Abstract: Background
Computer-aided drug design is still a state-of-the-art process in medicinal chemistry, and the main topics in this field have been extensively studied and well reviewed. These topics include compound databases, ligand-binding pocket prediction, protein-compound docking, virtual screening, target/off-target prediction, physical property prediction, molecular simulation and pharmacokinetics/pharmacodynamics (PK/PD) prediction. Message and Conclusion: However, there are also a number of secondary or miscellaneous topics that have been less well covered. For example, methods for synthesizing and predicting the synthetic accessibility (SA) of designed compounds are important in practical drug development, and hardware/software resources for performing the computations in computer-aided drug design are crucial. Cloud computing and general purpose graphics processing unit (GPGPU) computing have been used in virtual screening and molecular dynamics simulations. Not surprisingly, there is a growing demand for computer systems which combine these resources. In the present review, we summarize and discuss these various topics of drug design.
Keyword: Computer-aided drug design, Synthetic accessibility, Cloud computing, GPU computing, Virtual screening, Molecular dynamics simulation
Introduction
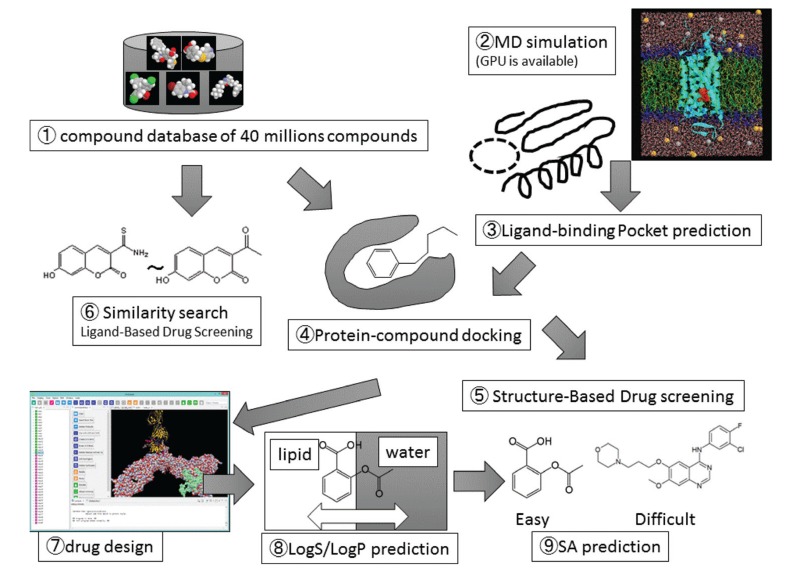
We review miscellaneous topics in computer-aided drug design (CADD), including non-scientific, technical, old and forgotten topics, since there have been a number of good reviews published already on the major topics in CADD. Computer-aided drug design is an assemblage of various computational methods and resources. These include compound databases, molecular dynamics simulations, ligand-binding pocket predictions, protein-compound dockings, structure-based drug screenings, ligand-based drug screenings, similarity searches, de-novo drug design, property predictions like LogS (aqueous solubility) and LogPow (water-octanol partitioning coefficient) prediction, target/off target predictions, and predictions of synthetic accessibility. Fig. (1) shows the relationship among these methods. They are based on the chemical compound structures, while the pharmacokinetics and pharmacodynamics studies are mainly based on the experimental data.
Fig. (1).
Schematic representation of methods in computer-aided drug design.
There have been many reviews reporting on compound databases, molecular dynamics simulation, ligand-binding pocket prediction, protein-compound docking, structure-based drug screening, ligand-based drug screening, similarity searches, and de-novo drug design. Thus, in the present review, we focus on a number of important but less-studied topics. We review the synthetic accessibility (SA) prediction SA is an important aspect of drug design, since in some cases computer-designed compounds cannot be synthesized. In addition, we briefly consider the correlation between the sales price of approved drugs and the SA values.
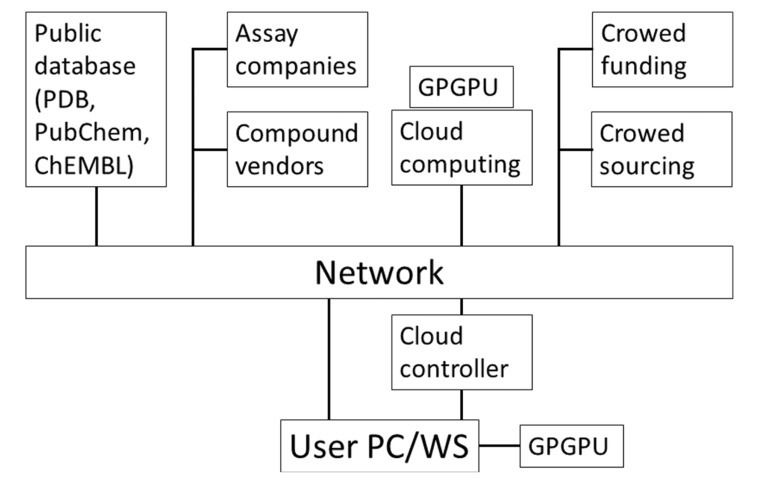
These methods and resources in CADD are supported by various computational technologies such as PCs, cluster machines, cloud computing, general purpose graphics processing unit (GPU) computing and ad-hoc specialized computers. Fig. (2) shows these resources used in CADD. Setting up a CADD system on a GPU or in the cloud is still a difficult task for users, while ordinary PCs and workstations are quite user-friendly. We also review how to set up a cloud and GPU computer environment.
Fig. (2).
Schematic representation of resources in computer-aided drug design.
Synthetic accessibility (SA) prediction using reaction databases
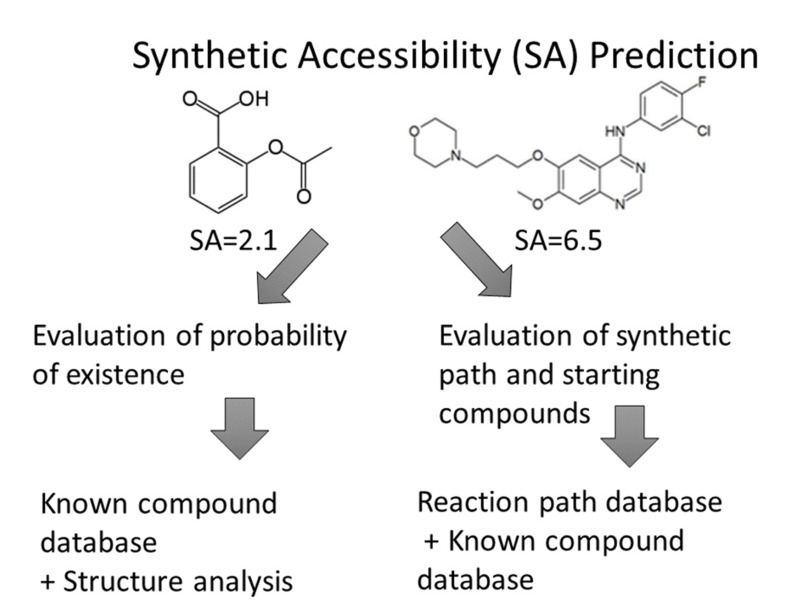
Synthetic accessibility (SA) is a well-known concept in organic chemistry, but calculation-based SA predictions have been studied only in the last 10 years [1-17]. On the other hand, researchers have been using software to predict synthetic reaction pathways since the 1960s [11, 16]. Computer-aided drug design, especially de-novo design, became popular in the 1980s, but SA estimation of the newly designed compounds have proven to be difficult. Automatic SA evaluation programs would be helpful in such cases and, indeed, several programs for the prediction of synthetic reaction pathways have been developed. However, these programs depend on the availability of reagents, the catalogs of which have changed every year, and they do not evaluate SA quantitatively (see Fig. 3).
Fig. (3).
Two major synthetic accessibility (SA) prediction procedures.
Several computational methods, including CAESA [3], RECAP [4], WODCA [5], LHASA [6], RASA [7], RSsvm [8], AIPHOS [9], and SYLVIA [10], have been developed to perform retro-synthesis [7, 11, 12, 16] and/or SA prediction for the compounds in question. In these methods, the synthetic reaction path is predicted based on reaction databases and the availability of reagents (starting materials). Parts of the available programs are summarized in Table 1. These methods work well in some projects and should be useful in drug design. However, the predicted reaction paths can be unrealistic in some cases, since steric effects (such as atom collisions and other inter-atomic interactions) are not taken into consideration in these approaches and the reaction databases include incorrect entries.
Table 1.
