR shifts.30 This area contains the catalytic site where the α-meso carbon of heme is hydroxylated and later removed as CO during the degradation of heme. The second site near Trp-158 suggests a small area appropriate for binding inhibitors; while this small site might be sampled by inhibitors, as also suggested by previous NMR studies showing perturbations of Phe-117 and Val-118,30 the utility for further inhibitor design is limited based on its small size. Another site of interest appears near Arg-188 and Trp-115 (Figure 2, right). The area of this potential alternative pocket suggests an alternative and previously unexplored mode of binding to HemO and modulation of HemO activity. The apolar FragMap highlights the high hydrophobic character of compounds that could bind to that site, and the nearby salt bridge between Arg-188 and Asp-99 suggests areas of hydrogen bonding or electrostatic interactions for polar moieties, such as the iminoguanidyl group present in compounds 1–25.
Figure 2.
Fragment maps from SILCS. Potential small-molecule binding sites of HemO are identified in silico using small molecule probes. Apolar, hydrogen bond donor and acceptor, negative and positive charged FragMaps are shown in green, blue, red, orange, and cyan colors and contoured at −1.2, −1.2, −1.2, −1.5, and −1.5 kcal/mol, respectively. The heme-binding face of HemO (indicated by His-26, panel a) and the back face of HemO (indicated by Asp-99 and Arg-188, panel b) show putative hydrophobic binding sites.
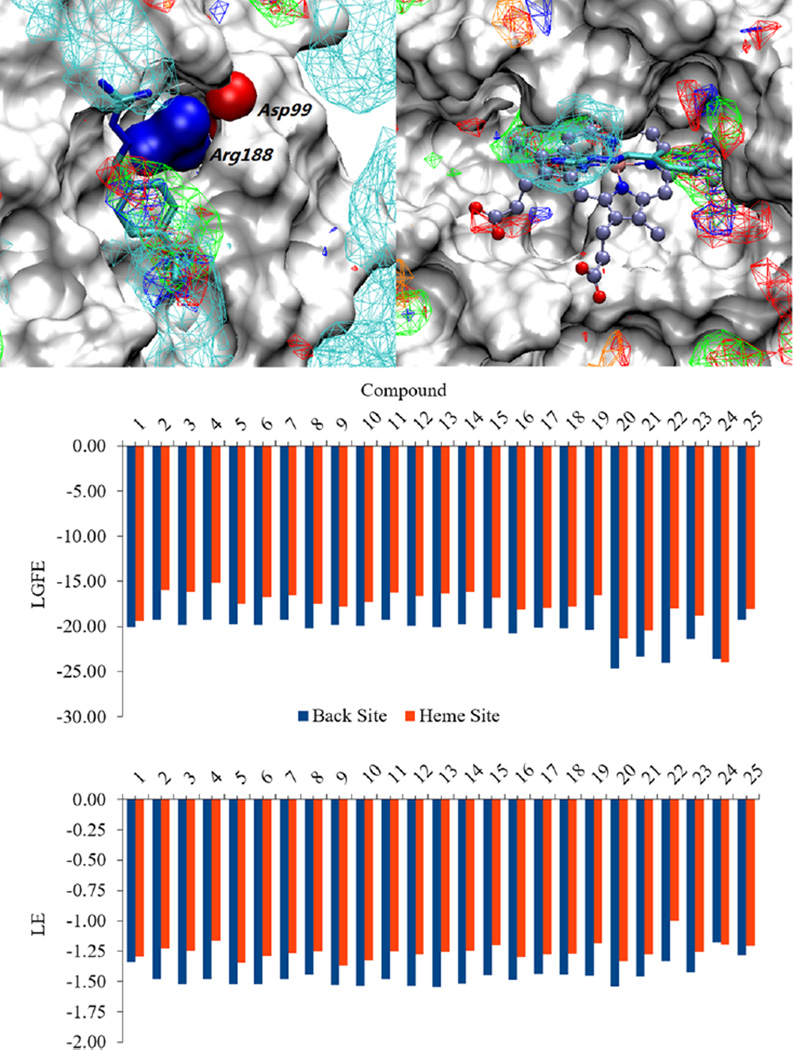
In order to further examine the ability of the compounds to bind to the proposed allosteric site of HemO, compounds 1– 25 were docked onto both the heme-binding site and the allosteric site using the Monte Carlo site identification by ligand competitive saturation (MC-SILCS) protocol.42 Figure 3 shows the predicted binding orientation of compound 23, as well as overlap with selected FragMaps for both the heme-binding site (upper right) and the allosteric site (upper left). Overlap with the FragMaps in the heme-binding pocket is poor, while strong overlap in the allosteric site indicates preferred binding at the putative site. Overlap of the aromatic ring and isopropyl groups with the apolar FragMaps and of the guanidinium moiety with the positive FragMaps show these moieties to make favorable contributions to binding in the site. The preference for the putative site is quantified by the ligand-grid free energy (LGFE) scores of the ligands calculated for both the heme-binding site and the allosteric site (Figure 3, center). All compounds except compound 24 are predicted to have greater affinity for the allosteric site over the heme-binding pocket. Additionally, when the ligand efficiencies (LE, the LGFE divided by the number of non-hydrogen atoms) are calculated for the ligands in both sites, all compounds except compound 24 are again predicted to interact more favorably with the allosteric site (Figure 3, bottom). Compound 24 also shows the least efficient binding, suggesting a lack of substantial contributions from its benzyloxy substitution. Validation of these computational results will provide additional evidence of an alternative interaction with HemO instead of competitive inhibition at the heme-binding pocket as previously proposed.
Figure 3.
Computational docking and scoring of compounds binding to HemO on the back site and the heme site. Compound 23 is shown docked to the proposed alternative binding site (upper left) as well as the heme binding site (upper right) overlaid with SILCS FragMaps generated for HemO. FragMaps are colored and contoured at −1.2 kcal/mol for the apolar and negative FragMaps and at −0.6 kcal/mol for the positive, hydrogen bond donor, and hydrogen bond acceptor FragMaps. LGFE scores (kcal/mol), based on the Monte Carlo docking of the ligands into the SILCS FragMaps, were calculated as an estimate of the affinity of the ligands for the two binding sites (middle). LE (kcal/mol/atom), which are calculated as the LGFE per non-hydrogen atom, are also shown (bottom).
Binding Studies by Fluorescence and STD NMR
Inhibitors 1–25 were tested for binding to HemO by intrinsic fluorescence quenching, as performed previously.29 Optical interference in these measurements due to high absorptivity of the inhibitors around 250–350 nm was corrected as suggested using experimentally determined extinction coefficients.43 As shown in Table 1, all compounds showed binding affinities between 4 and 20 µM regardless of aromatic substitution, except for compounds 17, 18, 21, and 24, which exhibited small-molecule fluorescence that interfered with the assay. Previous STD NMR studies30 suggested that aromatic protons contributed most to the inhibitor–HemO interaction; however all mono- and disubstituted compounds tested showed similar binding by fluorescence quenching. If the compounds bind in the active site cavity and given the relatively larger size of heme, the native substrate, it was predicted that substitutions allowing for more van der Waals contacts or aromatic stacking interactions (compounds 17–24) would show increased binding affinity compared to compound 1. Without any observed increase or decrease in binding affinity caused by aromatic substitution, it can be assumed that an alternative binding mode contributes to the binding of these compounds to HemO.
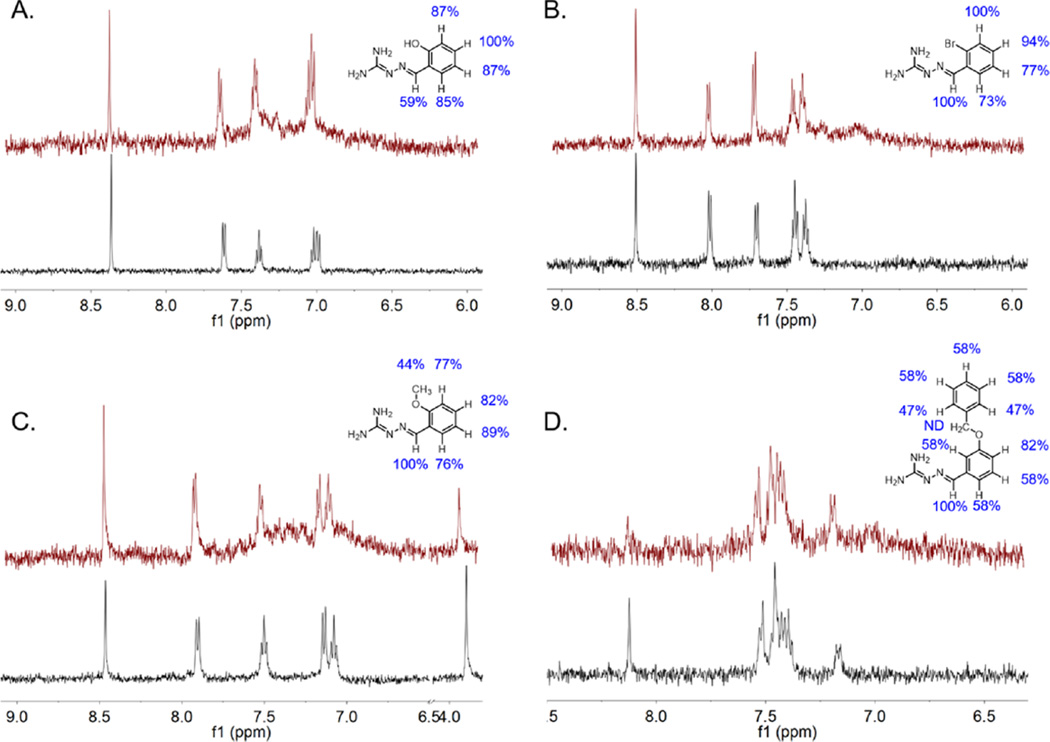
In order to further study this series of compounds with regard to binding mode, analysis of direct interaction between select compounds and HemO was performed by STD NMR.30 Selected compounds included those containing hydrophilic or lipophilic substitutions and individually resolved protons (9, 12, and 17) as well as the largest substitution, compound 24. Integration of the difference peaks allows for comparison of relative magnetization transfer, and therefore degree of interaction, of each proton with HemO (Figure 4). Compound 12 (part A) appears to interact most strongly at the paraposition of the benzyl ring, and the relative interaction decreases around the ring to the benzylimine proton at 59% that of the para-proton. However, compounds 9, 17, and 24 (parts B, C, and D, respectively) display the strongest interaction at the benzylimine proton. Compounds 9 and 17 both show good interactions with the aromatic protons, with equal relative interaction at the meta-position of compound 9 to the benzylimine proton. Additionally, the methoxy protons of compound 17 show much less interaction with HemO than the aromatic protons, much like the N-methyl protons of compound 1.30 The largest compound, 24, contains two sets of aromatic protons with overlapping signals that cannot be resolved. The two unique signals, for the benzylimine proton and the para-position proton, show the largest relative interactions at 100% and 82%, respectively. The methylene protons could not be detected due to the Watergate pulse sequence used to reduce the background water signal. Still, the available relative interactions suggest that the benzylimine ring interacts more directly with HemO than the benzyloxy ring, much like the other compounds studied in this series. Little change in both binding affinity and interaction via STD NMR for all compounds regardless of substitution(s) suggests that the compounds are not interacting in the heme-binding pocket, as previously proposed. The variety of functional groups in compounds 1–25, chosen for their hydrogen bonding, lipophilic, or steric properties, should interact differently with the mostly hydrophobic heme-binding pocket of HemO. If instead the iminoguanidyl moieties of the compounds contribute a majority of the interaction with HemO, then any sufficiently tolerated aromatic substitution will allow the compound to bind and give the results seen in Table 1, as well as the lack of substitution contributions to binding by STD NMR seen in Figure 3. The additional benzyloxy ring of compound 24 could also make contacts with the secondary binding area in the heme-binding site, increasing its preference for the heme-binding site over the allosteric site. These SILCS and docking studies propose an allosteric site of binding that matches the relatively flat SAR suggested by binding and STD NMR studies while also matching the NMR data previously shown upon binding of compound 1 to HemO in which residues Arg-188 and Trp-115 are significantly shifted.30
Figure 4.
