Abstract
Background
Genomic technologies are increasingly used to guide clinical decision-making in cancer control. Economic evidence about the cost-effectiveness of genomic technologies is limited, in part because of a lack of published comprehensive cost estimates. In the present micro-costing study, we used a time-and-motion approach to derive cost estimates for 3 genomic assays and processes—digital gene expression profiling (gep), fluorescence in situ hybridization (fish), and targeted capture sequencing, including bioinformatics analysis—in the context of lymphoma patient management.
Methods
The setting for the study was the Department of Lymphoid Cancer Research laboratory at the BC Cancer Agency in Vancouver, British Columbia. Mean per-case hands-on time and resource measurements were determined from a series of direct observations of each assay. Per-case cost estimates were calculated using a bottom-up costing approach, with labour, capital and equipment, supplies and reagents, and overhead costs included.
Results
The most labour-intensive assay was found to be fish at 258.2 minutes per case, followed by targeted capture sequencing (124.1 minutes per case) and digital gep (14.9 minutes per case). Based on a historical case throughput of 180 cases annually, the mean per-case cost (2014 Canadian dollars) was estimated to be $1,029.16 for targeted capture sequencing and bioinformatics analysis, $596.60 for fish, and $898.35 for digital gep with an 807-gene code set.
Conclusions
With the growing emphasis on personalized approaches to cancer management, the need for economic evaluations of high-throughput genomic assays is increasing. Through economic modelling and budget-impact analyses, the cost estimates presented here can be used to inform priority-setting decisions about the implementation of such assays in clinical practice.
Keywords: Personalized medicine, micro-costing, time-and-motion analyses, genomic technology, economic evaluations
INTRODUCTION
The use of molecular information is transforming the field of oncogenomics. Ongoing genomic discoveries in the area of cancer research have contributed to the potential for personalized approaches to cancer management1. Molecular characterization of tumours has already contributed to the discovery of targeted therapies such as trastuzumab for her2-positive breast cancer2 and rituximab for first-line chemotherapy in diffuse large B-cell lymphoma (dlbcl)3, both of which have transformed the standard of care for those disease sites. More recently, immunohistochemistry and cytogenetic analysis for the non-Hodgkin (nhl) and Hodgkin lymphomas have been used to identify prognostic biomarkers in the anaplastic large-cell lymphomas, to understand the prognostic impact of molecularly distinct cell-of-origin subgroups in cases of dlbcl, and to identify various factors that can be used to risk-stratify Hodgkin lymphoma patients with the aim of offering targeted treatment regimens4–6. In a fiscally conscious health care environment, there is great appeal in being able to identify patients for whom newer and potentially more costly treatments will be cost-effective.
Despite those advances in cancer research, economic evidence supporting the application of high-throughput genomic technologies in everyday practice is lacking7,8. The accelerated pace with which high-throughput genomic assays and next-generation sequencing technologies are being used in cancer diagnostics threatens to outpace an assessment of the economic implications of changes in practice9–11. A recent rapid review of the cost-effectiveness of next-generation sequencing technologies, including whole-genome, whole-exome, and targeted capture sequencing, conducted by the Canadian Agency for Drugs and Technologies in Health, found that the economic evidence is currently insufficient to make definitive claims about the cost-effectiveness of next-generation sequencing technologies12. Similarly, a review by Buchanan et al.7 of the literature on economic evaluations of genomic interventions identified a number of methodology challenges faced by health economists when undertaking evaluations of personalized medicine technologies, one of them being a lack of comprehensive cost estimates that are available to researchers. Some research institutions and manufacturers publish price lists of personalized medicine technologies, but because those services are often provided for profit, using the provided estimates to inform decision-making within a publicly funded health care system cannot be recommended. In addition, such estimates often neglect the multidisciplinary nature of the work (encompassing, for example, clinical researchers, bioinformatics analysis, laboratory oversight), and assumptions factored into the related calculations are not always made apparent, hence limiting their applicability12.
The foregoing challenges highlight a critical gap in the costing literature that can be addressed by the collection of comprehensive cost estimates for high-throughput genomic technologies. Use of micro-costing techniques13–16—in particular, those reflecting a bottom-up costing approach17,18—provides a rigorous method for directly measuring activities related to the relevant assays and assigning prices at a per-unit level to the resources used.
Various methods are available for measuring the quantities of resources used in costing studies, one of which is a direct observational (that is, time-and-motion) approach15,19,20. In a time-and-motion study, an external observer records, for a defined set of activities, the resources used and their quantities (that is, laboratory equipment, supplies) and the time taken in conducting the activities. Studies of this kind have been used in the health care setting to understand the workflow in surgical operating rooms21–23, the per-patient cost of disease screening24,25, and the implementation of health informatics and information technologies26,27.
The aim of the present study was to determine the per-case (that is, per-patient) cost of 3 commonly used high-throughput genomic assays—digital gene expression profiling (gep), fluorescence in situ hybridization (fish), and targeted capture sequencing—in the context of lymphoma cancer research in the province of British Columbia.
