Abstract
Shotgun metagenomics and computational analysis are used to compare the taxonomic and functional profiles of microbial communities. Leveraging this approach to understand roles of microbes in human biology and other environments requires quantitative data summaries whose values are comparable across samples and studies. Comparability is currently hampered by the use of abundance statistics that do not estimate a meaningful parameter of the microbial community and biases introduced by experimental protocols and data-cleaning approaches. Addressing these challenges, along with improving study design, data access, metadata standardization, and analysis tools, will enable accurate comparative metagenomics. We envision a future in which microbiome studies are replicable and new metagenomes are easily and rapidly integrated with existing data. Only then can the potential of metagenomics for predictive ecological modeling, well-powered association studies, and effective microbiome medicine be fully realized.
Shotgun sequencing is revolutionizing our understanding of microbiomes associated with humans and other environments by enabling in situ, culture-free genomic characterization of microbial communities. A shotgun metagenomic experiment involves sequencing a random sample of DNA fragments (to generate “reads”) from the pool of microbial genomes in a biological sample. Typically millions of reads, each on the order of 100 base pairs (bp), are obtained. Although complex and challenging to analyze, these metagenomic libraries can be used to identify and quantify microbial taxa and/or genes so that “who” is there and what they are doing can be compared across communities. This tool offers a powerful means for characterizing the immense microbial diversity on earth. However, there are a number of challenges standing in the way of ready comparisons across shotgun datasets. This Perspective seeks to outline the major issues and discuss how they might be overcome through application of current methods or development of new approaches.
Shotgun metagenomics has the potential to be highly quantitative, but it also presents many unique challenges. The genome from which each read comes and its position in that genome are unknown. Furthermore, the vast majority of microbial diversity is not represented in reference databases or otherwise characterized in most environments (Wu et al., 2009). Even for species with sequenced genomes, reference databases do not capture the full collection of genes present across different strains (Malmstrom et al., 2013). Leaving aside reads that cannot be confidently assigned to a taxon or gene, we are still faced with the challenge of converting the remaining reads to comparable estimates of abundance. This quantification is difficult due to a variety of experimental and bioinformatics biases that affect our ability to accurately estimate meaningful parameters of the underlying community. Another challenge is the size of shotgun metagenomes, which are typically much larger than data from individual genomes, targeted sequencing of specific genes from microbial communities (e.g., 16S, other taxonomic markers, biosynthetic genes), or other meta‘omic experiments (e.g., meta-proteomics, meta-metabolics).
Despite this complexity, metagenomic analyses have already revealed massive amounts of novel diversity, shed light on host-microbe interactions, explained cryptic health outcomes (Alivisatos et al., 2015; Dubilier et al., 2015), and been used for clinical diagnosis (Wilson et al., 2014b). Bioinformatics and statistics research has produced a first generation of tools for estimating the taxonomic and functional composition of a microbial community from shotgun metagenomics data (Box 1). Analysis strategies include mapping reads to reference databases using sequence homology, clustering reads to discover new taxa or protein families, assembling reads into genes or genomes, and various combinations of these approaches (Prakash and Taylor, 2012; Segata et al., 2013; Sharpton, 2014). The key data summaries are based on counts of reads assigned to taxa or functions. Studies examine different levels of taxonomic resolution, including individual strains (Box 2). As methods are rigorously benchmarked (Carr and Borenstein, 2014; Lindgreen et al., 2016; Nayfach et al., 2015a), iterative improvements and new approaches should soon enable accurate quantification of the abundances of individual taxa, genes, or pathways in a single metagenome.
Box 1. Taxonomic and Functional Profiling.
A common approach to quantifying organisms and functions represented in a shotgun metagenome is to first classify sequencing reads by using alignment to a reference database of genes and/or genomes to establish homology. The resulting counts of classified reads are used to compute statistics that estimate the abundance of taxonomic groups and gene families.
One promising extension of this approach is to generate a gene catalog by using metagenome assembly applied to samples from a similar environment (Li et al., 2014b; Sunagawa et al., 2015). In some environments, assembling complete or draft genomes may also be possible. The accuracy and efficiency of assembly algorithms can be improved by binning reads and/or assembled contigs based on features such as sequence composition, coverage, and co-variation (Alneberg et al., 2014; Cleary et al., 2015). Metagenome-derived sequences are then added to the reference database, which can increase the number of shotgun reads that can be classified by adding novel gene families and increasing the diversity of known gene families (Li et al., 2014b). Given that reads are expected to closely match one of the reference sequences, extremely fast sequence-alignment tools (e.g., Bowtie 2; Langmead and Salzberg, 2012) can then be used to map metagenomic reads to the gene catalog and quantify gene abundance.
A complementary approach, termed de novo profiling, involves applying unsupervised clustering to group shotgun reads into operational taxonomic units (OTUs) or gene families. This method does not rely on homology to known sequences and is therefore well suited to discovering novel taxa. De novo analysis has mostly been applied with targeted 16S gene sequencing, but the approach has been extended to shotgun metagenomes to identify OTUs (PhylOTU; Sharpton et al., 2011) and operational protein families (OPFs; Schloss and Handelsman, 2008), where longer reads will likely make the approach more useful. Although clustering can be performed without a reference database, the resulting OTUs and OPFs are typically assessed for homology to known sequences in order to infer annotations (Wang et al., 2007).
Further details of different approaches to taxonomic and functional profiling are given in the following: Prakash and Taylor, 2012; Segata et al., 2013; Sharpton, 2014.
Box 2. Strain-Level Variation.
The sequences in a shotgun metagenome contain information about nucleotide and copy-number variants carried by the specific strains of microbes in a community. Resolving metagenomics data at the sub-species level has great potential for understanding functional differences between communities, shedding light on the recent evolution of microbial populations (Kashtan et al., 2014; Shapiro et al., 2012), providing critical insight into pathogenicity (Rasko et al., 2011), and uncovering transmission between hosts (Nayfach et al., 2016). Strain analysis may also be able to shed light on processes occurring during host colonization, including niche competition and population bottlenecks (Lam and Monack, 2014). However, quantifying strain-level variation from metagenomes is challenging. For most species, directly detecting strains based on known strain-specific variants is impossible because very few (if any) genomes from the species have been sequenced.
One solution is to quantify genomic variation by aligning reads to reference genomes and identifying gene copy-number variants (Greenblum et al., 2015) and/or single-nucleotide variants (Nayfach et al., 2016; Schloissnig et al., 2013). Reference-based approaches work well for species from the human microbiome, which are well represented in genome databases, but may not be suitable in other environments, like soil, that are dominated by genes and genomes from microbial dark-matter (Rinke et al., 2013). These methods have the advantages of estimating patterns of variation genome-wide and being fast, automated tools that produce output that can be easily compared across samples. An alternative approach is to deconvolute mixtures of strains from patterns of genomic variation in metagenomes, either de novo (Luo et al., 2015) or using reference panels (Joseph et al., 2014). Other techniques include reference-free assembly from shotgun data (Cleary et al., 2015; Nielsen et al., 2014) and sub-species resolution analyses of 16S amplicons (Tikhonov et al., 2015).
These methods have revealed that different humans harbor quite distinct strains, and some of this variation is correlated with host phenotypes (Greenblum et al., 2015) and body sites (Oh et al., 2014). Levels of variation differ across bacterial species and genes (Schloissnig et al., 2013). Importantly, genes with copy-number variants are enriched for specific functions that may affect interactions with other microbiota and the host (Greenblum et al., 2015).
In the future, it will be important to evaluate the emerging approaches to strain-level analysis. One consideration is the data needed for good performance, such as dependence on reference genomes or a large number of samples (e.g., for covariation analysis). Other criteria for evaluation include sensitivity for low-abundance organisms, ability to resolve strains from high-diversity populations, ability to capture both core and variable genomic regions, and robustness to different levels of recombination.
The real power of metagenomics comes from comparing data across samples, either within a study or across studies. Detecting differences in abundance for individual microbes or microbial genes often requires larger sample sizes than are feasible within a project. Hence, meta-analyses and comparisons of new meta-genomes to existing cohorts are increasingly being done (Arumugam et al., 2011; Finucane et al., 2014; Koren et al., 2013; Li et al., 2014b; Lozupone et al., 2013; Yatsunenko et al., 2012), and large studies usually involve experiments conducted across multiple institutions (Ehrlich, 2011; Peterson et al., 2009). Databases of publicly available gene sequences, genomes, and metagenomes are growing exponentially (Kodama et al., 2012), providing rich information for contextualizing new experiments and fodder for computational studies that ask new questions of existing data. These investigations typically involve comparisons of the abundance of taxa and/or genes across samples or summary measures based on these, such as diversity metrics.