Binding epitopes of selected compounds [(A) 12; (B) 9; (C) 17; (D) 24] determined by STD NMR. For each compound, traditional pulse proton spectra are shown below in black and the difference spectra are shown above in red.
HXMS of HemO with Selected Compounds
We further investigated the interaction of compounds 1 and 22 by HXMS. Peptide coverage following hydrogen–deuterium exchange and pepsin digestion was approximately 98% with 7-fold redundancy (Figure S1 in Supporting Information). HXMS, in addition to providing confirmation of the ligand binding site on reduced hydrogen exchange, can also give insight on the global effects of ligand binding that may include deuteration enhancement.44 Observation of increased deuterium exchange is often a result of ligand binding causing unfavorable structural change in the protein. As previously observed by chemical shift perturbations of apo-HemO on binding of compound 1, the surface loop containing Asp-99, which forms a salt bridge with Arg-188 and the helices packing over the distal helix (residues 76–84 and 92–102), shows reduced hydrogen exchange indicative of protection upon compound binding (Figure S2). The data are consistent with the SILCS analysis and previous STD NMR that compound 1 most likely binds at an alternative hydrophobic site on the protein surface.
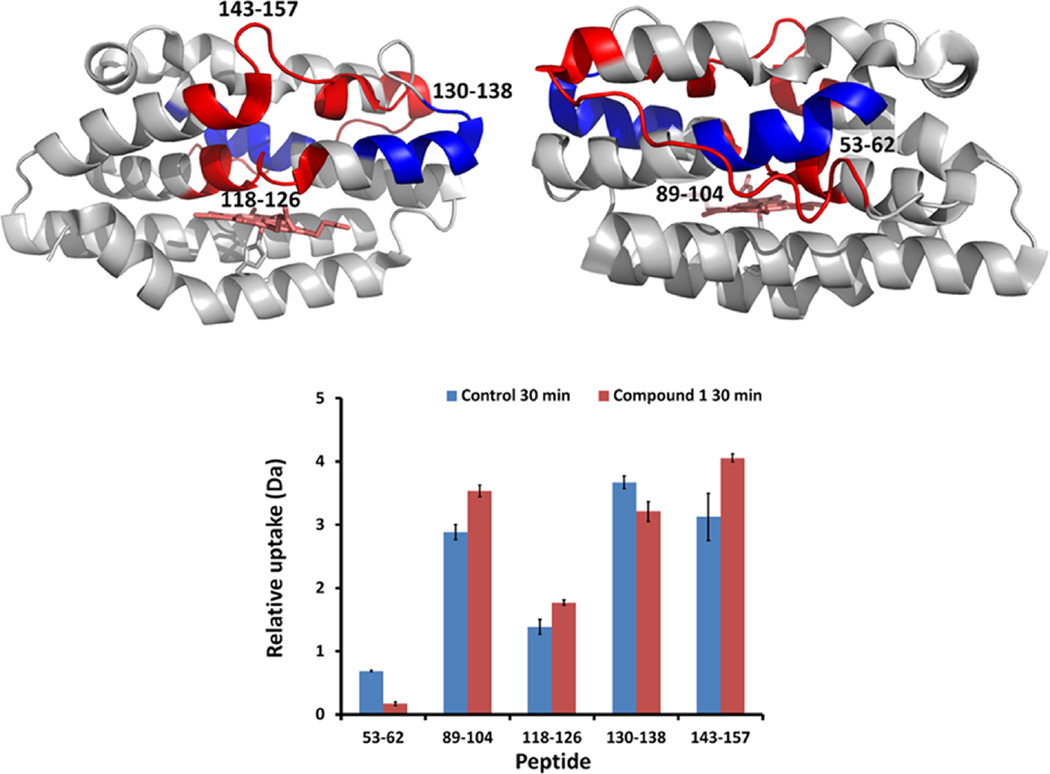
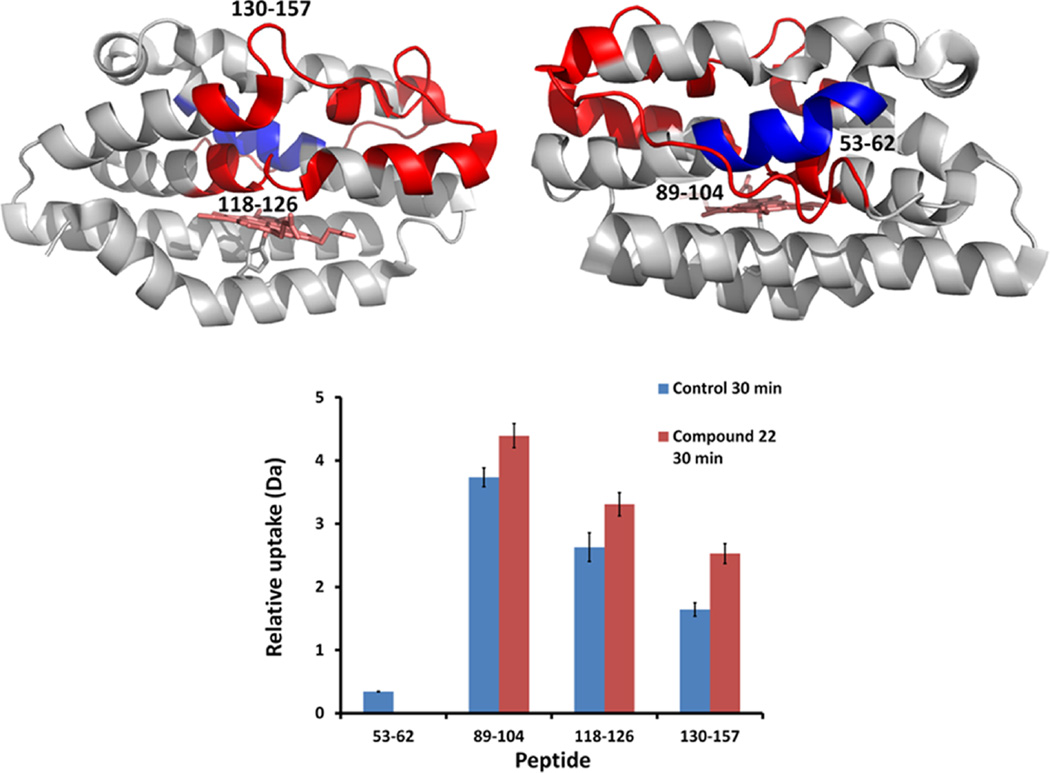
Interestingly, compound 1 in holo-HemO shows differences in hydrogen exchange within several of the same regions but where the overall effect is to increase hydrogen exchange within the distal helices (Figure 5). Much of the same hydrogen exchange profile on holo-HemO is also seen with compound 22 (Figure 6), whereas little-to-no change in hydrogen exchange is seen with compound 22 on apo-HemO. These data agree with the alternative binding site identified by SILCS and the compounds’ proposed preferences for binding to the allosteric site, marked by the signification protection of peptide 53–62. Further, the protection of this peptide compared to holo-HemO shows interactions with HemO when the heme site is occupied, pointing again to the allosteric binding site. The addition of lipophilic groups in compounds 20–24 showed an overall improvement across the spectrum of biological assays suggesting these inhibitors are causing global effects on protein stability. Extended interaction of the lipophilic substituents with the hydrophobic pocket may perturb the binding pocket adjacent to the salt bridge. Although we cannot be certain that the iminoguanidine group disrupts the salt bridge, mutation of either Asp-99 or Arg-188 results in a similar pattern of hydrogen exchange (data not shown), highlighting the importance of this region in maintaining structural integrity and enzymatic activity. The binding of ligands to the allosteric site appears to “loosen up” the distal pocket, which possibly results in disruption of the hydrogen bond network critical in stabilizing the activated oxygen intermediate required for initial hydroxylation of the heme.45
Figure 5.
Color coded protein structure of HemO representing deuterium uptake change upon binding of compound 1. Deuterium incorporation was mapped onto the HemO crystal structure (PDB code 1SK7) as viewed from front (heme-binding pocket, upper left) and from back (upper right). Peptide regions of holo-HemO that were more protected from deuterium exchange in the presence of 1 are colored in blue, whereas regions that became more prone to deuterium exchange are colored in red. Relative deuterium uptake of peptides showed differences between holo-HemO and holo-HemO in the presence of 1 at 30 min (bottom). Two peptides (53–62 and 130–138) were significantly more protected from deuterium exchange upon binding of 1 (Student’s t-test, p < 0.05n = 2), while other peptides showed significantly increased deuterium uptake (p < 0.05). Error bar indicates standard deviation.
Figure 6.
Color coded protein structure of HemO representing deuterium uptake change upon binding of compound 22. Deuterium incorporation perturbation data were mapped onto the HemO crystal structure (PDB code 1SK7) as viewed from front (heme-binding pocket, upper left) and from back (upper right). Peptide regions of 22-bound holo-HemO that were more protected from deuterium exchange are colored in blue, whereas regions that became more prone to deuterium exchange were colored in red. Relative deuterium uptake of peptides showed differences between holo-HemO and 22-bound holo-HemO at 30 min (bottom). One peptide (53–62) was significantly more protected from deuterium exchange upon binding of 22 (Student’s t-test, p < 0.05, n = 2), while other peptides showed significantly increased deuterium uptake (p < 0.05). Error bar indicates standard deviation.