METHODS
The study took the perspective of the BC Cancer Agency (bcca) and was conducted as part of a multi-year (2013–2017) research project in genomics and personalized health, one of its aims being to establish a province-wide system to guide the personalization of treatment for lymphoid cancers in British Columbia28. Enrolled patients were at least 16 years of age, had been diagnosed and were treated in British Columbia, were hiv-negative, and had 1 of 3 types of nhl (dlbcl, follicular lymphoma, chronic lymphocytic leukemia) or Hodgkin lymphoma. A separate set of procedures was being used for patients diagnosed with chronic lymphocytic leukemia, and hence those cases were excluded.
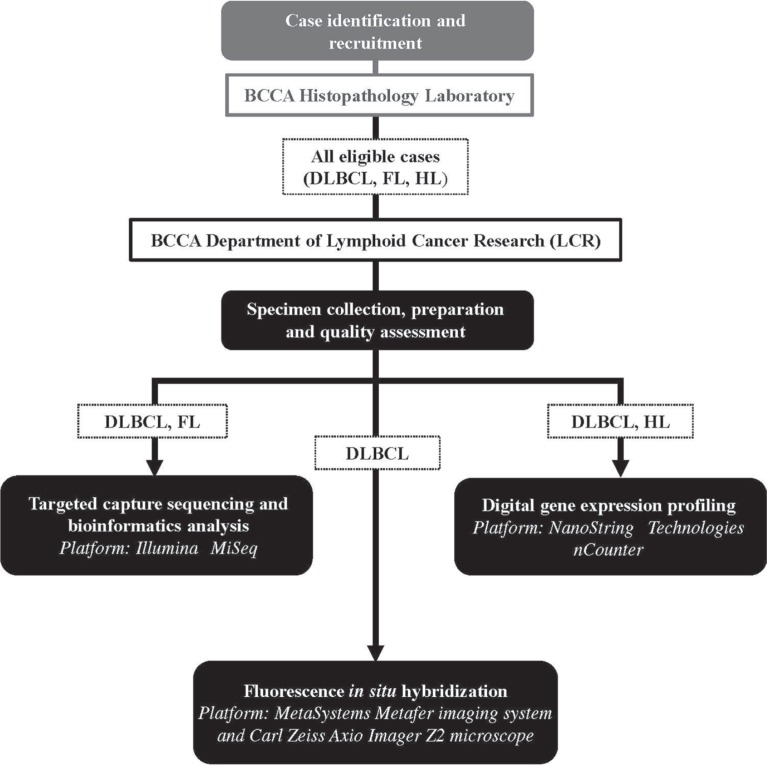
Formalin-fixed paraffin-embedded tissues from biopsies are delivered to the bcca for standard and molecular diagnostic analyses in the bcca histopathology laboratory (Figure 1). After analysis, leftover tissues from eligible dlbcl, follicular lymphoma, and Hodgkin lymphoma cases are transferred to the bcca Department of Lymphoid Cancer Research (lcr) laboratory for dna or rna extraction, quantification, quality assessment, and storage. Depending on the type of lymphoid cancer, specimens undergo targeted analysis to further characterize their unique genomic (fish, targeted capture sequencing) and transcriptomic (digital gep) features. The clinical assessment and genomic or transcriptomic profiles are then returned to the treating oncologist for evaluation of available treatment options.
FIGURE 1.
High-level overview of case-specific genomic analysis after entry into the research project. Formalin-fixed paraffin-embedded tissue from biopsies undergoes standard and molecular diagnostic analysis in the BC Cancer Agency (BCCA) histopathology laboratory. Leftover material from eligible diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), and Hodgkin lymphoma (HL) samples is transferred to the Department of Lymphoid Cancer Research for preparation, quality assessment, and storage. The DLBCL specimens undergo digital gene expression profiling, fluorescence in situ hybridization, targeted capture sequencing, and bioinformatics analysis. The HL specimens undergo gene expression profiling, and the FL specimens undergo targeted capture sequencing and bioinformatics analysis. The clinical assessment and genomic or transcriptomic profiles are then returned to the treating oncologist for evaluation of available treatment options. Trademarks: MiSeq: Illumina, San Diego, CA, U.S.A.; NanoString: NanoString Technologies, Seattle, WA, U.S.A.; Metafer: MetaSystems, Altlussheim, Germany; Axio Imager Z2: Carl Zeiss Microscopy, Oberkochen, Germany.
For the purposes of the broader research project, a set of 16 standard operating procedures (sops, see Table i) were developed to process individual cases through specimen collection, preparation, and quality assessment (sop-01 to sop-09), digital gep (sop-10 to sop-12), fish (sop-13 and sop-14), and targeted capture sequencing and bioinformatics analysis (sop-15 and sop-16). Digital gep is performed using the nCounter Digital Analyzer platform (NanoString Technologies, Seattle, WA, U.S.A.) with an 807-gene code set, which includes 20 genes to determine cell-of-origin for dlbcl cases29 and 26 genes to stratify Hodgkin lymphomas as either high- or low-risk30. Dual-colour fish is conducted using the Metafer imaging software (MetaSystems, Altlussheim, Germany) and a Carl Zeiss Axio Imager Z2 microscope (Carl Zeiss Microscopy, Oberkochen, Germany), with break-apart probes used to detect MYC, BCL2, and BCL6 rearrangements in dlbcl. Targeted capture sequencing is performed using Agilent SureSelect capture protocols (Agilent Technologies, Santa Clara, CA, U.S.A.) and the Illumina MiSeq platform (Illumina, San Diego, CA, U.S.A.) to detect somatic single nucleotide variants and insertions or deletions in 32 genes that are reported to be recurrently mutated in dlbcl, follicular lymphoma, and chronic lymphocytic leukemia, and to harbour clinically actionable potential. Each assay is conducted in the lcr laboratory. For each sop, resource use related to labour (staff), capital and equipment (for example, MiSeq desktop sequencer), supplies and reagents (for example, disposable gloves, commercial probes), and overhead was collected using a direct observational technique. A micro-costing approach was then applied to calculate the costs attributable to each resource.