Meaningful comparative metagenomics requires accurate quantification of taxon and gene abundances so that they can be numerically compared across samples. For example, a 3-fold higher gene abundance estimate should indicate roughly 3-fold more of the gene rather than a systematic difference in the scale of abundance estimates. This is unfortunately a much taller order than computing relative levels of genes or taxa within each sample because many aspects of study design, experimental protocols, and bioinformatics pipelines affect the relationship between true abundance in the community and the number of reads observed for a taxon or gene. Some of these issues have been previously described in the context of taxonomic profiling with 16S sequencing (Finucane et al., 2014; Goodrich et al., 2014), but others are specific to shotgun data. Our goal is to promote accurate comparative shotgun metagenomics by describing the primary hurdles and highlighting existing or potential solutions.
Experimental Protocols Affect Results and Should Be Tracked in Sample Metadata
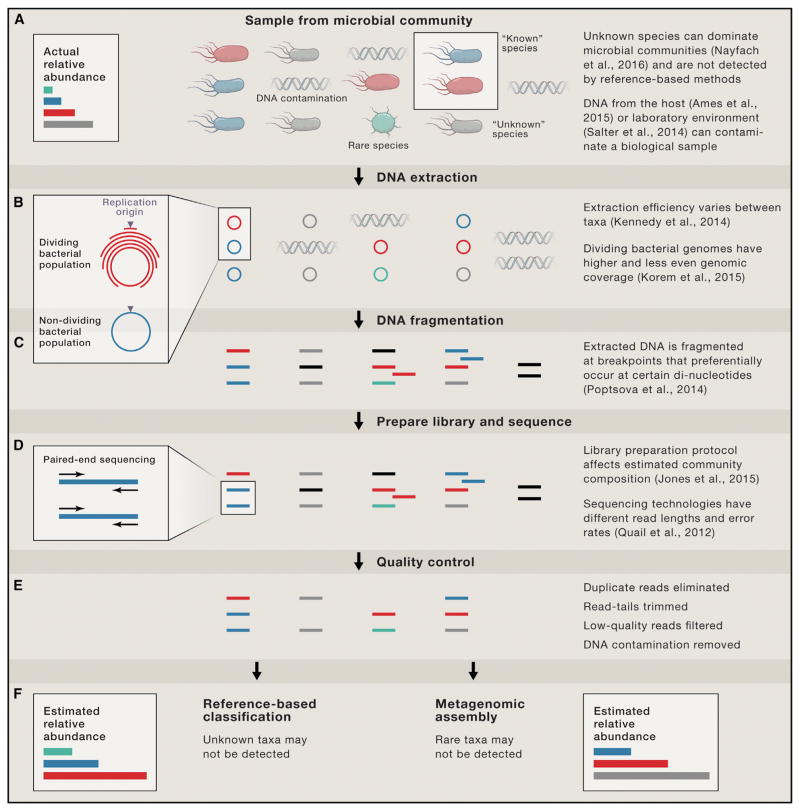
Experimental protocols influence the sequences obtained in a metagenomics experiment in a variety of ways (Figure 1). The International Human Microbiome Standards (IHMS) project (Voigt et al., 2015), Microbiome Quality Control (MBQC) project (Sinha et al., 2015), and numerous other studies are measuring the effects of different techniques on read distributions. Sample collection, storage (Voigt et al., 2015), DNA extraction (Kennedy et al., 2014), and library preparation (Jones et al., 2015) all influence the taxonomic composition of a metagenome and, by extension, the functional composition. Variable amounts of DNA from the host (Ames et al., 2015), reagents (Tanner et al., 1998), and post-sampling environment (Salter et al., 2014) will be sequenced along with microbial DNA and can strongly influence coverage and quantification of microbiota, especially in low biomass body sites and environments (Weiss et al., 2014). Additionally, the amount of DNA extracted per cell depends on growth rate of microbial populations—actively dividing cells will yield more genomic DNA, which accumulates at the origin of replication (Korem et al., 2015).
Figure 1. Challenges Associated with Estimating the Composition of a Microbial Community from Shotgun DNA Sequencing.
(A) A sample from a microbial community composed of four different microbial species. Colored cells (blue, red, green) indicate “known” species that have at least one genome sequence in reference databases. The green cell indicates a species that is rare within the microbial community. DNA contamination includes DNA from the host, laboratory environment, or experimental reagents.
(B) DNA is extracted from the microbial cells in the sample. Extraction efficiency varies for different taxa, depending on the experimental protocol. The amount of DNA extracted per cell depends on growth rate—actively dividing cells yield more genomic DNA, which accumulates at the origin of replication.
(C) Extracted DNA is broken into fragments by mechanical or enzymatic methods. Certain sequences are more likely to be breakpoints.
(D) A library is prepared from DNA fragments and sequenced. DNA fragments with high or low GC% are under-represented in the sequencing reads. Typically millions of short (e.g., 150 bp) reads are generated per sample.
(E) Bioinformatics quality-control steps may be performed to eliminate duplicate reads, trim low-quality bases from read ends, and remove reads from contamination sources or with low-quality scores.
(F) To infer the composition of the microbial community, high-quality reads are either compared to reference sequences or assembled de novo. Reference-based classification cannot account for unknown species and overestimates the abundances of known species. Metagenomic assembly may not detect rare species and overestimates abundance of abundant species.
DNA fragmentation (Poptsova et al., 2014) and PCR biases (Benjamini and Speed, 2012) introduced during library preparation result in a non-uniform sampling of possible sequencing reads and an under-representation of DNA with certain sequence features. Benjamini and Speed found that genomic fragments with high and low GC content are under-represented in Illumina libraries (Benjamini and Speed, 2012), and Manor and Borenstein found that intra-metagenome differences in coverage of different universal, single-copy genes can be explained by their GC content (Manor and Borenstein, 2015). Non-uniform coverage skews representation of both genomes and genes in shotgun data. Correction of these biases has yet to be incorporated into most analysis methods in part because solutions from other genomics applications (Benjamini and Speed, 2012; Roberts et al., 2011) require complete and high-quality reference genomes, which are unavailable for the vast majority of microbes in the environment.
Sequencing is another potential source of bias. Commonly used sequencers have different error rates and patterns (Quail et al., 2012), but their effects on taxonomic (Sinha et al., 2015) and functional (Nayfach et al., 2015a) composition are surprisingly minimal (O’Sullivan et al., 2014). Read length, on the other hand, is a source of bias on its own (Carr and Borenstein, 2014; Nayfach and Pollard, 2015), in large part because it is more difficult to detect homology for short reads, especially when sampled from a taxonomic group that is poorly represented in reference databases (Wommack et al., 2008). Long-read technologies help with homology detection but are currently much lower throughput and also prone to insertion and deletion (indel) errors (Carneiro et al., 2012) that can affect read-mapping accuracy (Nguyen et al., 2014). Further evaluations are needed to determine how different analysis methods perform across various length reads and indel rates.
With all of these biases, it is critical to ask whether the effects are comparable in scale to biological variation and therefore a threat to accurate comparisons. For example, the MBQC found that inter-laboratory variation (combined effects of many experimental differences) is on the same order as biological variation (Sinha et al., 2015). Voight et al. showed that technical variability from freezing fecal samples versus preserving them in solution is small compared to temporal and inter-subject variability. Lozupone et al. found that samples from different studies of Western adults clustered by study, whereas strong effect sizes such as age and lifestyle were great enough to outweigh technical variation (Lozupone et al., 2013). Thus, most experimental biases examined to date are not large enough to obscure biological effects. Quantifications of error from additional experimental protocols would enable the field to identify best practices.
There will always be some variability in experimental details, particularly when using old datasets. It is therefore imperative that all experimental methods be tracked in sample metadata so that protocol differences can be adjusted for in downstream comparisons. Currently, this is essentially never done. Ideally, experimental metadata would be required in order to upload sequence data to public repositories like the NCBI Sequence Read Archive and the European Nucleotide Archive. Capturing protocols at the time of data generation rather than months or years later at the time of a publication would improve the accuracy of the recorded information. On the computational side, tools that combine data across samples should be developed and benchmarked with various sources of experimental bias and their magnitudes in mind. Overall, we believe that these biases can be minimized through careful sample annotation and innovations in analysis.
