Abstract
The androgen receptor (AR)-signaling axis plays a key role in the pathogenesis of prostate cancer. The identification of AR targets contributing to prostate tumorigenesis is thus critical for the development of more effective therapies. Herein, we examined whether the AR could regulate classical protein tyrosine phosphatases, a family of enzymes increasingly associated with oncogenic processes. We found that protein tyrosine phosphatase 1B (PTP1B), a well-established regulator of metabolic signaling, was induced after androgenic stimulation of AR-expressing prostate cancer cells. This effect was observed both at the mRNA and protein levels, and translated into increased PTP1B activity. High-resolution location analyses on tiled array covering chromosome 20q revealed the recruitment of the AR to two response elements located within the first intron of the PTP1B gene (PTPN1) and correlated with an increase in RNA polymerase II recruitment to the transcriptional start site of PTPN1. Analysis of copy number alterations revealed that both PTPN1 and AR genes are co-amplified in metastatic tumors, and that PTPN1 amplification is associated with a subset of high-risk primary tumors. At the functional level, PTP1B depletion significantly delayed LNCaP tumor growth in vivo, and impaired androgen-induced cell migration and invasion in vitro. Importantly, androgen-independent cells also required PTP1B for optimal cell migration. Collectively, our results establish the AR as a transcriptional regulator of PTPN1 transcription, and suggest that PTP1B plays a tumor-promoting role in prostate cancer. This has important implications for prostate cancer biology, and supports the pre-clinical testing of PTP1B inhibitors for the treatment of the disease.
Keywords: PTP1B, Androgen Receptor, Prostate Cancer, Promoter Analysis, Xenograft
Introduction
Despite early diagnosis and efficient treatment of most early-stage tumors, prostate cancer remains the second leading cause of cancer-related deaths in North American men (1). This discrepancy is the result of the lack of biomarkers able to discriminate between indolent and aggressive tumors, as well as the absence of curative options for castration-resistant metastatic prostate cancer (CRMPC) (2). The androgen receptor (AR) plays a key role in the development and progression of prostate cancer, and androgen deprivation therapy (ADT) and/or AR blockade continues to be the standard treatment for advanced or recurrent tumors, even though the majority of patients eventually develop CRMPC (3).
Remarkably, most castration-resistant tumors still depend on the AR for their growth and survival, a feature that underscores the “addiction” of prostate cancer cells to AR signaling (4, 5). In primary tumors, this concept is further supported by the presence of common genetic alterations that directly and/or indirectly affect AR-dependent transcription. For instance, the loss of Nkx3.1 and PTEN tumor suppressor genes has been associated with Akt-mediated ligand-independent AR activity (6, 7), although recent findings challenge this concept and suggest that PI3K-Akt and AR signaling pathways regulate each other through reciprocal feedback (8, 9). Besides, the prostate-specific AR-induced TMPRSS2-ERG fusion protein has also been shown to regulate AR transcriptional activity (10). The characterization of signaling pathways acting upstream and downstream of the AR is therefore of paramount importance to identify new therapeutic targets that could interfere with AR signaling not only in CRMPC, but also at earlier stages of the disease.
One largely unexplored mechanism in prostate tumors is the regulation of tyrosine phosphorylation by classical protein tyrosine phosphatases (PTPs). Instead, the vast majority of studies have addressed the contribution of receptor and non-receptor tyrosine kinases, as important mediators of tumor-promoting signals responsible for the induction and/or enhancement of AR activity, as well as inducers of AR-independent survival mechanisms (11, 12). But abnormal regulation of tyrosine phosphorylation-dependent signaling in cancer cells can also be caused by altered PTP signaling. In fact, mutations and/or aberrant expression of several PTPs have been reported in different cancer types, and shown not only to counteract oncogenic tyrosine kinases, but as well to directly promote tumor development and progression (13). With respect to prostate cancer, however, only a limited number of classical PTPs have been investigated and their relationship with AR-dependent signaling remains largely unknown (13).
To address this issue, we first profiled the expression of classical PTPs in the context of AR-dependent signaling. Unexpectedly, we found that the protein tyrosine phosphatase 1B (PTP1B) gene PTPN1, previously reported as being induced in androgen-deprived conditions (14), is actually induced after androgen stimulation and is a direct transcriptional target of the AR. In primary prostate tumors, we observed a positive correlation between PTP1B expression, AR nuclear localization, and Ki-67 nuclear index. We also found that PTPN1 is frequently amplified in metastatic tumors and a subset of high-risk primary tumors. Finally, we provide evidence that PTP1B depletion decreases LNCaP tumor growth rates in vivo, and abrogates androgen-induced cell migration and invasion in vitro. These findings support a tumor-promoting role for PTP1B in prostate cancer and suggest that PTP1B inhibition may be of use in the treatment of the disease.
Materials and Methods
Cell culture
LNCaP, 22rv1, and DU145 cells were purchased from ATCC (Manassas, VA) and maintained in RPMI 1640 medium (Wisent, St-Bruno, Qc) supplemented with 10% FBS, L-glutamine, and 50µg/ml gentamycin. C4-2 cells were obtained from MD Anderson (Houston, TX) and maintained in the same conditions. The synthetic androgen analog R1881 was obtained for Perkin Elmer (Waltham, MA). For androgenic stimulation assays, cells were first androgen-deprived in phenol-free RPMI 1640 supplemented with 5% charcoal-stripped FBS, L-glutamine, and 50µg/ml gentamycin. After 48hrs, medium was refreshed and R1881 or ethanol (vehicle) was added for different time periods. For siRNA experiments, cells were incubated for 24hrs with 100nM siRNA or non-targeting control prior to androgen-deprivation.
Analysis of gene and protein expression
Total RNA extraction, reverse-transcription and quantitative real-time PCR (qPCR) were performed as already described (15). The primer sequences used can be found in Table S1. Threshold cycle numbers were calculated using the second derivative maximum obtain with the LightCycler®480 software version 3.5 (Roche, Laval, Qc). Data was normalized according to both PPIB and ARBP levels (Table S1).
Immunoblotting procedures were done as previously described (15). Membranes were probed with the following antibodies according to manufacturer’s instructions: mouse monoclonal anti-PTP1B (BD Transduction Laboratories, San Jose, CA), mouse monoclonal anti-AR (Lab Vision, Fremont, CA), rabbit polyclonal anti-calnexin (Cell Signaling Technology, Danver, MA), and mouse monoclonal anti-PSA (Lab Vision, Fremont, CA). Densitometry analyses were done with ImageJ (U.S. National Institutes of Health, Bethesda, MD; http://imagej.nih.gov/ij/).