TABLE I.
The standard operating procedures defined and observed for the study
| Assay | Procedure and description |
|---|---|
| Specimen collection, preparation, and quality assessment | |
| |
| Digital gene expression profiling | |
| |
| Fluorescence in situ hybridization (FISH) | |
| |
| Targeted capture sequencing and bioinformatics analysis | |
| |
NanoString Technologies, Seattle, WA, U.S.A.
MetaSystems, Heidelberg, Germany.
Illumina, San Diego, CA, U.S.A.
The research project obtained ethics approval from the University of British Columbia–bcca Research Ethics Board (H05-60103).
Time-and-Motion Study
Various approaches to conducting time-and-motion studies can be used, each of which has its own strengths and limitations16. For the present research study, a direct observer approach was used to avoid some of the input errors and inconsistencies that can affect other data collection techniques (work sampling, for instance)13,20.
The setting for the time-and-motion study was the lcr laboratory. An external observer (SC) conducted the observations during a 5-month observation period in 2014. The only exception to this observer-led approach occurred with respect to the bioinformatics analysis, whereby the mean per-case duration to analyze the targeted capture sequencing data (sop-16) was calculated based on the minimum and maximum time estimates provided by the bioinformatician. Depending on the sop, each observation lasted between 30 minutes and 5 hours. At the start of the observation period, approximately 150 cases had been enrolled in the genomics and personalized health research project. In the time-and-motion and micro-costing study, 99 individual cases were observed, with some cases being observed more than once across all sops (Table ii). The breakdown of unique cases by diagnostic type was 48 dlbcl cases (48.5%), 19 follicular lymphoma cases (19.2%), and 32 Hodgkin lymphoma cases (32.3%). The patients were predominately men (65%) and were less than 60 years of age at time of diagnosis (67%); all patients were diagnosed in 2014.
TABLE II.
Per-case hands-on time, with standard deviation, by standard operating procedure (SOP) and assay
| Assay | Procedure | Observations a [n samples (n batches)] | Mean per-case hands-on time [n minutes] |
|---|---|---|---|
| Specimen collection, preparation, and quality assessment | |||
| 1. Specimen collection and new case entry | 23 (5) | 11.8±4.5 | |
| 2. Purification of genomic DNA and total RNA from formalin-fixed paraffin-embedded tissue sections | 32 (4) | 34.1±10.7 | |
| 3. Quantifying, labelling, and storing RNA | 32 (4) | 4.0±2.7 | |
| 4. Quantifying, labelling, and storing DNA | 19 (2) | 3.2±0.9 | |
| 5. Preparing tumour DNA dilutions for targeted capture sequencing | 24 (3) | 2.9±1.5 | |
| 6. DNA quality assessment | 16 (2) | 12.3±3.3 | |
| 7. RNA quality assessment | 23 (2) | 11.8±5.4 | |
| 8. Identifying and tracking peripheral blood DNA cases | 8 (1) | 0.4 | |
| 9. Preparing peripheral blood DNA dilutions for targeted capture sequencing | 10 (2) | 5.4±1.0 | |
| TOTAL per-case mean hands-on time | 86.5±13.6 | ||
| Digital gene expression profiling | |||
| 10. NanoString Technologies nCounter Gene Expression Assayb hybridization procedure | 48 (3) | 5.5±0.9 | |
| 11. NanoString Technologies nCounter prep station operation | 48 (4) | 2.7±0.8 | |
| 12. NanoString Technologies nCounter Digital Analyzerb operation | 48 (4) | 6.7±8.8 | |
| TOTAL per-case mean hands-on time | 14.9±8.9 | ||
| Fluorescence in situ hybridization (FISH) | |||
| 13. FISH protocol for paraffin-embedded tissue | 5 (2) | 139.4±8.6 | |
| 14. FISH analysis | 5 (2) | 118.8±54.5 | |
| TOTAL per-case mean hands-on time | 258.2±55.2 | ||
| Targeted capture sequencing and bioinformatics analysis | |||
| 15. Illumina Multiplex Sequencingc | |||
| 16. Sequence analysis for mutation calling | 32 (1) | 124.1 | |
| TOTAL per-case mean hands-on time | 124.1 | ||
Refers to the number of times that the SOP was observed throughout the observation period. Per-case hands-on time was determined by summing the batch hands-on time estimates (multiple samples) and dividing by the number of samples within the batch. Total sample size adds to more than 99 because some cases were observed multiple times during the various SOPs.
NanoString Technologies, Seattle, WA, U.S.A.