Communities Should Be Profiled with Meaningful Parameters
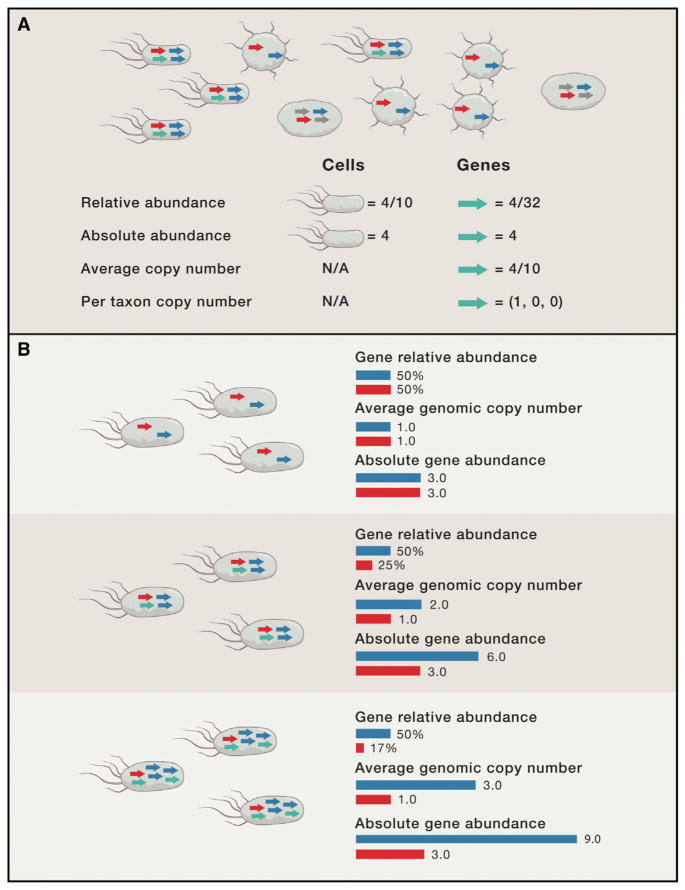
It is important to ask what aspect, or parameter, of the underlying community we wish to estimate in any metagenomic analysis. For example, defining what we mean by abundance in the community, which is distinct from any statistic computed from a sequencing library, clarifies the objectives of a metagenome analysis. Absolute abundance, relative abundance, and copy number are examples of parameters that capture different biological properties of a taxon or gene in a community (Figure 2A). These parameters vary in how they depend on abundances of other taxa and genes (Figure 2B). By detailing these differences, we aim to guide researchers toward appropriate parameter choices for their goals and provide insight into which analysis methods produce taxonomic and functional profiles that are comparable across samples.
Figure 2. Parameters Used for Taxonomic and Functional Profiling.
When computing the abundance of taxa and genes, it is important to think about what parameter of the underlying community one wishes to quantify.
(A) A community with ten cells composed of three taxa with different subsets of four different gene families (colored arrows). Two cellular abundance parameters and four gene abundance parameters are defined by examples.
(B) A comparison of gene relative abundance, average genomic copy number, and absolute abundance across three communities (top, middle, and bottom). The red gene is present at one copy per cell and has constant absolute abundance in all communities, but its relative abundance decreases with increasing genome size. The copy number of the blue gene increases with genome size, but its relative abundance is constant.
Most metagenomics studies have focused on community composition, meaning the relative amounts of different taxa and genes contained within a sample. A common parameter for taxonomic profiling is cellular relative abundance. This is the proportion of all cells in the community that belong to a taxonomic group or, equivalently, the probability that a randomly sampled cell is from that group. A functional analog of cellular relative abundance is gene relative abundance, which is also a compositional parameter. It can be conceptualized by combining the genomes from all cells in the community into a “bag of genes” and asking what proportion of genes belong to each gene family. Relative abundance is compositional, so an increase in the number of cells of a single taxon implies a decrease in the proportion accounted for by other taxa, even though their cell counts remain constant. Before using compositional parameters, it is important to note that they require specialized statistical methodology (Aitchison, 2003; Fisher and Mehta, 2014; Kurtz et al., 2015), without which spurious correlations and other errors are made (Faust and Raes, 2012). Also, the relative abundance of a taxon (or gene) is expected to differ between communities even when the absolute amount is constant because of changes in other taxa (or genes).
An alternative, and arguably more biologically meaningful, parameter for functional profiling (Beszteri et al., 2010; Manor and Borenstein, 2015; Nayfach and Pollard, 2015) is average genomic copy number, which is the expected number of copies of the gene (or pathway) per cell or, equivalently, in a randomly sampled cell. Average genomic copy number is less than one for many gene families, equal to one for universal, single-copy genes, and above one for gene families with multiple paralogs in a typical microbe from the community. In a microbial community with an average genome size of 1,000 genes, average genomic copy number will be three orders of magnitude greater than gene relative abundance. It is independent of genome size and the percentage of unknown genes and does not sum to one over all genes. It remains a relative parameter because adding cells with a particular set of genes to a community will alter average copy numbers. A related parameter is the copy number per taxon, which is a vector of expected copies per cell for each taxonomic group (e.g., species) in the community.
Another option is to consider absolute amounts of taxa or genes in a community (e.g., per unit of volume). Cellular absolute abundance of a taxon does not change with the addition or subtraction of cells from other taxa. Similarly, gene absolute abundance only changes if the cells that are added or subtracted carry that gene. Absolute abundances are quantitative parameters that can be compared across samples with standard statistical methods. On the other hand, they do not quantify composition or relative amounts of taxa or genes and cannot be estimated with sequence data alone.
By carefully defining different choices of taxonomic and functional parameters, we have highlighted the information that they capture and how their values will change across different communities (Figure 2B). The goals of a given analysis should drive the choice of taxonomic and/or functional parameters to investigate. For example, taxa or genes that vary by several orders of magnitude across communities will likely be detected as different using any of the abundance parameters, whereas those with small dynamic ranges might be detected using copy number, but not relative abundance.
Unbiased Estimation of Taxonomic and Functional Profiles from Shotgun Data
Once a community parameter is selected, one should consider how best to estimate it from shotgun data. Many computational tools have been designed for classifying metagenomic reads into taxonomic groups (reviewed in Lindgreen et al., 2016) or gene families (reviewed in Prakash and Taylor, 2012; Segata et al., 2013; Sharpton, 2014), but relatively few have been developed for accurately estimating abundance parameters.
Accurately estimating cellular or gene relative abundance parameters from shotgun metagenomes is difficult. The typical approach is to use the proportion of classified reads in a shotgun metagenome that map to a genome or gene. However, this statistic is biased and introduces unwanted variability into comparisons between communities. One important reason is variation in the proportion of unmapped reads across samples, which results in the overestimation of the relative abundance of known taxa or genes (Prakash and Taylor, 2012) (Figure 1). Two major sources of unmapped reads are novel taxa and taxa that are poorly characterized in reference databases because the sequenced representative(s) have different gene content from the strains in a sample. Even in well-studied environments like the human gut, it has been estimated that 43% of prokaryotic species abundance (Sunagawa et al., 2013) cannot be captured by current reference genome-based methods. Similarly, 64% of gene abundance in the human microbiome is not found in databases (Abubucker et al., 2012). The situation is far worse in other environments (e.g., soil, seawater), where it has been estimated that between 90% and 98% of microbes have no sequenced genome at the species level (Nayfach et al., 2016). A proposed solution is to estimate relative abundance with the proportion of total reads that map to a taxon or gene (i.e., include “unclassified” as a category, as in GRAMMy; Xia et al., 2011). However, this is still a poor estimate of cellular or gene relative abundance because unmapped reads derive from sources other than novel microbes, including host DNA, other contamination, sequencing errors, and read length (which affects sensitivity of alignment-based homology detection) (Wommack et al., 2008).
Statistics based on proportions of reads also suffer from difficulties related to the unknown sizes of genomes and genes in a metagenome. Because the probability of sequencing a read from a gene depends on its length, the number of mapped reads will vary between equally abundant genes with different lengths. For well-characterized genes, length can be estimated from gene-family models or database sequences to which reads from a metagenome map (Prakash and Taylor, 2012; Segata et al., 2013; Sharpton, 2014). Normalizing by length makes abundance estimates more comparable across genes, but it does not address comparability across samples. The problem is worse for estimating cellular relative abundance because genome sizes vary greatly, even within species. Additionally, the number of mapped reads to a genome will depend on the average length of other genomes in the community. As taxon-specific genome sizes in a metagenome are difficult to estimate for most communities, it is not feasible to normalize by this factor to get an unbiased estimate of cellular relative abundance.