In Cellulo HemO Inhibitory Activity of Compounds
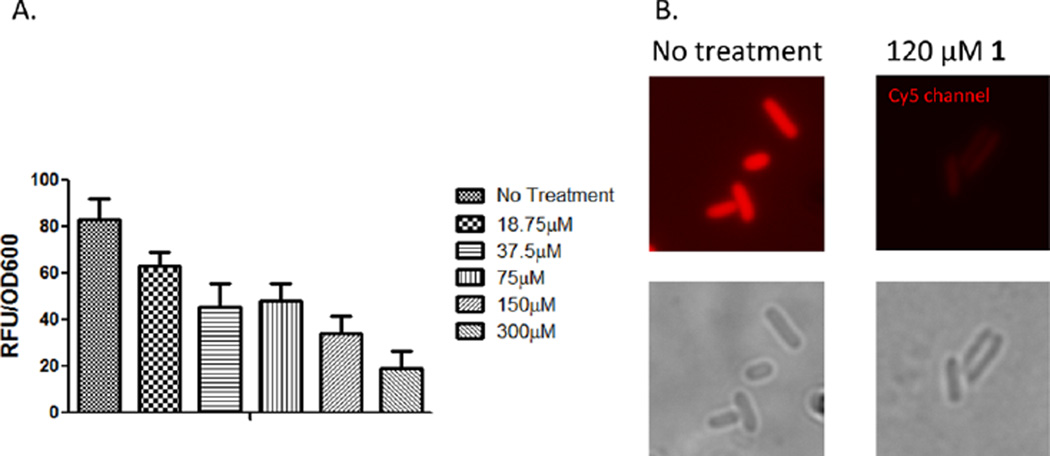
In order to assess the abilities of compounds 1–25 to inhibit the activity of HemO, it was necessary to develop a new, high-throughput assay. Current methods for measuring HO catalytic activity rely on spectroscopic measurements of bilirubin.46 As the Gram-negative HemO enzymes do not release biliverdin to eukaryotic biliverdin reductase enzymes, it is not possible to perform multiple turnover reactions in vitro. Instead, inspired by the work with bacterial phytochromes by Filonov47 and Shu,48 an in cellulo system coexpressing HemO and an engineered infrared fluorescent protein (IFP) was developed to assay inhibition of HemO activity in bacterial cells. The advantages of the assay are several including (i) a measure of the ability of the compounds to cross the bacterial cell membrane, (ii) ability to inhibit HemO activity, and (iii) lack of toxicity to E. coli cells that do not require a HemO for survival. Inhibition of HemO activity prevents the formation of biliverdin, which is measured spectrophotometrically as a decrease in fluorescence of the biliverdin-dependent IFP1.4. The in cellulo assay was optimized so that expression of IFP1.4 is not rate limiting (Figure S3A) and a linear range of fluorescence is obtained (Figure S3B). Utilizing the in cellulo assay, the concentration-dependent inhibition of HemO was measured for compound 1 as a function of decreased fluorescence over time, corrected for OD600 to account for differences in growth (Figure S3B). In addition the in cellulo assay provides a mechanism to screen out inhibitors with nonspecific cell toxicity versus HemO inhibitory activity, an important consideration for developing inhibitors specific to the invading pathogen versus the host microbial flora. We confirmed the concentration-dependent inhibition of HemO by compound 1 was not a consequence of cell toxicity (the cells grew at a similar rate when treated with compound 1) by microscopic analysis of the untreated and treated cultures (Figure 7). For routine screening of HemO inhibition the assay was adapted to a 96-well plate format and the fluorescence emission at 700 nm measured at mid log phase (16 h). The EC50 for the compounds was calculated as described in the methods, and the EC50 values for compounds 1–25 are shown in Table 1.
Figure 7.
In cellulo fluorescence quenching of HemO activity. (A) Concentration dependent inhibition of HemO activity by compound 1 as measured by biliverdin-dependent IFP1.4 fluorescence. Experiments were performed as described in Experimental Section. All experiments were performed in triplicate and averaged. Relative fluorescence was corrected for OD600, and values represent the average of three separate experiments. (B) In situ detection of the biliverdin-IFP1.4 within live bacteria imaged on the Cy5 channel of a fluorescence microscope in the absence (upper left panel) or presence of 120 µM compound 1 (upper right panel). Phase contrast images of the bacterial cells are shown in the lower panels.
Unlike the binding constants, the EC50 data comprise a wide range from 11.9 µM up to >200 µM, the highest concentration tested. Generally, more lipophilic compounds showed increased potency compared to the parent compound 1, while more hydrophilic compounds showed decreased potency. In addition to the change in polarity of the various functional groups tested, the position on the benzylidene ring also contributed to potency. For example, compounds 5 and 6 show potent inhibition with chloro substitutions on the 2′ and 3′ positions, whereas compound 7, the 4′-chloro substitution, shows inhibition similar to parent compound 1. Potency is restored with the 3′,4′ disubstituted compound 8. The reverse trend is seen in the hydroxyl substituted compounds. Compounds 13 and 14, substituted at the 3′ and 4′ positions, respectively, show poor potency (>200 µM), whereas the 2′ substituted compound 12 maintains activity similar to the parent compound 1. The inhibitory activity of compound 12 in contrast to 13 and 14 may be due to an internal hydrogen bond between the hydroxyl substituent and the imine nitrogen from the iminoguanidine moiety of 12, masking its hydrophilic character and increasing its ability to cross the Gram-negative cell membrane. Inhibitory activity is further increased in compound 16 with an additional bromo substitution at the 3′ position. Thus, the in cellulo assay allows a rapid ranking of compounds based on inhibitory activity and the ability to cross the Gram-negative cell membrane, confirming earlier data showing reduced biliverdin levels in HemO overexpressing E. coli treated with compound 1.29
Antimicrobial Activity of Compounds
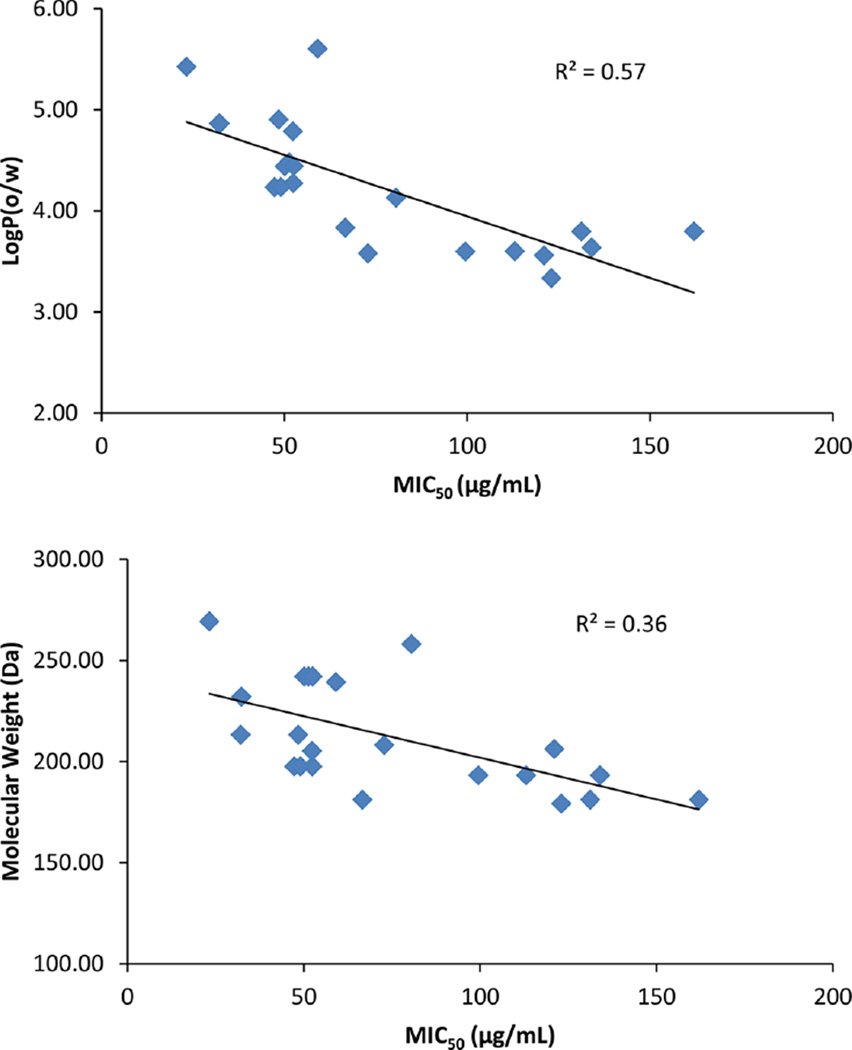
Compounds 1–25 were tested for antimicrobial activity against PAO1, a laboratory strain of P. aeruginosa, as well as JSRI-1 and JSRI-2, longitudinal clinical isolates of P. aeruginosa from a cystic fibrosis patient.26 The minimum inhibitory concentration for 50% growth (MIC50) showed great variation depending upon the substitutions of the aromatic ring as shown in Table 3. Generally, increased lipophilicity led to more potent inhibition of growth, as exemplified by compounds 20–24, whereas increased hydrophilicity (13–15) greatly reduced the compounds’ abilities to inhibit growth. Compounds 12 and 16 showed moderate activity, much like in the in cellulo assay shown in Table 1. Halogen substitutions showed mildly increased activity, and the dichloro-substituted inhibitor 8 showed equal potency to the lipophilic compounds 20–24. The relationship between MIC50 and lipophilicity (as determined by log P(o/w)) for these compounds is shown in Figure 8. Correlations are seen indicating that physical properties of the compounds related to membrane permeability affecting MIC50, as well as EC50, are more important for activity than the in vitro binding affinities given the weak correlation between KD and MIC50.
Table 3.
MIC50 and MBC of Compounds 1–25 in PAO1 and Clinical Isolates
| MIC50 (µg/mL)a |
MBC (µg/mL)b |
|||||
|---|---|---|---|---|---|---|
| compd | PAO1 | JSRI 1 | JSRI 2 | PAO1 | JSRI 1 | JSRI 2 |
| 1 | 121 | 32.5 | 15.5 | 1000 | 1000 | ≤250 |
| 2 | 131.2 | 28.6 | 72.9 | 1000 | >1000 | ≤250 |
| 3 | 66.6 | 70 | 68.2 | 1000 | 1000 | 250 |
| 4 | 162 | 87.4 | 69.4 | 1000 | 1000 | 500 |
| 5 | 47.3 | 16.3 | 35 | 500 | 500 | 250 |
| 6 | 52.4 | 19.2 | 33.5 | 250 | 500 | 250 |
| 7 | 49 | 10.7 | 41.6 | 500 | 250 | ≤62.5 |
| 8 | 32.3 | 24.1 | 16.4 | 125 | 125 | 125 |
| 9 | 50.1 | 62.9 | 33.4 | 500 | 500 | 250 |
| 10 | 51.3 | 23 | 19.4 | 250 | 500 | ≤125 |
| 11 | 52.5 | 19.7 | 13.4 | 250 | 250 | ≤62.5 |
| 12 | 123 | 58.5 | 79.1 | >1000 | >1000 | 250 |
| 13 | 750 | 391 | 617 | NDc | NDc | NDc |
| 14 | 408 | 443 | 121 | >1000 | >1000 | >1000 |
| 15 | 1140 | 338 | 564 | NDc | NDc | NDc |
| 16 | 80.5 | 145 | 45.6 | 500 | 1000 | 250 |
| 17 | 99.5 | 38.4 | 125 | 1000 | >1000 | 500 |
| 18 | 134 | 48.6 | 93.4 | >1000 | 1000 | 500 |
| 19 | 113 | 30.5 | 38 | 500 | 1000 | 500 |
| 20 | 32.1 | 30.4 | 36.1 | 250 | 250 | 125 |
| 21 | 48.4 | 31.7 | 75.6 | 250 | 250 | 125 |
| 22 | 59.1 | 34.6 | 38 | 125 | 125 | ≤62.5 |
| 23 | 52.3 | 20.6 | 81 | 125 | 125 | ≤125 |
| 24 | 23.2 | 25.5 | 33.9 | 125 | 125 | 62.5 |
| 25 | 72.8 | 61.4 | 47.2 | 1000 | 1000 | ≤125 |
Minimum inhibitory concentration for 50% growth (MIC50).
Minimum bactericidal concentration (MBC).
Not determined.
Figure 8.
