Abstract
The combination of protein display technologies and noncanonical amino acids (ncAAs) offers unprecedented opportunities for the high throughput discovery and characterization of molecules suitable for addressing fundamental and applied problems in biological systems. Here we demonstrate that ncAA-compatible yeast display facilitates evaluations of conjugation chemistry and stability that would be challenging or impossible to perform with existing mRNA, phage, or E. coli platforms. Our approach enables site-specific introduction of ncAAs into displayed proteins, robust modification at azide-containing residues, and quantitative evaluation of conjugates directly on the yeast surface. Moreover, screening allows for the selective enrichment of chemically modified constructs while maintaining a genotype–phenotype linkage with encoded azide functionalities. Thus, this platform is suitable for the high throughput characterization and screening of libraries of chemically modified polypeptides for therapeutic lead discovery and other biological applications.
Keywords: yeast display, noncanonical amino acids, click chemistry, antibodies, high throughput screening
Introduction
The biosynthetic production of proteins containing noncanonical amino acids (ncAAs) provides powerful approaches to solving many outstanding problems in biology and biomolecular engineering (Johnson et al., 2010; Sun et al., 2014) including the atomic-level dissection of protein biophysical properties (Kwon et al., 2010; Ye et al., 2010) and the construction of therapeutic leads (Frost et al., 2015; Horiya et al., 2014; Ng et al., 2015). Emerging efforts combining ncAAs with mRNA (Hayashi et al., 2012; Horiya et al., 2014), phage (Liu et al., 2008), or E. coli (Van Deventer et al., 2014) display show promise in the discovery of proteins with an expanded range of chemical functionality exhibiting improved biophysical and therapeutic characteristics. However, existing approaches utilizing ncAAs in display formats remain limited in scope. mRNA and phage display are restricted to indirect assessments of protein function and chemical functionalization of proteins containing reactive ncAA side chains. While E. coli display allows for some quantitative on-cell characterizations, only a narrow range of proteins can be presented on the bacterial cell surface. Thus, versatile ncAA-compatible display technologies that overcome these limitations are highly desirable.
Here we present a yeast-based approach to the construction, evaluation, and screening of bioconjugates. Using Saccharomyces cerevisiae-based yeast display (Boder and Wittrup, 1997; Van Deventer and Wittrup, 2014) and amber suppression (Chin et al., 2003; Van Deventer et al., 2015; Wang and Wang, 2008), we constructed variants of the human IgG1 antibody constant region (Fc) (Traxlmayr et al., 2012) and antigen-binding Fc fragments (Fcabs) (Hasenhindl et al., 2013; Traxlmayr et al., 2013) containing reactive ncAAs. These well-characterized proteins serve as an ideal model system for extending our previously described ncAA-compatible yeast display/secretion system (Van Deventer et al., 2015) to bioconjugate evaluation. Stable Fcs and Fcabs tolerate a range of ncAA substitutions, and azide functionality is selectively addressable using copper-catalyzed or strain-promoted azide–alkyne cycloadditions. Bioconjugate thermal stability and generalizable approaches to the assessment of protein modifications are readily carried out on the yeast surface. Model screening experiments indicate that ncAA-compatible yeast display maintains genotype–phenotype linkages during flow cytometry-based screening.
Materials and methods
Media preparation, yeast electroporation, and yeast strain construction
Procedures for the preparation of liquid media, solid media, and electrocompetent yeast were performed as described previously (Van Deventer and Wittrup, 2014; Van Deventer et al., 2015). The strain RJY100 was constructed using standard homologous recombination approaches and has been described in detail elsewhere (Van Deventer et al., 2015).
Plasmid construction
The reporter constructs, which encode antibody-like scFv-Fcs fused at their C-termini to the display anchor Aga2p, were assembled in several steps as detailed by Van Deventer et al. and verified by Sanger sequencing (Van Deventer et al., 2015). All sequencing in this work was submitted to the Koch Institute Biopolymers Facility (Cambridge, MA) or to Quintara Biosciences (Cambridge, MA). The plasmid pRS315-OmeRS was constructed from the SacI-PstI fragment of pSNR-OmeRS (Wang and Wang, 2008) and similarly digested pRS315 as described (Van Deventer et al., 2015). The resulting construct encodes a constitutively expressed E. coli tyrosyl-tRNA synthetase (TyrRS) variant capable of performing aminoacylation with O-methyl-L-tyrosine and a constitutively expressed E. coli tRNATyr variant containing a CUA anticodon. Plasmids encoding E. coli TyrRS variants capable of performing aminoacylation with p-azido-L-phenylalanine and p-acetyl-L-phenylalanine were produced by using gBlock DNA fragments encoding the amino acid mutations of p-acetylPheRS-1 and p-azidoPheRS-1 reported by Chin et al. (2003) in PCR mutagenesis of pRS315-OmeRS. After PCR amplification of pRS315-OmeRS with the gBlocks, the PCR mixtures were digested with DpnI, transformed into E. coli, and plated on selective media (LB containing ampicillin). Colonies were inoculated in selective liquid media, grown to saturation, and miniprepped. The resulting plasmid DNA was sequenced in order to identify correctly mutated plasmids, which were named pRS315-AzFRS and pRS315-AcFRS.
The constructs pYD1-Fc, pYD-H10-03-6, and pYD-Stab19 were subjected to site-directed mutagenesis in order to produce constructs containing amber codons at amino acid positions 282, 375, and 400 (EU numbering). PCR reactions with plasmids were digested with DpnI, transformed into E. coli, and plated on selective media. Colonies were inoculated in selective liquid media, grown to saturation, and miniprepped. The resulting plasmid DNA was sequenced in order to identify correctly mutated plasmids, which were designated with the names of their parental plasmids followed by the position of the amber codon and ‘TAG’ (e.g. the plasmid encoding the Fc containing an amber codon at position 282 is designated ‘pYD1-Fc-282TAG’).
Yeast transformations, propagation, and induction
The reporter construct plasmid pCHA-FcSup-TAG-FAPA (TRP marker) and aaRS variant plasmids (LEU marker) were transformed simultaneously into Zymo competent RJY100 cells, plated on solid SD-SCAA media (−LEU −TRP −URA), and grown at 30°C until colonies appeared (2−3 days). Transformation of RJY100 cells with Fc display constructs containing amber codons and pRS315-AcFRS proceeded similarly. Control reporter and Fc or Fcab constructs encoding only canonical amino acids were transformed into RJY100 cells, plated on solid SD-CAA media (−TRP −URA), and grown at 30°C until colonies appeared (2−3 days).