Illumina, San Diego, CA, U.S.A.
The amount of staff “hands-on” time, capital and equipment, and supplies and reagents used to process the batch of samples for every observation during the sop was recorded—that is, “fixed” times (the extended periods during which samples are on the equipment with minimal-to-no staff oversight, such as overnight incubation periods) were not used. Per-case estimates of hands-on time reflect the average time required for a batch of cases to be processed through the sop across multiple observations of the procedure or procedures (Table ii). In that way, the mean per-case estimate reflects an average across all cases observed, and not the total time required to process a single case in isolation.
The hands-on time for each sop was recorded using a stopwatch, and the number of resources used was recorded in a data collection template; the resulting data were later transcribed. When a set of cases was batched for processing together (for example, digital gep), total resource use was divided by the number of cases in the batch.
Micro-Costing Analysis
A cost model was developed to calculate the monetary value of the 4 resource categories: labour, capital and equipment, supplies and reagents, and overhead. The mean hands-on time for each sop and the annual case throughput—that is, the number of cases that can feasibly be processed through the sop, given current resources—were used to estimate the proportion of the unit cost for each resource that could be attributed to the 16 sops. Case throughput was based on historical referral patterns to the bcca for newly diagnosed nhl (dlbcl and follicular lymphoma) and Hodgkin lymphoma cases31,32, together with the project-specific average case accrual rate of 3–4 specimens per week. The result was an estimated case throughput of 180 cases annually. Although that estimate is conservative, it was considered reasonable, given the resource capacity of the study setting and the assumption in the cost model that resources (for example, number of staff) remained constant over time.
All costs are reported in 2014 Canadian dollars using the health care component of Statistics Canada’s Consumer Price Index33. The valuations of each of the 4 resource categories are described in the subsections that follow. To respect the privacy of staff and contracts and purchasing agreements, actual source costs are not reported here.
Labour
The mean per-case hands-on time derived from the time-and-motion study was used to calculate the proportion of total working hours devoted to conducting the sop. A per-minute salary rate was calculated based on a typical 7.5-hour workday, taking into consideration statutory holidays and vacation days25. The resulting per-minute rate was multiplied by the mean hands-on time to derive a per-case labour cost estimate. Minimum and maximum salary ranges were used in the calculation of upper and lower labour cost estimates.
Capital and Equipment
The unit cost for each type of capital resource and equipment used in a sop reflects the purchase price of the item. Because most of the capital resources and equipment used for the assays are shared between projects and laboratory staff, the proportion specific to the genomics research study was calculated based on mean per-case hands-on and fixed times for all sops. The annual capital resource and equipment costs were then divided by the estimated case throughput to establish a per-case capital and equipment cost. Straight-line amortization was used to calculate the annual cost of each unit during its useful lifetime (in years)14, which was informed by laboratory staff. When that information was not known, the duration of the service contract was used as a conservative estimate of the useful duration of the unit. Upper and lower per-case cost estimates were calculated based on manufacturer prices, where available (for example, manufacturer Web site).
Supplies and Reagents
The unit prices for supplies and reagents reflect the local purchase prices of the items. Each purchase price was divided by the number of units contained within the purchased item to establish a per-unit cost, which was then multiplied by the units required to process a single case through the sop. Maximum and minimum values for major drivers of supply and reagent costs were based on manufacturer prices, where available, and were used to calculate upper and lower per-case cost estimates.
Overhead
The 2014 overhead costs were provided by the bcca’s operations management division and included costs related to information technology, security, building operation, and maintenance. To adjust for cost-sharing within the laboratory, the annual overhead cost (per square foot) was distributed between the laboratory directors so as to reflect the proportion attributed to a research project14. The cost per square foot was multiplied by the size of the lab bay in which each assay took place and was then divided by the estimated annual case throughput to establish a per-case overhead cost. To avoid double counting, overhead was applied only once in the calculation of assay costs.
Overall Cost
Once costs were derived for each resource category, a total per-case cost estimate for each assay was calculated by summing the per-case costs for labour, capital and equipment, supplies and reagents, and overhead across all relevant sops. Maximum and minimum cost estimates for each sop and assay were also calculated by summing the upper and lower cost estimates.
RESULTS
The present study provided a baseline understanding of the time and costs associated with processing some commonly used genomic assays in a research setting. Time estimates and per-case cost estimates are reported in the subsections that follow.
Estimates of SOP Hands-On Time
Table ii presents mean per-case hands-on time (with standard deviation) for each sop. For some sops (specifically, sop-08 and sop-15), only 1 full observation of the procedure was conducted; hence, a standard deviation is not available. The fewest cases were observed for the fish and targeted capture sequencing assays, in part because of staff and observer availability, but also because of the requirement for the larger batch size needed to process the assay (such as for targeted capture sequencing). The hands-on time of the sops related to specimen collection, preparation, and quality assessment was estimated at 86.5 ± 13.6 minutes per case. The assay that was found to be the most labour-intensive was fish, at 258.2 ± 55.2 minutes per case, followed by targeted capture sequencing with bioinformatics analysis at 124.1 minutes (just one observation). The assay with the lowest hands-on time was digital gep with an 807-gene code set: 14.9 ± 8.9 minutes per case. Bioinformatics analysis for gep was not observed.