A selection of software programs for synthetic accessibility.
| Software Name | Company | URL |
|---|---|---|
| CAESA | Keymodule Ltd | http://www.keymodule.co.uk/products/caesa/index.html |
| SYLVIA | Molecular Networks GmbH | https://www.molecular-networks.com/products/sylvia |
| AIPHOS | ChemInfoNavi | http://www.cheminfonavi.co.jp/main/product/aiphos.html |
| LHASA | Radboud University Nijmegen | http://cheminf.cmbi.ru.nl/cheminf/lhasa/ |
| WODCA | Universität Erlangen-Nürnberg | http://www2.chemie.uni-erlangen.de/software/wodca/index.html |
| MolDesk | Information and Mathematical Science Bio Inc. | http://www.moldesk.com/ |
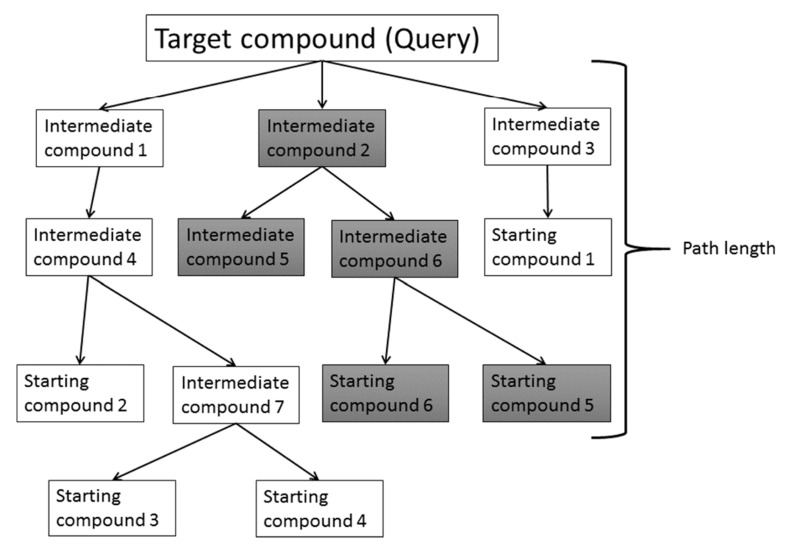
Fig. (4) shows one example of an SA prediction based on reaction data [7, 11, 12, 16]. The compound in question is decomposed into fragments by breaking the relatively unstable bonds. This process is repeated until these fragments reach the starting chemicals included in the database. The bond breaking is performed based on the reaction database. This decomposition process generates a number of reaction paths. Then the difficulty of each reaction is estimated and the total SA of the compound in question is calculated. There are several ways to estimate the SA. One method evaluates the depth of the shortest path among all the predicted paths, since the SA depends on the reaction steps from the starting materials to the final product [7]. Generally speaking, if the compound is synthesized within 5 steps, the synthesis is easy. The other method evaluates the SA by considering the number of possible reaction paths, the difficulty of each step in the predicted path, and LogP on which the difficulty of the purification process depends [7]. The difficulty of synthesis will be decreased if the number of possible reaction paths increases. The difficulties of the reactions are estimated by chemists a priori. LogP prediction is more precise than LogS prediction and the descriptor-based LogP prediction is fast enough for practical use.
Fig. (4).
Reaction path-based synthetic accessibility prediction.
These retro-synthesis approaches depend on the reaction database. Satoh et al. examined the 329 data entries of the ChemInform (ISIS-ChemInform MDL-Information systems Inc.) database [18]. They reported that 46% of the data contained some error. Namely, they found wrong numbers of reaction steps in 25% of the data, and wrong reactions in 21%, wrong reaction sites in 19%, wrong chemical structures in 15%, wrong reaction conditions in 9% and wrong yield constants in 5% of the data. Most of the errors were human input errors. The SA and reaction path prediction theory should be improved in the future, as same as the theory, the quality check of various databases should be important.
Synthetic accessibility prediction without a reaction database
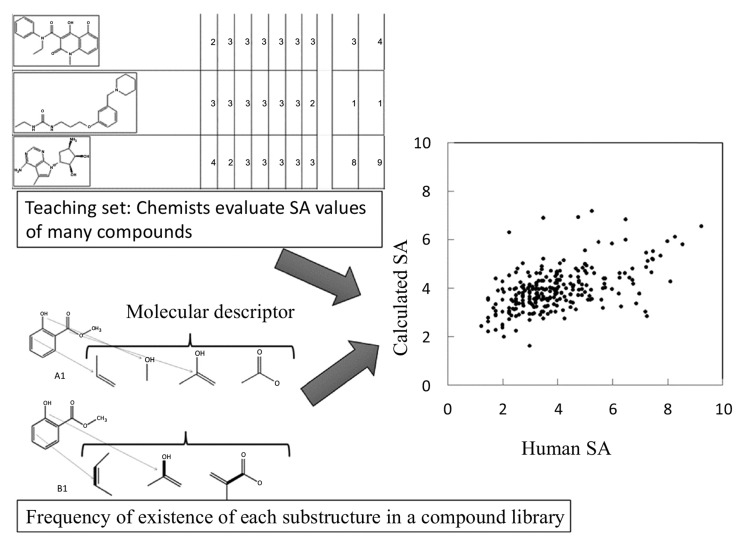
A quantitative SA prediction method was reported by Takaoka et al. (2003) [13]. This method had two important features. First, it is able to predict SA values based on only the chemical structure of the compound in question and thus does not require a reaction database. Second, the method has already been assessed in a study in which expert manual assessment was used to evaluate the SA (in the original paper, “synthetic easiness”) of compounds in a teaching data set. The compounds in the teaching data set were described by molecular descriptors, and the weights of the descriptors were determined to reproduce the SA. This approach worked well, suggesting that SA is useful.
Of course, SA is not a well-defined concept but a fuzzy idea [14]. Many years ago, people thought that organic compounds could not be synthesized by humans but only by God. In the 19th century, organic compounds were made from charcoal and organic polymers could not be synthesized. In the middle of the 20th century, many reactions were developed based on oil. Thus, the SA values of compounds would have changed in each epoch along with the available organic chemistry.
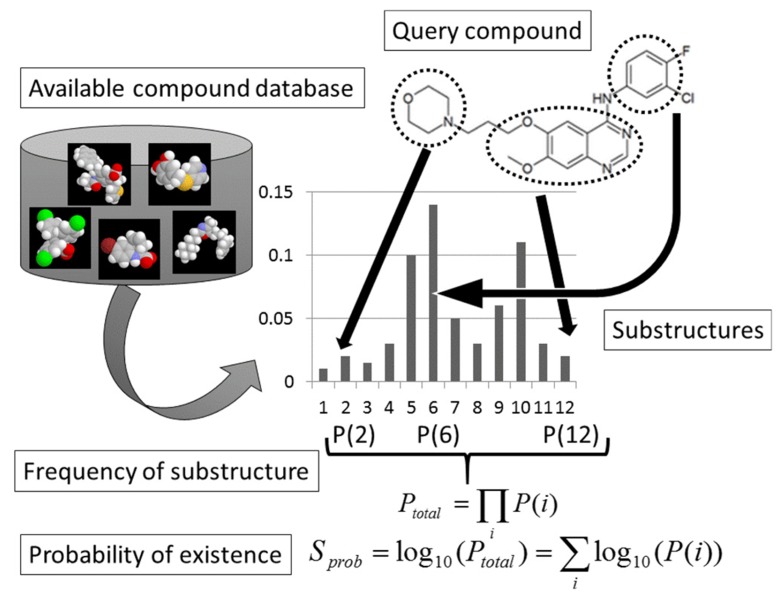
The latest methods are based on a commercially available compound database and molecular descriptors. In these methods [13, 15], reaction databases and retro-synthesis analyses are not necessary (see Fig. 5). Instead, SA is estimated from the probability of the existence of substructures of the compound calculated based on a compound library, the number of symmetry atoms, and the number of chiral centers in the compound. A steric effect, such as that of atomic collision, could be partially taken into consideration by the probability of the existence of substructures of the compound. It has recently become easier to access free compound databases, such as PUBChem [19], ChEMBL [20], ZINC [21, 22], and LigandBOX [23, 24], and many vendors have made the catalogs of their compounds (see Table 2). In general, the databases of compound structures are more reliable than the reaction databases.
Fig. (5).
Substructure-based synthetic accessibility prediction.
Table 2.