To propagate samples and prepare them for induction, colonies containing the appropriate plasmids were inoculated in selective media (5 mL; cells containing control constructs only: SD-CAA (−TRP −URA); cells containing both constructs with amber codons and suppression constructs: SD-SCAA (−LEU −TRP −URA)) and allowed to grow to saturation at 30°C (2−3 days). Alternatively, samples from saturated cultures stored at 4°C were pelleted and resuspended to an OD600 of ~0.5 to 1.0 in 5 mL fresh media and allowed to grow to saturation overnight. Following saturation, the cultures were diluted to an OD600 of 1 in fresh media and grown at 30°C until reaching mid log phase (OD 2−5; 4−8 h). Cells were pelleted (30 s at 12 000 rpm and 4°C) and resuspended to an OD600 of 1 in induction media (cells containing reporter constructs only: SG-CAA (−TRP −URA); cells containing both reporter constructs and suppression constructs: SG-SCAA (−LEU −TRP −URA)). To enable site-specific incorporation of ncAAs, SG-SCAA was supplemented with p-azido-L-phenylalanine, p-acetylphenylalanine, or O-methyl-L-tyrosine (1 mM final concentration of L amino acid) and grown at 20°C for at least 16 h.
Flow cytometry (analysis)
Freshly induced samples (see above), samples subjected to click chemistry reactions (see below), or thermally denatured samples (see below) were labeled in 1.7 mL microcentrifuge tubes or 96-well V-bottom plates. Flow cytometry was performed on an Accuri/BD C6 flow cytometer (BD) within the Wittrup Laboratory, on FACSCanto II, LSR II HTS, or LSR Fortessa machines (BD) at the Koch Institute Flow Cytometry Core, or on an Attune NxT flow cytometer within the Tufts University Science and Technology Center.
To label samples in tubes, 2 million cells per sample (1 mL of culture at an OD600 of 1 contains approximately 10 million cells) were transferred to microcentrifuge tubes and pelleted (30 s at 12 000 rpm and 4°C). The sample was washed twice in 1 mL PBSA (PBS, pH 7.4, with 0.1% BSA) and then resuspended in 50 μL PBSA. Primary labeling was performed at room temperature on a rotary wheel for 30 min using antibodies or other ligands in concentrations as presented in Supplementary Table SVIII; labeling reagents varied depending on the experiment type in question. Cells were then pelleted and transferred to ice. All remaining labeling steps were performed on ice with ice-cold PBSA and a chilled microcentrifuge. Cells were washed once and resuspended in 50 µL ice-cold PBSA. Secondary labeling was performed for 15 min on ice in the dark using labeling dilutions indicated in Supplementary Table SVIII. Cells were pelleted and washed with 1 mL ice-cold PBSA and placed on ice in the dark. Immediately prior to flow cytometric analysis, each cell sample was resuspended in 500 μL ice-cold PBSA and filtered through a 12 × 75 mm tube with cell strainer cap.
To label samples in plates, at least 200 000 cells per sample were transferred into a 96-well V-bottom plate and pelleted (3 min at 2400 rpm and 4°C). Samples were resuspended in PBSA (50 μL well−1) containing primary detection reagents as outlined in Supplementary Table SVIII. Plates were incubated with rotation (150 rpm) at room temperature on a Southwest Science digital orbital shaker for 30 min. Cells were then pelleted and washed in ice-cold PBSA (200 μL well−1). The washed samples were resuspended in PBSA (50 μL) containing secondary labels as described in Supplementary Table SVIII and incubated in the dark on a plate shaker at 4°C for 15 min. After incubation, cells were pelleted and washed in ice-cold PBSA (200 μL well−1). Samples were then pelleted and left covered on ice. Immediately prior to flow cytometry analysis, samples were resuspended in ice-cold PBSA (200 μL well−1).
Click chemistry
All click chemistry reactions were performed in microcentrifuge tubes using empirically determined reaction conditions giving robust and selective modifications. For both copper-catalyzed azide–alkyne cycloaddition (CuAAC) and strain-promoted azide–alkyne cycloaddition (SPAAC) reactions, 2 million freshly induced cells were used per condition in most analytical experiments, 25 million freshly induced cells were used in the CuAAC dithiothreitol (DTT) reduction analysis experiments, and all sorting experiments used 50 million freshly induced cells per condition. Biotin-alkyne 3, propargylamine (4), and DBCO-biotin 6 were dissolved in DMSO and diluted to appropriate stock concentrations for reactions. DBCO-PEG 7 was dissolved in distilled, deionized water immediately prior to reactions. Each cell sample was pelleted and washed twice with PBSA prior to a final resuspension in PBS before the reactions. CuAAC was performed using the general protocol of Hong et al. (2009). Except for the recommended alkyne and azide concentrations, all other aspects of the protocol were followed, including the order of reagent addition. Samples were vortexed after addition of each reagent. Biotin-alkyne 3 was added to all reactions at a final concentration of 20 μM except in sorting experiments, where it was added at 100 μM. Propargylamine (4) was added a concentration of 20 μM, 100 μM, or 400 μM. All reactions proceeded for 15 min at room temperature in sealed microcentrifuge tubes, after which time samples were pelleted and washed three times in ice-cold PBSA. SPAAC reactions on cells resuspended in PBS were also performed for 15 min. For the case of DBCO-biotin 6, these reactions were carried out at 4°C after addition of 6 (10 μM final concentration) and vortexing of the reaction tube. For PEG conjugations, DBCO-PEG 7 was added to final concentrations of either 200 μM or 800 μM and vortexed. These reactions were incubated at room temperature for 15 min, after which time cells were pelleted and washed three times in ice-cold PBSA. For the two-step detection of reaction products, CuAAC or SPAAC reactions with the alkyne- or DBCO-containing molecule of interest were conducted, followed by washing three times in ice-cold PBSA and resuspension in PBS. A second reaction using the appropriate biotin-alkyne or DBCO-biotin reagent was then performed on the once-clicked samples. Two-step CuAAC results described here are the result of triplicate experiments. These results and all other data reported here are representative of experiments performed on multiple days. After completion of click chemistry reactions, samples were subjected to thermal denaturation (described below) and/or flow cytometry analysis (described above) and sorting (described below). Biotin labeling was assessed only on cells staining positively for the Xpress antibody.