Cost Estimates
Table iii gives per-case cost estimates for each sop and assay, including the upper and lower estimates. Preparation and quality assessment was estimated to cost $146.30 per case (minimum: $123.74; maximum: $153.38). Digital gep with an 807-gene code set was estimated to cost $898.35 per case (minimum: $798.43; maximum: $936.95); fish was estimated at $596.60 per case (minimum: $377.03; maximum: $760.48); and targeted capture sequencing, including bioinformatics analysis, was estimated at $1,029.16 per case (minimum: $765.02; maximum: $1,233.43). An 807-gene code set—which is typically used in research settings for broad panel discoveries, including cell-of-origin set—was used for the gep cases observed during the present study. In clinical application of gep, a subset of that comprehensive gene panel is more likely to be used if the main application is cell-of-origin discovery. For that reason, the cost for digital gep with a 20-gene code set was also calculated: $419.49 per case (minimum: $320.89; maximum: $457.39).
TABLE III.
Estimated per-case cost of each standard operating procedure (SOP) and assay, overall and by resource category
| Assay | Procedure | Estimated per-case cost (2014 CA$) | |
|---|---|---|---|
|
|
|||
| Average | Upper, lower | ||
| Specimen collection, preparation, and quality assessment | |||
| 1. Specimen collection and new case entry | 16.11 | 16.82, 13.56 | |
| 2. Purification of genomic DNA and total RNA from formalin-fixed paraffin-embedded tissue sections | 49.31 | 52.29, 42.00 | |
| 3. Quantifying, labelling, and storing RNA | 8.11 | 8.79, 6.12 | |
| 4. Quantifying, labelling, and storing DNA | 7.98 | 8.66, 6.03 | |
| 5. Preparing tumour DNA dilutions for targeted capture sequencing | 7.44 | 7.57, 6.63 | |
| 6. DNA quality assessment | 19.00 | 19.76, 16.13 | |
| 7. RNA quality assessment | 20.14 | 20.85, 17.59 | |
| 8. Identifying and tracking peripheral blood DNA cases | 4.15 | 4.50, 3.14 | |
| 9. Preparing peripheral blood DNA dilutions for targeted capture sequencing | 14.06 | 14.14, 12.54 | |
| TOTAL estimated per-case cost | 146.30 | 153.38, 123.74 | |
| Digital gene expression profiling (807-gene code set) | |||
| 10. NanoString Technologies nCounter Gene Expression Assaya hybridization procedure | 408.40 | 408.85, 407.50 | |
| 11. NanoString Technologies nCounter prep station operation | 110.90 | 111.12, 110.45 | |
| 12. NanoString Technologies nCounter Digital Analyzera operation | 379.05 | 416.98, 280.48 | |
| TOTAL estimated per-case cost | 898.35 | 936.95, 798.43 | |
| Fluorescence in situ hybridization (FISH) | |||
| 13. FISH protocol for paraffin-embedded tissue | 363.20 | 448.14, 292.40 | |
| 14. FISH analysis | 233.40 | 312.34, 84.63 | |
| TOTAL estimated per-case cost | 596.60 | 760.48, 377.03 | |
| Targeted capture sequencing and bioinformatics analysis | |||
| 15. Illumina Multiplex Sequencingb | 1,011.75 | 1,213.64, 751.86 | |
| 16. Sequence analysis for mutation calling | 17.41 | 19.79, 13.16 | |
| TOTAL estimated per-case cost | 1,029.16 | 1,233.43, 765.02 | |
NanoString Technologies, Seattle, WA, U.S.A.
Illumina, San Diego, CA, U.S.A.
The total cost to analyze a typical dlbcl, follicular lymphoma, and Hodgkin lymphoma case was calculated using the per-case assay cost estimates (Table iii). As depicted in Figure 1, all eligible cases undergo specimen collection, preparation, and quality assessment at a mean cost of $146.30 per case. In addition, a typical dlbcl undergoing digital gep with an 807-gene code set, fish, and targeted capture sequencing is estimated to cost $2,670.41 per case. If the gep with a 20-gene code set was to be used instead, the cost declines to $2,191.55 per case. A typical follicular lymphoma case undergoing only targeted capture sequencing and bioinformatics analysis is estimated to cost $1,175.46 per case, and a typical Hodgkin lymphoma case undergoing digital gep with an 807-gene code set is estimated to cost $1,044.65 per case ($565.79 per case using a 20-gene code set).
DISCUSSION
Faced with priority-setting decisions between various treatment and therapeutic options, decision-makers in cancer control look to clinical, cost-effectiveness, and budget-impact analyses to inform the evidence base from which such decisions and policies can be made34–37. To the best of our knowledge, the present study is the first to report per-case cost estimates for 3 high-throughput genomic as-says frequently used in cancer diagnostics, including fish, targeted capture sequencing, and digital gep. Used in economic decision models and budget-impact analyses, those cost estimates can help to determine the cost-effectiveness of the associated technologies and can inform decisions related to their implementation in clinical practice. The costing model developed in this study provides a platform from which future cost analyses of other genomic technologies, or of other settings, can be built.