Correlation between MIC50 and log P(o/w) (top) as well as molecular weights (bottom) for compounds with MIC50 lower than 200 µg/mL for PAO1. Correlation coefficients R2 are also shown.
Following MIC50 growth conditions, new cultures were inoculated to classify growth inhibition as bacteriostatic or bactericidal. Compounds with the best activity against growth via MIC50 were also shown to act in a bactericidal manner, as growth was not recovered upon greater than 1000-fold dilution of culture into new media. Again, compounds 8 and 20–24 showed the most potent bactericidal activity. Further, most compounds showed higher potency against the JSRI-2 isolate, which was previously shown to more efficiently utilize heme as an iron source,26 as well as being more resistant to antibiotics such as tobramycin and gentamycin. While bactericidal activity was not anticipated from these compounds, increased activity against the JSRI isolates versus PAO1 supports their targeting the heme utilization pathway and shows sustained activity in otherwise resistant strains.
CONCLUSIONS
Various analogues of iminoguanidine 1 were explored computationally, synthesized, tested for binding to HemO, and tested for antimicrobial activity. Although little differences were seen in binding by fluorescence and STD NMR for many of the compounds, structure–activity relationships for this series were drawn from a combination of SAR methods with in cellulo activity and antimicrobial assays. From this series, compound 23 showed the most improved overall activity, with a binding affinity of 5.7 µM, an EC50 of 11.3 µM, and an MIC50 of 52.3 µg/mL against PAO1. Unfortunately the pulses used in STD NMR do not allow for analysis of compound 23 due to its upfield isopropyl group. Compound 23 should demonstrate good selectivity for HemO over α-2 adrenergic receptors according to the known structure–activity relationship of the α-2 adrenergic agonists.49 Nguyen and co-workers report that α-2 adrenergic activity is maintained only with the 2′,6′-dicholoro substitution found in guanabenz (see Figure 1) and altered chloro substitutions abrogate agonistic activity.49 It was also seen that more lipophilic compounds 20–24 proved more active against all strains of P. aeruginosa than the hydrophilic compounds 13–15, which correlates well with their inhibitory activity in the E. coli IFP assay. Correlations shown in Figure 8 confirm the greater role of lipophilicity and cell membrane permeability than binding affinity for in vivo activity. Additionally, many compounds displayed increased activity against the clinical isolates JSRI-1 and JSRI-2, which were previously characterized as highly efficient heme-utilizing strains.
A somewhat unexpected result was the relative insensitivity of the KD values to the different functional groups, with the maximum and minimum affinities of 4.8 and 21 µM obtained, respectively. Consistent with the KD values, the LGFE scores were also relatively insensitive to the studied modifications as seen in Figure 3. To understand the molecular origins of this phenomenon, the LGFE scores determined for the allosteric site were partitioned into the contributions of different moieties in the ligands, based on the sum of the atom-GFE scores for each moiety. The allosteric-site analysis is shown in Table 2 for selected compounds. Results show that the sum of the GFEs for the phenyl and guanidinium groups becomes less favorable as the GFE contribution of the substituent becomes more favorable. This indicates a situation in which favorable interactions of the substituents lead to less favorable interactions of the parent phenyl and guanidinium moieties. With compound 20, the LGFE score is significantly more favorable; however, when the ligand efficiencies are extracted from the LGFE by dividing by the number of non-hydrogen atoms, the ligand efficiency of compound 20 falls in the range of the other compounds. This indicates that the more favorable LGFE is an artifact of the SILCS method associated with the large size of the substituent. Thus, the similarity of the affinities is indicated to be due to a “push–pull” mechanism where favorable interactions of the substituents lead to less favorable interactions of the parent compound moieties, yielding a flat SAR profile.
Table 2.
Contributions of Selected Moieties to the LGFE Scores for Selected Compounds
| compd | substituent | KD (µM) | LGFEa contributions (kcal/mol) |
total |
||||
|---|---|---|---|---|---|---|---|---|
| Phb | Guanc | Ph + Guan | Subst | LGFE | LE | |||
| 8 | 3,4-dichloro | 8.4 ± 0.7 | −13.2 | −4.9 | −18.1 | −2.1 | −20.2 | −1.44 |
| 12 | 2-hydroxyl | 9.2 ± 0.8 | −12.7 | −6.4 | −19.1 | −0.8 | −19.9 | −1.53 |
| 17 | 2-methoxyl | ND | −12.3 | −6.3 | −18.7 | −1.5 | −20.1 | −1.44 |
| 20 | 1,2-naphthyl | 12 ± 1.8 | −12.3 | −5.5 | −17.9 | −6.8 | −24.7 | −1.54 |
| 23 | 4-isopropyl | 5.7 ± 1.0 | −11.9 | −4.4 | −16.3 | −5.1 | −21.4 | −1.43 |
Ligand grid free energy.
Phenyl.
Guanidinium.
Structure–activity relationships from chemical shift perturbation NMR and HXMS studies, together with the computational SILCS methodology, suggested a novel allosteric binding site from the active site for compounds 1–25. This new binding site comprises a shallow hydrophobic cleft formed by the two helices running along the backside of the distal helix, which are further stabilized by a salt bridge formed between Asp-99 and Arg-188. The lack of improvement in binding affinities of the various compounds is consistent with a push-pull mechanism in the binding site. However, as shown by HXMS, it is possible to expand on the inhibitory activity of 1 by generating back side instability of HemO upon binding to the pocket adjacent to the Asp-99/Arg-188 salt bridge, as demonstrated by compound 22. Consistent with the proposed binding of the inhibitors to the newly identified allosteric site, we have recently shown that mutation of either Asp-99 or Arg-188 to an Ala results in poorly active HemO mutants when overexpressed in E. coli (data not shown). Finally, the alternative binding site is unique to the bacterial HemO providing for selectivity against the human HO-1 and HO-2 enzymes.
EXPERIMENTAL SECTION
General Procedures
All reagents and solvents were of analytical grade and used without further purification. NMR spectra were obtained on a Varian INOVA 400 MHz NMR spectrometer at 25 °C. Chemical shifts are reported as δ values (parts per million) using the residual solvent peak as an internal reference. Data for 1H NMR are reported in the following order: chemical shift, multiplicity (s, singlet; d, doublet; t, triplet; sept, septuplet; dd, double doublet; dt, double triplet; m, multiplet), number of protons, coupling constant (Hz). Data for 13C NMR are reported as δ values (parts per million). UV spectra were obtained on a Nanodrop 2000c spectrophotometer. High-resolution mass spectra (HRMS) were obtained on a JEOL AccuTOF with ESI/APCI ion sources coupled to an Agilent 1100 HPLC system. HPLC analysis was performed on a Shimazdu HPLC fitted with a C-18 reversed-phase column (Phenomenex, 4.6 mm × 250 mm) with a flow rate of 0.5 mL/min using CH3OH–H2O 3:1 with 0.1% NH4OAc mobile phase. The purity of final products are >95%.
General Procedure for the Synthesis of Compounds 1–25
To a solution of aminoguanidine hydrochloride (110 mg, 1.0 mmol) in methanol (5 mL) was added the corresponding benzaldehyde (1.1 mmol). Solution was heated to reflux and stirred for 6 h. Solvent was evaporated. Residue was stirred in CH2Cl2 (20 mL) and filtered.
Precipitate was washed with additional CH2Cl2 (50 mL) to generate the desired product.
(E)-2-(4-(Dimethylamino)benzylidene)hydrazine-carboximidamide Hydrochloride (1)
Yield, 80%; mp 216–217 °C. 1H NMR (DMSO-d6) δ 2.93 (s, 6H), 6.68 (d, 2H, J = 8.8 Hz), 7.60 (d, 2H, J = 8.0 Hz), 7.96 (s, 1H), 11.56 (s, 1H). 13C NMR (DMSO-d6): 111.4, 120.5, 128.8, 147.4, 151.6, 155.0. UV λmax (log ε) 346 (4.47). HRMS (ESI) m/z: calculated 206.1400 for [C10H16N5]+, found 206.1404. HPLC analysis: retention time = 7.82 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(2-Fluorobenzylidene)hydrazinecarboximidamide Hydrochloride (2)
Yield, 86%; mp 193–195 °C. 1H NMR (DMSO-d6) δ 7.29 (m, 2H), 7.52 (dt, 1H, J1 = 6.8 Hz, J2 = 6.4 Hz), 8.23 (t, 1H, J = 7.6 Hz), 8.39 (s, 1H), 12.17 (s, 1H). 13C NMR (DMSO-d6): 115.8, 116.0, 124.7, 127.0, 132.4, 132.5, 139.3, 155.3, 174.8. UV λmax (log ε) 273 (4.45). HRMS (ESI) m/z: calculated 181.0884 for [C8H10FN4]+, found 181.0891. HPLC analysis: retention time = 7.70 min; peak area, 98.8%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3-Fluorobenzylidene)hydrazinecarboximidamide Hydrochloride (3)