List of major chemical vendors and number of compounds supplied by them.
| Vender Name | Year | ||||
|---|---|---|---|---|---|
| 2015 | 2014 | 2013 | 2008 | 2006 | |
| Ambinter | 120764 | 101720 | |||
| AMRI | 195420 | ||||
| ACB Blocks(Fragment) | 1291 | 1292 | 1292 | ||
| Alinda | 186836 | 254092 | |||
| AnalytiCon(Fragment) | 222 | 218 | 204 | ||
| AnalytiConMACROx | 1118 | 1027 | |||
| AnalytiConMEGx | 5130 | 4579 | 4855 | 1092 | 522 |
| AnalytiConMEGxp | 2799 | 2066 | |||
| AnalytiConNATx | 26282 | 25498 | 24264 | 19080 | 9640 |
| Aronis | 45746 | ||||
| ART-CHEM | 185637 | 164136 | |||
| ASDI | 105587 | 42578 | |||
| Asinex | 402046 | 426345 | 426345 | 233540 | 236765 |
| Asinex(Fragment) | 23191 | 10412 | 3479 | 9315 | |
| AsinexPlatinum | 16240 | 126615 | 132155 | ||
| AsinexSynergy | 10740 | ||||
| Aurora | 24647 | 59164 | |||
| Bahrain | 998 | 998 | |||
| Bellen(Fragment) | 2911 | 2911 | |||
| BioBioPha(Natural) | 3283 | 2910 | 2675 | ||
| BioMar | 128 | ||||
| Bionet | 52025 | 47191 | 41618 | 46384 | 43410 |
| Bionet(Fragment) | 25906 | 20584 | 13127 | ||
| ButtPark | 21926 | 21926 | 21926 | 17273 | 14212 |
| CBI | 125594 | 125230 | |||
| ChemOvation | 3121 | 3143 | 3143 | 2922 | |
| Chem-X-Infinity | 11650 | 11825 | 13148 | 332 | |
| Chem-X-Infinity(Fragment) | 1156 | 727 | 700 | 2400 | |
| Cerep | 29230 | ||||
| Chemstar | 60081 | 60081 | |||
| ChemT&I | 844248 | 641842 | |||
| Enamine | 2089786 | 2056024 | 1883713 | 952314 | 506809 |
| Enamine(Fragment) | 42044 | 157022 | 153444 | ||
| EvoBlocks(Fragment) | 944 | 944 | 700 | ||
| Florida | 30910 | 30419 | |||
| FSSI | 102509 | ||||
| He.Co. | 7087 | 6136 | |||
| Innovapharm | 324568 | ||||
| Intermed | 32037 | 50295 | |||
| Labotest | 80096 | 81805 | 82296 | 107737 | 97794 |
| LifeChemicals | 392633 | 378162 | 333920 | 267020 | 147680 |
| LifeChemicals(Fragment) | 32877 | 30042 | |||
| Maybridge | 54228 | 54575 | 54768 | 56839 | 58854 |
| Maybridge(Fragment) | 8956 | 8915 | 9273 | ||
| MDD | 31254 | 31255 | |||
| MDPI | 11189 | 19749 | 10649 | 10649 | |
| Menai | 5005 | 5005 | 5210 | 5017 | |
| Menai(Fragment) | 402 | 402 | 402 | ||
| MolMall | 14415 | ||||
| OTAVA | 279563 | 263086 | 254048 | 118173 | 62091 |
| OTAVA(Fragment) | 11719 | 8748 | 7810 | ||
| Peakdale | 14636 | 6072 | 8548 | ||
| Peakdale | 6257 | 6572 | |||
| Pharmeks | 396620 | 396620 | 376899 | 276367 | 228630 |
| Pharmeks(Natural) | 49587 | 49587 | 44840 | 31720 | 29038 |
| Princeton | 1160008 | 1012682 | 416068 | 873993 | 517484 |
| RareChemicals | 11279 | 11279 | 11279 | 15245 | 9375 |
| SALOR | 174978 | 142206 | 186899 | 142876 | 137428 |
| ScientificExchange | 47833 | 47843 | 47733 | 43964 | 34569 |
| SPECS | 207973 | 199969 | 199969 | 199270 | 151570 |
| SPECS(Natural) | 866 | 456 | 456 | 335 | |
| Synthon Labs | 32706 | 49727 | |||
| TimTec | 130361 | 128078 | 1066286 | 308111 | 198599 |
| UOS | 565990 | 540678 | 457256 | 700730 | 331710 |
| VillaPharma | 4612 | ||||
| Vitas-M | 1319150 | 1321860 | 1163246 | 311813 | 220970 |
| Vitas-M(Natural) | 24694 | 24694 | |||
| Vitas-M(Fragment) | 18909 | 18909 | 8315 | ||
| WuxiAppTec | 104245 | 93892 | 81983 | ||
| WuxiAppTec(Fragment) | 1155 | 908 | |||
| WuxiAppTec(Natural) | 1417 | 4565 | 4689 | ||
| Zelinsky | 386127 | 386127 | 385559 | ||
| Toal number of entities | 8497227 | 8884648 | 7908899 | 6542933 | 4435139 |
| Nonredundant Total Number | 5,150,322 | 5,026,965 | 4,740,609 | 4,206,460 | |
| Number of New chemical entiries | 229,015 | 157,820 | 195,377 | ||
Table 2 shows trends in the number of compounds provided by some of the major compound vendors available on the LigandBOX database (URL http://ligandbox.protein.osaka-u.ac.jp/ligandbox//cgi-bin/index.cgi?LANG=en). As shown in this table, the total number of commercially available compounds (stocks) has not changed for several years. Every year, several hundred-thousand newly designed compounds appear and almost the same numbers of compounds are sold out. Thus the total number of compounds in stock has not changed substantially. Also, the number of natural compounds has been increasing gradually. On the other hand, the number of fragments has been increasing recently, since fragment-based drug discovery became a trend in medicinal chemistry.
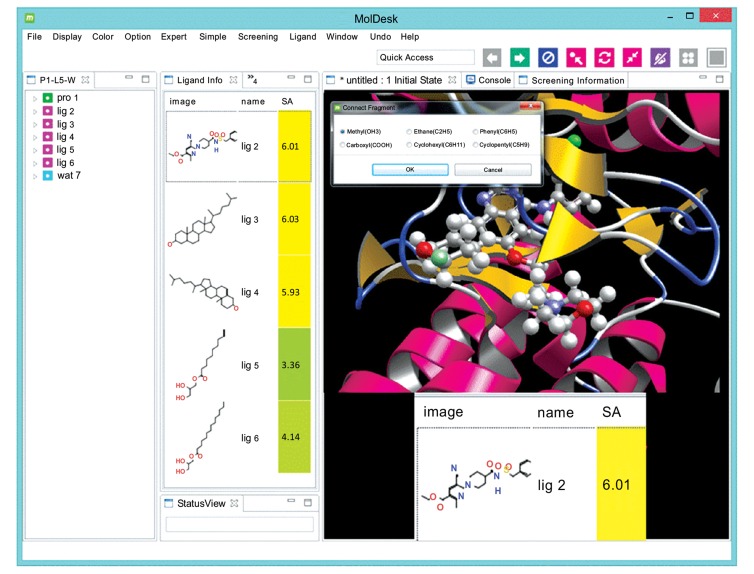
Fig. (6) shows one example of an SA prediction algorithm included in the MolDesk software package [17] (URL: http://www.moldesk.com/). This routine performs the SA prediction within about 0.1 seconds for a given compound with one click manipulation.
Fig. (6).
An example of SA prediction by a commercial program (MolDesk GUI software. Information and Mathematical Science Bio Inc., Tokyo Japan).
In the MolDesk software package, SA is calculated as follows:
| Eq. 1 |
| EQ2. |
where c0, c1, c2, c3, and c4 are fitting parameters. Parameters c0-c4 are optimized to reproduce the SA determined by expert manual assessment. In this review, the SA determined by expert manual assessment is designated the “human SA” and the SA calculated by Eq. 1 is called the “calculated SA.” SA is estimated from the probability of existence (Sprob) calculated based on a compound library, the number of symmetry atoms (Nsym), the total number of atoms (Ntot) of the compound, the number of chiral centers of the compound (Nchiral), and the graph complexity (Sgraph-complexity) [25]. The Nsym is the number of chemically (topologically) equivalent atoms.
If a compound consists of fragments frequently found in an available-compound database, it should be easy to synthesize. On the other hand, if a compound consists of fragments rarely or never found in an available-compound database, it would be difficult to synthesize. The probability of existence (Sprob) was calculated on the basis of the decomposition of the compound into fragments, and the probability of existence of each fragment was estimated according to the compound library. Any kind of substructure descriptor (the extended connectivity fingerprint (ECFP) descriptors developed by SciTegic [15], MACCS key, Dragon, etc.) could be used for the SA prediction.