A potential solution for accurately estimating cellular relative abundance is to use only shotgun reads that map to a set of taxonomically informative marker genes, whose length and copy number are constant or known across taxa (e.g., certain ribosomal proteins). The rationale for this approach is that genome size is not an issue, as in targeted 16S sequencing. It may also address issues with unmapped reads if the marker genes are able to recruit reads from novel taxa. Identifying accurate marker genes can be challenging, particularly for environments with limited genomes sequenced. Several tools implement the marker gene approach. MetaPhyler (Liu et al., 2011) maps reads to a set of universal, single-copy genes using alignment parameters that are tuned for specific taxonomic levels (Liu et al., 2011). mOTU (Sunagawa et al., 2013) also maps reads to a set of universal, single-copy genes but uses a reference database that contains genes from “novel” taxa identified from human gut metagenomic assemblies (Sunagawa et al., 2013). Meta-PhlAn (Segata et al., 2012) utilizes a hierarchical framework to assign reads to genes that are unique to taxonomic groups (Segata et al., 2012). MicrobeCensus (Nayfach and Pollard, 2015) estimates the total read-depth of all cellular microbes (Bacteria, Archaea, Eukaryotes) in a metagenome, which can be used to normalize the read-depth of known taxonomic groups (Nayfach et al., 2016). All of these methods enable detection of novel organisms at lower levels (e.g., species) because they estimate organism abundance at higher taxonomic levels (e.g., domain). Similar approaches could be used to estimate the relative abundance of viruses in metagenomes, although in reality this will be a much more difficult task due to the lack of universal marker genes in viruses and because viral diversity in many environments is poorly represented in reference databases (Dutilh et al., 2014).
For gene abundance, a solution is to estimate average genomic copy number, which is straightforward to estimate and easy to interpret. MUSiCC does so by normalizing gene relative abundances by the median relative abundance of universal single-copy genes whose genomic copy number is very close to one in all sequenced microbes (Manor and Borenstein, 2015). MicrobeCensus also uses reads mapped to single-copy genes and estimates the total coverage of genomes from cellular organisms, which can be used to compute reads per kilobase per genome (RPKG), which is closely related to average genomic copy number (Nayfach and Pollard, 2015). Because these metrics are normalized using a relatively small subset of genes in the community, estimating them with low error may require deeper sequencing than is needed for gene relative abundance. Copy number per taxon can be estimated directly by mapping reads to reference genomes (Greenblum et al., 2015; Nayfach et al., 2016) or indirectly using methods that deconvolute read counts across taxa (Carr et al., 2013).
Another appealing strategy is to focus on absolute taxon and gene abundances, which avoids challenges associated with compositional statistics. Unfortunately it is impossible to estimate absolute abundance parameters from shotgun metagenomes alone. However, progress has been made toward doing so by combining sequencing with density measurements from flow cytometry (Hingamp et al., 2013) or quantitative PCR (Liu et al., 2012) and by incorporating DNA or mRNA standards (Satinsky et al., 2013). Good standards capture technical variation in sample preparation and sequencing, and hence they may also be useful for estimating biases and normalizing read counts to compare relative abundance across samples. On the other hand, standards depend on recovery rate being constant and do not quantify variation from obtaining or handling the sample. As density estimation techniques and standards improve in accuracy and cost, absolute abundance estimation may be a promising direction.
We conclude that the most biologically meaningful and quantifiable parameters for metagenome profiling are cellular relative abundance and average genomic copy number. For cellular relative abundance, the best estimation approach appears to be using marker genes. However, the different marker gene methods have not yet been thoroughly benchmarked and compared to genome coverage approaches using realistic datasets. One approach would be to create in silico or in vitro metagenomes with different read lengths, novel taxa, host contamination, sequencing biases, and PCR artifacts. An ideal method should produce accurate estimates of cellular relative abundance that are not biased by these factors. An additional important goal is to detect the presence and identity of novel taxonomic groups, just as 16S sequencing is able to detect novel operational taxonomic units (OTUs). Even after benchmarking has been done, it will be critical to rigorously evaluate and periodically update marker gene sets as more genomes are sequenced. In the future, it also may be possible to accurately estimate gene relative abundance as reference databases continue to expand. Database bias can be reduced by building comprehensive gene catalogs for different environments, as has been done for the human gut microbiome with metagenome assembly methods (Li et al., 2014b).
Meta-analysis Benefits from Uniform Bioinformatics Processing of Raw Sequence Data
Analysis of a shotgun metagenome typically begins with bioinformatics processing of sequencing reads to ensure high-quality data for read mapping or assembly. These steps include but are not limited to trimming bases with low quality, filtering low-quality reads, removing sequencing adaptors, removing host sequences, and removing duplicated reads. Data in public repositories are a mix of raw data (e.g., Consortium, 2012; Sunagawa et al., 2015) and quality-controlled data (e.g., Li et al., 2014b) that differ in sequence quality, read length, and library size. It is unclear whether these differences prevent accurate meta-analyses. Furthermore, it is not known what data-processing methods are optimal for various types of analysis.
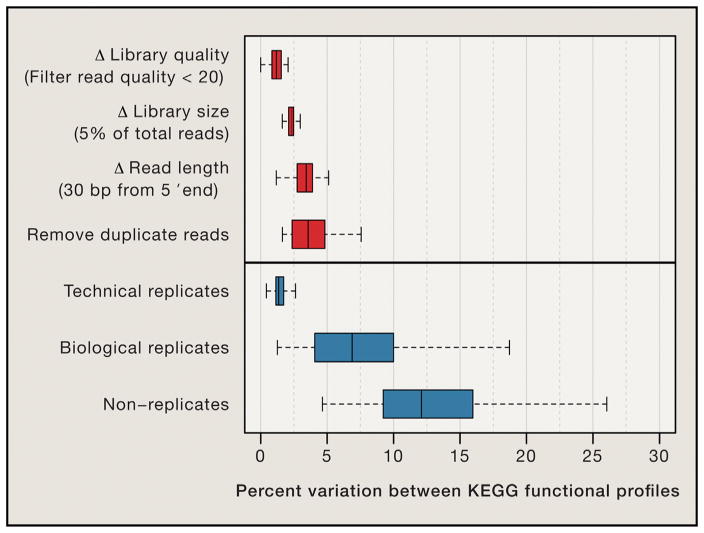
To quantify the magnitude of technical differences introduced by data processing, we took 26 human gut metagenomes of varying quality, processed them using different quality-control methods, and used the resulting reads to estimate the average genomic copy number and relative abundance of KEGG Orthology Groups. Similar trends were observed for both abundance parameters, so we emphasize results for average genomic copy number. We compared the variation introduced by data processing to the variation observed between a large set of gut metagenomes from the Human Microbiome Project (Consortium, 2012), including technical replicates (N = 1,474; median = 1.3% variation), biological replicates from the same host at different times (N = 144; median = 6.8%), and non-replicates from different hosts (N = 179; median = 12.1%). A separate cohort of European individuals from the MetaHIT Consortium (Nielsen et al., 2014) showed similar levels of variability for technical replicates, biological replicates, and non-replicates (medians = 2.3%, 5.9%, 10.9%, respectively).
Surprisingly, most of the data-processing methods had relatively little impact on the functional profiles of the metagenomes (Figure 3). For example, sequence quality filtering rarely altered copy-number estimates by more than the variation observed between technical replicates, and differences in read length introduced only slightly more variation. Metagenomes naturally differ in their sequencing depth and are sometimes rarefied (i.e., downsampled to the smallest library). We found that average genomic copy-number estimates were surprisingly robust to library-size differences, with libraries only 5% of their original size introducing <2.5% variation overall, although effects differ for common versus rare genes. Although rarefaction may improve accuracy of tests for taxonomic differences in 16S studies when library sizes differ by more than 2-fold (Weiss et al., 2015), our results suggest that large library-size differences do not significantly confound biological effects in gene profiling with shotgun metagenomics. We therefore suggest that rarefaction not be used on shotgun metagenomes, except perhaps for statistics that are correlated with sequencing effort (e.g., richness) and those that are biased (e.g., nucleotide diversity, beta-diversity; Nayfach et al., 2015a) or have high variance at low coverage. Explicit normalization methods that use all reads (e.g., mixed models; McMurdie and Holmes, 2014) may be more effective.
Figure 3. Differences in Functional Profiles due to Read Length, Library Size, and Quality Control Are Small Compared to Biological Variation.