Yield, 77%; mp 202–203 °C. 1H NMR (DMSO-d6) δ 7.28 (dd, 1H, J1 = 8.4 Hz, J2 = 1.6 Hz), 7.49 (q, 1H, J = 6.4 Hz), 7.61 (d, 1H, J = 7.6 Hz), 7.85 (d, 1H, J = 10 Hz), 8.19 (s, 1H), 11.87 (s, 1H). 13C NMR (DMSO-d6): 113.4, 113.6, 117.6, 117.9, 125.0, 131.2, 131.3, 136.3, 136.3, 146.0, 155.6, 161.6, 164.1. UV λmax (log ε) 278 (4.56). HRMS (ESI) m/z: calculated 181.0884 for [C8H10FN4]+, found 181.0898. HPLC analysis: retention time = 7.86 min; peak area, 98.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(4-Fluorobenzylidene)hydrazinecarboximidamide Hydrochloride (4)
Yield, 49%; mp 195–197 °C. 1H NMR (DMSO-d6) δ 7.26 (t, 2H, J = 8.8 Hz), 7.91 (dt, 2H, J1 = 5.6 Hz, J2 = 2.8 Hz), 8.14 (s, 1H), 12.03 (s, 1H). 13C NMR (DMSO-d6): 116.3, 130.4, 146.0, 155.9, 162.6, 165.0. UV λmax (log ε) 277 (4.50). HRMS (ESI) m/z: calculated 181.0884 for [C8H10FN4]+, found 181.0894. HPLC analysis: retention time = 7.91 min; peak area, 98.2%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(2-Chlorobenzylidene)hydrazinecarboximidamide Hydrochloride (5)
Yield, 72%; mp 198–200 °C. 1H NMR (DMSO-d6) δ 7.42 (dt, 2H, J1 = 20.4 Hz, J2 = 1.2 Hz), 7.50 (d, 1H, J = 6.8 Hz), 8.27 (d, 1H, J = 7.2 Hz), 8.52 (s, 1H), 12.26 (s, 1H). 13C NMR (DMSO-d6): 127.9, 128.1, 130.3, 131.1, 132.4, 133.7, 143.1, 155.7. UV λmax (log ε) 279 (4.56). HRMS (ESI) m/z: calculated 197.0589 for [C8H10ClN4]+, found 197.0596. HPLC analysis: retention time = 8.61 min; peak area, 97.6%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3-Chlorobenzylidene)hydrazinecarboximidamide Hydrochloride (6)
Yield, 73%; mp 227–229 °C. 1H NMR (DMSO-d6) δ 7.49 (m, 2H), 7.77 (d, 1H, J = 7.6 Hz), 8.10 (s, 1H), 8.18 (s, 1H), 12.18 (s, 1H). 13C NMR (DMSO-d6): 126.8, 127.3, 130.5, 131.0, 134.2, 136.1, 145.6, 155.9. UV λmax (log ε) 277 (4.57). HRMS (ESI) m/z: calculated 197.0589 for [C8H10ClN4]+, found 197.0602. HPLC analysis: retention time = 9.23 min; peak area, 98.3%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(4-Chlorobenzylidene)hydrazinecarboximidamide Hydrochloride (7)
Yield, 21%; mp 146–149 °C. 1H NMR (DMSO-d6) δ 7.53 (d, 2H, J = 8.4 Hz), 7.91 (d, 2H, J = 9.2 Hz), 8.16 (s, 1H), 11.86 (s, 1H). 13C NMR (DMSO-d6): 128.7, 129.2, 132.3, 134.9, 145.4, 155.3. UV λmax (log ε) 285 (4.53). HRMS (ESI) m/z: calculated 197.0589 for [C8H10ClN4]+, found 197.0593. HPLC analysis: retention time = 9.42 min; peak area, 96.0%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3,4-Dichlorobenzylidene)hydrazinecarboximidamide Hydrochloride (8)
Yield, 58%; mp 221–223 °C. 1H NMR (DMSO-d6) δ 7.84 (dd, 1H, J1 = 8.4 Hz, J2 = 1.6 Hz), 8.17 (s, 1H), 8.29 (d, 1H, J = 0.8 Hz), 12.27 (s, 1H). 13C NMR (DMSO-d6): 128.0, 128.6, 130.9, 131.8, 132.6, 134.3, 144.1, 155.5, 159.1. UV λmax (log ε) 286 (4.47). HRMS (ESI) m/z: calculated 231.0199 for [C8H9Cl2N4]+, found 231.0206. HPLC analysis: retention time = 12.86 min; peak area, 95.0%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(2-Bromobenzylidene)hydrazinecarboximidamide Hydrochloride (9)
Yield, 52%; mp 220–221 °C. 1H NMR (DMSO-d6) δ 7.39 (t, 1H, J = 7.6 Hz), 7.46 (t, 1H, J = 8.0 Hz), 7.69 (d, 1H, J = 8.0 Hz), 8.26 (d, 1H, J = 7.6 Hz), 8.52 (s, 1H), 12.12 (s, 1H). 13C NMR (DMSO-d6): 124.1, 125.5, 128.4, 132.4, 132.7, 133.5, 145.7, 155.5. UV λmax (log ε) 281 (4.56). HRMS (ESI) m/z: calculated 241.0083 for [C8H10BrN4]+, found 241.0087. HPLC analysis: retention time = 9.31 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3-Bromobenzylidene)hydrazinecarboximidamide Hydrochloride (10)
Yield, 81%; mp 225–229 °C. 1H NMR (DMSO-d6) δ 7.39 (t, 1H, J = 7.2 Hz), 7.62 (d, 1H, J = 8.0 Hz), 7.79 (d, 1H, J = 8.0 Hz), 8.14 (s, 1H), 8.22 (s, 1H), 12.10 (s, 1H). 13C NMR (DMSO-d6): 122.7, 127.7, 129.7, 131.2, 133.4, 136.3, 145.6, 155.9. UV λmax (log ε) 281 (4.56). HRMS (ESI) m/z: calculated 241.0083 for [C8H10BrN4]+, found 241.0080. HPLC analysis: retention time = 9.61 min; peak area, 99.0%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(4-Bromobenzylidene)hydrazinecarboximidamide Hydrochloride (11)
Yield, 60%; mp 156–159 °C. 1H NMR (DMSO-d6) δ 7.61 (d, 2H, J = 8.0 Hz), 7.79 (d, 2H, J = 8.8 Hz), 8.11 (s, 1H), 8.74 (s, 1H), 12.06 (s, 1H). 13C NMR (DMSO-d6): 129.9, 132.1, 133.2, 146.0, 155.8, 159.5. UV λmax (log ε) 285 (4.53). HRMS (ESI) m/z: calculated 241.0083 for [C8H10BrN4]+, found 241.0088. HPLC analysis: retention time = 9.84 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(2-Hydroxybenzylidene)hydrazinecarboximidamide Hydrochloride (12)
Yield, 69%; mp 226–228 °C. 1H NMR (DMSO-d6) δ 6.82 (t, 1H, J = 7.2 Hz), 6.91 (d, 1H, J = 8.4 Hz), 7.23 (t, 1H, J = 7.2 Hz), 7.93 (d, 1H, J = 7.6 Hz), 8.41 (s, 1H), 10.14 (s, 1H), 11.94 (s, 1H). 13C NMR (DMSO-d6): 116.6, 119.6, 120.0, 127.1, 132.3, 143.8, 155.6, 157.0. UV λmax (log ε) 276 (4.50). HRMS (ESI) m/z: calculated 179.0927 for [C8H11N4O]+, found 179.0930. HPLC analysis: retention time = 10.24 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3-Hydroxybenzylidene)hydrazinecarboximidamide Hydrochloride (13)
Yield, 35%; mp 153–156 °C. 1H NMR (DMSO-d6) δ 6.88 (t, 1H, J = 8.0 Hz), 7.24 (m, 3H), 8.08 (d, 1H, J = 2.4 Hz), 9.71 (d, 1H, J = 3.2 Hz), 11.99 (s, 1H). 13C NMR (DMSO-d6): 114.5, 118.1, 119.0, 130.1, 135.1, 147.5, 155.8, 158.1, 159.5. UV λmax (log ε) 277 (4.41). HRMS (ESI) m/z: calculated 179.0927 for [C8H11N4O]+, found 179.0929. HPLC analysis: retention time = 6.69 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(4-Hydroxybenzylidene)hydrazinecarboximidamide Hydrochloride (14)
Yield, 83%; mp 251–253 °C. 1H NMR (DMSO-d6) δ 6.79 (d, 2H, J = 8.0 Hz), 7.64 (d, 2H, J = 8.4 Hz), 8.01 (s, 1H), 10.03 (s, 1H), 11.68 (s, 1H). 13C NMR (DMSO-d6): 116.0, 124.8, 129.8, 147.5, 155.6, 160.3. UV λmax (log ε) 287 (4.58). HRMS (ESI) m/z: calculated 179.0927 for [C8H11N4O]+, found 179.0933. HPLC analysis: retention time = 6.52 min; peak area, 99.3%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3,5-Dihydroxybenzylidene)hydrazine-carboximidamide Hydrochloride (15)
Yield, 45%; mp 258–260 °C. 1H NMR (DMSO-d6) δ 6.40 (s, 1H), 6.68 (d, 2H, J = 1.6 Hz), 7.98 (s, 1H), 9.52 (s, 2H), 11.89 (s, 1H). 13C NMR (DMSO-d6): 105.4, 106.2, 135.4, 148.0, 155.7, 159.0, 159.5. UV λmax (log ε) 286 (4.50). HRMS (ESI) m/z: calculated 195.0877 for [C8H11N4O2]+, found 195.0878. HPLC analysis: retention time = 6.25 min; peak area, 99.4%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3-Bromo-2-hydroxybenzylidene)hydrazine-carboximidamide Hydrochloride (16)
Yield, 74%; mp 263–265 °C. 1H NMR (DMSO-d6) δ 6.89 (t, 1H, J = 7.6 Hz), 7.62 (d, 1H, J = 8.0 Hz), 7.83 (d, 2H, J = 8.0 Hz), 8.43 (s, 1H), 9.94 (s, 1H), 12.13 (s, 1H). 13C NMR (DMSO-d6): 111.8, 121.4, 121.8, 128.2, 134.8, 145.4, 152.6, 155.1. UV λmax (log ε) 282 (4.55). HRMS (ESI) m/z: calculated 257.0032 for [C8H10BrN4O]+, found 257.0030. HPLC analysis: retention time = 8.81 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(2-Methoxybenzylidene)hydrazinecarboximidamide Hydrochloride (17)
Yield, 85%; mp 244–246 °C. 1H NMR (DMSO-d6) δ 3.86 (s, 3H), 7.01 (t, 1H, J = 6.8 Hz), 7.11 (d, 1H, J = 8.4 Hz), 7.44 (t, 1H, J = 8.0 Hz), 8.09 (d, 1H, J = 7.2 Hz), 8.47 (s, 1H), 12.08 (s, 1H). 13C NMR (DMSO-d6): 55.7, 111.8, 120.6, 121.2, 126.3, 132.1, 142.0, 155.3, 157.7. UV λmax (log ε) 276 (4.52). HRMS (ESI) m/z: calculated 193.1084 for [C9H13N4O]+, found 193.1095. HPLC analysis: retention time = 7.75 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3-Methoxybenzylidene)hydrazinecarboximidamide Hydrochloride (18)
Yield, 99%; mp 135–138 °C. 1H NMR (DMSO-d6) δ 3.80 (s, 3H), 7.03 (t, 1H, J = 6.8 Hz), 7.37 (d, 2H, J = 4.8 Hz), 7.48 (s, 1H), 8.14 (s, 1H), 11.70 (s, 1H). 13C NMR (DMSO-d6): 55.8, 112.1, 117.1, 121.2, 130.3, 135.1, 147.4, 155.5, 160.0. UV λmax (log ε) 278 (4.57). HRMS (ESI) m/z: calculated 193.1084 for [C9H13N4O]+, found 193.1093. HPLC analysis: retention time = 7.88 min; peak area, 97.8%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(4-Methoxybenzylidene)hydrazinecarboximidamide Hydrochloride (19)
Yield, 79%; mp 79–81 °C. 1H NMR (DMSO-d6) δ 3.80 (s, 3H), 7.00 (d, 2H, J = 8.8 Hz), 7.79 (d, 2H, J = 8.4 Hz), 8.11 (s, 1H), 11.60 (s, 1H). 13C NMR (DMSO-d6): 55.8, 114.6, 126.3, 129.7, 147.3, 155.5, 161.6. UV λmax (log ε) 287 (4.56). HRMS (ESI) m/z: calculated 193.1084 for [C9H13N4O]+, found 193.1090. HPLC analysis: retention time = 7.56 min; peak area, 99.9%.