Thermal denaturation
To perform irreversible thermal denaturation experiments on the yeast surface (Hasenhindl et al., 2013; Traxlmayr et al., 2012), freshly induced cells or cells subjected to click chemistry were resuspended in PBS, added to thin-walled PCR tubes (≥50 000 cells in 50 μL tube−1), and placed on ice for at least 10 min prior to incubation at elevated temperatures. Tubes were then loaded into a BioRad MyCycler and incubated at 55°C, 60°C, 65°C, 70°C, 75°C, or 80°C for 10 min. After incubation, samples were transferred back to ice for at least 10 min prior to labeling for the detection of folded Fc domains and the Xpress epitope (Supplementary Table SVIII). After the completion of flow cytometry, all data was analyzed using methods described by Hasenhindl et al. (2013) with the following modifications: (1) background subtraction of the lowest median FcγRI detection levels was performed for all denaturation series; (2) background-subtracted data were normalized to the highest median FcγRI detection level for a given series in the range of 55–80°C (resulting in the ‘fraction folded’ data presented here); and (3) data fits were constrained so that the upper limit of the ‘fraction folded’ was equal to a value of 1. All data presented here are the result of denaturations performed in triplicate on a single freshly induced sample or sample subjected to click chemistry.
Dithiothreitol reduction
Cells were grown, induced, and subjected to CuAAC or SPAAC as described above. To remove display constructs from the yeast surface (Boder and Wittrup, 1997; Lim et al., 2010), cells were treated with 100 mM DTT in deionized water for 2 h at 37°C with occasional mixing to ensure complete reduction of displayed constructs. DTT-treated cells were then pelleted, washed twice in PBSA, and subjected to flow cytometry labeling and analysis as described above.
Flow cytometry (sorting)
Cells encoding wild-type Fcs or TAG-containing Fcs and amber suppression machinery were induced in 5–100 mL volumes using the methods described above. Cells displaying wild-type Fcs or Fcs containing AzF substitutions in numbers sufficient to perform all sorting and control experiments were washed twice in PBSA, mixed in desired ratios (100 Fc: 1 Fc 400 AzF; 1000 Fc: 1 Fc 400 AzF; 10 000 Fc: 1 Fc 400 AzF) or aliquoted in appropriate numbers for controls (50 million cells displaying wild-type Fcs, 10 million cells displaying AzF-substituted Fcs), pelleted, and resuspended in PBS. CuAAC reactions and all subsequent washes were performed as above. Samples were sorted using either an Aria III (BD Biosciences) or MoFlo Legacy (Beckman Coulter) flow cytometer at the Koch Institute Flow Cytometry Core. Sorted cells were collected in glass tubes containing 1 mL of SDCAA (which contains leucine and leads to loss of the suppression plasmid in cells containing the Fc 400 AzF/TAG construct). The sides of these tubes were washed with 4 mL additional SDCAA and grown at 30°C until the cultures reached saturation. Analysis of sorted samples was performed after induction in SGCAA and labeling with FcγRI and anti-Xpress as described above. All sorting results reported here were obtained from 3 to 5 replicates in which all CuAAC and sorting was performed independently. Fold enrichments were calculated by determining the fraction of truncated clones in sorting outputs (i.e. the percentage of clones in the ‘Fc 400 TAG’ population divided by the sum of the percentages of clones in the ‘Fc 400 TAG’ and ‘Fc’ populations) and dividing by the initial ratio of Fc: Fc 400 AzF used in the experiment.
DNA sequencing of sorted clones
One replicate of the sorted output of 100 Fc: 1 Fc 400 AzF was expanded in SDCAA to allow for flow cytometry analysis (see above) and DNA sequencing in parallel. A portion of the sorted culture was inoculated in SDCAA at an OD600 of 1 and grown for 3−4 h. The culture was then pelleted and resuspended in 200 μL Zymo Research plasmid extraction buffer 1 supplemented with 8 μL zymolyase. The resulting mixture was incubated at 37°C overnight, followed by plasmid extraction with the Zymo Research buffers and an Epoch Biosciences DNA column. Eluted DNA was subjected to digestion with HpaI and NotI overnight (to remove the suppression plasmid), transformed into competent Mach1 E. coli, and grown at 37°C overnight on LB plates containing ampicillin. Individual colonies were miniprepped and submitted for sequencing.
Viability experiments
Six aliquots (washed twice in PBSA) of 2 million induced yeast displaying Fcs containing AzF at position 400 were pelleted and resuspended in 220 μL PBSA. Three of these samples were subjected to CuAAC with 20 μM 3 as described above. All six aliquots were then pelleted and washed three times in ice-cold PBSA and resuspended to a final volume of 1 mL PBSA. 1:10, 1:100, and 1:1000 dilutions of the samples were plated on SD-SCAA plates and grown at 30°C for 3−4 days. The reduction in viability reported here is the ratio of the number colonies that appeared in plated samples subjected to CuAAC divided by the number of colonies that appeared in plated samples that were left untreated.