The variability in per-case cost estimates for each assay is not surprising given the variability in the proportion of the cost contributed by each resource category to the performance of each sop. Targeted capture sequencing and digital gep are examples of assays that process cases in batches, with a relatively lower reliance on labour than is required for fish, whose case processing is much more individualized. Supplies and reagents accounted for most of the per-case cost for fish, likely because multiple break-apart probes are used to detect MYC, BCL2, and BCL6 rearrangements in every dlbcl case. The capital and equipment category was a major contributor to the costs of gep and targeted capture sequencing, which could in part be attributable to the conservative estimate of “useful lifetime” used in the depreciation calculations.
External validation of the cost estimates derived in this study is challenging, given that estimates for the relevant technologies are not generally available from traditional costing resources such as administrative data-sets and reimbursement systems12,38. That being said, the BC Medical Services Plan laboratory fee schedule includes a reimbursement rate of $466.46 for complex cytogenetic analysis (fish, fee item 93050), with additional billing codes applied if multiple probes are used on the same sample (that is, fee item P93051 at $192.68, up to a maximum of 3 times)38. In one cost-effectiveness study39 that evaluated the use of digital gep to determine tumour-site origin, a per-case cost of US$4,400 (CA$4,338) was used for tissue-of-origin gep with a 2000-gene code set. Other institutional price lists suggest that the cost of gep with a gene code set of 700 or more lies somewhere between US$315 per sample (CA$369)40 and CA$500 per assay for catalogued code sets41. Some references to targeted capture sequencing suggest that the cost for a large disease-targeted multigene sequencing test range as high as US$2,000–US$10,000 (CA$2,120–CA$10,600)42, with one study using a base-case estimate of US$2,400 (CA$2,544) for a 34-gene sequencing panel test43. That variability for cost estimates highlights the importance of transparency in the calculation of estimates so as to ensure their representativeness for the setting or disease group of interest, an aspect of micro-costing that we have aimed to achieve in the present study.
A number of limitations of our study should be noted. The setting for the study was one research laboratory situated within an established cancer centre in British Columbia. The nature of the setting has implications for the applicability of the resulting estimates outside a research-based setting. It is reasonable to expect that per-case costs could change once the assays are moved into clinical practice and become part of a regulated, provincially funded system. Specifically, additional costs would have to be considered for items related to data storage, additional quality control or assurance protocols, expanded informatics support, and lab management and oversight7. Furthermore, compared with the research-grade reagents used in the present study, clinical-grade reagents are likely to come at an increased cost.
One of the strengths of a direct observational approach is that, compared with self-reported estimates, the data collected tend to be more precise. However, a direct observer approach also has a number of limitations, including the cost associated with training staff (acting as observers) to reliably collect information during a continuous observation period. For that reason, other forms of data collection such as activity logs tend to be used more frequently in practice13. One of the unintended consequences of using a direct observational approach could be the tendency for the performance of an activity to be altered to seem more favourable to the observer. To the extent possible, mitigation of that effect was attempted through the collection of multiple observations of each sop over an extended period of time, consultation with laboratory managers to ensure validity, and where possible, comparison of the results with available estimates from the broader grey literature. Estimates of time for each assay were based on observations from an individual staff member who was responsible for the procedure and experienced in its execution. The efficiency with which cases were processed through each sop could differ in other laboratory settings. It is also important to note that the time estimates reported here exclude “fixed” times, and hence the total duration of each sop (that is, the sum of the fixed and hands-on times) is longer in practice than is expressed here.
The small sample sizes observed for each sop (Table ii) also somewhat limit the level of analysis that could be conducted for the study, including calculation of the standard deviations for some assays. Future research studies might consider expanding the study criteria or increasing the observation period to increase sample size.
For some of the capital resources and equipment, “useful lifetime” is not widely known, because the technologies are relatively new and continuously changing. For those items, the depreciation calculations used a conservative estimate that reflected the length of the service contracts, a choice that could have overrepresented the “true” annual cost of the items. This potential overrepresentation therefore poses a limitation, in that the cost estimates are highly sensitive to changes in sample throughput.
CONCLUSIONS
The demand for high-throughput genomic assays—including cytogenetic analyses (for example, fish), digital gep, and targeted capture sequencing—for application to cancer diagnostics and personalization of cancer-specific interventions has risen dramatically over time. Pressure to apply such technologies outside the traditional research settings and into standard clinical practice is increasing. The challenge for health economists and funders of the technologies at the health care system level is to understand the economic impact and to allocate resources wisely. Our study provides a set of reliable cost estimates that can be used to inform economic evaluations of personalized approaches to cancer care. Future work can continue to build on this approach and to focus on the generalizability of the results to other cancer sites and populations.
ACKNOWLEDGMENTS
This research was supported by Genome British Columbia and Genome Canada (141LYM), the Canadian Institutes of Health Research (cihr), the BC Cancer Foundation, and the Canadian Centre for Applied Research in Cancer Control. The Canadian Centre for Applied Research in Cancer Control is funded by the Canadian Cancer Society Research Institute (019789).
The authors express their gratitude to the staff of the bcca lcr, the bcca finance and research facilitation divisions, and the Michael Smith Genome Sciences Centre, bcca, for their contributions to the work reported here. CS is the recipient of a Career Investigator Award from the Michael Smith Foundation for Health Research and a New Investigator award from cihr. DWS is supported by the BC Cancer Foundation.