(E)-2-(Naphthalen-1-ylmethylene)hydrazine-carboximidamide Hydrochloride (20)
Yield, 46%; mp 153–154 °C. 1H NMR (DMSO-d6) δ 7.65 (m, 3H), 8.04 (m, 2H), 8.24 (d, 1H, J = 6.8 Hz), 8.45 (d, 1H, J = 8.4 Hz), 9.01 (s, 1H), 12.05 (s, 1H). 13C NMR (DMSO-d6): 123.6, 125.9, 126.7, 127.1, 127.6, 127.9, 129.0, 129.3, 130.8, 131.4, 133.8, 146.2, 155.7, 159.5. UV λmax (log ε) 320 (4.23). HRMS (ESI) m/z: calculated 213.1135 for [C12H13N4]+, found 213.1139. HPLC analysis: retention time = 10.22 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(Naphthalen-2-ylmethylene)hydrazine-carboximidamide Hydrochloride (21)
Yield, 74%; mp 237–241 °C. 1H NMR (DMSO-d6) δ 7.58 (m, 2H), 7.96 (m, 3H), 8.20 (m, 2H), 8.34 (s, 1H), 12.13 (s, 1H). 13C NMR (DMSO-d6): 123.0, 126.8, 127.4, 127.8, 128.3, 128.4, 129.5, 131.2, 132.7, 133.9, 146.8, 155.4, 159.1. UV λmax (log ε) 270 (4.55). HRMS (ESI) m/z: calculated 213.1135 for [C12H13N4]+, found 213.1136. HPLC analysis: retention time = 10.26 min; peak area 95.3%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-([1,1’-Biphenyl]-4-ylmethylene)hydrazine-carboximidamide Hydrochloride (22)
Yield, 40%; mp 179–182 °C. 1H NMR (DMSO-d6) δ 7.41 (m, 2H), 7.50 (t, 2H, J = 8.0 Hz), 7.61 (t, 3H, J = 8.0 Hz), 7.97 (d, 2H, J = 8.0 Hz), 8.24 (s, 1H), 12.13 (s, 1H). 13C NMR (DMSO-d6): 126.4, 126.7, 126.8, 126.9, 127.1, 128.0, 128.2, 129.0, 129.0, 132.6, 141.9, 146.3, 155.4, 159.1. UV λmax (log ε) 302 (4.42). HRMS (ESI) m/z: calculated 213.1291 for [C14H15N4]+, found 213.1296. HPLC analysis: retention time = 11.79 min; peak area, 97.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(4-Isopropylbenzylidene)hydrazinecarboximidamide Hydrochloride (23)
Yield, 20%; mp 116–121 °C. 1H NMR (DMSO-d6) δ 1.21 (d, 7H, J = 6.8 Hz), 2.92 (sept, 1 H, J = 6.8 Hz), 7.32 (d, 2H, J = 8.0 Hz), 7.77 (d, 2H, J = 8.0 Hz), 8.13 (s, 1H), 11.88 (s, 1H). 13C NMR (DMSO-d6): 23.6, 33.4, 126.7, 127.7, 131.1, 146.9, 151.2, 155.3. UV λmax (log ε) 284 (4.67). HRMS (ESI) m/z: calculated 205.1448 for [C11H17N4]+, found 205.1457. HPLC analysis: retention time = 11.98 min; peak area, 96.0%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(3-(Benzyloxy)benzylidene)hydrazinecarboximidamide Hydrochloride (24)
Yield, 37%; mp 170–172. 1H NMR (DMSO-d6) δ 5.15 (s, 2H), 7.10 (m, 1H), 7.39 (m, 4H), 7.48 (m, 2H), 7.63 (s, 1H), 8.14 (s, 1H), 8.77 (s, 1H), 12.00 (s, 1H). 13C NMR (DMSO-d6): 69.4, 112.6, 117.2, 121.0, 128.0, 128.5, 129.8, 134.8, 136.8, 146.6, 155.4, 158.7. UV λmax (log ε) 278 (4.35). HRMS (ESI) m/z: calculated 269.1397 for [C15H17N4O]+, found 269.1401. HPLC analysis: retention time = 12.37 min; peak area, 99.9%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
(E)-2-(4-Nitrobenzylidene)hydrazinecarboximidamide Hydrochloride (25)
Yield, 34%; mp 243–247 °C. 1H NMR (DMSO-d6) δ 8.12 (d, 2H, J = 8.8 Hz), 8.25 (m, 3H), 12.35 (s, 1H). 13C NMR (DMSO-d6): 124.2, 129.0, 140.2, 144.9, 148.5, 156.0. UV λmax (log ε) 315 (4.46). HRMS (ESI) m/z: calculated 208.0829 for [C8H10N5O2]+, found 208.0835. HPLC analysis: retention time = 7.82 min; peak area, 99.8%; eluent A, CH3OH; eluent B, NH4OAc solution (0.1%); isocratic (3:1) over 60 min with a flow rate of 0.5 mL/min and detection at 254 nm.
Protein Purification
P. aeruginosa HemO was purified as previously reported with slight modification.50 Protein was bound to a Q Sepharose Fast Flow column (2.5 cm × 6 cm) equilibrated with 20 mM tris (pH 8.0), 50 mM NaCl, and 50 mM 1,3-diaminopropane (DAP). Residual biliverdin was retained on the column, and apoprotein was eluted with a 50–250 mM NaCl gradient. Apo-HemO fractions (determined by SDS–PAGE) were pooled and dialyzed against 20 mM Tris-HCl (pH 8.0).
Determining Binding Affinities (KD) by Intrinsic Fluorescence Quenching
Fluorescence experiments were performed on a Synergy H1 hybrid microplate reader using flat-bottom black 96-well plates. Protein concentration was 1 µM HemO in 20 mM Tris-HCl (pH 8.0). Compounds were dissolved in DMSO, and final concentrations ranged from 0.5 to 1000 µM. Solutions were excited at 295 nm, and emission spectra were recorded from 300 to 400 nm. Decrease in maximum emission (332 nm) was corrected using experimentally determined extinction coefficients for each compound.43 Binding was determined using eq 1 and fit to a one-site binding model.
| (1) |
STD NMR. Preparation of samples and collection of STD spectra were conducted as previously reported with slight modifications.30 Briefly, to a 950 µL solution of 50 mM sodium phosphate (pH = 7.4) in D2O was added 50 µL of desired compound dissolved in DMSO-d6 and 10 µL of purified HemO such that final concentration of HemO was between 3 and 5 µM. After 4 h incubation at room temperature the mixture was centrifuged at 6000g for 3 min to remove residual precipitate. After preparation of 600 µL sample was added 3 µL of 50 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) dissolved in D2O as internal standard.
In Cellulo Activity Assay
We have developed an E. coli cell based assay for the measurement of HemO activity. The assay is based on the coexpression of a HemO mutant with biliverdin-IXα regioselectivity and the IFP1.4.48 E. coli BL21 (DE3) cells were transformed with pET28a expressing IFP 1.4 and pBAD expressing HemO. Cells were plated on LB-agar containing 30 µg/mL kanamycin and 15 µg/mL tetracycline. A single colony was used to inoculate a 25 mL culture of LB medium containing 30 µg/mL kanamycin and 15 µg/mL tetracycline. The culture was incubated with shaking (200 rpm) at 37 °C overnight and used to inoculate a fresh culture treated to a final OD600 of 0.1. Cultures were further incubated until reaching an OD600 = 0.7 at which point IFP 1.4 expression was induced with IPTG (final concentration 1 mM). The HemO and IFP 1.4 proteins were allowed to express at 25 °C induced with 0.02% arabinose (uninduced wells were used for negative control) and aliquoted (200 µL) in 96-well plates. Wells were treated with 0.5–200 µM compound (in a final concentration 1% DMSO) or 1% DMSO in the positive control. Cultures were incubated at 25 °C for 16 h following which the OD600 and fluorescence (ex 630, em 700) were measured on a Synergy H1 hybrid microplate reader. Background fluorescence in the absence of HemO expression was measured as 0% HemO activity (negative control). Maximum fluorescence on expression of HemO was considered 100% activity (positive control). The percent inhibition was calculated using eq 2 following correction of the fluorescence intensity as a function of OD600.
| (2) |
The EC50 for each compound was calculated from the concentration-dependent curve at 50% fluorescence inhibition. Fluorescence images of live bacteria were obtained on a Nikon eclipse Ti fluorescence microscope using NIS-Elements. Cells were imaged through a 100× objective by phase contrast and through a Cy5 filter for detection of near-infrared emissions from the biliverdin-IFP1.4 chromophore.
Growth Inhibition Assay
P. aeruginosa growth inhibition was conducted as previously described. Briefly, a culture (25 mL) of each strain was grown from a freshly plated colony for 8–12 h at 37 °C in LB medium. Cultures were diluted back to an OD600 of 0.05 and aliquoted into 96-well plates (200 µL) in either LB medium alone or containing 8–1000 µg/mL of compound. The OD600 was measured following 12 h of growth on a Synergy H1 hybrid microplate reader and subtracted from LB media containing 50 µg/mL gentamycin to prevent bacterial growth. MIC50 values were calculated from the average of three experiments as previously described. Cultures from wells containing 62.5–1000 µg/mL of compound were used to inoculate fresh cultures in 5 mL of LB medium. Cultures were grown for 12 h, and the lowest concentration that showed no growth by measurement of OD600 was considered to be the minimum bactericidal concentration (MBC).
SILCS
SILCS is a novel computer-aided drug design (CADD) protocol that uses all-atom explicit-solvent molecular dynamics (MD) simulations that include small organic solutes, such as benzene, propane, methanol, and others, to identify spatial functional-group binding patterns on the target. Benefiting from the use of explicit solvent MD, SILCS intrinsically incorporates target flexibility and desolvation effects and has more advantages over other CADD methods which usually only consider limited target flexibility and desolvation effects.
The current SILCS run was performed using the new grand canonical Monte Carlo (GCMC)/MD protocol for SILCS. Compared to the MD-alone technique, the GCMC/MD protocol has the benefit of quickly targeting occluded pockets and decreasing the false binding pattern caused by low fragment exchange rate at specific positions.52 The protein structure in the complex crystal structure (PDB code 1SK7) was used to initialize the SILCS simulation with removal of cofactors such as heme and sulfate ions. The protein structure was processed by Reduce53 to determine the most physical protonation state of histidine residues and optimal side chain orientations of asparagine and glutamine residues. The protein was solvated in a water box, the size of which is determined to have the protein extrema separated from the box edge by 12 Å on all sides. Eight representative solutes with different chemical functionalities (benzene, propane, acetaldehyde, methanol, formamide, imidazole, acetate, and methylammonium) were added into the system at ~0.25 M concentration, to probe the functional group requirements of the protein.
Ten such systems with different initial fragment positions were created to expedite the convergence of the simulations. Each system was minimized for 5000 steps with the steepest descent (SD) algorithm54 in the presence of periodic boundary conditions (PBC) and was followed by a 250 ps MD equilibration. During SILCS simulations, weak restraints were applied on the backbone Ca carbon atoms with a force constant (k in ½ kδx2) of 0.12 kcal mol−1 Å−1 to prevent the rotation of the protein in the simulation box. Ten GCMC/ MD simulations were run for 100 cycles where each cycle has 200 000 steps of GCMC and 0.5 ns of MD, yielding a cumulative 200 million steps of GCMC and 500 ns of MD. During GCMC, solutes and water are exchanged between their gas-phase reservoirs; the simulation system and the excess chemical potential used to drive such exchange are periodically fluctuated over every 3 cycles to yield the average value to be close to the hydration free energy of that fragment. The configuration at the end of each GCMC run is used as the starting configuration for the following MD. During MD, the Nosé–Hoover method55,56 was used to maintain the temperature at 298 K, and pressure was maintained at 1 bar using the Parrinello–Rahman barostat.57 CHARMM36 protein force field,58 CHARMM general force field (CGenFF),59,60 and modified TIP3P water model61 were used to describe protein, fragments, and water during the simulation, respectively. GCMC was performed by in-house code, and MD was conducted using GROMACS program.62
FragMaps Preparation and Ligand Docking
3D probability distributions of the selected atoms from the small molecules, called “FragMaps”, from the SILCS simulations were constructed and combined to obtain both specific and generic FragMap types as previously described.41 Atoms from snapshot outputs every 10 ps from each SILCS simulation trajectory were binned into 1Å×1Å×1Å cubic volume elements (voxels) of a grid spanning the entire system to acquire the voxel occupancy for each FragMap atom type being counted. The voxel occupancies computed in the presence of the protein were divided by the value in bulk to obtain a normalized probability. Normalized distributions were then converted to GFE based on a Boltzmann transformation for quantitative use.