Publicly available metagenomes often differ in their library sizes, read lengths, and quality-control measures, which leads one to ask, how comparable are metagenomes from different studies? Twenty-six human gut metagenomes of varying quality were processed using different quality-control methods, and the resulting reads were used to estimate the relative abundance of KEGG Orthology Groups (KOs). We compared the variation introduced by these factors (top) with the variation observed between a large set of technical (N = 1,474), biological (N = 144), and non-replicate gut metagenomes (N = 179) from the Human Microbiome Project (Consortium, 2012) that contained at least one million reads (bottom). Trimming reads from their 5′ ends was done to simulate libraries of different read length; downsampling metagenomes by 95% was done to simulate libraries of different size; fastq-mcf (Aronesty, 2011) was used for de-duplication and quality filtering. To estimate the average genomic copy number of functional groups, reads were mapped to the integrated catalog of reference genes in the human gut microbiome (Li et al., 2014a, 2014b) using bowtie2 (Langmead and Salzberg, 2012) and normalized by the median coverage of 30 universal single-copy genes (Wu et al., 2013). The percent variation between two metagenomes was measured by the following: (1) taking the sum of absolute deviations across KOs, (2) dividing this by the total abundance of KOs in both metagenomes, and (3) multiplying this by 100.
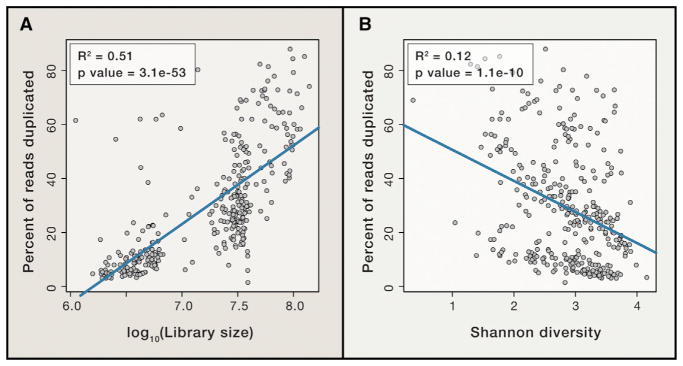
Filtering duplicate metagenomic reads had the greatest effect on estimates of gene copy number (median = 3.6%) with some samples changing by up to 7.6%. This procedure is commonly employed based on the hypothesis that duplicate reads arise as a result of experimental biases from PCR (Gomez-Alvarez et al., 2009). However, biological duplicates can also arise from abundant organisms, especially in deeply sequenced libraries. In these cases, de-duplication can lead to underestimation of the abundance of common taxa and genes. Supporting this hypothesis, 63% of the variation in duplication rates across HMP gut metagenomes was explained by library size (R2 = 0.51, p = 3e-53) and species-level alpha diversity based on the Shan-non diversity index (Keylock, 2005) (R2 = 0.12, p = 1e-10) (Figure 4). For these reasons, we recommend against duplicate filtering for quantitative metagenomic analysis and instead suggest minimizing experimental biases, for example by using a PCR-free library preparation (Jones et al., 2015) or employing statistical models designed to correct these issues (Benjamini and Speed, 2012; Roberts et al., 2011).
Figure 4. The Presence of Duplicated Reads Is Largely a Function of Library Size and Microbial Diversity.
FASTQC was used to estimate the percent of duplicated reads across 181 human gut meta-genomes from the Human Microbiome Project and compared to (A) library size and (B) species-level alpha diversity using the Shannon diversity index (Keylock, 2005). Species abundance of bacteria and archaea was estimated with mOTU (Sunagawa et al., 2013). Together, library size and Shannon diversity explain 63% of the variation in sequence duplication rates.
Although the effect of the quality-control methods we evaluated was small compared with biological variation, we expect that these differences can add up, especially when combined together or with other technical differences, such as DNA extraction and library preparation. Likewise, we expect that larger differences in read length, sequence quality, and/or library size will have a greater impact on comparability. We will only be able to fully assess these effects through cross-study comparisons, which will require analysis of unprocessed datasets. Deposition of such datasets into public repositories has not been consistently practiced, and we strongly encourage it to enable uniform processing of the raw sequences to avoid bioinformatics bias in meta-analyses and to allow researchers to choose the optimal quality-control methods for their specific goals.
Additionally, we recommend that bioinformatics tools build in quality-control features needed for their specific goals. For example, metagenomic assembly clearly benefits from using sequence quality scores to trim and filter reads (Mende et al., 2012), as does 16S-based diversity estimation (Bokulich et al., 2013) and polymorphism quantification. Likewise, methods that reduce redundancy of high coverage reads (Howe et al., 2014) can increase the efficiency of metagenomic assembly by reducing library complexity. On the other hand, functional profiles (Nayfach et al., 2015a) and taxonomic profiles (Nayfach et al., 2016) can be estimated with only a few million reads, reducing computational burden. Deposition of raw data and integration of tunable quality processing into bioinformatics tools are important steps for improving the accuracy of comparative metagenomics.
Study Design and Statistical Modeling
When designing a metagenome study, we advocate using sufficient replication and controlling or randomizing over variables that affect microbiome composition. But this is not always possible, especially in meta-analyses. Biological effects (e.g., disease status, drug sensitivity) will frequently be confounded with technical biases (Knight et al., 2012), as well as population differences in variables associated with microbiome shifts, such as diet, geography, animal facility, antibiotics (Goodrich et al., 2014), or other aspects of medical treatment (Forslund et al., 2015). Furthermore, metagenomes are often convenience samples that are not representative of the underlying population of interest (Knight et al., 2012). For example, ocean expedition routes are understandably determined by weather, so analyses of marine metagenomes must account for sampling date when estimating microbial distributions (Ladau et al., 2013). Similarly, failure to account for metformin treatment led to discordant findings regarding gut microbiome dysbiosis in type 2 diabetes (Forslund et al., 2015).
There is nonetheless hope for metagenomics meta-analysis. If experimental protocols and sample characteristics are well documented in metadata and raw data are available for uniform processing, many biases can be accounted for in statistical models and tests, especially when confounding is not complete (e.g., cases are not all from one study and controls from another) and replication is sufficient (McMurdie and Holmes, 2014). Tara Oceans (Sunagawa et al., 2015) is an example of a project that made highly structured experimental and environmental meta-data publicly accessible. Tools from the RNA-seq literature (e.g., edgeR [Robinson et al., 2010] and DESeq [Anders and Huber, 2010]) can be employed to adjust for sources of bias through statistical modeling, and extensions specific to metagenomics are being developed (e.g., metagenomeSeq [Paulson et al., 2013] and phyloseq [McMurdie and Holmes, 2014]). As samples frequently are not a random draw from the underlying population of interest, it would be appropriate to control for any measured confounders, being cognizant of unmeasured confounders when interpreting results, and avoiding extrapolation beyond the range of the observed data.
The Sequence Data Deluge Is an Opportunity and a Challenge
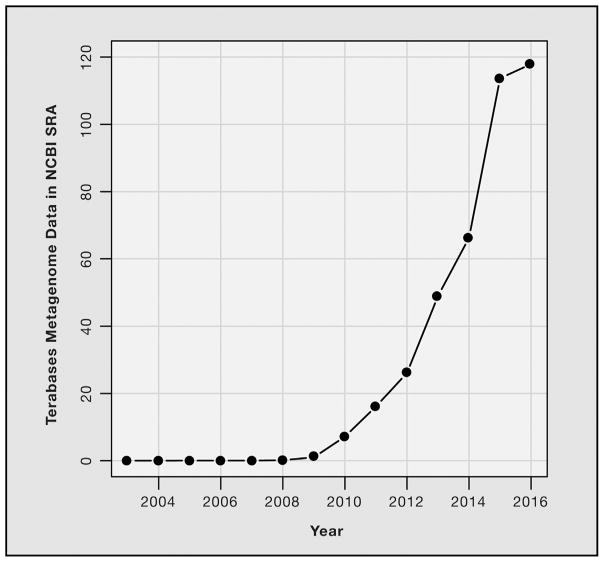
There are massive amounts of publicly available data from genes, genomes, and metagenomes that can be leveraged for meta-analyses or interpretation of new metagenomics experiments. Repositories, such as those hosted by the National Center for Biotechnology Information (NCBI), European Bioinformatics Institute (EBI), and Joint Genome Institute (JGI), contain many terabytes of gene and genome sequence that are used to build reference databases for metagenomics studies. Reference sequences are annotated in a variety of ways, including their taxonomy and functions when known (examples in Segata et al., 2013). Metagenomes are hosted in the NCBI Sequence Read Archive (SRA), EBI European Nucleotide Archive (ENA), and other repositories like MG-RAST (Meyer et al., 2008). The growth of available data is staggering. For example, an order of magnitude more metagenomics data is now publicly available in the SRA compared to just 5 years ago (Figure 5). By hosting tera-bytes of publicly available shotgun data, these repositories enable meta-analyses and comparisons of new data to other populations.