All compounds in the library were decomposed into small fragments. Let N and N(i) be the total number of fragments found in the library and the total number of fragments found in the library that were exactly the same as the i-th fragment of the compound in question, respectively. The probability of existence of the i-th fragment in library (P(i)) is given by
| Eq3. |
The total probability of existence of the compound in question is then given as
| EQ4. |
and
| EQ5. |
As shown in Fig. (7), the compound library should consist of already-synthesized available compounds.
Fig. (7).
Substructure-based synthetic accessibility (SA) prediction. SA is estimated from the probability of existence (Sprob) calculated based on a compound library, the number of symmetry atoms (Nsym), the total number of atoms (Ntot) of the compound, the number of chiral centers of the compound (Nchiral), and the graph complexity (Sgraph-complexity).
The correlation coefficients between the predicted SA values and the human SAs are about 0.5-0.8 for these prediction methods. The R value (0.56) and the average error (1.2) obtained by MolDesk are similar to those between the human SAs (R=0.59 with a standard deviation of 0.22 and average error=1.1).
4. Correlation among human SAS
Takaoka et al. reported that the human SA values (synthetic accessibility (or synthetic easiness in the original text) values of visual inspection by individuals) were slightly dependent on the individual chemists, even within the same company. Nonetheless, while the concept of SA remains somewhat fuzzy and poorly defined in the manner of concepts like “good” or “bad,” such abstractions are often quite useful.
Tables 3 -6 show the correlations among human SAs reported in previous articles [7, 10, 17]. The human SAs were strongly dependent on the skill and experience of the individual chemist. The SA values estimated by the 2 chemists who belonged to the same company were still similar to each other. The SA values estimated by the chemists from different companies were different from each other and showed almost no correlation. SA would be expected to depend on the equipment available at the individual company, as well as on the training and experience of the company chemists.
Table 3.
Correlation coefficient between human SAs evaluated by two chemists.
| FMP1 | FMP2 | NARD1 | NARD2 | RASA | SYLVIA | Average | |
|---|---|---|---|---|---|---|---|
| FMP1 | 1.00 | 0.93 | 0.57 | 0.56 | 0.35 | 0.37 | 0.56 |
| FMP2 | 0.93 | 1.00 | 0.55 | 0.53 | 0.28 | 0.41 | 0.54 |
| NARD1 | 0.57 | 0.55 | 1.00 | 0.86 | 0.47 | 0.94 | 0.68 |
| NARD2 | 0.56 | 0.53 | 0.86 | 1.00 | 0.44 | 0.92 | 0.66 |
| RASA | 0.35 | 0.28 | 0.47 | 0.44 | 1.00 | N.D. | 0.39 |
| SYLVIA | 0.37 | 0.41 | 0.94 | 0.92 | N.D. | 1.00 | 0.66 |
FMP: Fujimoto Chemicals. NARD: Nard Institute.
Table 6.
Correlation coefficient between human SAs evaluated by five chemists reported by Taisho Pharmaceutical Co., Ltd.
| Taisho | Chemist 1 | Chemist 2 | Chemist 3 | Chemist 4 | Chemist 5 | Average |
|---|---|---|---|---|---|---|
| chemist 1 | 1.00 | 0.50 | 0.40 | 0.40 | 0.56 | 0.47 |
| chemist 2 | 0.50 | 1.00 | 0.42 | 0.47 | 0.52 | 0.48 |
| chemist 3 | 0.40 | 0.42 | 1.00 | 0.40 | 0.48 | 0.43 |
| chemist 4 | 0.40 | 0.47 | 0.40 | 1.00 | 0.48 | 0.44 |
| chemist 5 | 0.56 | 0.52 | 0.48 | 0.48 | 1.00 | 0.51 |
Relationship between the sales price of approved drugs and SA
In Japan, about 10% of approved drugs are not sold in the market, since the drug prices do not meet the drug development costs. Thus the drug price is an important factor in drug development. We examined the relationship between the sales price and SA of approved drugs in the United States and Japan (US) [17]. Of course, the price of a drug is critical to its practical adoption by patients. For this reason, the ability to predict the price of a new drug in the drug-design process would be highly useful.
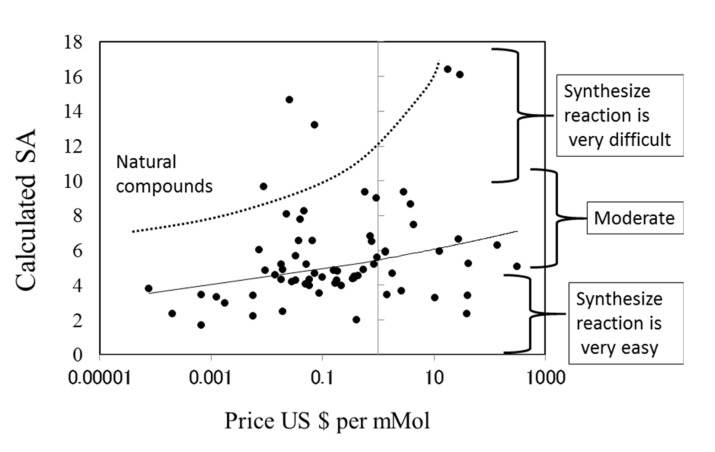
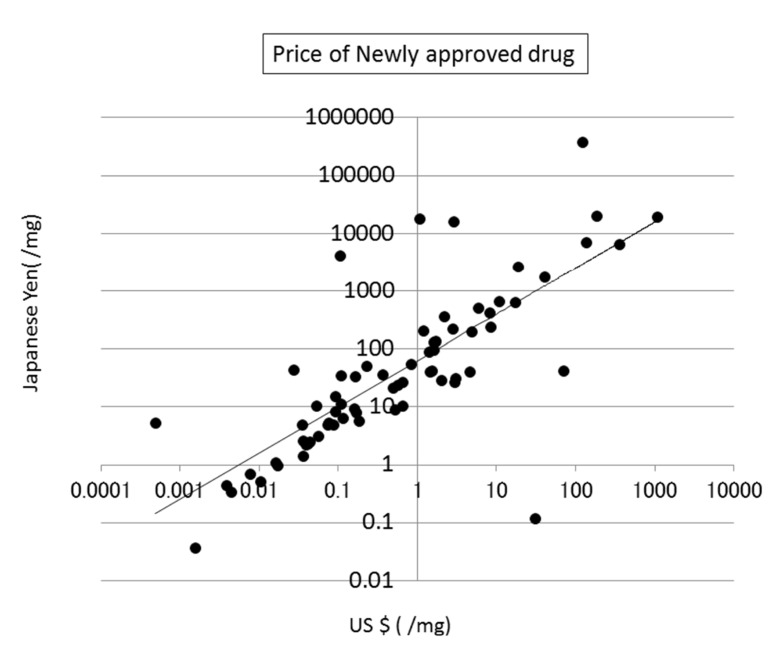
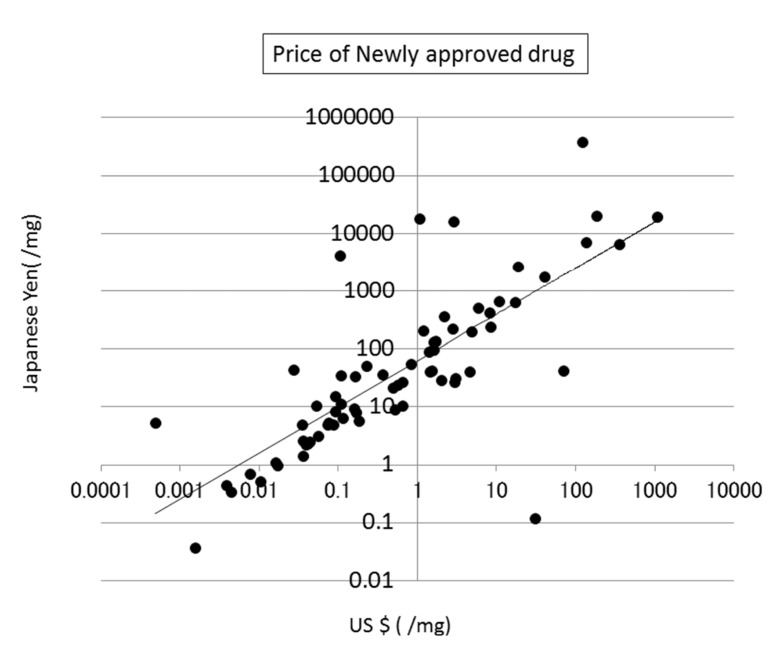
The prices of new drugs in the United States and Japan showed a weak correlation to the calculated SA values. Their correlation coefficients were 0.32 and 0.29, respectively (see Fig. (8)). The prices of generic drugs showed the same trends as the new drugs. The reason for this should be that the price of a generic drug follows the price of the original drug.