Phosphatase assay
Cells were lysed in ice-cold RIPA buffer supplemented with 3mM DTT and EDTA-free complete protease inhibitor cocktail (Roche, Laval, Qc). PTP1B was immunoprecipitated 2hrs at 4°C using 200µg of protein lysate, 1ug of mouse monoclonal anti-PTP1B clone AE4-2J (EMD4Biosciences, Mississauga, ON), and 30µl of Protein G agarose beads (Fisher Canada, Nepean, ON). Beads were then washed three times in RIPA buffer and once in assay buffer (50mM HEPES, 3mM DTT, 0.1mg/ml BSA). The phosphatase assay was performed as previously described (16) using DIFMUP (Invitrogen) as the PTP1B substrate and, where indicated, 50µM of a PTP1B inhibitor (kind gift from Brian Kennedy, Merck Frosst, Pointe-Claire, Qc).
siRNA and stable miRNA expression systems
Detailed experimental procedures are reported in SI Materials and Methods and in Table S1.
ChIP assays and ChIP-on-chip on chr.20 tiled array
Chromatin was prepared from LNCaP cells exposed to 1nM R1881 or vehicle for 4hrs following pre-treatment with 20µM bicalutamide (BIC) or vehicle for 30min. Chromatin-immunoprecipitation (ChIP) was performed as described previously (17) using antibodies specific to AR (mouse monoclonal anti-AR from Lab Vision, Fremont, CA and BD Biosciences, San Jose, CA). Quantification of ChIP enrichment by real-time Q-PCR was carried out using the LightCycler®480 instrument (Roche, Laval, Qc). Amplification and labeling of AR-bound ChIP fragments was performed as described previously (18). Hybridization was carried out on custom designed chr.20 Agilent tiled arrays (150 bp resolution) and analyzed using Feature Extraction 10 and ChIP Analytics 3.1 (Agilent). The primers used for standard ChIP quantification and validation are listed in Table S3.
Computational motif discovery
De novo and known motif discovery was performed using the MEME Suite (http://meme.sdsc.edu/meme4_4_0/intro.html). Motif discovery and chromosomal location mapping were performed using the Genomatix Software Suite (http://www.genomatix.de/en/produkte/genomatix-software-suite.html).
Immunohistochemistry
Detailed experimental procedures and PTP1B scoring scale are reported in SI Material and Methods and in Table S2 respectively.
PTPN1 copy number alteration in prostate cancer
Analysis of copy number alteration at the PTPN1 locus is based on the published copy number profiles from 181 primary and 37 metastatic prostate tumors (19) and mRNA expression profiling of a subset of 150 of these cases (131 primary and 19 metastatic tumors) using the Nexus Copy Number software v6.0 (Biodiscovery, Inc.). The significance of association between amplification of the PTPN1 locus or its increased RNA expression and the indicated events (Figure 4E) was identified based on the two-tailed Fisher’s Exact test. For RNA expression data, a z-score threshold of 1.5 standard deviations was used to separate low (≤ −1.5) or high (≥ 1.5) from normal expression.
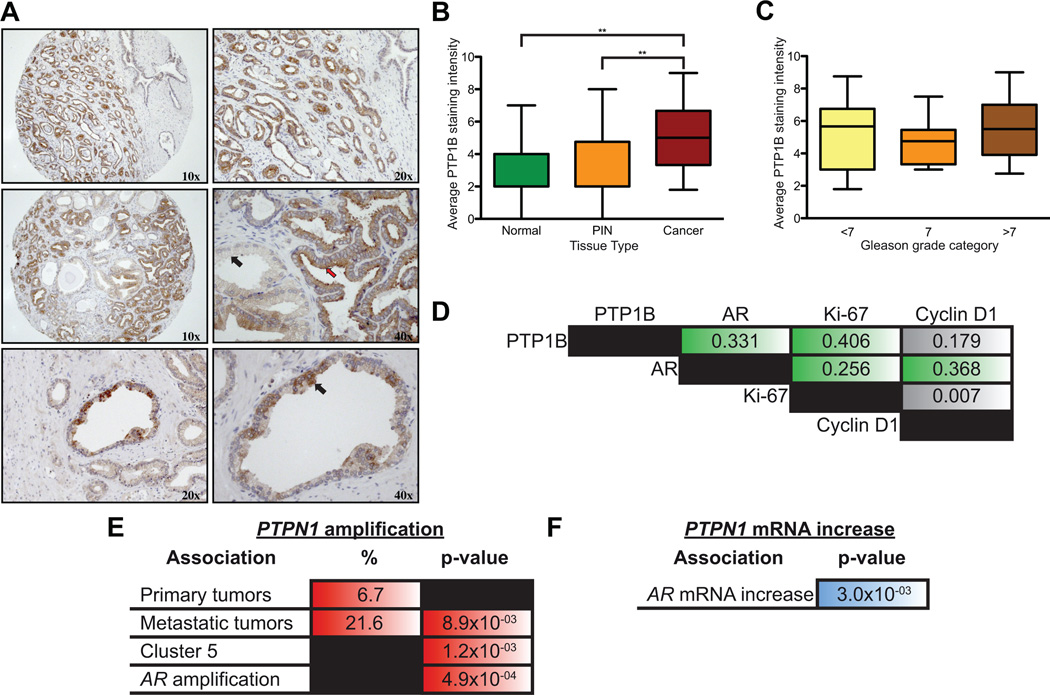
Figure 4.
PTP1B and AR are co-expressed and co-amplified in prostate tumors. (A) Immunohistochemical analyses of a prostate tumor tissue microarray (TMA) demonstrated a strong PTP1B expression in tumor tissue (upper panel), a strong PTP1B expression in tumor cells (middle panel, red arrow), and weak expression in normal glands (middle panel, black arrow). Punctate staining was often observed in normal gland (lower panels). (B and C) Expression of PTP1B is (B) elevated in cancer tissues compared to normal or PIN glands (Mann-Whitney, **p<0.01; Median +/− SD), (C) but does not vary according to the degree of histological differentiation (Gleason score). (D) Spearman correlations between PTP1B expression and the nuclear localization of AR, Ki-67, and cyclin D1 (green boxes represent p<0.01). (E) PTPN1 amplification is significantly associated with metastatic (N=37) and not primary prostate tumors (N=181). It significantly correlates with AR amplification (all samples, N=217) an event exclusively observed in metastatic samples. PTPN1 amplification is also associated with a subset of genomic alterations (Cluster 5) which displays early recurrence after radical prostatectomy (19). (F) PTPN1 mRNA expression in prostate cancer tumors positively correlates with AR mRNA expression (samples with available mRNA expression data: N=150).