Figure 5. Growth of Shotgun Metagenome Data in the NCBI Sequence Read Archive.
Cumulative size in terabases of publicly available shotgun metagenomic data in the NCBI Sequence Read Archive (SRA). Sequencing runs were identified using the SRAdb database (Zhu et al., 2013) by the following: library_source = “META-GENOMIC,” study_type = “Metagenomics,” and library_strategy = “WGS.”
Despite having access to growing amounts of information, researchers still face many challenges when leveraging these resources. The sequence and annotation databases used to taxonomically and functionally annotate shotgun reads can affect estimates of abundance, diversity, and other community properties. This can even be an issue with de novo discovery of operational taxonomic units or protein families from shotgun data because these are frequently annotated or interpreted in the context of reference data. Taxonomy can be inconsistently described across databases, only resolved to the genus level (8.6% of bacterial genomes), or in disagreement with average nucleotide identity (9.8% of bacterial genomes) (Mende et al., 2013; Nayfach et al., 2016). This unevenness could be addressed by adopting a shared taxonomy based on an operational, sequence-based definition. The issue continues at the gene level as well. Functional annotations of genes are sparse, frequently inconsistent between databases, and differentially mapped to pathways and other higher-order functional categories, which can lead to discordant conclusions about the biological capabilities of a community (Nayfach et al., 2015a). Additionally, there are major phylogenetic and functional biases in what has been sequenced and annotated (Wu et al., 2009). Metagenome assembly produces gene catalogs that help to fill these gaps (Li et al., 2014b; Sunagawa et al., 2015). These enable faster and more accurate homology search when the catalog is assembled from an environment resembling the shotgun data being annotated.
Keeping pace with gene and genome sequencing is another major issue. Many metagenome bioinformatics tools are developed and distributed with static reference databases that run the risk of rapidly becoming out of date. For example, the number of genomes we clustered into species in 2015 (Nayfach et al., 2016) was an order of magnitude more than were analyzed with a similar approach 2 years earlier (Mende et al., 2013), and our database in 2016 is already missing new genomes. A simple solution is to say that software-associated databases should be perpetually updated. But this is a tall order for an individual lab that is focused on methods development rather than information management and whose projects are not funded for ongoing data curation (Knight et al., 2012). An alternative solution is therefore to promote development of software that allows users to provide their own reference database (e.g., Nayfach et al., 2015a). This approach puts the burden of database creation and updating on the user, which may be realistic if tools for querying and storing public sequence data continue to improve and/or perpetually updated databases in formats utilized by metagenomics tools are hosted in reliable central repositories. An even better solution would be for bioinformatics labs to design software to directly query centralized databases, avoiding the need for users to manage large databases themselves while also improving reproducibility by ensuring that different researchers are working with the same data.
Other bioinformatics challenges in utilizing public sequence resources include storage and manipulation of big data, tracking quality and completeness of sequencing projects, and translating between databases. Although tools for easily querying databases for sequences based on their metadata (e.g., read length, environment) help to address access and organization (Börnigen et al., 2015; Zhu et al., 2013), a specific obstacle to using public shotgun data is linking accession numbers of metagenomes in public repositories (e.g., NCBI SRA) to the sample identifiers used in most publications. Adoption of globally unique sample identifiers, an effort underway (Chase et al., 2015), should improve upon error-prone and laborious manual solutions. We conclude that widespread utilization of publicly available shotgun metagenomes will benefit from better interfaces to read data and perpetually updated reference sequence databases.
Sample Metadata Is Hard to Access and Link to Sequence Libraries
Metadata is critical for comparative metagenomics. Knowledge of how a metagenome was produced allows researchers to adjust for technical biases, whereas annotation of the environment from which it was sampled enables statistical adjustment for confounders and tests of associations with host or ecosystem characteristics. Unfortunately, metadata is currently both incomplete and difficult to access for most metagenomics experiments, jeopardizing the reproducibility of individual studies (Ravel and Wommack, 2014) and preventing data integration across studies. The Genomic Standards Consortium’s MIxS standard (Yilmaz et al., 2011) (MIGS, MIMS, MIMARKS, MIENS) provides a solution that has been adopted by several large projects (Gilbert et al., 2010) and extended for specific environments (e.g., MIxS-BE for built environments; Glass et al., 2014). In addition to adopting a shared standard, it is important that meta-data be freely and easily accessible whenever approved by ethics committees (Huttenhower et al., 2014). Parsing metadata from files in journal supplements, lab websites, or dbGaP (Mailman et al., 2007) and manually linking it to sequence data is error prone and untenable for large meta-analyses. The Bio-Project and BioSample databases at NCBI are searchable, centralized solutions (Barrett et al., 2012) that tools like SRAdb can parse (Zhu et al., 2013). For example, the Tara Oceans (Sunagawa et al., 2015) expedition used these databases to link publicly accessible metadata for 243 globally distributed seawater samples directly to raw sequencing reads. We would like to see this approach adopted more widely as an alternative to putting meta-data in supplemental files or other difficult-to-access locations.
Conclusions and Future Prospects
A future in which metagenomes are quantitatively compared across studies is within our grasp. One important step will be adopting statistics that estimate meaningful properties of a microbial community. Cellular relative abundance and average genomic copy number are biologically motivated parameters that can be estimated with minimal bias using appropriate normalization techniques. Experimental protocols and data-analysis methods can influence taxonomic and functional pro-files estimated from shotgun metagenomes, as well as downstream results. However, the magnitude of these effects is often small relative to biological variation, and confounders that are recorded in accessible metadata can be included in statistical models to adjust for bias.
Data access and standardization are therefore imperative. Accurate comparative metagenomics requires raw sequence reads so that bioinformatics bias can be eliminated through processing and modeling data from different studies in a uniform manner. Because most analyses use reference databases of genes and genomes, expanding the functional and phylogenetic diversity of data resources is a high priority. For example, microbial eukaryotes (Keeling et al., 2014) and viruses (Mizuno et al., 2013) present unique challenges but need to be represented because they are important players in many microbial communities. Solutions to database bias may include assembling novel sequences from metagenomes (Box 1) as well as efforts to sequence unculturable genomes (Rinke et al., 2013) and new genomes from specific environments (Fodor et al., 2012) and neglected clades (Wu et al., 2009). Accessing metagenome metadata is currently a major hurdle, which can be overcome through broader utilization of public repositories for sharing metadata in formats that are easily parsed and queried. To make robust meta-analytic approaches broadly available will require further development of computational tools that empower scientists with limited programming experience to rapidly and easily query the abundance of specific taxa and genes across publicly available datasets (Börnigen et al., 2015; Nayfach et al., 2015b; Pesant et al., 2015).
One of the most promising directions for microbiome research is integration (Franzosa et al., 2015; Segata et al., 2013) and systems-level modeling (Greenblum et al., 2013) of data from multiple different meta’omics platforms. To do this accurately and quantitatively will require careful consideration of sources of bias affecting each technology, including how methods perform on communities with different compositions. We have focused here on barriers to comparing shotgun DNA-sequencing libraries. However, similar questions should be asked of other modalities, starting with defining a meaningful parameter of the underlying community. For example, should metatranscriptomic analyses aim to estimate transcripts per cell and, if so, how should read counts be normalized to estimate this quantity without bias? Universal, single-copy genes may not be as useful as they are for normalizing DNA metagenomes due to differences in the overall transcriptional activity between cells (Maurice et al., 2013). Absolute transcript numbers of some genes may be estimable with mRNA standards (Satinsky et al., 2013), but this approach requires further benchmarking. An interesting alternative is high-throughput targeted sequencing of known taxonomic and functional markers from single cells (Spencer et al., 2016). As good quantitative statistics are developed for each meta’omics approach, it will be important to additionally consider how measurements will be compared across technologies.