Results
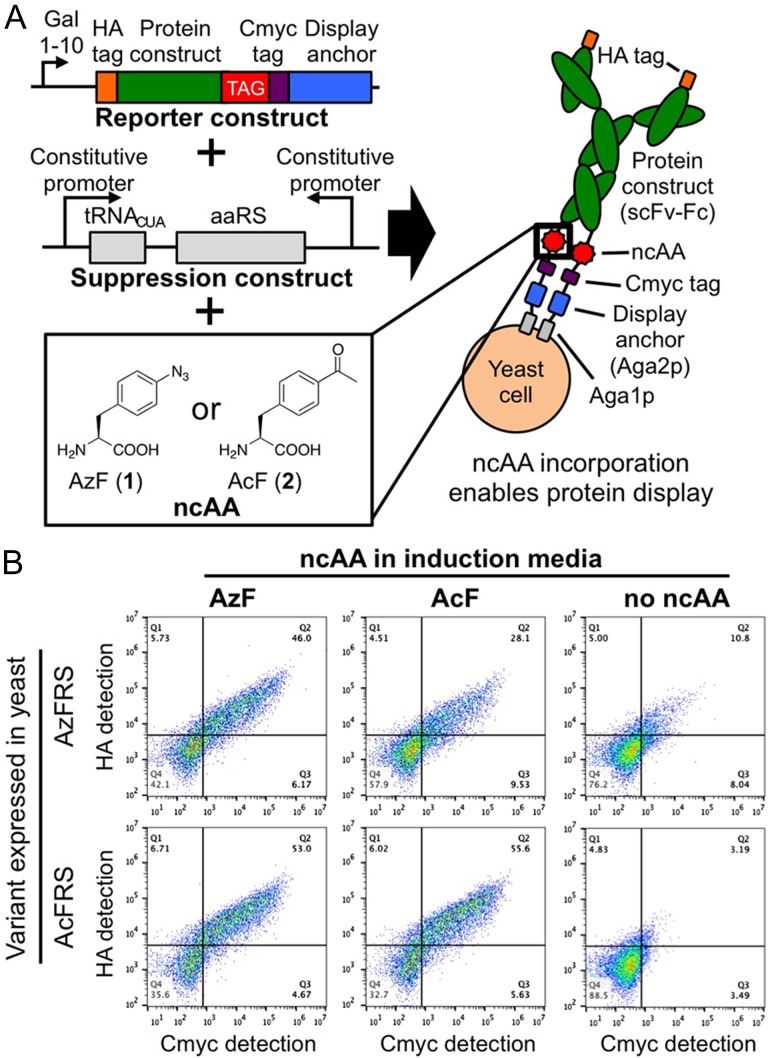
We first investigated site-specific ncAA incorporation in proteins displayed on the yeast surface using a previously reported single chain variable fragment-Fc (scFv-Fc) construct that is only displayed if an amber (TAG) codon is suppressed (Fig. 1A). A separate plasmid enables the constitutive expression of orthogonal suppressor tRNA/aminoacyl-tRNA synthetase (aaRS) pairs (Chin et al., 2003; Van Deventer et al., 2015; Wang and Wang, 2008). Figure 1B depicts display levels of reporter constructs on cells expressing aaRS variants evolved for the recognition of the ncAAs p-azido-L-phenylalanine (AzF; 1) and p-acetyl-L-phenylalanine (AcF; 2) after induction in media supplemented with 1, 2, or no ncAA. Full-length proteins are detected when the medium of cells expressing an aaRS variant is supplemented with its ‘cognate’ ncAA. Display levels observed here are somewhat lower than the display levels of a reporter construct containing only canonical amino acids, but are similar among all aaRSs and ‘cognate’ ncAAs tested (Supplementary Figure S1, Supplementary Table SI). When the induction media is supplemented with a ‘non-cognate’ ncAA, display levels remain elevated, indicating that the aaRSs used here are somewhat promiscuous (see also Supplementary Figure S1 and Supplementary Table SI) (Antonczak et al., 2011).
Fig. 1.
Reporter system for the detection of proteins containing ncAAs on the yeast surface. (A) Yeast transformed with reporter (single chain variable fragment-Fc) and suppression constructs produce full-length proteins when induced in media containing a ncAA recognized by the orthogonal aaRS. (B) Flow cytometry analysis of reporter protein display levels detected by labeling the cmyc and HA epitope tags. Prior to detection, cultures were induced in media containing 1 mM p-azido-L-phenylalanine (AzF; 1), 1 mM p-acetyl-L-phenylalanine (AcF; 2), or no ncAA.
Induction of cells in media lacking ncAAs results in reduced display levels, suggesting that ncAAs are present in the majority of constructs displayed on the yeast surface. However, with the AzFRS and OmeRS variants tested here, more than 10% of cells induced in the absence of a ncAA display the reporter construct to some extent, indicating canonical amino acid incorporation in the displayed construct in response to the amber codon (Supplementary Table SI). These findings suggest that even when inducing yeast in the presence of a ncAA, some displayed constructs may contain a canonical amino acid in place of the desired ncAA. We conservatively estimated the fraction of displayed constructs likely to contain a canonical amino acid after induction in the presence of a ncAA by taking the ratio of the percentage of displaying cells after induction in the absence of ncAA to the percentage of displaying cells in the presence of ncAA for a given aaRS (Supplementary Table SI). For the ‘cognate’ pairs examined here, these ratios are 0.235, 0.0574, and 0.230 for AzFRS-AzF, AcFRS-AcF, and OmeRS-OmeY, respectively. These values represent ‘worst-case’ ratios because, in reality, the presence of ncAA in the induction media leads to higher reporter construct display levels than in the absence of ncAA (Fig. 1, Supplementary Figure S1), and the high concentration of ncAA in the media reduces competition by canonical amino acids for ncAA incorporation (Tanrikulu et al., 2009; Wiltschi et al., 2008). However, even when using these conservative calculations, the AcFRS variant appears to perform very well, with the fraction of constructs containing canonical amino acids estimated to be no greater than 0.0574, 0.0602, or 0.0628 for AcF, AzF, and OmeY, respectively. Moreover, the AcFRS variant supports higher display levels and percentages of displaying cells than the other variants examined here (Fig. 1, Supplementary Figure S1, Supplementary Table SI). Because AcFRS appears to support higher levels of displayed constructs and exhibits greater discrimination against canonical amino acid incorporation during amber suppresion, we performed all subsequent suppression experiments with this variant.