Fig. (8).
Correlation between SA and the price of newly approved drugs in the US.
The price of a drug may depend on many kinds of costs, including the costs of development, phase trials, and patents, as described in the next section (see Fig. (9)). In addition, the market size and efficacy of the drug should be considered to decide the price. This means that a drug that is difficult to synthesize is not always expensive. The average SA of a drug is about 6, meaning that the average difficulty of drug synthesis is not extremely high.
Fig. (9).
Setting of drug prices in the US and Japan. The setting of drug prices depends on the country, type of disease and type of drug.
Drug prices in the US and Japan
Because different countries have different methods of setting the prices of drugs prices, these prices vary widely around the world. In the US, the pharmaceutical companies determine drug prices based on a capitalist paradigm. On the other hand, in Japan the government sets the price of drugs based on their estimated importance to the healthcare system. These differences also reflect the different health insurance systems in each country. Thus a comparison between the drug prices in the US and Japan should reveal differences between a free market-based and government-based healthcare system.
There is almost no correlation between the sales prices of drugs in the US and Japan [17]. On the other hand, Fig. (10) shows the correlation between the logarithm of the price of drugs in the US and the price in Japan. The correlation is very high (R = about 0.8). This means that the prices of drugs are roughly estimated and the values of drugs could change depending on each society.
Fig. (10).
Correlation between the sales prices of drugs in the US and Japan.
Cloud computing in CADD
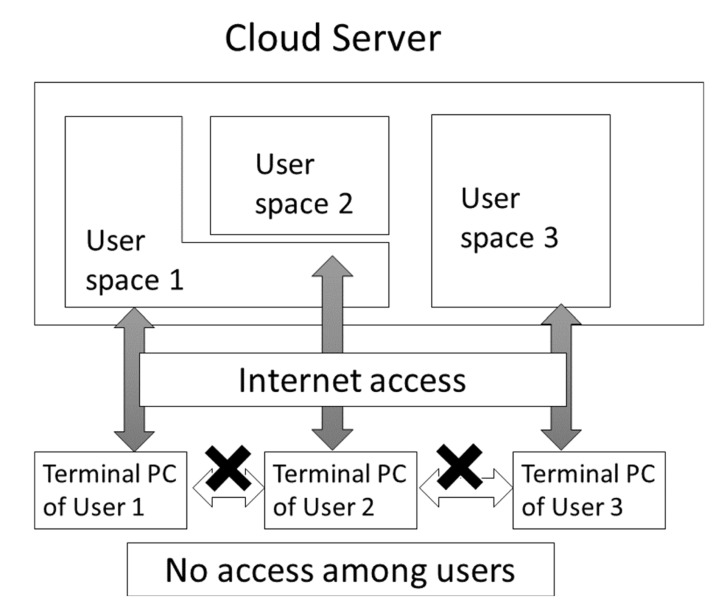
Cloud computing has become a quite popular technology. Usually, the cloud computer is a large-scale computer and computer resources such as CPU and disk space are served on demand (see Fig. (11)). Two of the major cloud computer services are the Amazon Web Service (AWS: URL https://aws.amazon.com/?nc1=h_ls)
Fig. (11).
Rough sketch of access control in cloud computing.
and AZURE by Microsoft (URL: https://azure.microsoft.com/ja-jp/). The cloud computer is very similar to the conventional server computer. The difference between the cloud and conventional server is that the cloud is a virtual machine on which the OS, middleware, programs and computational environment must be prepared for application computing by users in general, and the virtual machine disappears when the users deallocates the virtual machine. Although this procedure (allocation and deallocation of the virtual machine) enables computers to be used flexibly, it is also complicated and time-consuming. The other important aspect of cloud computing is the requirement of high security or privacy. On a conventional server, we can see the status of other users’ jobs. Conversely, we cannot see any information on the other users of a virtual machine. The scalability and high security have enabled many pharmaceutical companies to perform their virtual screenings, similarity searches and MD simulations on cloud computers.
Cloud computing also enables us to access specialized hardware. The general purpose graphics processing unit (GPU) computational resource is somewhat troublesome for many users. Since the evolution of the GPU hardware is very fast, the GPU software is strongly dependent on the hardware, and the CUDA software (URL: https://developer.nvidia.com/cuda-zone) depends on the GPU hardware. Every year, new GPU hardware has appeared and the CUDA version has been updated. The GPU program should be tuned up for each GPU, since the performance of GPU programs depends on the balance of the number of GPU cores and memory-band width. Also the application of GPU is quite limited. This means that the GPU is used only when the GPU programs are available. In contrast, CPUs are always used for all application programs.
One of the problems of cloud computing is that the cloud machine is a virtual machine that basically begins as an empty box in cases where we allocate our own resources. We must prepare our own computational environment before starting target computations such as virtual screenings or MD simulations. This is almost equivalent to the procedure of setting up a brand new machine, and it must be done each time a new connection is made to the cloud computer. The other problem is that we must pay for the number of computations and in many cases it is difficult to predict the computational cost before the computations have actually been run. Finally, it is necessary to retrieve the data from the cloud computer before the close of service on each use.
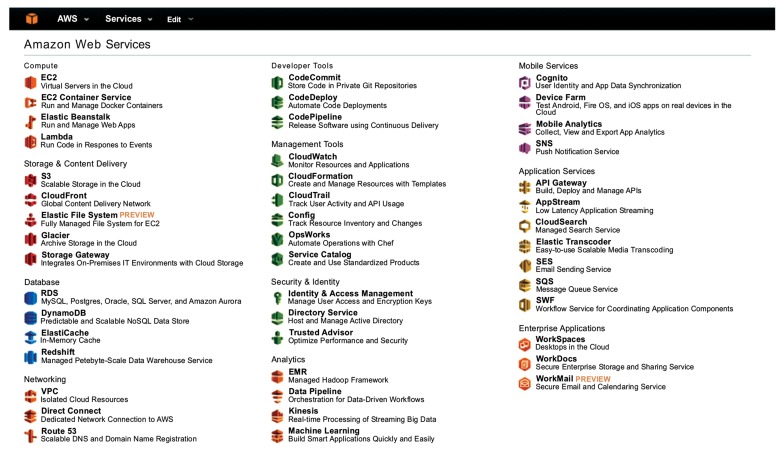
AWS provides more than 40 services (see Fig. (12)), including CPU, disk, database, and security resources. Most of the charges for these services are proportional to the amount of usage of the cloud services. To realize a scientific calculation, the users must integrate and combine these cloud services adequately. Because there are so many cloud services, selecting the best combination for each scientific calculation can be hard work for the users. In addition, the users need resources on demand. Instead of asking the system providers, in some cases the users can manage their cloud computing using the available cloud services.
Fig. (12).
Service menu of Amazon cloud computing.
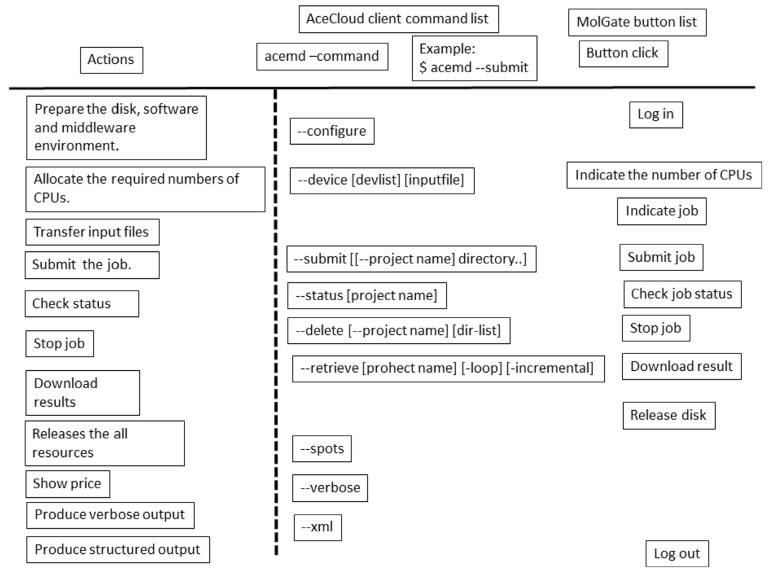
The cloud controller software assists in the process of preparing the computational environment and retrieval of data. There have been several reports on this software. Since the explanation of the mechanism of the cloud controller software is beyond the scope of this review, we will simply explain how to use such software. The controller software depends on the kind of computations to be made. Thus, we must select the best controller software for the computer programs that we want to use. AceCloud is a command-line cloud controller [26]. The users can submit their jobs, check the job status, abort them, and retrieve the results through AceCloud (see Fig. 13).