Ligand docking was conducted under the Monte Carlo based SILCS (MC-SILCS) framework for both the heme binding site and the allosteric site.41 The MC sampling was guided by LGFE, which is the summation of the GFE contributions of the classified ligand atoms in a molecule, and it represents the overlap of the functional groups in a molecule with corresponding types of FragMaps. Five independent MC-SILCS runs were performed for each ligand and each run was repeated in two-step cycles. The first step of each cycle is 10 000 Metropolis MC steps that sample a wide range of different binding poses, and the second step is 40 000 steps of MC simulated annealing (SA) slow cooling designed to find a local minimum on the LGFE surface. The initial ligand conformation is randomized by randomly adjusting the torsion angels around all single bonds in the molecule, and its potential energy was checked to ensure that unphysical conformations are rejected. CGenFF was used to describe the ligands during the MC-SILCS simulations. The generated conformation was then placed at the desired binding site with a randomized orientation, and its LGFE was checked to remove bad starting orientations. The five MC SILCS runs involved repeated MC cycles that were repeated until the best three LGFE scores were with 0.5 kcal/mol, with a minimum of 50 cycles performed. The docking pose with the most favorable LGFE was reported as the predicted binding mode.
HXMS
Deuterium exchange reactions were performed as described by Chen et al. (2007).62 HemO protein samples were prepared by diluting the protein to a final concentration of 100 µM with 20 mM HEPES buffer in H2O (pH 7.4). Deuterium exchange was initiated by diluting the HemO protein 50-fold with 20 mM HEPES buffer in D2O (pD 7.4, deuteration buffer) at 23 °C. 100 pmol of protein was removed from the reaction at 0 s, 10 min, and 30 min, and the deuteration reaction was immediately quenched by lowering the pH to 2.5 with ice-cold HCl. Quenched samples were frozen on dry ice prior to analysis.
For reactions containing holo-HemO, the heme required for 95% saturation was estimated using a KD of 1 µM. 100 µM apo-HemO was reconstituted with 114 µM heme during the sample preparation, and heme was added to the deuteration buffer to a final concentration of 21 µM. For reactions of inhibitor bound HemO, the inhibitor (1 or 22) required for 95% binding was estimated using a KD of 10 µM. Apo- or holo-HemO was incubated with 285 µM inhibitor during the sample preparation, and the inhibitor was added to the deuteration buffer to a final concentration of 192 µM.
Quenched samples were thawed and immediately injected into a nanoACQUITY UPLC system with HDX manager (Waters). The protein was digested online at 10 °C using an Enzymate BEH pepsin column (2.1 mm × 30 mm, Waters) in 1 min. The digest was trapped and desalted online on an ACQUITY Vanguard BEH C18 precolumn (2.1 mm × 5 mm, Waters) at 0 °C for 4 min at a flow rate of 125 µL/ min in 0.1% formic acid. Peptides were separated on an ACQUITY UPLC BEH C18 column (1.7 µm, 1 mm × 100 mm, Waters) at 0 °C by a 15 min linear acetonitrile gradient (5–50%) with 0.1% formic acid at a flow rate of 40 µL/min. The eluent was directed into the ion source of a coupled SYNAPT G2 HDMS mass spectrometer (Waters). Mass spectra were acquired in the MSE mode over the m/z range of 50–2000. Mass spectrometer parameters were as follows: electrospray ionization positive (ESI+) mode, capillary voltage 3 kV, collision energy 20–30 eV, sampling cone voltage 30 V, source temperature 80 °C, desolvation temperature 175 °C, and desolvation gas flow 500 L/ h. To generate a peptide list for ion search, 100 pmol of undeuterated protein in 2 mM HCl in H2O was injected. The undeuterated peptides were identified using Waters ProteinLynx Global Server software. The peptide list generated was imported into Waters DynamX software to guide the search of deuterated peptides, and the relative deuterium incorporation levels for each deuterated peptide were calculated using the 0 s sample as reference.
Supplementary Material
Acknowledgments
The authors thank Angela Nguyen and Amanda Oglesby-Sherrouse for their generous donations of the JSRI-1 andJSRI-2 strains and guidance for their handling. This research was funded by a predoctoral fellowship from the ACS Division of Medicinal Chemistry and a predoctoral fellowship from the American Foundation for Pharmaceutical Education to G.A.H.; NIH Grant T32GM066706 to G.A.H.; and NIH Grant AI102883 to A.W.
ABBREVIATIONS USED
- CHARMM
chemistry at Harvard molecular mechanics
- GCMC
grand canonical Monte Carlo
- GFE
grid free energy
- HemO
heme oxygenase
- HEPES
(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HO
human heme oxygenase
- HXMS
hydrogen–deuterium exchange mass spectrometry
- IFP
infrared fluorescent protein
- LB
Luria–Bertani
- LGFE
ligand-grid free energy
- MBC
minimum bactericidal concentration
- MIC50
minimum inhibitory concentration for 50% growth
- PBC
periodic boundary conditions
- SA
simulated annealing
- SD
steepest descent
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SILCS
site identification by ligand competitive saturation
- STD NMR
saturation transfer difference nuclear magnetic resonance
- UPLC
ultra-performance liquid chromatography
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.6b00757.
Additional figures depicting the HXMS peptide coverage following pepsin digestion, HXMS profile of apo-HemO with compound 1, and Western blot and fluorescence emission profiles of the E. coli HemO IFP expression system (PDF)
Molecular formula strings (CSV)
Author Contributions
All authors contributed to the writing of this manuscript. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): A.D.M., Jr., is cofounder and CSO of SilcsBio LLC.
REFERENCES
- 1.Clatworthy AE, Pierson E, Hung DT. Targeting Virulence: A New Paradigm for Antimicrobial Therapy. Nat. Chem. Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 2.Page MG, Heim J. New Molecules from Old Classes: Revisiting the Development of Beta-Lactams. IDrugs. 2009;12:561–565. [PubMed] [Google Scholar]
- 3.Fair RJ, Hensler ME, Thienphrapa W, Dam QN, Nizet V, Tor Y. Selectively Guanidinylated Aminoglycosides as Antibiotics. ChemMedChem. 2012;7:1237–1244. doi: 10.1002/cmdc.201200150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fàbrega A, Madurga S, Giralt E, Vila J. Mechanism of Action of and Resistance to Quinolones. Microb. Biotechnol. 2009;2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guénard S, Muller C, Monlezun L, Benas P, Broutin I, Jeannot K, Plésiat P. Multiple Mutations Lead to MexXY-OprM-Dependent Aminoglycoside Resistance in Clinical Strains of Pseudomonas aeruginosa . Antimicrob. Agents Chemother. 2014;58:221–228. doi: 10.1128/AAC.01252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voor AF, Severin JA, Lesaffre EM, Vos MC. Carbapenem Use and Medical Devices Highest Risk Factors for Carbapenem-Resistant Pseudomonas aeruginosa: A Systematic Review and Meta-Analyses. Antimicrob. Agents Chemother. 2014;58:2626–2637. doi: 10.1128/AAC.01758-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costabile G, d’ Angelo I, Rampioni G, Bondí R, Pompili B, Ascenzioni F, Mitidieri E, d’ Emmanuele di Villa Bianca R, Sorrentino R, Miro A, et al. Toward Repositioning Niclosamide for Antivirulence Therapy of Pseudomonas aeruginosa Lung Infections: Development of Inhalable Formulations through Nanosuspension Technology. Mol. Pharmaceutics. 2015;12:2604. doi: 10.1021/acs.molpharmaceut.5b00098. [DOI] [PubMed] [Google Scholar]
- 8.Alekshun MN, Levy SB. Targeting Virulence to Prevent Infection: To Kill or Not to Kill? Drug Discovery Today: Ther Strategies. 2004;1:483–489. [Google Scholar]
- 9.Rasko DA, Sperandio V. Anti-Virulence Strategies to Combat Bacteria-Mediated Disease. Nat. Rev. Drug Discovery. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 10.Limoli DH, Rockel AB, Host KM, Jha A, Kopp BT, Hollis T, Wozniak DJ. Cationic Antimicrobial Peptides Promote Microbial Mutagenesis and Pathoadaptation in Chronic Infections. PLoS Pathog. 2014;10:e1004083. doi: 10.1371/journal.ppat.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeukens J, Boyle B, Kukavica-Ibrulj I, Ouellet MM, Aaron SD, Charette SJ, Fothergill JL, Tucker NP, Winstanley C, Levesque RC. Comparative Genomics of Isolates of a Pseudomonas aeruginosa Epidemic Strain Associated with Chronic Lung Infections of Cystic Fibrosis Patients. PLoS One. 2014;9:e87611. doi: 10.1371/journal.pone.0087611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balasubramanian D, Kumari H, Mathee K. Pseudomonas aeruginosa AmpR: An Acute-Chronic Switch Regulator. Pathog. Dis. 2015;73:1–14. doi: 10.1111/2049-632X.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux D, Danilchanka O, Guillard T, Cattoir V, Aschard H, Fu Y, Angoulvant F, Messika J, Ricard J-D, Mekalanos JJ. Fitness Cost of Antibiotic Susceptibility during Bacterial Infection. Sci. Transl. Med. 2015;7:297ra114. doi: 10.1126/scitranslmed.aab1621. [DOI] [PubMed] [Google Scholar]
- 14.Mittal R, Sharma S, Chhibber S, Harjai K. Iron Dictates the Virulence of Pseudomonas aeruginosa in Urinary Tract Infections. J. Biomed. Sci. 2008;15:731–741. doi: 10.1007/s11373-008-9274-7. [DOI] [PubMed] [Google Scholar]
- 15.Hauser AR. Pseudomonas aeruginosa Virulence and Antimicrobial Resistance. Crit. Care Med. 2014;42:201–202. doi: 10.1097/CCM.0b013e3182a120cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fillat MF. The FUR (ferric Uptake Regulator) Superfamily: Diversity and Versatility of Key Transcriptional Regulators. Arch. Biochem. Biophys. 2014;546:41–52. doi: 10.1016/j.abb.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 17.Lowy FD, Pollack S, Fadl-Allah N, Steigbigel NH. Susceptibilities of Bacterial and Fungal Urinary Tract Isolates to Desferrioxamine. Antimicrob. Agents Chemother. 1984;25:375–376. doi: 10.1128/aac.25.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins HL, Kaufmann SH, Schaible UE. Iron Chelation via Deferoxamine Exacerbates Experimental Salmonellosis via Inhibition of the Nicotinamide Adenine Dinucleotide Phosphate Oxidase-Dependent Respiratory Burst. J. Immunol. 2002;168:3458–3463. doi: 10.4049/jimmunol.168.7.3458. [DOI] [PubMed] [Google Scholar]