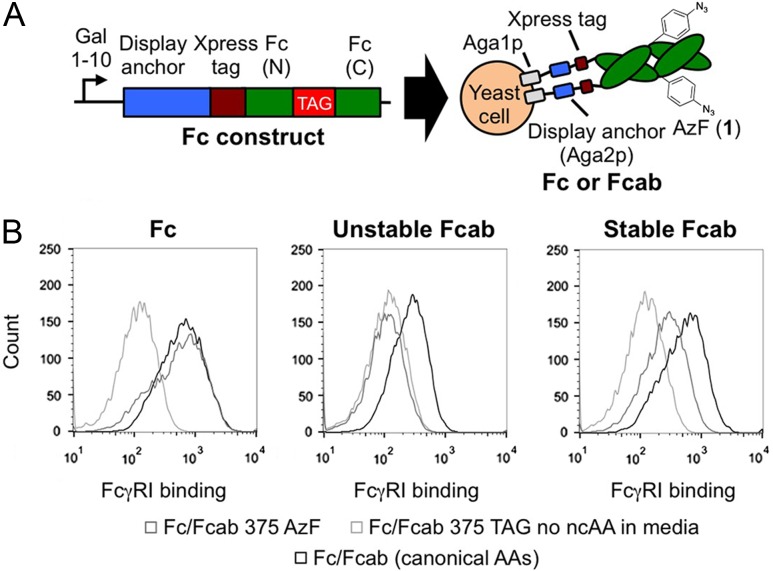
The modular nature of the suppression construct enabled us to incorporate ncAAs into Fcs and Fcabs on the yeast surface (Fig. 2A) and study the effects of ncAA substitution on protein conformation. Amber codons were positioned at amino acids 282, 375, and 400 of the Fc (EU numbering), positions known to tolerate amino acid substitutions and enable quantitative chemical modification (Shen et al., 2012). The Fc gamma receptor FcγRI, which only recognizes folded Fc domains (Hasenhindl et al., 2013), was used to determine if ncAA substitutions affect protein conformation. Figure 2B indicates that FcγRI binds to wild-type Fc and to an Fc variant containing AzF at position 375 (Fc 375 AzF) similarly, confirming that Fc 375 AzF retains its native fold. In contrast, FcγRI does not bind to cells encoding an Fc with an amber codon at position 375 but induced in media lacking AzF, confirming that full-length Fc is required for FcγRI binding. Similar trends are observed with Fc 282 AzF, Fc 400 AzF, and with other ncAA substitutions (Supplementary Figure S2). Studying the FcγRI binding properties of AzF-substituted Fcabs revealed that ncAA substitutions can disrupt Fcab folding in unstable constructs (Fig. 2B, Supplementary Figure S2). The Fcab H10-03-6 contains an engineered binding site to Her2 at the C-terminus of the Fc domain, but reduced stability, while the Stab19 variant of H10-03-6 exhibits stability close to that of the wild-type Fc (Traxlmayr et al., 2013). Figure 2B indicates that H10-03-6 375 AzF loses its ability to bind FcγRI, while Stab19 375 AzF retains FcγRI binding. Thus, ncAA-compatible yeast display enables sensitive detection of conformational changes resulting from ncAA substitutions in constructs of interest.
Fig. 2.
Study of the properties of ncAA-containing Fcs and Fcabs. (A) Schematic representation of Fc or Fcab display constructs. Fc (N) and Fc (C) refer to the N- and C-terminal portions of the Fc, respectively. (B) Detection of folded Fc or Fcab domains on the yeast surface with the conformation-specific Fc gamma receptor I (FcγRI) ligand. FcγRI binds similarly to cells displaying Fc 375 AzF and cells displaying wild-type Fcs. FcγRI does not bind to cells displaying H10-03-6 375 AzF (H10-03-6: unstable Fcab), but binds to cells displaying Stab19 375 AzF (Stab19: stabilized Fcab). The histograms depicted here are restricted to cells exhibiting display of the Xpress tag.
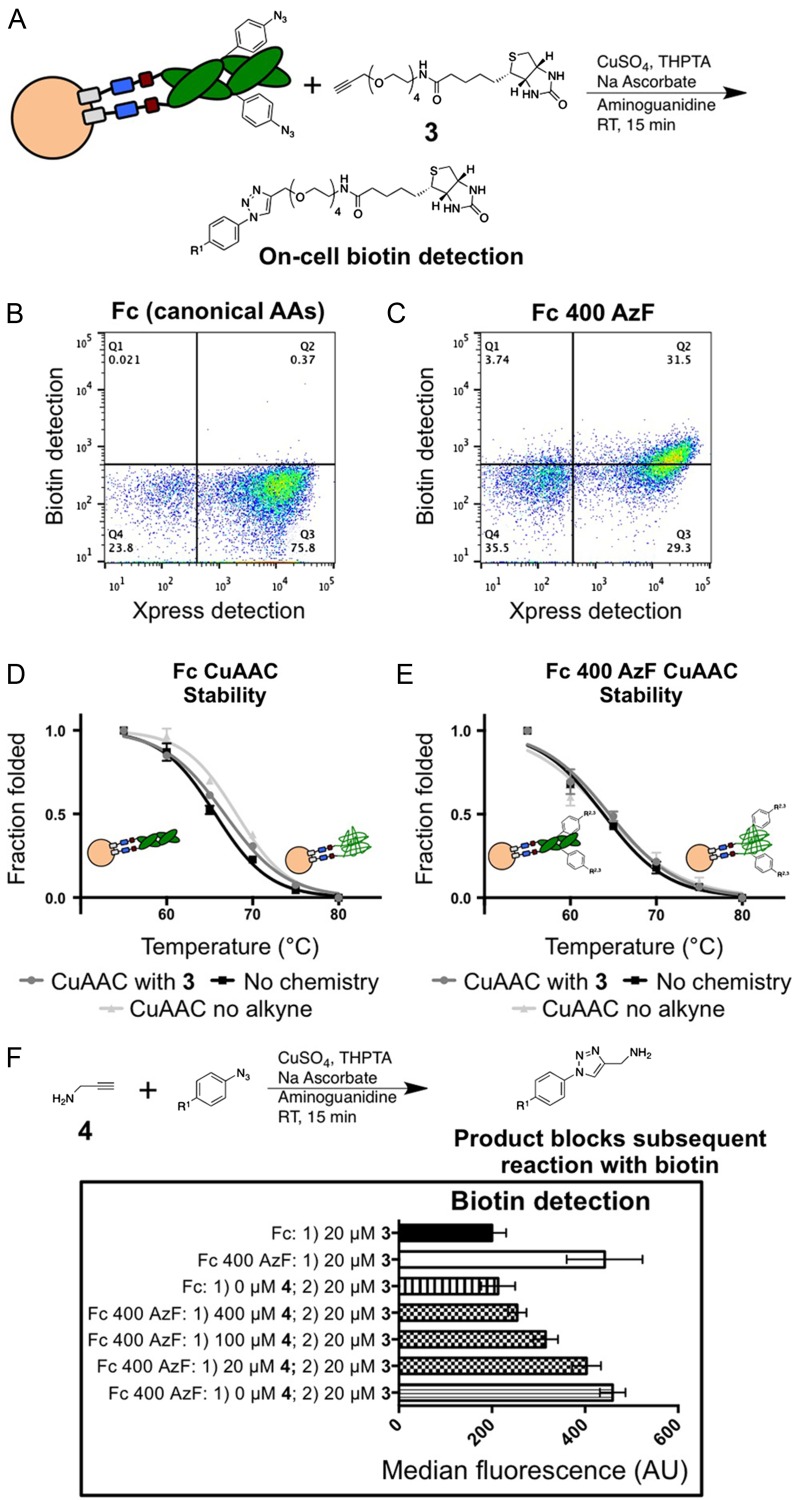
The incorporation of AzF into Fcs enabled us to perform click chemistry on the yeast surface and evaluate conjugate thermal stability. Figure 3A–C illustrates CuAAC (Hong et al., 2009) modifications with biotin-alkyne derivative 3. Biotin detection remains at background levels when cells displaying wild-type Fcs undergo CuAAC (Fig. 3B), while the fluorescence levels of cells displaying Fc 400 AzF increase after CuAAC (Fig. 3C), confirming biotinylation. SPAAC (Kuzmin et al., 2010) reactions exhibited similar selectivities on the yeast surface (Supplementary Figure S3). We used DTT treatments to further investigate CuAAC and SPAAC reaction specificity on the yeast surface (Supplementary Figure S4, Supplementary Table SII) by removing disulfide-bonded display constructs from the yeast surface (Boder and Wittrup, 1997; Lim et al., 2010). We found that following DTT treatments, the levels of biotin detected on cells previously subjected to either CuAAC or SPAAC was greatly decreased, although not fully to background levels. The incomplete decrease in detected biotin levels following DTT treatment raises the possibility that side reactions are occurring on the yeast surface; this possibility will be considered further in the ‘Discussion’ section. Taken as a whole, though, CuAAC and SPAAC experiments indicate that AzF-containing display constructs readily react to form conjugates on the yeast surface.