Migration and invasion assays
LNCaP, C4-2, and DU145 cells were treated for siRNA experiments as described in the cell culture section, except for DU145 cells that were not androgen-deprived prior to the experiment. Details regarding the migration procedure are reported in SI Materials and Methods.
Mouse experimental procedures
Six-week old male SCID mice (Jackson Laboratories, Bar Harbor, ME) were subcutaneously injected with 2×106 shCTRL1a, shCTRL1b, shPTP1B1a, or shPTP1B2 cells resuspended in a 200µl 70% PBS/30% Matrigel solution. Ten mice per group were monitored daily for their physical health and once a week for tumor occurrence. Tumor volume was measured using the following equation: length × width × depth × 0.5236 (20). The animal protocol followed the Canadian Council on Animal Care ethical regulations and was approved by the McGill University Research and Ethics Animal Committee.
Statistics
Statistical analyses were performed with the Prism 5.0 GraphPad Software (La Jolla, CA). The Mann-Whitney or the Kruskal-Wallis tests were used to compare the distributions between two or more groups, respectively. The Spearman’s rho correlation test was used to measure correlation coefficients between clinicopathological and molecular variables. Kaplan-Meier and Log-Rank analyses were used to compare time of tumor onset between shCTRL and shPTP1B mice.
Results
Androgen treatment induces PTP1B expression in AR-expressing prostate cancer cells
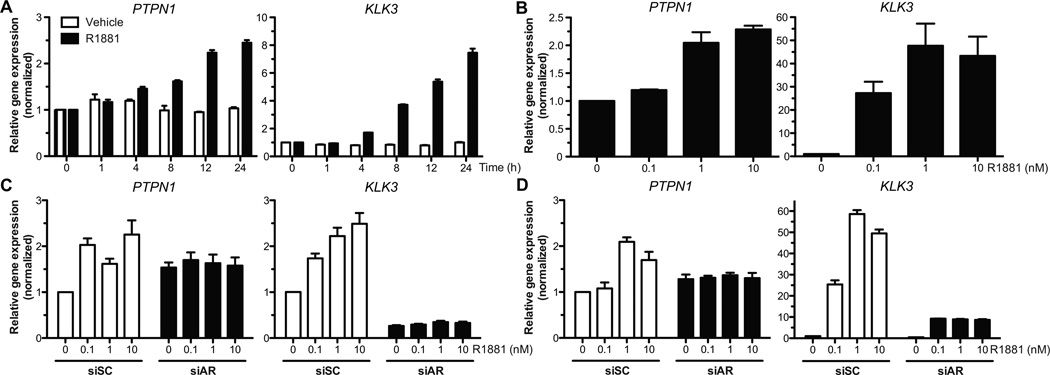
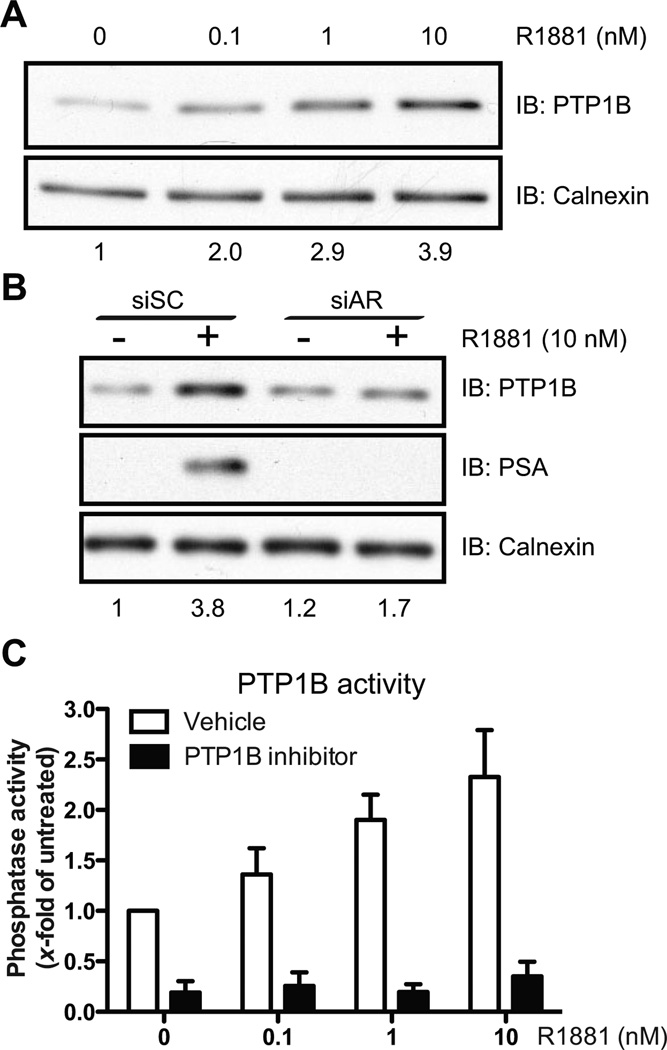
We screened for classical PTP transcripts that could be modulated by androgen stimulation. Androgen-sensitive LNCaP cells were maintained in steroid-depleted medium for 48hrs before stimulation with the synthetic androgen analog R1881. We performed gene expression profiling after treating for 24hrs of treatment, and detected changes in mRNA levels for 11 classical PTPs (Fig. S1). Surprisingly, we noted an AR-dependent increase in the levels of the PTPN1 transcript, despite previous findings linking the induction of PTPN1 expression to androgen deprivation (14). Using qPCR, we confirmed a time- and dose-dependent increase in PTPN1 mRNA levels in LNCaP cells treated with R1881 (Fig. 1A–B). In AR-expressing androgen-independent C4-2 and 22rv1 cells, the up-regulation of PTPN1 mRNA was also significant (Fig. 1C and S2A). In contrast, PTPN1 levels were not affected by R1881 treatment in AR-depleted or AR-negative cells (Fig. 1C–D and S2B–D). Notably, basal PTPN1 mRNA levels did not decrease in untreated AR-depleted cells (Fig. 1C–D) despite a PTPN1 mRNA half-life of about 8hrs (Fig. S3), indicating that other transcription factors can also regulate basal PTPN1 expression. Ultimately, the AR-mediated activation of PTPN1 led to an increase in PTP1B protein levels (Fig. 2A–B), which translated into a dose-dependent increase in total PTP1B phosphatase activity (Fig. 2C). Interestingly, substrate-trapping experiments revealed differential binding of PTP1B to phosphotyrosine substrates in androgen-depleted and R1881-treated cells (Fig. S4). Taken together, these results support a connection between AR-dependent signaling and PTP1B activity in prostate cancer cells.