CONFLICT OF INTEREST DISCLOSURES
We have read and understood Current Oncology’s policy on disclosing conflicts of interest, and we declare the following interests: DWS has received honoraria from Celgene in a consulting and advisory role. He is also a named inventor on a pending patent describing gep for prognostication in classical Hodgkin lymphoma and potentially a named inventor on a pending patent on the use of gep to assign cell-of-origin in dlbcl that has been licensed to NanoString Technologies.
REFERENCES
- 1.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 2.Genentech . Herceptin (Trastuzumab) Development Timeline [Web page] San Francisco, CA: Genentech; n.d. [Available at: http://www.gene.com/media/product-information/herceptin-development-timeline; cited 31 August 2015] [Google Scholar]
- 3.Sehn L. Diffuse large B-cell lymphoma: one treatment no longer fits all. Oncology (Williston Park) 2014;28:334–6. [PubMed] [Google Scholar]
- 4.Scott DW, Mottok A, Ennishi D, et al. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol. 2015;33:2848–56. doi: 10.1200/JCO.2014.60.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derenzini E, Younes A. Predicting treatment outcome in classical Hodgkin lymphoma: genomic advances. Genome Med. 2011;3:26. doi: 10.1186/gm240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parrilla Castellar ER, Jaffe ES, Said JW, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124:1473–80. doi: 10.1182/blood-2014-04-571091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchanan J, Wordsworth S, Schuh A. Issues surrounding the health economic evaluation of genomic technologies. Pharmacogenomics. 2013;14:1833–47. doi: 10.2217/pgs.13.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips KA, Trosman JR, Kelley RK, Pletcher MJ, Douglas MP, Weldon CB. Genomic sequencing: assessing the health care system, policy, and big-data implications. Health Aff (Millwood) 2014;33:1246–53. doi: 10.1377/hlthaff.2014.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caulfield T, Evans J, McGuire A, et al. Reflections on the cost of “low-cost” whole genome sequencing: framing the health policy debate. PLoS Biol. 2013;11:e1001699. doi: 10.1371/journal.pbio.1001699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayer R, Galea S. Public health in the precision-medicine era. N Engl J Med. 2015;373:499–501. doi: 10.1056/NEJMp1506241. [DOI] [PubMed] [Google Scholar]
- 11.Rogowski W, Payne K, Schnell-Inderst P, et al. Concepts of “personalization” in personalized medicine: implications for economic evaluation. Pharmacoeconomics. 2014;33:49–59. doi: 10.1007/s40273-014-0211-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canadian Agency for Drugs and Technology in Health (cadth) Next Generation DNA Sequencing: A Review of the Cost Effectiveness and Guidelines. Ottawa, ON: CADTH; 2014. [PubMed] [Google Scholar]
- 13.Mogyorosy Z, Smith P. The Main Methodological Issues in Costing Health Care Services: A Literature Review. York, UK: Centre for Health Economics, University of York; 2005. [Google Scholar]
- 14.Drummond MF, Sculpher MJ, Torrance GW, O’Brien BJ, Stoddart GL. Methods for the Economic Evaluation of Health Care Programmes. 3rd ed. New York, NY: Oxford University Press; 2005. [Google Scholar]
- 15.Frick KD. Microcosting quantity data collection methods. Med Care. 2009;47(suppl 1):S76–81. doi: 10.1097/MLR.0b013e31819bc064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baladi JF. A Guidance Document for the Costing Process. Ver. 1.0. Ottawa, ON: Canadian Coordinating Office for Health Technology Assessment; 1996. [Google Scholar]
- 17.Chapko MK, Liu CF, Perkins M, Li YF, Fortney JC, Maciejewski ML. Equivalence of two healthcare costing methods: bottom-up and top-dow n. Health Econ. 2009;18:1188–201. doi: 10.1002/hec.1422. [DOI] [PubMed] [Google Scholar]
- 18.Wordsworth S, Ludbrook A, Caskey F, Macleod A. Collecting unit cost data in multicentre studies. Eur J Health Econ. 2005;6:38–44. doi: 10.1007/s10198-004-0259-9. [DOI] [PubMed] [Google Scholar]
- 19.Lopetegui M, Yen PY, Lai A, Jeffries J, Embi P, Payne P. Time motion studies in healthcare: what are we talking about? J Biomed Inform. 2014;49:292–9. doi: 10.1016/j.jbi.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkler SA, Knickman JR, Hendrickson G, Lipkin M, Jr, Thompson WG. A comparison of work-sampling and time-and-motion techniques for studies in health services research. Health Serv Res. 1993;28:577–97. [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrich A, Chow MP, Skierczynski BA, Lu Z. A 36-hospital time and motion study: how do medical-surgical nurses spend their time? Perm J. 2008;12:25–34. doi: 10.7812/TPP/08-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallidou AA, Cummings GG, Schalm C, Estabrooks CA. Health care aides use of time in a residential long-term care unit: a time and motion study. Int J Nurs Stud. 2013;50:1229–39. doi: 10.1016/j.ijnurstu.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Ampt A, Westbrook J, Creswick N, Mallock N. A comparison of self-reported and observational work sampling techniques for measuring time in nursing tasks. J Health Serv Res Policy. 2007;12:18–24. doi: 10.1258/135581907779497576. [DOI] [PubMed] [Google Scholar]