Fig. 3.
Copper-catalyzed azide–alkyne cycloaddition (CuAAC) and thermal denaturation on the yeast surface. (A) Schematic representation of CuAAC reaction between yeast displaying AzF-containing Fcs and biotin-alkyne 3. (B), (C) Flow cytometric detection of biotin after reactions with yeast displaying wild-type Fcs (B) or Fc 400 AzF (C). (D), (E) CuAAC thermal stability studies. Wild-type Fcs (D) or Fc 400 AzF (E) exhibits the same thermal denaturation behaviors irrespective of exposure to CuAAC reaction conditions or covalent modification with 3. (F) Two-step detection of reaction products on the yeast surface. CuAAC reactions were used to functionalize proteins on the yeast surface with propargylamine (4). A second CuAAC reaction with 3 and subsequent biotin detection revealed that reactions with 400 μM 4 result in cells exhibiting background levels of fluorescence, while lowering the concentration of 4 in solution leads to increased levels of fluorescence, suggesting incomplete reactions. R1: Fc-yeast; R2: azide; R3: triazole-biotin; THPTA: Tris(3-hydroxypropyltriazolylmethyl)amine; RT: room temperature.
We used thermal denaturation experiments (Hasenhindl et al., 2013; Traxlmayr et al., 2013) to assess the stability of displayed bioconjugates. Figure 3D–E depicts FcγRI binding as a function of denaturation temperature in samples left untreated, samples subjected to CuAAC with 3, or samples subjected to CuAAC conditions without 3. The data indicate that wild-type Fc (Fig. 3D) and Fc 400 AzF (Fig. 3E) are unaffected by either the CuAAC conditions or covalent modification with 3 (verification of conjugation with 3 can be seen in Supplementary Figure S5; midpoints of thermal denaturation are reported in Supplementary Table SIII). Similar trends were observed with SPAAC-mediated reactions at several positions (Supplementary Figure S6, Supplementary Table SIV). These findings indicate that azides in proteins displayed on the yeast surface are selectively addressable using CuAAC and SPAAC, and that the resulting bioconjugates are (1) amenable to stability evaluations that are well-correlated with measurements in solution (Hasenhindl et al., 2013); and (2) stable after biotinylation, consistent with previous investigations (Acchione et al., 2012).
We used reactions with 3 to establish a general approach to detecting CuAAC reaction products on the yeast surface (Dirksen et al., 2014; Ng et al., 2012). In the two-step reaction scheme, an alkyne-containing molecule of interest is used in the initial reaction, followed by reaction with 3 and subsequent biotin detection to evaluate the success of the first reaction. This strategy enabled identification of reaction conditions with the model compound propargylamine (4) resulting in modification levels similar to those achieved with a single-step biotinylation reaction (Fig. 3C and F, Supplementary Table SV). Importantly, omission of 4 in the first CuAAC reaction followed by reaction with 3 in the second reaction results in fluorescence levels comparable to a single reaction with 3, demonstrating that the azide functionality is only consumed when an alkyne is present during the reaction. This two-step detection process was also successfully extended to SPAAC reactions (Supplementary Figure S3, Table SVI). Although all alkyne reactants (≥10 μM) were added in vast excess of displayed azide-containing proteins (≤10 nM), the concentrations of alkyne required for efficient reaction varied from compound to compound in both CuAAC and SPAAC reactions, suggesting that empirical determination of conditions leading to complete reactions is crucial (Dirksen et al., 2014). These two-step experiments demonstrate generalizable approaches to the detection of reaction products on the yeast surface and the determination of high-yielding reaction conditions.