Figure 1.
PTP1B mRNA expression is induced upon R1881 treatment. (A and B) PTPN1 expression in LNCaP cells was quantified as described in Materials and Methods after treatment with R1881 (10nM or the specified concentration) or the vehicle. PTPN1 mRNA levels were significantly increased in a (A) time- and (B) dose-dependent (24hrs treatment) manner (Kruskal-Wallis test, p<0.01 and p<0.05 respectively; representative experiment, N=3, +/− SEM). Relative PSA gene (KLK3) expression levels were monitored as treatment control. (C and D) PTPN1 mRNA levels were quantified after 24hrs of R1881 treatment in cells transfected with siRNAs against the AR (siAR) or control sequences (siSC). The PTPN1 mRNA increase in siSC transfected (C) C4-2 or (D) LNCaP cells was significant (Kruskal-Wallis test, p<0.05 for both cell lines; representative experiment, N=3, +/− SEM). No R1881-dependent induction was observed in siAR conditions.
Figure 2.
PTP1B protein and phosphatase activity is increased following R1881 treatment. (A and B) Cell lysates from LNCaP cells treated for 36hrs with (A) different R1881 concentrations or (B) with 10nM R1881 after transfection with siSC or siAR were resolved by SDS/PAGE followed by Western immunoblotting against PTP1B, PSA, and Calnexin.. Numbers below represent PTP1B levels relative to Calnexin (Representative experiments, (A) N=3 and (B) N=2). (C) PTP1B phosphatase activity in LNCaP cells was assessed after 36hrs of R1881 treatment, and a dose-dependent increase in phosphatase activity was observed (Kruskal-Wallis test, p<0.05; N=3, +/− SEM). A PTP1B inhibitor (50 µM) was used to address the specificity of the assay.
The PTPN1 gene is a direct AR transcriptional target
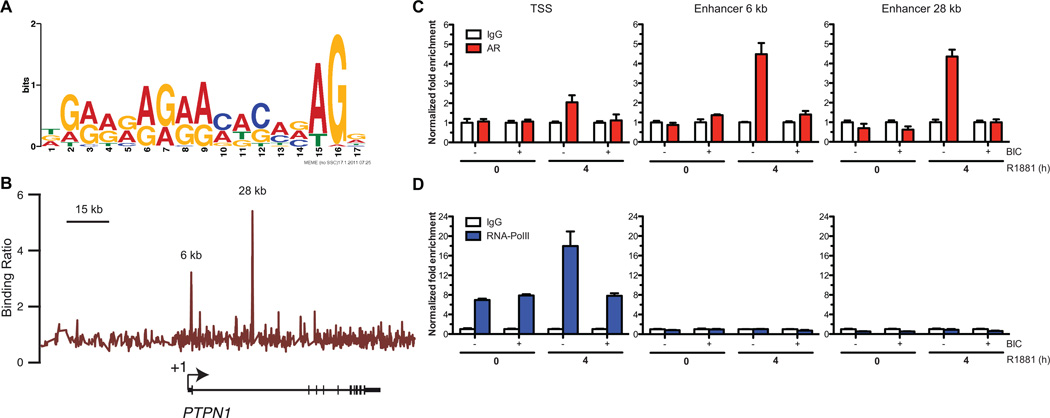
We assessed whether the AR could regulate PTPN1 expression through direct recruitment on androgen-responsive elements (ARE) located in the vicinity of the PTPN1. Since the location of AREs is not restricted to gene promoter regions and often contains degenerated motifs (21, 22), we designed a high-resolution tiled microarray spanning the entire human 20q chromosome enclosing PTPN1 to perform location analysis of AR by chromatin immunoprecipitation hybridized to the array (ChIP-on-chip) in R1881-treated LNCaP cells. The AR-bound segments identified by ChIP-on-chip were submitted to de novo binding site identification. The most enriched sequence (p-value 4.3×10−3) (Fig. 3A) was significantly associated to the known glucocorticoid-responsive element (GRE/ARE) (p-value 1.7×10−4). No significant AR recruitment was detected within the PTPN1 promoter while two AR-associated segments were identified in the first intron of PTPN1 located at +6.10 kb and +28.22 kb downstream of the transcription start site (TSS) respectively (Fig. 3B). The AR-bound region located at +28.22 kb harbors a putative GRE/ARE binding site. To test whether the recruitment of AR at these sites was modulated by androgen, we pre-treated LNCaP cells with the AR antagonist bicalutamide (BIC) for 30min prior to R1881 stimulation. BIC treatment prevented the R1881-dependent recruitment of AR at the +6.10 kb and +28.22 kb enhancer sites (Fig. 3C), suggesting that the recruitment of AR at these enhancers is directed by androgen. Interestingly, modest AR occupancy was also detected at the PTPN1 TSS when cells were treated with R1881 (Fig. 3C) concomitant with a significant R1881-dependent increase in RNA polymerase II (RNAPol II) recruitment (Fig. 3D). BIC pretreatment also abrogated the R1881-dependent AR and RNAPol II recruitment at the PTPN1 TSS (Fig. 3C–D), suggesting that activation of AR is necessary in order to contribute to the recruitment of RNApol II for transcriptional activation of the gene. Collectively, these findings validate AR as a transcriptional regulator of PTPN1 expression in androgen-dependent prostate cancer cells.
Figure 3.
PTPN1 is a direct target of the AR. (A) Sequence logo depicting a GRE sequence enriched in AR-bound segments identified by ChIP-on-chip obtained following a de novo motif discovery. (B) Binding profile of AR from the ChIP-on-chip performed with LNCaP cells on a high-resolution tiled array covering PTPN1 and its vicinity (N=2). (C and D) Standard ChIP in LNCaP cells shows that (C) AR is recruited to the 6 kb (Kruskal-Wallis test, p<0.05) and 28 kb enhancers (Kruskal-Wallis test, near to significance, p=0.075) upon R1881 treatment. We also observed a slight but non-significant increase in AR recruitment at the TSS (Kruskal-Wallis test, p<0.05). AR binding at these sites was abrogated when cells were pretreated with BIC (Representative experiment, N=2 +/− SEM). (D) RNAPol II occupancy at the TSS is increased after treatment with R1881 (Kruskal-Wallis test, p<0.05) but not when cells where pretreated with BIC (Representative experiment, N=2 +/− SEM).