With the development of unbiased, quantitative methods for comparative meta’omics, many exciting new questions are emerging. For example, it is now possible to reconstruct the pathways, modules, and metabolic potential of microbiomes (Abubucker et al., 2012). With accurate functional profiles, we can infer microbial interactions (Levy and Borenstein, 2013) and metabolic dependencies (Zelezniak et al., 2015), identify microbial metabolites that affect host biology (Donia and Fisch-bach, 2015), and inform bioprospecting efforts (Wilson et al., 2014a). Abundance estimates that are comparable across samples also enable ecological and evolutionary investigations of co-variation in taxonomic groups and gene families. An interesting example is the network of correlations between abundances of different antibiotic resistance genes across diverse environments, which highlights the role of specific human behaviors in the spread of drug resistance (Li et al., 2015). Patterns of co-variation are also improving OTU identification (Preheim et al., 2013), metagenome assembly (Nielsen et al., 2014), and detection of interacting taxa (Fisher and Mehta, 2014). One particularly exciting development is the ability to characterize strain-level diversity in shotgun metagenomes, which has revealed massive differences in strain composition and gene content within and between human hosts (Box 2). Our understanding of this genomic variation and its consequences will no doubt be revolutionized by the incorporation of long-read sequencing (Kuleshov et al., 2016), chromatin-capture technology (Marbouty and Koszul, 2015), and single-cell genomics (Stepanauskas, 2015) into the metagenomics toolbox.
References
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J. The Statistical Analysis of Compositional Data. Caldwell, N.J: Blackburn Press; 2003. [Google Scholar]
- Alivisatos AP, Blaser MJ, Brodie EL, Chun M, Dangl JL, Donohue TJ, Dorrestein PC, Gilbert JA, Green JL, Jansson JK, et al. MICROBIOME. A unified initiative to harness Earth’s microbiomes. Science. 2015;350:507–508. doi: 10.1126/science.aac8480. [DOI] [PubMed] [Google Scholar]
- Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. Binning meta-genomic contigs by coverage and composition. Nat Methods. 2014;11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- Ames SK, Gardner SN, Marti JM, Slezak TR, Gokhale MB, Allen JE. Using populations of human and microbial genomes for organism detection in metagenomes. Genome Res. 2015;25:1056–1067. doi: 10.1101/gr.184879.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. Command-line tools for processing biological sequencing data. 2011 ea-utils, https://expressionanalysis.github.io/ea-utils/
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beszteri B, Temperton B, Frickenhaus S, Giovannoni SJ. Average genome size: a potential source of bias in comparative metagenomics. ISME J. 2010;4:1075–1077. doi: 10.1038/ismej.2010.29. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börnigen D, Moon YS, Rahnavard G, Waldron L, McIver L, Shafquat A, Franzosa EA, Miropolsky L, Sweeney C, Morgan XC, et al. A reproducible approach to high-throughput biological data acquisition and integration. PeerJ. 2015;3:e791. doi: 10.7717/peerj.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, De-Pristo MA. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics. 2012;13:375. doi: 10.1186/1471-2164-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr R, Borenstein E. Comparative analysis of functional metagenomic annotation and the mappability of short reads. PLoS ONE. 2014;9:e105776. doi: 10.1371/journal.pone.0105776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr R, Shen-Orr SS, Borenstein E. Reconstructing the genomic content of microbiome taxa through shotgun metagenomic deconvolution. PLoS Comput Biol. 2013;9:e1003292. doi: 10.1371/journal.pcbi.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JH, Bolyen ET, Rideout JR, Caporaso JG. cualid: globally unique, correctable, and human-friendly sample identifiers for comparative omics studies. mSystems. 2015;1:e00010–e00015. doi: 10.1128/mSystems.00010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary B, Brito IL, Huang K, Gevers D, Shea T, Young S, Alm EJ. Detection of low-abundance bacterial strains in metagenomic datasets by eigengenome partitioning. Nat Biotechnol. 2015;33:1053–1060. doi: 10.1038/nbt.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T.H.M.P.; Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Fischbach MA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N, McFall-Ngai M, Zhao L. Microbiology: Create a global microbiome effort. Nature. 2015;526:631–634. doi: 10.1038/526631a. [DOI] [PubMed] [Google Scholar]
- Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, Boling L, Barr JJ, Speth DR, Seguritan V, Aziz RK, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal meta-genomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich SD. MetaHIT: The European Union Project on Metagenomics of the Human Intestinal Tract. In: Nelson EK, editor. Metagenomics of the Human Body. New York: Springer New York; 2011. pp. 307–316. [Google Scholar]
- Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS ONE. 2014;9:e84689. doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CK, Mehta P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS ONE. 2014;9:e102451. doi: 10.1371/journal.pone.0102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor AA, DeSantis TZ, Wylie KM, Badger JH, Ye Y, Hepburn T, Hu P, Sodergren E, Liolios K, Huot-Creasy H, et al. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLoS ONE. 2012;7:e41294. doi: 10.1371/journal.pone.0041294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Krogh Pedersen H, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, Huttenhower C. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol. 2015;13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Meyer F, Jansson J, Gordon J, Pace N, Tiedje J, Ley R, Fierer N, Field D, Kyrpides N, et al. The Earth Microbiome Project: meeting report of the “EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6 2010. Stand Genomic Sci. 2010;3:249–253. doi: 10.4056/aigs.1443528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass EM, Dribinsky Y, Yilmaz P, Levin H, Van Pelt R, Wendel D, Wilke A, Eisen JA, Huse S, Shipanova A, et al. MIxS-BE: a MIxS extension defining a minimum information standard for sequence data from the built environment. ISME J. 2014;8:1–3. doi: 10.1038/ismej.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, Ley RE. Conducting a microbiome study. Cell. 2014;158:250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum S, Chiu HC, Levy R, Carr R, Borenstein E. Towards a predictive systems-level model of the human microbiome: progress, challenges, and opportunities. Curr Opin Biotechnol. 2013;24:810–820. doi: 10.1016/j.copbio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum S, Carr R, Borenstein E. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 2015;160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingamp P, Grimsley N, Acinas SG, Clerissi C, Subirana L, Poulain J, Ferrera I, Sarmento H, Villar E, Lima-Mendez G, et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 2013;7:1678–1695. doi: 10.1038/ismej.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe AC, Jansson JK, Malfatti SA, Tringe SG, Tiedje JM, Brown CT. Tackling soil diversity with the assembly of large, complex metagenomes. Proc Natl Acad Sci USA. 2014;111:4904–4909. doi: 10.1073/pnas.1402564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Knight R, Brown CT, Caporaso JG, Clemente JC, Gevers D, Franzosa EA, Kelley ST, Knights D, Ley RE, et al. Advancing the microbiome research community. Cell. 2014;159:227–230. doi: 10.1016/j.cell.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MB, Highlander SK, Anderson EL, Li W, Dayrit M, Klitgord N, Fabani MM, Seguritan V, Green J, Pride DT, et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proc Natl Acad Sci USA. 2015;112:14024–14029. doi: 10.1073/pnas.1519288112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Li B, Ghonasgi T, Haase CP, Qin ZS, Dean D, Read TD. Direct amplification, sequencing and profiling of Chlamydia trachomatis strains in single and mixed infection clinical samples. PLoS ONE. 2014;9:e99290. doi: 10.1371/journal.pone.0099290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, Ding H, Marttinen P, Malmstrom RR, Stocker R, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy NA, Walker AW, Berry SH, Duncan SH, Farquarson FM, Louis P, Thomson JM, Satsangi J, Flint HJ, Parkhill J, et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS ONE. 2014;9:e88982. doi: 10.1371/journal.pone.0088982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keylock CJ. Simpson diversity and the Shannon–Wiener index as special cases of a generalized entropy. Oikos. 2005;109:203–207. [Google Scholar]
- Knight R, Jansson J, Field D, Fierer N, Desai N, Fuhrman JA, Hugenholtz P, van der Lelie D, Meyer F, Stevens R, et al. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol. 2012;30:513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen R International Nucleotide Sequence Database Collaboration. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, Matot E, Jona G, Harmelin A, Cohen N, et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015;349:1101–1106. doi: 10.1126/science.aac4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov V, Jiang C, Zhou W, Jahanbani F, Batzoglou S, Snyder M. Synthetic long-read sequencing reveals intraspecies diversity in the human microbiome. Nat Biotechnol. 2016;34:64–69. doi: 10.1038/nbt.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11:e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladau J, Sharpton TJ, Finucane MM, Jospin G, Kembel SW, O’Dwyer J, Koeppel AF, Green JL, Pollard KS. Global marine bacterial diversity peaks at high latitudes in winter. ISME J. 2013;7:1669–1677. doi: 10.1038/ismej.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LH, Monack DM. Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection. PLoS Pathog. 2014;10:e1004527. doi: 10.1371/journal.ppat.1004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Borenstein E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc Natl Acad Sci USA. 2013;110:12804–12809. doi: 10.1073/pnas.1300926110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Yang Y, Ma L, Ju F, Guo F, Tiedje JM, Zhang T. Meta-genomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015;9:2490–2502. doi: 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. Data from: An integrated catalog of reference genes in the human gut microbiome. GigaScience Database. 2014a doi: 10.1038/nbt.2942. http://gigadb.org/dataset/100064. [DOI] [PubMed]