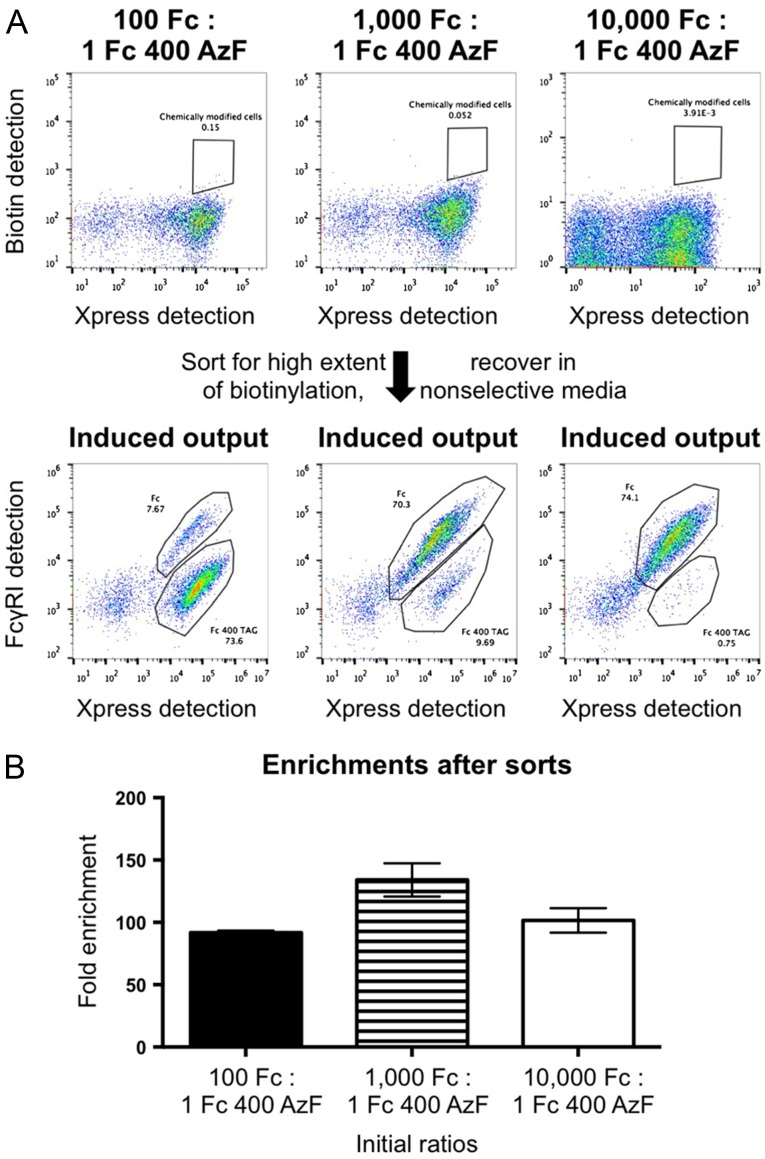
Having established robust approaches to constructing and evaluating bioconjugates on the yeast surface, we explored the feasibility of performing high throughput screening with conjugates. In these experiments, yeast displaying wild-type Fcs were mixed with yeast displaying Fc 400 AzF in varying ratios, subjected to CuAAC with 3, and sorted via fluorescence-activated cell sorting for cells exhibiting high levels of biotinylation (Fig. 4A, Supplementary Figure S7). Although cells displaying AzF-containing Fcs exhibited reduced viability after CuAAC (approximately two thirds reduction by colony counting assays), sorted cells were readily recovered using growth-based recoveries, in line with findings for chemically modified phage (Heinis et al., 2009). Enriched populations were propagated, induced in media lacking AzF, and labeled with FcγRI and anti-Xpress to identify full-length (labeled ‘Fc’ in Fig. 4A) and truncated (labeled ‘Fc 400 TAG’ in Fig. 4A) clones. (Sorted populations were recovered in yeast media containing leucine, leading to retention of display plasmids only; cells displaying wild-type Fcs do not contain the suppression plasmid and thus cannot be propagated in the media used for retention of the suppression plasmid. See the section ‘Flow cytometry (sorting)’ in the Materials and methods section for additional details.)) Figure 4B depicts enrichment ratios calculated based on the fraction of truncated constructs observed in sorting outputs (see also ‘Flow cytometry (sorting)’ section in Materials and methods and Supplementary Table SVII). Enrichments are between 90- and 130-fold over a range of starting ratios. We attribute incomplete single-pass enrichments to the overlapping fluorescence profiles of biotinylated and nonbiotinylated cells and inherent noise in fluorescence-activated cell sorting, both of which are known to limit enrichment efficiency (VanAntwerp and Wittrup, 2000). To confirm retention of the genotype–phenotype linkage of the covalently modified azide functionality, DNA from cells recovered in a ‘100:1’ sort was isolated and subjected to sequence analysis. 16/18 sequenced clones contained the amber codon at position 400 of the Fc, consistent with flow cytometric analysis. These model sorting experiments demonstrate the high throughput enrichment of chemically modified conjugates while maintaining a genotype–phenotype linkage with reactive ncAAs.
Fig. 4.
Fluorescence-activated cell sorting of yeast-displayed bioconjugates. (A) Representative flow cytometry plots before and after sorting. Mixtures of yeast displaying wild-type Fcs and yeast displaying Fc 400 AzF were subjected to CuAAC with 3, labeled for biotin detection, and sorted. Enriched populations were recovered in media containing leucine, which resulted in retention of only the encoded display constructs, in order to avoid growth biases from the suppression plasmid. After outgrowth, populations were induced and labeled with FcγRI in order to distinguish between full-length Fc constructs (indicated by ‘Fc’) and truncated Fc constructs (indicated by ‘Fc 400 TAG’). Differences in flow cytometry plot appearances are from the use of different instruments on different days. (B) Enrichment factors from sorts. Fold enrichments were calculated by dividing the observed fraction of truncated Fcs by the initial ratio of Fc to Fc 400 AzF. Values reported here are the results of three or more independent sorting experiments at the indicated initial ratios.
Discussion
This work demonstrates the power and versatility of ncAA-compatible yeast display in the area of bioconjugate construction and evaluation. ncAA incorporation can be evaluated on the yeast surface using reporter proteins such as scFv-Fcs (Fig. 1). In our experiments, the sensitivity of flow cytometry enabled us to evaluate display levels supported by various aaRS–ncAA combinations as well as residual display levels observed in the absence of a ncAA during induction; both of these considerations are important for the analysis and directed evolution of more efficient suppression systems for ncAA incorporation (Des Soye et al., 2015; O'Donoghue et al., 2013). We found that some of the aaRS mutants we evaluated here appear to support reporter construct display in the absence of ncAA (Fig. 1, Supplementary Figure S1, Supplementary Table SI), raising the possibility of highly heterogeneous mixtures of full-length constructs on the yeast surface that may confound analysis. However, our use of the AcFRS variant for the bulk of our experiments resulted in what we expect to be very low fractions of constructs containing canonical amino acids (approximate fractions of 0.06 for ‘worst-case’ display scenarios calculated as described in ‘Results’). Thus, more than 90 percent of the full-length Fcs and Fcabs we displayed on the yeast surface in this work contained the ncAA of interest, suggesting that observed properties are almost entirely attributable to ncAA-containing proteins. Evaluating potential protein heterogeneity with other aaRS–ncAA pairs will be important as efforts are made to expand the range of chemical functionality that can be displayed on the yeast surface.