Elevated PTP1B expression in prostate primary tumors correlates with AR nuclear localization and the tumor proliferative index
The AR-dependent regulation of PTP1B in androgen-dependent cells prompted us to characterize PTP1B expression in androgen-dependent primary prostate tumors. We analyzed PTP1B expression by immunohistochemistry on a tissue microarray (TMA) regrouping >300 cores from 62 patients that had undergone radical prostatectomy. We detected PTP1B expression in both normal adjacent and cancerous luminal cells, as well as in prostatic intra-epithelial neoplasia lesions (PIN). Most normal and PIN tissues displayed weak cytoplasmic PTP1B levels with occasional strong punctate staining of luminal cells (Fig. 4A). In contrast, tumor cells showed a marked increase in PTP1B expression (Fig. 4A–B). The over-expression of PTP1B in primary tumors was not related to the degree of histological differentiation (Fig. 4C and Table S3), or to other clinicopathological parameters except for a negative correlation with the surgical margin status (Table S3). At the molecular level, however, PTP1B expression positively correlated with AR nuclear localization (Fig. 4D), thus providing in vivo evidence for a link between AR activation and PTP1B expression. Interestingly, both nuclear AR and PTP1B also correlated with the tumor proliferative index, as determined by Ki-67 staining (Fig. 4D). This suggests that PTP1B may not only be regulated by the AR, but may also exert tumor-promoting effects in prostate tumors.
Amplification of the PTPN1 gene is associated with metastatic prostate cancer and a subgroup of high-risk primary tumors
To gather additional evidence for a role of PTP1B in prostate cancer pathogenesis, we examined the status of the PTPN1 gene in an independent cohort of 218 patients (19). Interestingly, PTPN1 amplification was found in 6.6% (12/181) of primary tumors and enriched in 21.6% (8/37) of the metastatic tumor samples (Fig. 4E). In addition, the PTPN1 amplification in primary tumors was significantly associated with a previously identified pattern of copy-number alterations (Cluster 5 as defined by Taylor et al. (19)), found predictive of early recurrence after radical prostatectomy. PTPN1 amplification and mRNA expression also significantly correlated with AR gene amplification and transcript levels, respectively (Figs. 4E–F). These findings complement our ChIP-on-chip and immunohistochemical results and are consistent with the AR-mediated induction of PTPN1 in human prostate cancer. Moreover, given the special role of AR signaling in prostate cancer and its exclusive amplification in late stage disease, our identification of a positive correlation between AR and PTPN1 amplification and expression strongly supports a role for PTP1B in prostate cancer progression.
PTP1B plays a tumor-promoting role in prostate cancer cells
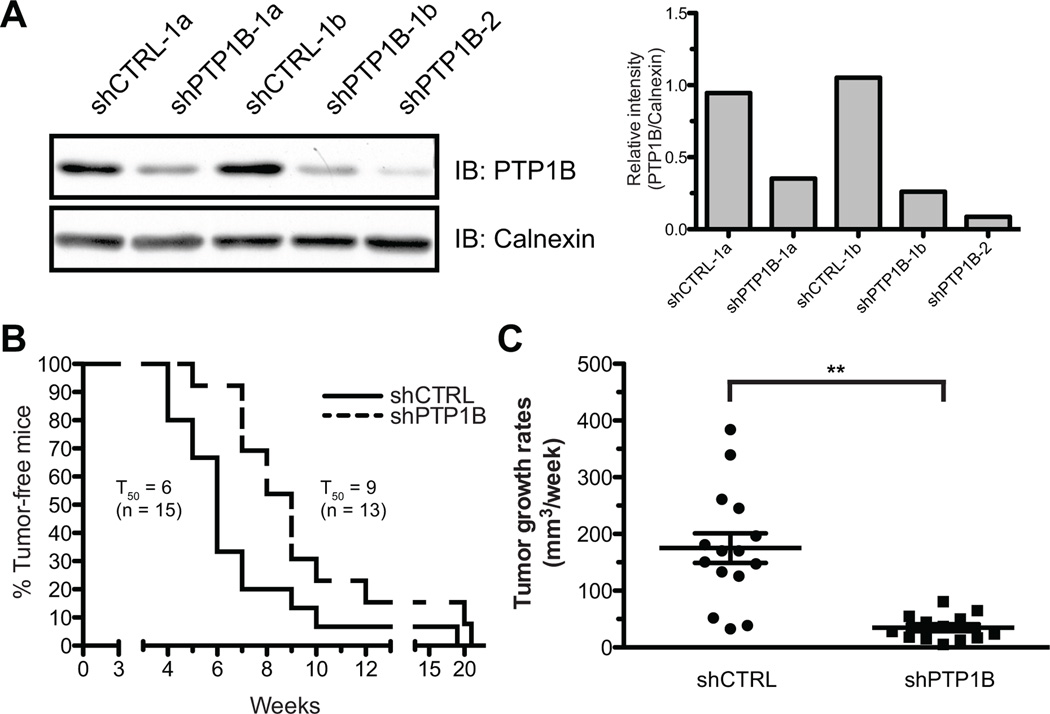
To address the role of PTP1B in vivo, stable LNCaP-derived clones expressing shRNAs against PTP1B (shPTP1B), or a non-targeting control shRNA (shCTRL), were injected in the right flank of male SCID mice (Fig. 5A). Subsequent weekly monitoring revealed that PTP1B depletion significantly delayed tumor occurrence, with a median time to onset of 9 weeks for shPTP1B tumors, compared to 6 weeks for shCTRL xenografts (Fig. 5B). On average, shPTP1B tumors also grew more slowly at a rate of ~35mm3 per week, compared to ~175mm3 a week for shCTRL tumors (Fig. 5C). Importantly, analysis of PTP1B expression in endpoint tumors showed no significant difference between shPTP1B and shCTRL specimens (Fig. S5), suggesting that the restoration of endogenous PTP1B levels was required for the growth of shPTP1B clones.
Figure 5.
PTP1B down-regulation delays tumor occurrence and decreases tumor growth rates in vivo. (A) Western immunoblotting showing PTP1B levels in stable shCTRL and shPTP1B clones (left panel) relative to Calnexin (right panel). For the xenograft assays, a total of 40 mice were injected with either shCTRL-1a (n=10), shCTRL-1b (n=10), shPTP1B-1a (n=10), and shPTP1B-2 (n=10) clones. For data analysis, shCTRL and shPTP1B mice were pooled together (B) Tumor development occurred in 15 shCTRL mice, and 13 shPTP1B animals. Kaplan-Meier analyses of tumor occurence shows that shPTP1B tumors at a median time of onset (TD50) of 9 weeks compared to 6 weeks for shCTRL clones (Log-rank test, p<0.05). (C) Weekly monitoring of tumor growth revealed that shPTP1B tumors growth significantly slower than the shCTRL tumors (Mann-Whitney test, p<0.01).