We show one example of the commercial cloud controller software “MolGate” (BY-HEX LLP, Tokyo Japan. URL http://by-hex.com/ 2015) in Fig. (13). MolGate is a web-based program with SSH communication and the users can use both the CPU and GPU resources. When input files are prepared for MD simulation a priori, MolGate can prepare the environment on the cloud and start the calculation within 10 minutes. The example shows how to perform MD simulation on AWS. Fig. (13) shows the procedures followed by the user. The user must allocate and deallocate the computational resources manually in addition to performing the login process. The procedure consists of the following four steps.
Fig. (13).
Command list for cloud computing.
Step 1. Users open “MolGate” and connect to AWS. They then indicate what kind of server and how many servers are needed for the computation. There are about 40 types of servers available. These include m1.large, m1.xlarge, m2.4xlarge, cg1.4xlarge, cc2.8xlarge, c4.8xlarge, c4.2xlarge, etc. (https://aws.amazon.com/ec2/pricing/?nc1=h_ls. Last access: 2015/09/26). Each server includes CPU cores, memories and hard disks.
Step 2. Users select the data that should be calculated on the local machine. MolGate prepares the computational environment for this calculation as a background job. The simulation programs, analysis programs and MPI environment are prepared on AWS automatically. Users select the data that should be calculated on the local machine. MolGate sends or unzips the zipped files.
Step 3. Users indicate the number of servers and estimate the CPU cost of the job. MolGate calculates the expected CPU time for the job. Users should change the server type and/or modify the input file for the simulation if necessary. Then users perform their job.
Step 4. Users retrieve the resulting data after the job is finished.
On AWS, users can see only their virtual machine. Once the CPU resources are allocated, users will not be bothered by other users. Most of the costs of cloud computing are due to the CPU time, while the disk space usage is very inexpensive or even free in many cases.
To use the cloud computer, we should estimate the cost of the job a priori [27]. The computational cost depends on the size of the simulation system, the duration of the simulation, what kind of program will be used, and which service (type of server machine) of the cloud will be used. BY-HEX LLP provides a service for predicting the cost (URL: http://by-hex.com/). The total number of atoms is entered into the simulation system, and the site estimates the price per one MD simulation step. This prediction is still primitive, but the service is useful.
Machine setup: GPU and compound database
GPU computation has also become very popular [28-36]. GPU computing is particularly suitable for performing molecular dynamics simulation programs like AMBER [33], Gromacs [34], NAMD [35] and psygene-G/myPresto [36]. The computational details of these programs are described in detail elsewhere. Some of these GPU programs are freely available. One of the most serious problems for end users is how to set up the GPU machine for these MD programs. The other problem is that the system size for the GPU computation must be larger than the minimum size that is determined by the program. Since most GPU programs adopt a space-decomposition method for parallel computing, the system must be decomposed into sub systems. This means that the MD of a small system (like a single molecule) is not suitable for GPU computing.
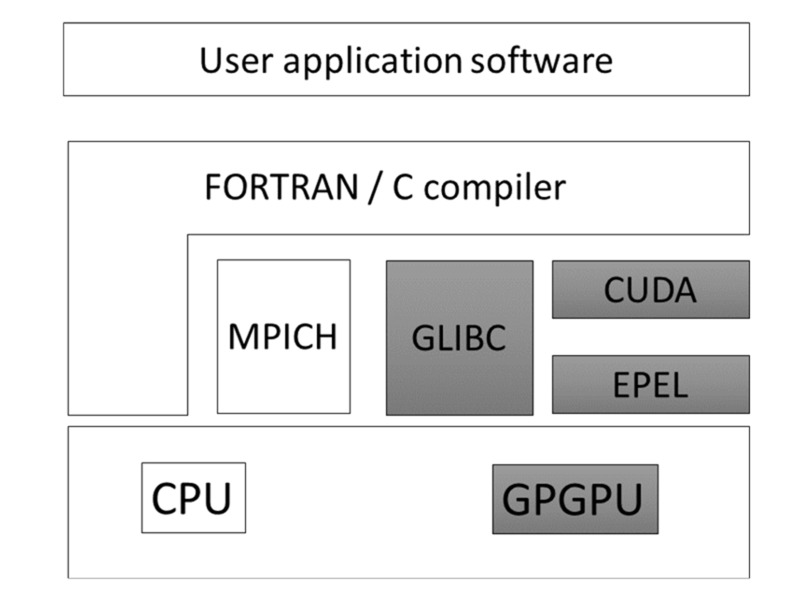
Fig. (14) shows how to set up the MD program for the GPU. Many computers have GPU boards that are used mainly for graphics. To utilize the GPU for an application program, we must uninstall the GPU graphics driver software and install CUDA, a computer language for use with GPU. Most of the GPU-MD programs adopt CUDA for GPU computations. The slot number of each GPU board in the computer must be explicitly indicated in CUDA. Suppose our machine has three GPU boards. In some cases, a poor GPU is allocated to slot 1 for graphics. The additional two expensive GPU boards have slots 0 and 2 for the MD simulation. In some cases, a poor GPU is allocated to slot 0 for graphics. The additional two expensive GPU boards have slots 1 and 2 for the
Fig. (14).
Software and hardware GPU framework.
MD simulation. This allocation depends on the machine. If the GPU programs use a slow and a fast GPU board, the computation speed depends on the speed of the slow GPU board. This setup job is somewhat time-consuming work. The user can request that the computer vendor set up the machine, or in some cases, the vendor will provide a pre-installed machine such as MolSpace (LEVEL FIVE Co., Ltd. Tokyo; URL: http://www.level-five.jp/).
In this section, we explain how to start using the GPU computation, since the set up the environment for the GPU computation is a much more time-consuming and complex process than the conventional CPU. Most of the GPU programs run on the CUDA language. This means that the GPU of the Intel Core-i series is not suitable for GPU computing, since the GPU of core-i is not designed for CUDA. Many computers utilize the GPU card for graphics. To use the GPU card for a scientific calculation program, we must stop using the GPU for graphics and change the status of the GPU card for application only. Our example is designed for CUDA7.0 running on Linux x86_64 (RedHat6 (RHEL6) and CentOS6). The RPM package of CUDA for RHEL6/CentOS6 is available from the NVIDIA CUDA download website (https://developer.nvidia.com/cuda-downloads).
In order to use GPUs for running CUDA application programs, we must check the versions of the software and hardware. For example, CUDA for CentOS6 runs on the combination of kernel 2.6.32, GCC version 4.4.7, and GLIBC version 2.12. Each CUDA version runs on each GPU card. We must check what kind of GPU card is available on the CUDA GPUs site (https://developer.nvidia.com/cuda-gpus). These points can be checked using the following 5 steps.
Step 1. Check the version of the OS (obtained by “$ cat /etc/*release”).
Step 2. Check the version of the kernel (obtained by “$ uname -a”).
Step 3. Check the version of GCC (obtained by “$ gcc --version”).
Step 4. Check the version of GLIBC (obtained by “$ rpm -q glibc”).
Step 5. Check the GPU card (obtained by “$ lspci | grep -i nvidia”).
In the present review, we explain how to set up the GPU for MD simulation mainly for an NVIDIA CUDA environment for RedHat6/CentOS6, since CUDA is now popular for GPU simulations. In order to use the NVIDIA driver PRM package, an EPEL package must be installed. This process should be done by following 7 steps as follows.
Step 1. Download a suitable EPEL, such as # yum install (http://dl.fedoraproject.org/pub/epel/epel-release-latest-6.noarch.rpm).
Step 2. Install the CUDA on the command mode (run level 3), then set the run level using the command “# /sbin/init 3.”
Step 3. Install the CUDA repository (”# rpm --install cuda-repo-rhel6-7-0-local-7.0-28.x86_64.rpm”).
Step 4. Clear the cache of yum (“# yum clean expire-cache”).
Step 5. Install CUDA by yum (“# yum install cuda”). Answer yes (“y”) to all the questions.
Step 6. Restart the computer system by typing the command ”# /sbin/shutdown -r now.”
Step 7. Change the default parameter of EPEL. Change “enable=1” to “enable=0” for [epel] in the file “/etc/yum.repos.d/epel.repo”.