- 24.Robinson S, Roberts T, Barton P, et al. Healthcare and patient costs of a proactive chlamydia screening programme: the Chlamydia Screening Studies project. Sex Transm Infect. 2007;83:276–81. doi: 10.1136/sti.2006.023374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry SG, Ness RM, Stiles RA, Shintani AK, Dittus RS. A cost analysis of colonoscopy using microcosting and time-and-motion techniques. J Gen Intern Med. 2007;22:1415–21. doi: 10.1007/s11606-007-0281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poissant L, Pereira J, Tamblyn R, Kawasumi Y. The impact of electronic health records on time efficiency of physicians and nurses: a systematic review. J Am Med Inform Assoc. 2005;12:505–16. doi: 10.1197/jamia.M1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng K, Guo MH, Hanauer DA. Using the time and motion method to study clinical work processes and workflow: methodological inconsistencies and a call for standardized research. J Am Med Inform Assoc. 2011;18:704–10. doi: 10.1136/amiajnl-2011-000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connors J, Marra M, Gascoyne R. Personalized Treatment of Lymphoid Cancer: British Columbia As Model Province. Ottawa, ON: Genome Canada; 2012. [Google Scholar]
- 29.Scott DW, Wright GW, Williams PM, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–17. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott DW, Chan FC, Hong F, et al. Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical Hodgkin lymphoma. J Clin Oncol. 2013;31:692–700. doi: 10.1200/JCO.2012.43.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.BC Cancer Agency (bcca) Referral Rates for Cancer Cases Diagnosed in 2012. Vancouver, BC: BCCA; 2014. [Available online at: http://www.bccancer.bc.ca/statistics-and-reports-site/Documents/referral_rates_2012.pdf; cited 5 January 2015] [Google Scholar]
- 32.Canadian Cancer Society . Types of non-Hodgkin lymphoma [Web page] Toronto, ON: Canadian Cancer Society; n.d. [Current version available at: http://www.cancer.ca/en/cancer-information/cancer-type/non-hodgkin-lymphoma/non-hodgkin-lymphoma/types-ofnhl/?region=bc; cited 5 January 2015] [Google Scholar]
- 33.Statistics Canada . Consumer Price Index, by province (Canada) [Web resource] Ottawa, ON: Statistics Canada; 2015. [Current version available at: http://www.statcan.gc.ca/tables-tableaux/sum-som/l01/cst01/econ09a-eng.htm; cited 5 September 2015] [Google Scholar]
- 34.Ferrusi IL, Leighl NB, Kulin NA, Marshall DA. Do economic evaluations of targeted therapy provide support for decision makers? Am J Manag Care. 2011;17(suppl 5):SP61–70. [PMC free article] [PubMed] [Google Scholar]
- 35.Elshaug AG, Moss JR, Littlejohns P, Karnon J, Merlin TL, Jiller JE. Identifying existing health care services that do not provide value for money. Med J Aust. 2009;190:269–73. doi: 10.5694/j.1326-5377.2009.tb02394.x. [DOI] [PubMed] [Google Scholar]
- 36.Dowie J. Why cost-effectiveness should trump (clinical) effectiveness: the ethical economics of the South West quadrant. Health Econ. 2004;13:453–9. doi: 10.1002/hec.861. [DOI] [PubMed] [Google Scholar]
- 37.Regier DA, Bentley C, Mitton C, et al. Public engagement in priority-setting: results from a pan-Canadian survey of decision-makers in cancer control. Soc Sci Med. 2014;122:130–9. doi: 10.1016/j.socscimed.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 38.BC Medical Services Plan Laboratory Medicine Fee Schedule. Victoria, BC: Medical Services Commission; 2013. [Available online at: http://www2.gov.bc.ca/assets/gov/health/practitioner-pro/medical-services-plan/40-laboratory-medicine.pdf; cited 10 September 2015] [Google Scholar]
- 39.Hornberger J, Degtiar I, Gutierrez H, et al. Cost-effectiveness of gene-expression profiling for tumor-site origin. Value Health. 2013;16:46–56. doi: 10.1016/j.jval.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Molecular Biology Core Facilities (mbcf), Dana Farber Cancer Institute . NanoString nCounter [Web resource] Boston, MA: MBCF; 2015. [Available at: http://mbcf.dfci.harvard.edu/genomics-core/nanostring; cited 30 August 2015] [Google Scholar]
- 41.Princess Margaret Genomics Centre (pmgc) NanoString Services Price List. Toronto, ON: PMGC; 2015. [Available online at: https://www.pmgenomics.ca/pmgenomics/services/pdf/NanoString_Services_PriceList.pdf; cited 30 August 2015] [Google Scholar]
- 42.Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yonghong L, Bare LA, Bender RA, et al. Cost effectiveness of sequencing 34 cancer-associated genes as an aid for treatment selection in patients with metastatic melanoma. Mol Diagn Ther. 2015;19:169–77. doi: 10.1007/s40291-015-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]