We next attempted to identify the mechanism(s) by which PTP1B promotes tumor growth. While PTP1B silencing did not affect cell proliferation (Fig. S6), it significantly impaired the migratory and invasive properties of LNCaP cells (Fig. 6A and S7–8). Importantly, this was exclusively observed in androgen-stimulated conditions, which provides a functional link between AR-induced PTP1B expression and PTP1B function. Combined with our observations in primary tumors, these results clearly support an AR-dependent tumor-promoting role for PTP1B in androgen-dependent prostate cancer cells.
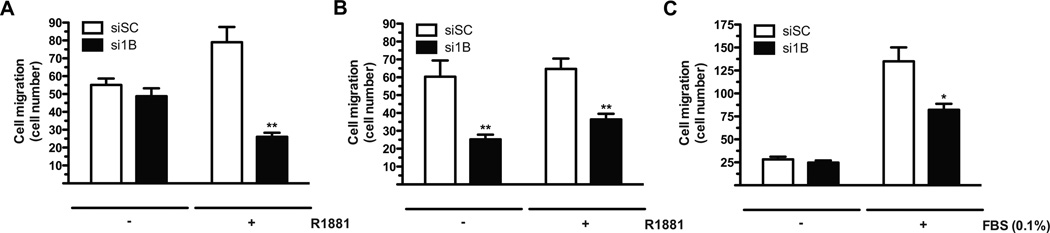
Figure 6.
PTP1B down-regulation impedes cell migration in vitro. (A and B) (A) LNCaP and (B) C4-2 cells transiently transfected with an siRNA against PTP1B (si1B) or a non-targeting sequence (siSC) were androgen-deprived for 48hrs. Cells were then treated with R1881 or the vehicle for 36hrs before FBS-induced cell migration as described in Materials and Methods. (C) DU145 cells were transiently transfected with si1B or siSC for 72hrs before FBS-induced cell migration (Mann-Whitney test; * p<0.05, ** p<0.01 compared to the corresponding siSC; N=3 +/− SEM).
We also examined the role of PTP1B in androgen-independent prostate cancer cells. In contrast to parental LNCaP cells, PTP1B silencing in C4-2 cells affected migration both in the presence and absence of androgen stimulation (Fig. 6B and S8). Importantly, the AR remains transcriptionally active in androgen-deprived C4-2 cells (23), which suggests that the AR-dependent regulation of PTP1B activity is maintained in AR-expressing androgen-independent cells. Besides, AR depletion in C4-2 cells also triggered an R1881-independent increase in PTP1B levels, an event that was not observed for the PSA gene (Fig. 1C). This indicates that in the context of AR down-regulation, other transcription factors can substitute for the AR to promote PTP1B expression and activity in androgen-independent cells. In line with this, PTP1B is highly expressed in AR-negative DU145 cells and PTP1B knockdown also impaired cell migration (Fig. 6C and S8). Overall, these results demonstrate that PTP1B exerts tumor-promoting functions in both androgen-dependent and -independent prostate cancer cells.
Discussion
PTP1B is a well-established regulator of metabolic signaling and has become an attractive drug target for the treatment of diabetes and obesity (24). In these diseases, PTP1B counteracts the action of tyrosine kinases such as the insulin receptor and Jak2, leading to insulin resistance and attenuation of leptin signaling, respectively (25, 26). With regards to cancer, this inhibitory function should intuitively suppress oncogenic signaling, yet accumulating evidence suggests that PTP1B can exert the opposite effect (27). Our results corroborate this trend and are in line with previous studies in breast (28–30), colon (31), and gastric cancer (32) where PTP1B ablation and/or inhibition abrogated the tumorigenic properties of malignant cells. More specifically, we show here that PTP1B promotes prostate cancer cell migration and invasion in vitro and is associated with tumor growth and metastasis in vivo (Figs. 3–4), which is not unexpected given the number of PTP1B substrates that play a direct role in these processes such as FAK, CrkII, p130Cas, cortactin, and p62dok (27). We have yet to identify the prostate-specific PTP1B substrate(s) responsible for the observed phenotype, but proteomic analyses are underway to characterize the phosphotyrosine substrates that differentially interact with PTP1B in androgen-deprived and androgen-supplemented conditions (Fig. S4).
The mechanisms that modulate PTP1B activity in tumor cells are still largely undefined, but the transcriptional up-regulation of PTPN1 has long been suspected. Indeed, previous characterization of the PTPN1 promoter has identified multiple binding sites for transcription factors that are aberrantly regulated not only in metabolic disorders, but also in malignant disease. These include Egr-1, Sp1, Sp3 (33), YB-1 (34), NF-κB (35), and HIF (36), that can be activated by a variety of stimuli such as glucose, insulin, pro-inflammatory cytokines, oncoproteins, or tumor-suppressor loss. In addition, an increase in PTP1B levels and/or stability has been reported in breast cancer cells expressing a constitutively active ErbB2 mutant (28, 37), as well as in a subset of B-cell lymphoma cell lines following IL4 treatment (38). Here, we demonstrate that PTPN1 transcription can also be modulated by the AR-signaling axis, a vital pathway in prostate cancer biology. We also show that PTPN1 and AR are co-amplified in metastatic tumors (Fig. 4E), and that PTP1B expression positively correlates with both AR expression (Fig. 4F) and nuclear localization (Fig 4D). All together, these results clearly establish PTPN1 as an AR-regulated gene, and corroborate findings of a previous study that reported the co-amplification of PTPN1 and AR in a human prostate cancer xenograft model (39, 40). Interestingly, PTPN1 is located on chromosome 20q13, a region frequently amplified in several tumor types (40). Despite conflicting results that question the relevance of this amplified region in hereditary prostate cancer (41), the fact that PTPN1 is frequently amplified in metastatic tumors as well as in a subset of high-risk primary tumors (Fig. 4E) supports a tumor-promoting role for this region in sporadic prostate cancer development and/or progression.