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. MetaHIT Consortium. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014b;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- Lindgreen S, Adair KL, Gardner PP. An evaluation of the accuracy and speed of metagenome analysis tools. Sci Rep. 2016;6:19233. doi: 10.1038/srep19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Gibbons T, Ghodsi M, Treangen T, Pop M. Accurate and fast estimation of taxonomic profiles from metagenomic shotgun sequences. BMC Genomics. 2011;12(2):S4. doi: 10.1186/1471-2164-12-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Aziz M, Kachur S, Hsueh PR, Huang YT, Keim P, Price LB. BactQuant: an enhanced broad-coverage bacterial quantitative real-time PCR assay. BMC Microbiol. 2012;12:56. doi: 10.1186/1471-2180-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vázquez-Baeza Y, Jansson JK, Gordon JI, Knight R. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D. ConStrains identifies microbial strains in metagenomic datasets. Nat Biotechnol. 2015;33:1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom RR, Rodrigue S, Huang KH, Kelly L, Kern SE, Thompson A, Roggensack S, Berube PM, Henn MR, Chisholm SW. Ecology of uncultured Prochlorococcus clades revealed through single-cell genomics and biogeographic analysis. ISME J. 2013;7:184–198. doi: 10.1038/ismej.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor O, Borenstein E. MUSiCC: a marker genes based framework for metagenomic normalization and accurate profiling of gene abundances in the microbiome. Genome Biol. 2015;16:53. doi: 10.1186/s13059-015-0610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M, Koszul R. Metagenome analysis exploiting high-throughput chromosome conformation capture (3C) data. Trends Genet. 2015;31:673–682. doi: 10.1016/j.tig.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende DR, Waller AS, Sunagawa S, Järvelin AI, Chan MM, Arumugam M, Raes J, Bork P. Assessment of metagenomic assembly using simulated next generation sequencing data. PLoS ONE. 2012;7:e31386. doi: 10.1371/journal.pone.0031386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende DR, Sunagawa S, Zeller G, Bork P. Accurate and universal delineation of prokaryotic species. Nat Methods. 2013;10:881–884. doi: 10.1038/nmeth.2575. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013;9:e1003987. doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Pollard KS. Average genome size estimation improves comparative metagenomics and sheds light on the functional ecology of the human microbiome. Genome Biol. 2015;16:51. doi: 10.1186/s13059-015-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Bradley PH, Wyman SK, Laurent TJ, Williams A, Eisen JA, Pollard KS, Sharpton TJ. Automated and accurate estimation of gene family abundance from shotgun metagenomes. PLoS Comput Biol. 2015a;11:e1004573. doi: 10.1371/journal.pcbi.1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Fischbach MA, Pollard KS. MetaQuery: a web server for rapid annotation and quantitative analysis of specific genes in the human gut microbiome. Bioinformatics. 2015b;31:3368–3370. doi: 10.1093/bioinformatics/btv382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of transmission and global biogeography of bacteria. BioRxiv. 2016 doi: 10.1101/gr.201863.115. http://dx.doi.org/10.1101/031757. [DOI] [PMC free article] [PubMed]
- Nguyen NP, Mirarab S, Liu B, Pop M, Warnow T. TIPP: taxonomic identification and phylogenetic profiling. Bioinformatics. 2014;30:3548–3555. doi: 10.1093/bioinformatics/btu721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E, et al. MetaHIT Consortium. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- O’Sullivan DM, Laver T, Temisak S, Redshaw N, Harris KA, Foy CA, Studholme DJ, Huggett JF. Assessing the accuracy of quantitative molecular microbial profiling. Int J Mol Sci. 2014;15:21476–21491. doi: 10.3390/ijms151121476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Deming C, Conlan S, Kong HH, Segre JA, Segre JA NISC Comparative Sequencing Program. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesant S, Not F, Picheral M, Kandels-Lewis S, Le Bescot N, Gorsky G, Iudicone D, Karsenti E, Speich S, Troublé R, et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci Data. 2015;2:150023. doi: 10.1038/sdata.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, et al. NIH HMP Working Group. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poptsova MS, Il’icheva IA, Nechipurenko DY, Panchenko LA, Khodikov MV, Oparina NY, Polozov RV, Nechipurenko YD, Grokhovsky SL. Non-random DNA fragmentation in next-generation sequencing. Sci Rep. 2014;4:4532. doi: 10.1038/srep04532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash T, Taylor TD. Functional assignment of metagenomic data: challenges and applications. Brief Bioinform. 2012;13:711–727. doi: 10.1093/bib/bbs033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preheim SP, Perrotta AR, Martin-Platero AM, Gupta A, Alm EJ. Distribution-based clustering: using ecology to refine the operational taxonomic unit. Appl Environ Microbiol. 2013;79:6593–6603. doi: 10.1128/AEM.00342-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Il-lumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Wommack KE. All hail reproducibility in microbiome research. Microbiome. 2014;2:8. doi: 10.1186/2049-2618-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satinsky BM, Gifford SM, Crump BC, Moran MA. Use of internal standards for quantitative metatranscriptome and metagenome analysis. Methods Enzymol. 2013;531:237–250. doi: 10.1016/B978-0-12-407863-5.00012-5. [DOI] [PubMed] [Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics. 2008;9:34. doi: 10.1186/1471-2105-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. Computational meta’omics for microbial community studies. Mol Syst Biol. 2013;9:666. doi: 10.1038/msb.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, Polz MF, Alm EJ. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton TJ. An introduction to the analysis of shotgun metagenomic data. Front Plant Sci. 2014;5:209. doi: 10.3389/fpls.2014.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton TJ, Riesenfeld SJ, Kembel SW, Ladau J, O’Dwyer JP, Green JL, Eisen JA, Pollard KS. PhylOTU: a high-throughput procedure quantifies microbial community diversity and resolves novel taxa from metagenomic data. PLoS Comput Biol. 2011;7:e1001061. doi: 10.1371/journal.pcbi.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 2015;16:276. doi: 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SJ, Tamminen MV, Preheim SP, Guo MT, Briggs AW, Brito IL, Weitz AD, Pitkänen LK, Vigneault F, Juhani Virta MP, Alm EJ. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 2016;10:427–436. doi: 10.1038/ismej.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas R. Re-defining microbial diversity from its single-celled building blocks. Environ Microbiol Rep. 2015;7:36–37. doi: 10.1111/1758-2229.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR, Coelho LP, Arumugam M, Tap J, Nielsen HB, et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods. 2013;10:1196–1199. doi: 10.1038/nmeth.2693. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, et al. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- Tanner MA, Goebel BM, Dojka MA, Pace NR. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov M, Leach RW, Wingreen NS. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2015;9:68–80. doi: 10.1038/ismej.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt AY, Costea PI, Kultima JR, Li SS, Zeller G, Sunagawa S, Bork P. Temporal and technical variability of human gut metagenomes. Genome Biol. 2015;16:73. doi: 10.1186/s13059-015-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Amir A, Hyde ER, Metcalf JL, Song SJ, Knight R. Tracking down the sources of experimental contamination in microbiome studies. Genome Biol. 2014;15:564. doi: 10.1186/s13059-014-0564-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SJ, Xu Z, Amir A, Peddada S, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vazquez-Baeza Y, Birmingham A, et al. Effects of library size variance, sparsity, and compositionality on the analysis of microbiome data. PeerJ. 2015;3:e1408. [Google Scholar]
- Wilson MC, Mori T, Rückert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UA, Heycke N, Schmitt S, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014a;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014b;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Bhavsar J, Ravel J. Metagenomics: read length matters. Appl Environ Microbiol. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Jospin G, Eisen JA. Systematic identification of gene families for use as “markers” for phylogenetic and phylogeny-driven ecological studies of bacteria and archaea and their major subgroups. PLoS ONE. 2013;8:e77033. doi: 10.1371/journal.pone.0077033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia LC, Cram JA, Chen T, Fuhrman JA, Sun F. Accurate genome relative abundance estimation based on shotgun metagenomic reads. PLoS ONE. 2011;6:e27992. doi: 10.1371/journal.pone.0027992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, Gilbert JA, Karsch-Mizrachi I, Johnston A, Cochrane G, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci USA. 2015;112:6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Stephens RM, Meltzer PS, Davis SR. SRAdb: query and use public next-generation sequencing data from within R. BMC Bioinformatics. 2013;14:19. doi: 10.1186/1471-2105-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]