- 19.Kontoghiorghes GJ, Kolnagou A, Skiada A, Petrikkos G. The Role of Iron and Chelators on Infections in Iron Overload and Non Iron Loaded Conditions: Prospects for the Design of New Antimicrobial Therapies. Hemoglobin. 2010;34:227–239. doi: 10.3109/03630269.2010.483662. [DOI] [PubMed] [Google Scholar]
- 20.Ji C, Miller PA, Miller MJ. Iron Transport-Mediated Drug Delivery: Practical Syntheses and In Vitro Antibacterial Studies of Tris-Catecholate Siderophore-Aminopenicillin Conjugates Reveals Selectively Potent Antipseudomonal Activity. J. Am. Chem. Soc. 2012;134:9898–9901. doi: 10.1021/ja303446w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baco E, Hoegy F, Schalk IJ, Mislin GLA. Diphenyl-benzo[1,3]dioxole-4-Carboxylic Acid Pentafluorophenyl Ester: A Convenient Catechol Precursor in the Synthesis of Siderophore Vectors Suitable for Antibiotic Trojan Horse Strategies. Org. Biomol. Chem. 2014;12:749–757. doi: 10.1039/c3ob41990h. [DOI] [PubMed] [Google Scholar]
- 22.Ji C, Juárez-Hernández RE, Miller MJ. Exploiting Bacterial Iron Acquisition: Siderophore Conjugates. Future Med. Chem. 2012;4:297–313. doi: 10.4155/fmc.11.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benson DR, Rivera M. Metallomics and the Cell. Springer; 2013. Heme Uptake and Metabolism in Bacteria; pp. 279–332. [DOI] [PubMed] [Google Scholar]
- 24.Wilks A, Barker KD. Mechanism of Heme Uptake and Utilization in Bacterial Pathogens. Handb. Porphyrin Sci. 2011;15:357–398. [Google Scholar]
- 25.Wilks A, Heinzl G. Heme Oxygenation and the Widening Paradigm of Heme Degradation. Arch. Biochem. Biophys. 2014;544:87–95. doi: 10.1016/j.abb.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen AT, O’Neill MJ, Watts AM, Robson CL, Lamont IL, Wilks A, Oglesby-Sherrouse AG. Adaptation of Iron Homeostasis Pathways by a Pseudomonas aeruginosa Pyoverdine Mutant in the Cystic Fibrosis Lung. J. Bacteriol. 2014;196:2265–2276. doi: 10.1128/JB.01491-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marvig RL, Damkiær S, Khademi SH, Markussen TM, Molin S, Jelsbak L. Within-Host Evolution of Pseudomonas aeruginosa Reveals Adaptation toward Iron Acquisition from Hemoglobin. mBio. 2014;5:e00966–e00914. doi: 10.1128/mBio.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Neill MJ, Wilks A. The P. aeruginosa Heme Binding Protein PhuS Is a Heme Oxygenase Titratable Regulator of Heme Uptake. ACS Chem. Biol. 2013;8:1794–1802. doi: 10.1021/cb400165b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furci LM, Lopes P, Eakanunkul S, Zhong S, MacKerell AD, Wilks A. Inhibition of the Bacterial Heme Oxygenases from Pseudomonas aeruginosa and Neisseria meningitidis: Novel Antimicrobial Targets. J. Med. Chem. 2007;50:3804–3813. doi: 10.1021/jm0700969. [DOI] [PubMed] [Google Scholar]
- 30.Hom K, Heinzl GA, Eakanunkul S, Lopes PE, Xue F, MacKerell AD, Jr, Wilks A. Small Molecule Antivirulents Targeting the Iron-Regulated Heme Oxygenase (HemO) of P. aeruginosa. J. Med. Chem. 2013;56:2097–2109. doi: 10.1021/jm301819k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baum T, Eckfeld DK, Metz N, Dinish JL, Rowles G, Van Pelt R, Shropshire AT, Fernandez SP, Gluckman MI. Bruce WF2 6-Dichlorobenzylidene Amino Guanidine Acetate (Wy-8678). A New Hypotensive Agent. Experientia. 1969;25:1066–1067. doi: 10.1007/BF01901433. [DOI] [PubMed] [Google Scholar]
- 32.Specht S, Sarite SR, Hauber I, Hauber J, Gorbig UF, Meier C, Bevec D, Hoerauf A, Kaiser A. The Guanylhydrazone CNI-1493: An Inhibitor with Dual Activity against Malaria—inhibition of Host Cell pro-Inflammatory Cytokine Release and Parasitic Deoxyhypusine Synthase. Parasitol. Res. 2008;102:1177–1184. doi: 10.1007/s00436-008-0891-x. [DOI] [PubMed] [Google Scholar]
- 33.Löwenberg M, Verhaar A, van den Blink B, ten Kate F, van Deventer S, Peppelenbosch M, Hommes D. Specific Inhibition of c-Raf Activity by Semapimod Induces Clinical Remission in Severe Crohn’s Disease. J. Immunol. 2005;175:2293–2300. doi: 10.4049/jimmunol.175.4.2293. [DOI] [PubMed] [Google Scholar]
- 34.Ulloa L. The Vagus Nerve and the Nicotinic Anti-Inflammatory Pathway. Nat. Rev. Drug Discovery. 2005;4:673–684. doi: 10.1038/nrd1797. [DOI] [PubMed] [Google Scholar]
- 35.Soll RM, Lu T, Tomczuk B, Illig CR, Fedde C, Eisennagel S, Bone R, Murphy L, Spurlino J, Salemme F. R Amidinohydrazones as Guanidine Bioisosteres: Application to a New Class of Potent, Selective and Orally Bioavailable, Non-Amide-Based Small-Molecule Thrombin Inhibitors. Bioorg. Med. Chem. Lett. 2000;10:1–4. doi: 10.1016/s0960-894x(99)00632-0. [DOI] [PubMed] [Google Scholar]
- 36.Gaubert G, Bertozzi F, Kelly NM, Pawlas J, Scully AL, Nash NR, Gardell LR, Lameh L, Olsson R. Discovery of Selective Nonpeptidergic Neuropeptide FF2 Receptor Agonists. J. Med. Chem. 2009;52:6511–6514. doi: 10.1021/jm9011998. [DOI] [PubMed] [Google Scholar]
- 37.Carniato D, Gourvest J-F, Ruxer J-M, Knolle J, Peyman A, Bodary SC, Gadek T. R Iminoguanidine derivatives, preparation method, use as medicines. U.S. Patent 6,391,904. 2002 May;21 [Google Scholar]
- 38.Barron S, Lewis B, Wellmann K, Carter M, Farook J, Rng J, Trent Rogers D, Holley R, Crooks P, Littleton J. Polyamine Modulation of NMDARs as a Mechanism to Reduce Effects of Alcohol Dependence. Recent Pat. CNS Drug Discovery. 2012;7:129–144. doi: 10.2174/157488912800673128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conard VA, Shriner RL. Aminoguanidine Derivatives. J. Am. Chem. Soc. 1933;55:2867–2870. [Google Scholar]
- 40.Rng J. Ph.D. Dissertation. Lexington, KY: University of Kentucky; 2006. R Synthetic Aromatic Agmatine Analogs as Allosteric Modulators of the N-Methyl-D-Aspartate (NMDA) Receptor Channel. [Google Scholar]
- 41.Raman EP, Yu W, Lakkaraju SK, MacKerell AD. Inclusion of Multiple Fragment Types in the Site Identification by Ligand Competitive Saturation (SILCS) Approach. J. Chem. Inf. Model. 2013;53:3384–3398. doi: 10.1021/ci4005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu W, Lakkaraju SK, Raman EP, Fang L, MacKerell AD., Jr Pharmacophore Modeling Using Site-Identification by Ligand Competitive Saturation (SILCS) with Multiple Probe Molecules. J. Chem. Inf. Model. 2015;55:407–420. doi: 10.1021/ci500691p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epps DE, Raub TJ, Caiolfa V, Chiari A, Zamai M. Determination of the Affinity of Drugs toward Serum Albumin by Measurement of the Quenching of the Intrinsic Tryptophan Fluorescence of the Protein. J. Pharm. Pharmacol. 1999;51:41–48. doi: 10.1211/0022357991772079. [DOI] [PubMed] [Google Scholar]
- 44.Sowole MA, Konermann L. Effects of Protein-ligand Interactions on Hydrogen/deuterium Exchange Kinetics: Canonical and Noncanonical Scenarios. Anal. Chem. 2014;86:6715–6722. doi: 10.1021/ac501849n. [DOI] [PubMed] [Google Scholar]
- 45.Friedman J, Lad L, Li H, Wilks A, Poulos TL. Structural Basis for Novel Δ-Regioselective Heme Oxygenation in the Opportunistic Pathogen Pseudomonas aeruginosa. Biochemistry. 2004;43:5239–5245. doi: 10.1021/bi049687g. [DOI] [PubMed] [Google Scholar]
- 46.Rücker H, Amslinger S. Identification of Heme Oxygenase-1 Stimulators by a Convenient ELISA-Based Bilirubin Quantification Assay. Free Radical Biol. Med. 2015;78:135–146. doi: 10.1016/j.freeradbiomed.2014.10.506. [DOI] [PubMed] [Google Scholar]
- 47.Filonov GS, Piatkevich KD, Ting L-M, Zhang J, Kim K, Verkhusha VV. Bright and Stable near-Infrared Fluorescent Protein for in Vivo Imaging. Nat. Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian Expression of Infrared Fluorescent Proteins Engineered from a Bacterial Phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ratliff M, Zhu W, Deshmukh R, Wilks A, Stojiljkovic I. Homologues of Neisserial Heme Oxygenase in Gram-Negative Bacteria: Degradation of Heme by the Product of the pigA Gene of Pseudomonas aeruginosa . J. Bacteriol. 2001;183:6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rawel HM, Frey SK, Meidtner K, Kroll J, Schweigert FJ. Determining the Binding Affinities of Phenolic Compounds to Proteins by Quenching of the Intrinsic Tryptophan Fluorescence. Mol. Nutr. Food Res. 2006;50:705–713. doi: 10.1002/mnfr.200600013. [DOI] [PubMed] [Google Scholar]
- 51.Lakkaraju SK, Raman EP, Yu W, MacKerell AD., Jr Sampling of Organic Solutes in Aqueous and Heterogeneous Environments Using Oscillating Excess Chemical Potentials in Grand Canonical-like Monte Carlo-Molecular Dynamics Simulations. J. Chem. Theory Comput. 2014;10:2281–2290. doi: 10.1021/ct500201y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and Glutamine: Using Hydrogen Atom Contacts in the Choice of Side-Chain Amide Orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 53.Levitt M, Lifson S. Refinement of Protein Conformations Using a Macromolecular Energy Minimization Procedure. J. Mol. Biol. 1969;46:269–279. doi: 10.1016/0022-2836(69)90421-5. [DOI] [PubMed] [Google Scholar]
- 54.Nosé S. A Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 55.Hoover WG. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A: At., Mol., Opt. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 56.Parrinello M, Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 57.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, MacKerell A. D., Jr Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone Φ, Ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu W, He X, Vanommeslaeghe K, MacKerell AD. Extension of the CHARMM General Force Field to Sulfonyl-Containing Compounds and Its Utility in Biomolecular Simulations. J. Comput. Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durell SR, Brooks BR, Ben-Naim A. Solvent-Induced Forces between Two Hydrophilic Groups. J. Phys. Chem. 1994;98:2198–2202. [Google Scholar]
- 61.Hess B, Kutzner C, Van Der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 62.Chen S, Brier S, Smithgall TE, Engen JR. The Abl SH2-Kinase Linker Naturally Adopts a Conformation Competent for SH3 Domain Binding. Protein Sci. 2007;16:572–581. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.