Our high-resolution analysis of AR binding to PTPN1 is the first to demonstrate that PTP1B expression is not solely regulated by transcription factors binding to the classical PTPN1 promoter region. In R1881-stimulated LNCaP cells, direct binding of the AR to PTPN1 occurs through two enhancers embedded into the large 50.77 kb first intron. In fact, intronic AR binding is quite common: Hu et al. recently reported in a genome-wide study that 49% of the AR binding sites from mouse epididymis were intronic, most of them located within the first intron (42), which is also consistent with other studies in LNCaP (22, 43), HPr-1AR (44) and primary human muscle cells (45). Our results also indicate that other transcription factors regulate PTP1B expression in androgen-deprived LNCaP cells, as well as in androgen-independent cells. In fact, AR gene expression programs have been shown to differ between androgen-dependent and -independent cells (46), thus raising the possibility that alternative factors can activate a subset of critical tumor-promoting genes no longer regulated by the AR. This is particularly relevant given recent discoveries showing that frequent prostate cancer alterations, i.e. constitutive PI3K signaling and TMPRSS2-ERG expression, can down-regulate AR transcriptional activity (8–10). Among potential candidates, the aforementioned NF-κB and YB-1 factors are particularly attractive given their established roles in prostate cancer progression. For instance, the regulation of PTP1B by NF-κB (35) could reconcile our findings with a previous report linking PTP1B expression to androgen withdrawal-induced neuroendocrine differentiation (14), an aggressive tumor phenotype that has been associated with NF-κB activation (47, 48). Similarly, YB-1 has been shown to be activated in LNCaP tumors following castration (49), and a strong correlation with elevated PTP1B expression has also been reported in androgen-independent DU145 cells (34). Finally, given the location of their respective response elements in the classical PTPN1 promoter region, it is also conceivable that these factors cooperate with the AR to regulate PTPN1 transcription in low androgen conditions. In line with this, a significant increase in PTP1B levels was observed in C4-2 cells stimulated with low androgen levels (0.1nM; Fig. 1C), a phenomenon that was not observed in parental LNCaP cells (Fig. 1D).
In summary, our study uncovers a previously unsuspected regulation of PTP1B by the AR, and provides direct evidence for a tumor-promoting role of PTP1B in prostate cancer. The combination of our cellular and in vivo findings advocates for the future preclinical testing of existing antisense- or small molecules inhibitors of PTP1B for the treatment of the disease (50). Moreover, our initial profiling of androgen-induced PTPs (Fig. S1) justifies further investigation to address the role and significance of other classical PTPs in prostate cancer.
Supplementary Material
Acknowledgments
We thank Matthew Feldhammer for critical review of the manuscript. We are also grateful to Marianne Filion, Ailsa Leloy, Robert Nadon, and Joseph James Bowden for their technical assistance and/or helpful discussions. L.L. is a recipient of a Canadian Institutes for Health Research (CHIR) fellowship award. D. P. L. is a recipient of a CIHR Frederick Banting and Charles Best doctoral research award. G.D. is supported by pre-doctoral traineeship award (W81XWH-10-1-0489) from the U.S. Department of Defense Breast Cancer Research Program. F.S. is the recipient of the Université de Montréal Chair in Prostate Cancer Research. M.L.T. is the holder of the Jeanne and Jean-Louis Levesque Chair in Cancer Research.
Grant Support
This work was supported by CIHR operating grants to V.G. (MOP-64275) and M.L.T (MOP-62887).
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damber JE, Aus G. Prostate cancer. Lancet. 2008;371:1710–1721. doi: 10.1016/S0140-6736(08)60729-1. [DOI] [PubMed] [Google Scholar]
- 4.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 5.Gurel B, Iwata T, Koh CM, Yegnasubramanian S, Nelson WG, De Marzo AM. Molecular alterations in prostate cancer as diagnostic, prognostic, and therapeutic targets. Adv Anat Pathol. 2008;15:319–331. doi: 10.1097/PAP.0b013e31818a5c19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao H, Ouyang X, Banach-Petrosky WA, Shen MM, Abate-Shen C. Emergence of androgen independence at early stages of prostate cancer progression in Nkx3.1; Pten mice. Cancer Res. 2006;66:7929–7933. doi: 10.1158/0008-5472.CAN-06-1637. [DOI] [PubMed] [Google Scholar]
- 7.Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, et al. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367–378. doi: 10.1016/j.ccr.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 8.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang YM, Kung HJ, Evans CP. Nonreceptor tyrosine kinases in prostate cancer. Neoplasia. 2007;9:90–100. doi: 10.1593/neo.06694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julien SG, Dube N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer. 2011;11:35–49. doi: 10.1038/nrc2980. [DOI] [PubMed] [Google Scholar]
- 14.Wu C, Zhang L, Bourne PA, Reeder JE, di Sant'Agnese PA, Yao JL, et al. Protein tyrosine phosphatase PTP1B is involved in neuroendocrine differentiation of prostate cancer. Prostate. 2006;66:1125–1135. doi: 10.1002/pros.20412. [DOI] [PubMed] [Google Scholar]
- 15.Hardy S, Wong NN, Muller WJ, Park M, Tremblay ML. Overexpression of the protein tyrosine phosphatase PRL-2 correlates with breast tumor formation and progression. Cancer Res. 2010;70:8959–8967. doi: 10.1158/0008-5472.CAN-10-2041. [DOI] [PubMed] [Google Scholar]
- 16.Stuible M, Zhao L, Aubry I, Schmidt-Arras D, Bohmer FD, Li CJ, et al. Cellular inhibition of protein tyrosine phosphatase 1B by uncharged thioxothiazolidinone derivatives. Chembiochem. 2007;8:179–186. doi: 10.1002/cbic.200600287. [DOI] [PubMed] [Google Scholar]
- 17.Laganiere J, Deblois G, Giguere V. Functional genomics identifies a mechanism for estrogen activation of the retinoic acid receptor alpha1 gene in breast cancer cells. Mol Endocrinol. 2005;19:1584–1592. doi: 10.1210/me.2005-0040. [DOI] [PubMed] [Google Scholar]
- 18.Deblois G, Hall JA, Perry MC, Laganiere J, Ghahremani M, Park M, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–6157. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- 19.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato N, Gleave ME, Bruchovsky N, Rennie PS, Beraldi E, Sullivan LD. A metastatic and androgen-sensitive human prostate cancer model using intraprostatic inoculation of LNCaP cells in SCID mice. Cancer Res. 1997;57:1584–1589. [PubMed] [Google Scholar]