In order to compile our GPU application on our computer, we must set the path for the CUDA library. We add the following two lines in ~/.cshrc for c-shell or ~/.bashrc for bash.
export PATH=/usr/local/cuda/bin:$PATH
export LD_LIBRARY_PATH=/usr/local/cuda/lib64:$LD_LIBRARY_PATH
Before starting our own application calculations, we check the software and hardware environment.
We download the sample program for CUDA and compile it using the following two steps.
Step 1. Put the CUDA sample on an suitable directory. For example, to put the sample on the current directory (.), we use the command “$ cuda-install-samples-7.0.sh .”.
Step 2. Compile the sample using the command (”$ cuda-install-samples-7.0.sh .”).
In the above section, we have shown how to prepare the CUDA environment for GPU applications. Now we can start the GPU applications. There are several GPU programs available on the Internet, and most of these are MD simulation programs. We will explain how to use psygene-G of the myPresto program suit developed by our group, since most of the other application programs should follow a similar procedure. Psygene-G is available on the myPresto download site (http://presto.protein.osaka-u.ac.jp/myPresto4/); after downloading, unzip the file using any suitable directory.
Psygene-G is an MPI parallel GPU program [36]. To compile this program, we must modify Makefile for our software environment. There are two Makefiles. One is for the MPI parallel program part (src/Makefile) and the other is for the CUDA program part (src/cuda/Makefile). Users should modify the Makefile of each application program in a manner similar to the following example.
Modification of Makefile for the MPI program (src/Makefile) to GPU in this example:
The CUDA version is indicated as “CUDA=cuda.x.y” for CUDA version x.y.
Activate the “#GPU Lib settings” block.
Psygene-G is written in FORTRAN90, and the MPI library for FORTRAN90 is indicated by “FC=mpif90”.
This is psygene-G specific indication. The memory size for a million atoms is indicated by “OPTIONS = -D_LARGE_SYSTEM”

Modification of Makefile for the CUDA program (src/cuda/Makefile) to GPU in this example
The CUDA version is indicated as “CUDA=cuda.x.y” for CUDA version x.y.
Compute capability that is the version of GPU card as described on the CUDA GPUs site (https://developer.nvidia.com/cuda-gpus). For Tesla K20, the compute capability is indicated as “GEN_SM = -arch=sm_35”.
The option of CUB library (for CUDA5.0 and more recent version) is indicated. “USE_CUB=1” for psygene-G, and USE_CUB=0 means the use of Thrust library that is a supplement of CUDA.
[ src/cuda/Makefile ].

If both Makefiles are correctly modified, psygene-G should be compiled by the make command in the form “$ make”.
In the case of psygene-G/myPresto, the control file (input file) for MD simulation is almost the same as that for the conventional MD simulation program for CPU. The following explanation is myPresto-specific, but a similar modification would be necessary in order to use the other MD programs. The recent MD simulation program adopts the space division technique with cut-off interaction calculations like the reaction field. We explain this to help understand the input file.
The coordinate of the center of the system is indicated. “CENTRX= 0.0D0 CENTRY= 0.0D0 CENTRZ= 0.0D0”
The size of the system is indicated. “LXCELL= 0.75D+02 LYCELL= 0.75D+02 LZCELL= 0.65D+02”
How to divide the system. In this example, the system cell is divided into 2x2x2 sub-cells. “DXCELL= 2 DYCELL= 2 DZCELL= 2”
[ md.inp-EXE> SPACEINPUT ]
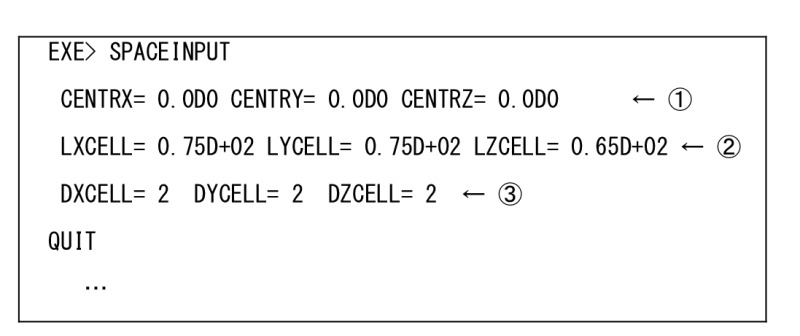
Input for GPU for psygene-G
How to allocate the GPU device. Multiple GPU cards are available by the MPI. “GPUALC=MPI” means the GPU cards are allocated by the MPI automatically. “GPUALC=RNK” means the GPU-allocation is defined by the machinefile.
The GPU-device numbers are indicated. “USEDEV=0,2” means that the GPU devices 0 and 2 are used for the calculation, In many cases, GPU device 1 is used for the graphics (depending on the hardware vendors).
“NBDSRV= GPU” means that the GPU cards are activated.
The GPU kernel is indicated by option NBDKNL. “NBDKNL= GRID” means that the space-decomposition method is used for GPU calculation.
The precision of the GPU kernel is indicated. The recent GPU cards can calculate in both single precision and double precision; however, the single precision calculation is much faster than the double precision calculation. Thus, we must use the single precision calculation on GPU boards whenever possible. In psygene-G, each pairwise interaction calculation between atoms is calculated in single precision, but the summing-up of these results is performed in double precision..
[ md.inp-EXE> GPU ]

[ machines ]

In this machine file example, ranks 0-3, ranks 4-5, and ranks 6-7 are allocated to host0, host1 and host2, respectively.
Finally, we can perform the GPU calculation.
Psygene-G calculation is performed by “$ psygene-G < md.inp.
Recently, an integrated solution system for CADD appeared to reduce the above procedure for setting up the GPU environment. MolSeries is a system that includes MolSpace (workstation that provide GPU and execution environment for myPresto computer-aided drug design program suit etc.) (LEVEL FIVE Co., Ltd. Tokyo Japan), MolDesk (pre-installed CADD software) (Information and Mathematical Science Bio Inc., Tokyo Japan), and MolGate (cloud controller) (BY-HEX LLP. Tokyo Japan). Users can perform the MD simulation with GPU computing, protein-compound docking, virtual screening, drug design, and synthetic-accessibility prediction using both the local machine and the cloud system without the set-up procedure.
Conclusion
In the present work, we reviewed the concept of synthetic accessibility prediction as a minor part of the CADD programs, and we reviewed the software / hardware environment supporting the CADD in greater depth, such as cloud computers and GPU.
Synthetic accessibility (SA) prediction has become a practical tool in CADD. There are two types of SA predictions. One is based on prediction of the reaction path and the other is based on the chemical structure only. These programs are now available even for commercial use. The predicted SA showed good agreement with the SA estimated by chemists, with a correlation coefficient of 0.5-0.8. Considering that the correlation coefficient between the SAs estimated by chemists is around 0.6, the computational prediction works well. Since SA is a fuzzy idea, different chemists can propose different SA values for the same given compound. The drug prices are only weakly dependent on the SA values (R=0.3). Thus the evaluation of drug prices is difficult work.
The use of cloud computing is still limited compared to the wide application of conventional computing on server computers. Currently, chemical computations on the cloud are relatively harder than those on local machines or ordinary server computers, although some cloud controllers are now available and these programs should help laypeople to perform various computations on the cloud. GPU computing has been popular. But setting up the GPU for chemical computation is still a difficult task. To use cloud and GPU computing, a support service and supporting software would be helpful. In the future, these relatively new methods should be actively studied.
Table 4.
Correlation coefficient between human SAs evaluated by five chemists reported in RASA software.
| RASA | Chemist 1 | Chemist 2 | Chemist 3 | Chemist 4 | Chemist 5 | Average |
|---|---|---|---|---|---|---|
| chemist 1 | 1.00 | 0.83 | 0.76 | 0.76 | 0.74 | 0.77 |
| chemist 2 | 0.83 | 1.00 | 0.84 | 0.78 | 0.74 | 0.79 |
| chemist 3 | 0.76 | 0.84 | 1.00 | 0.84 | 0.81 | 0.81 |
| chemist 4 | 0.76 | 0.78 | 0.84 | 1.00 | 0.82 | 0.80 |
| chemist 5 | 0.74 | 0.74 | 0.81 | 0.82 | 1.00 | 0.78 |
Table 5.
Correlation coefficient between human SAs evaluated by five chemists reported in SYLVIA software.
| SYLVIA | Chemist 1 | Chemist 2 | Chemist 3 | Chemist 4 | Chemist 5 | Average |
|---|---|---|---|---|---|---|
| chemist 1 | 1.00 | 0.75 | 0.77 | 0.84 | 0.74 | 0.78 |
| chemist 2 | 0.75 | 1.00 | 0.78 | 0.73 | 0.74 | 0.75 |
| chemist 3 | 0.77 | 0.78 | 1.00 | 0.82 | 0.75 | 0.78 |
| chemist 4 | 0.84 | 0.73 | 0.82 | 1.00 | 0.81 | 0.80 |
| chemist 5 | 0.74 | 0.74 | 0.75 | 0.81 | 1.00 | 0.76 |
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institute of Advanced Industrial Science and Technology (AIST), the New Energy and Industrial Technology Development Organization of Japan (NEDO), the Japan Agency for Medical Research and Development (AMED) and the Ministry of Economy, Trade, and Industry (METI) of Japan.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Schneider G., Fechner U. Computer-based de novo design of drug like molecules. Nat. Rev. Drug Discov. 2005;4:49–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- 2.Baber J.C., Feher M. Predicting synthetic accessibility: Application in drug discovery and development. Mini Rev. Med. Chem. 2004;4:681–692. doi: 10.2174/1389557043403765. [DOI] [PubMed] [Google Scholar]
- 3.Gillet V., Myatt G., Zsoldos Z., Johnson A. SPROUT, HIPPO and CAESA: Tool for de novo structure generation and estimation of synthetic accessibility. Perspect. Drug Discov. Des. 1995;3:34–50. [Google Scholar]
- 4.Lewell X.Q., Judd D.B., Watson S.P., Hann M. RECAP-retrosynthetic combinatorial analysis procedure: A powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 1998;38:511–522. doi: 10.1021/ci970429i. [DOI] [PubMed] [Google Scholar]
- 5.Pförtner M., Sitzmann M., Gasteiger J., editors. Handbook of Cheminformatics. Weinheim: Wiley-VCH; 2003. pp. 1457–1507. [Google Scholar]
- 6.Johnson A., Marshall C., Judson P. Starting material oriented retrosynthetic analysis in the lhasa program. 1. general description. J. Chem. Inf. Comput. Sci. 1992;32:411–417. [Google Scholar]
- 7.Huang Q., Li L.L., Yang S.Y. RASA: A rapid retrosynthesis-based scoring method for the assessment of synthetic accessibility of drug-like molecules. J. Chem. Inf. Model. 2011;51:2768–2777. doi: 10.1021/ci100216g. [DOI] [PubMed] [Google Scholar]
- 8.Podolyan Y., Walters M.A., Karypis G. Assessing synthetic accessibility of chemical compounds using machine learning methods. J. Chem. Inf. Model. 2010;50:979–991. doi: 10.1021/ci900301v. [DOI] [PubMed] [Google Scholar]
- 9.Funatsu K., Sasaki S. Computer-assisted organic synthesis design and reaction prediction system, “AIPHOS. Tetrahedr Comput Methodol. 1988;1:27–38. [Google Scholar]
- 10.Boda K., Seidel T., Gasteiger J. Structure and reaction based evaluation of synthetic accessibility. J. Comput. Aided Mol. Des. 2007;21:311–325. doi: 10.1007/s10822-006-9099-2. [DOI] [PubMed] [Google Scholar]
- 11.Corey E.J., Wipke W.T. Computer-assisted design of complex organic syntheses. Science. 1969;166:178–192. doi: 10.1126/science.166.3902.178. [DOI] [PubMed] [Google Scholar]
- 12.Timothy D., Salatin T.D., Jorgensen W.L. Computer-assisted mechanistic evaluation of organic reactions. 1. overview. J. Org. Chem. 1980;45:2043–2057. doi: 10.1021/jo00246a020. [DOI] [PubMed] [Google Scholar]
- 13.Takaoka Y., Endo Y., Yamanobe S., et al. Development of a method for evaluating drug-likeness and ease of synthesis using a data set in which compounds are assigned scores based on chemists’ Intuition. J. Chem. Inf. Comput. Sci. 2003;43:1269–1275. doi: 10.1021/ci034043l. [DOI] [PubMed] [Google Scholar]
- 14.Lajiness M.S., Maggiora G.M., Shanmugasundaram V. Assessment of the consistency of medicinal chemists in reviewing sets of compounds. J. Med. Chem. 2004;47:4891–4896. doi: 10.1021/jm049740z. [DOI] [PubMed] [Google Scholar]
- 15.Ertl P., Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 2009;1:8. doi: 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bersohn M. Automatic problem solving applied to synthetic chemistry. Bull. Chem. Soc. Jpn. 1972;45:1897–1903. [Google Scholar]
- 17.Fukunishi Y., Kurosawa T., Mikami Y., Nakamura H. Prediction of synthetic accessibility based on commercially available compound databases. J. Chem. Inf. Model. 2014;54:3259–3267. doi: 10.1021/ci500568d. [DOI] [PubMed] [Google Scholar]
- 18.Satoh H., Tanaka T. Verification of a chemical reaction database- is it sufficient for practical use in chemical research? J. Comput. Chem. Jpn. 2003;2:87–94. [Google Scholar]
- 19.Wang Y., Xiao J., Suzek T.O., Zhang J., Bryant S.H. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623-33. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaulton A., Bellis L.J., Bentro A.P., et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irwin J.I., Shoichet B.K. ZINC - a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irwin J.I., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC - a free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukunishi Y., Sugihara Y., Mikami Y., Sakai K., Kusudo H., Nakamura H. Advanced in-silico drug screening to achieve high hit ratio - Development of 3D-compound database. Synthesiology. 2008;2:64–72. [Google Scholar]
- 24.Kawabata T., Sugihara Y., Fukunishi Y., Nakamura H. LigandBox - a Database for 3D structures of chemical compounds. Biophysics (Oxf.) 2013;9:113–121. doi: 10.2142/biophysics.9.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertz S.H. The first general index of molecular complexity. J. Am. Chem. Soc. 1981;103:3599–3601. [Google Scholar]
- 26.Harvey M.J., De Fabritiis G. AceCloud: Molecular dynamics simulations in the Cloud. J. Chem. Inf. Model. 2015;55:909–914. doi: 10.1021/acs.jcim.5b00086. [DOI] [PubMed] [Google Scholar]
- 27.Moghadam B.T., Alvarsson J., Holm M., Eklund M., Carlsson L., Spjuth O. Scaling predictive modeling in drug development with cloud computing. J. Chem. Inf. Model. 2015;55:19–25. doi: 10.1021/ci500580y. [DOI] [PubMed] [Google Scholar]
- 28.Mortier J., Rakers C., Bermudez M. Murgueitio1 MS, Riniker S, Wolber G. The impact of molecular dynamics on drug design: applications for the characterization of ligand-macromolecule complexes. Drug Discov. Today. 2015;20:686–702. doi: 10.1016/j.drudis.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Elsen E., Houston M., Vishal V., Darve E., Hanrahan P., Pande V. N-body simulation on GPUs.; SC '06: Proceedings of the 2006 ACM/IEEE conference on Supercomputing; New York, NY. 2006. [Google Scholar]
- 30.Stone J.E., Phillips J.C., Freddolino P.L., Hardy D.J., Trabuco L.G., Schulten K.J. Accelerating molecular modeling applications with graphics processors. Comput. Chem. 2007;28:2618–2640. doi: 10.1002/jcc.20829. [DOI] [PubMed] [Google Scholar]
- 31.Owens J.D., Luebke D., Govindaraju N., et al. A survey of general‐purpose computation on graphics hardware. Comput. Graph. Forum. 2007;26:80–113. [Google Scholar]
- 32.Anderson J.A., Lorenz C.D., Travesset A.J. General purpose molecular dynamics simulations fully implemented on graphics processing units. Comput. Phys. 2008;227:5342–5359. [Google Scholar]
- 33.Goetz A.W., Williamson M.J., Xu D., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goga N., Marrink S., Cioromela R., Moldoveanu F. GPU-SD and DPD parallelization for Gromacs tools for molecular dynamics simulations.; 2012. [Google Scholar]
- 35.Phillips J.C., Stone J.E., Schulten K. Adapting a Message-Driven Parallel Application to GPU-Accelerated Clusters. Piscataway, NJ; SC '08: Proceedings of the 2008 ACM/IEEE Conference on Supercomputing; 2008. [Google Scholar]
- 36.Mashimo T., Fukunishi Y., Kamiya N., Takano Y., Fukuda I., Nakamura H. Molecular dynamics simulations accelerated by GPU for biological macromolecules with a non-Ewald scheme for electrostatic interactions. J. Chem. Theory Comput. 2013;9:5599–5609. doi: 10.1021/ct400342e. [DOI] [PubMed] [Google Scholar]