- 21.Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, et al. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 2007;8:871–878. doi: 10.1038/sj.embor.7401046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, et al. A Hierarchical Network of Transcription Factors Governs Androgen Receptor-Dependent Prostate Cancer Growth. Molecular Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dehm SM, Tindall DJ. Ligand-independent androgen receptor activity is activation function-2-independent and resistant to antiandrogens in androgen refractory prostate cancer cells. J Biol Chem. 2006;281:27882–27893. doi: 10.1074/jbc.M605002200. [DOI] [PubMed] [Google Scholar]
- 24.Dube N, Tremblay ML. Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: from diabetes, obesity to cell cycle, and cancer. Biochim Biophys Acta. 2005;1754:108–117. doi: 10.1016/j.bbapap.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 25.Cheng A, Uetani N, Simoncic PD, Chaubey VP, Lee-Loy A, McGlade CJ, et al. Attenuation of leptin action and regulation of obesity by protein tyrosine phosphatase 1B. Dev Cell. 2002;2:497–503. doi: 10.1016/s1534-5807(02)00149-1. [DOI] [PubMed] [Google Scholar]
- 26.Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 27.Lessard L, Stuible M, Tremblay ML. The two faces of PTP1B in cancer. Biochim Biophys Acta. 2010;1804:613–619. doi: 10.1016/j.bbapap.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 28.Arias-Romero LE, Saha S, Villamar-Cruz O, Yip SC, Ethier SP, Zhang ZY, et al. Activation of Src by protein tyrosine phosphatase 1B Is required for ErbB2 transformation of human breast epithelial cells. Cancer Res. 2009;69:4582–4588. doi: 10.1158/0008-5472.CAN-08-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentires-Alj M, Neel BG. Protein-tyrosine phosphatase 1B is required for HER2/Neu-induced breast cancer. Cancer Res. 2007;67:2420–2424. doi: 10.1158/0008-5472.CAN-06-4610. [DOI] [PubMed] [Google Scholar]
- 30.Julien SG, Dube N, Read M, Penney J, Paquet M, Han Y, et al. Protein tyrosine phosphatase 1B deficiency or inhibition delays ErbB2-induced mammary tumorigenesis and protects from lung metastasis. Nat Genet. 2007;39:338–346. doi: 10.1038/ng1963. [DOI] [PubMed] [Google Scholar]
- 31.Zhu S, Bjorge JD, Fujita DJ. PTP1B contributes to the oncogenic properties of colon cancer cells through Src activation. Cancer Res. 2007;67:10129–10137. doi: 10.1158/0008-5472.CAN-06-4338. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Liu B, Chen X, Su L, Wu P, Wu J, et al. PTP1B expression contributes to gastric cancer progression. Med Oncol. 2011 doi: 10.1007/s12032-011-9911-2. [DOI] [PubMed] [Google Scholar]
- 33.Fukada T, Tonks NK. The reciprocal role of Egr-1 and Sp family proteins in regulation of the PTP1B promoter in response to the p210 Bcr-Abl oncoprotein-tyrosine kinase. J Biol Chem. 2001;276:25512–25519. doi: 10.1074/jbc.M101354200. [DOI] [PubMed] [Google Scholar]
- 34.Fukada T, Tonks NK. Identification of YB-1 as a regulator of PTP1B expression: implications for regulation of insulin and cytokine signaling. Embo J. 2003;22:479–493. doi: 10.1093/emboj/cdg067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zabolotny JM, Kim YB, Welsh LA, Kershaw EE, Neel BG, Kahn BB. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J Biol Chem. 2008;283:14230–14241. doi: 10.1074/jbc.M800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suwaki N, Vanhecke E, Atkins KM, Graf M, Swabey K, Huang P, et al. A HIF-Regulated VHL-PTP1B-Src Signaling Axis Identifies a Therapeutic Target in Renal Cell Carcinoma. Sci Transl Med. 2011;3:85ra47. doi: 10.1126/scitranslmed.3002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiener JR, Kerns BJ, Harvey EL, Conaway MR, Iglehart JD, Berchuck A, et al. Overexpression of the protein tyrosine phosphatase PTP1B in human breast cancer: association with p185c-erbB-2 protein expression. J Natl Cancer Inst. 1994;86:372–378. doi: 10.1093/jnci/86.5.372. [DOI] [PubMed] [Google Scholar]
- 38.Lu X, Malumbres R, Shields B, Jiang X, Sarosiek KA, Natkunam Y, et al. PTP1B is a negative regulator of interleukin 4-induced STAT6 signaling. Blood. 2008;112:4098–4108. doi: 10.1182/blood-2008-03-148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bar-Shira A, Pinthus JH, Rozovsky U, Goldstein M, Sellers WR, Yaron Y, et al. Multiple genes in human 20q13 chromosomal region are involved in an advanced prostate cancer xenograft. Cancer Res. 2002;62:6803–6807. [PubMed] [Google Scholar]
- 40.Tabach Y, Kogan-Sakin I, Buganim Y, Solomon H, Goldfinger N, Hovland R, et al. Amplification of the 20q chromosomal arm occurs early in tumorigenic transformation and may initiate cancer. PLoS ONE. 2011;6:e14632. doi: 10.1371/journal.pone.0014632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaid DJ, Chang BL. Description of the International Consortium For Prostate Cancer Genetics, and failure to replicate linkage of hereditary prostate cancer to 20q13. Prostate. 2005;63:276–290. doi: 10.1002/pros.20198. [DOI] [PubMed] [Google Scholar]
- 42.Hu S, Yao G, Guan X, Ni Z, Ma W, Wilson EM, et al. Research Resource: Genome-Wide Mapping of in Vivo Androgen Receptor Binding Sites in Mouse Epididymis. Mol Endocrinol. 2010;24:2392–2405. doi: 10.1210/me.2010-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takayama K, Kaneshiro K, Tsutsumi S, Horie-Inoue K, Ikeda K, Urano T, et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene. 2007;26:4453–4463. doi: 10.1038/sj.onc.1210229. [DOI] [PubMed] [Google Scholar]
- 44.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes & Development. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyce A, Bai Y, Nagpal S, Thompson CC. Research Resource: The Androgen Receptor Modulates Expression of Genes with Critical Roles in Muscle Development and Function. Mol Endocrinol. 2010;24:1665–1674. doi: 10.1210/me.2010-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen Receptor Regulates a Distinct Transcription Program in Androgen-Independent Prostate Cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin RJ, Lho Y, Connelly L, Wang Y, Yu X, Saint Jean L, et al. The nuclear factor-kappaB pathway controls the progression of prostate cancer to androgen-independent growth. Cancer Res. 2008;68:6762–6769. doi: 10.1158/0008-5472.CAN-08-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan TC, Veeramani S, Lin MF. Neuroendocrine-like prostate cancer cells: neuroendocrine transdifferentiation of prostate adenocarcinoma cells. Endocr Relat Cancer. 2007;14:531–547. doi: 10.1677/ERC-07-0061. [DOI] [PubMed] [Google Scholar]
- 49.Gimenez-Bonafe P, Fedoruk MN, Whitmore TG, Akbari M, Ralph JL, Ettinger S, et al. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate. 2004;59:337–349. doi: 10.1002/pros.20023. [DOI] [PubMed] [Google Scholar]
- 50.Liu G. Technology evaluation: ISIS-113715, Isis. Curr Opin Mol Ther. 2004;6:331–336. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.