Similar to the evaluation of ncAA incorporation events, the sensitivity of flow cytometry enabled us to evaluate chemical reactions on the yeast surface in detail. Some of our flow cytometry results raise the possibility of side reactions after performing CuAAC or SPAAC (Fig. 3, Supplementary Figure S4, Supplementary Table SII). Most notably, display construct removal with DTT following chemical reactions did not fully decrease biotin detection levels to background levels. These findings may be explained in part by the enhanced permeability of yeast after click chemistry and DTT treatment (Klis et al., 2007), which could increase staining during flow cytometry preparations. However, these results could also indicate background incorporation of AzF into other proteins on the yeast surface (and subsequent chemical modification) or a more general set of side reactions that can occur during CuAAC or SPAAC (Agard et al., 2006; Gutmann et al., 2016; Link et al., 2004). Our attempts to characterize display constructs removed from the yeast surface with Western blotting were unsuccessful, mirroring current challenges faced by researchers evaluating chemical reactions on phage (Ng et al., 2012). Off-target reactions may impact the performance of some applications of yeast display with ncAAs, such as the detection of low levels of a particular functional group on the yeast surface during the directed evolution of aaRSs (Tanrikulu et al., 2009). Future work will consider strategies for increasing reaction specificity on the yeast surface, developing analytical approaches for evaluating chemical reactions on the yeast surface, and mitigating potential effects of off-target reactions on bioconjugate construction, evaluation, and screening.
The use of Fcs and Fcabs in combination with previously established modification sites enabled the development of generalizable strategies for characterizing chemical reactions, evaluating thermal stability, and performing conjugate screening on the yeast surface. These experiments also highlight the advantages of ncAA-compatible yeast display over other display technologies: bivalent, glycosylated Fcs and Fcabs are challenging to display using mRNA, phage, or E. coli display. Moreover, flow cytometry-based evaluations are not possible with mRNA or phage, and the fragile membrane structure of E. coli prevents thermal stability evaluations. Previous work has shown that binding and stability measurements made on the yeast surface are highly correlated with solution-based measurements (Hasenhindl et al., 2013; Lipovsek et al., 2007), suggesting that the approaches described here will be valuable for generating lead conjugates. ncAA-compatible yeast display will enable high throughput identification of conjugates with desirable reactivity and stability (Junutula et al., 2008; Traxlmayr et al., 2013) prior to assessing more nuanced properties in vivo (Chari et al., 2014). The approaches described here will also be applicable to the identification of chemically modified polypeptides such as protein-small molecule hybrids (Horiya et al., 2014; Ng et al., 2015) and constrained polypeptides (Frost et al., 2015; Sako et al., 2008) exhibiting desirable properties. ncAA-compatible yeast display will be a valuable addition to early-stage reagent and therapeutic discovery efforts.
Supplementary material
Supplementary material is available at PEDS online.
Acknowledgements
We thank Prof. K.D. Wittrup of MIT for providing laboratory space and funding during the initial stages of this work. We thank Prof. L. Wang of the Salk Institute for Biological Studies, Dr V.K. Vyas and Prof. G.R. Fink of the Whitehead Institute, Dr M.W. Traxlmayr, Dr G.E. Whitworth, Prof. B. Imperiali, J. Liu, and Prof. J.A. Johnson of MIT for providing materials. We thank the Koch Institute Flow Cytometry Core for flow cytometry assistance.
Funding
J.A.V. was supported in part by startup funds from Tufts University and in part by postdoctoral fellowships from the Ludwig Institute for Cancer Research and the National Institutes of Health [grant number F32 CA168057]; D.N.L. and J.Z. were supported in part by the MIT UROP Office; H.P.K. was supported in part by a Stern's Fellowship at Tufts University; R.L.K. was supported by the National Institutes of Health Biotechnology Predoctoral Research Training Program at MIT [grant number T32 GM008334].
References
- Acchione M., Kwon H., Jochheim C.M. and Atkins W.M. (2012) mAbs, 4, 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard N.J., Baskin J.M., Prescher J.A., Lo A. and Bertozzi C.R. (2006) ACS Chem. Biol., 1, 644–648. [DOI] [PubMed] [Google Scholar]
- Antonczak A.K., Simova Z., Yonemoto I.T., Bochtler M., Piasecka A., Czapinska H., Brancale A. and Tippmann E.M. (2011) Proc. Natl. Acad. Sci. U S A, 108, 1320–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder E.T. and Wittrup K.D. (1997) Nat. Biotechnol., 15, 553–557. [DOI] [PubMed] [Google Scholar]
- Chari R.V., Miller M.L. and Widdison W.C. (2014) Angew. Chem. Int. Ed. Engl., 53, 3796–3827. [DOI] [PubMed] [Google Scholar]
- Chin J.W., Cropp T.A., Anderson J.C., Mukherji M., Zhang Z. and Schultz P.G. (2003) Science, 301, 964–967. [DOI] [PubMed] [Google Scholar]
- Des Soye B.J., Patel J.R., Isaacs F.J. and Jewett M.C. (2015) Curr. Opin. Chem. Biol., 28, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen A., Madsen M., Dello Iacono G., Matin M.J., Bacica M., Stankovic N., Callans S. and Bhat A. (2014) Bioconjug. Chem., 25, 1052–1060. [DOI] [PubMed] [Google Scholar]
- Frost J.R., Jacob N.T., Papa L.J., Owens A.E. and Fasan R. (2015) ACS Chem. Biol., 10, 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann M., Memmel E., Braun A.C., Seibel J., Meinel L. and Luhmann T. (2016) Chembiochem, 17, 866–875. [DOI] [PubMed] [Google Scholar]
- Hasenhindl C., Traxlmayr M.W., Wozniak-Knopp G., Jones P.C., Stadlmayr G., Ruker F. and Obinger C. (2013) Protein Eng. Des. Sel., 26, 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Morimoto J. and Suga H. (2012) ACS Chem. Biol., 7, 607–613. [DOI] [PubMed] [Google Scholar]
- Heinis C., Rutherford T., Freund S. and Winter G. (2009) Nat. Chem. Biol., 5, 502–507. [DOI] [PubMed] [Google Scholar]
- Hong V., Presolski S.I., Ma C. and Finn M.G. (2009) Angew. Chem. Int. Ed. Engl., 48, 9879–9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiya S., Bailey J.K., Temme J.S., Guillen Schlippe Y.V. and Krauss I.J. (2014) J. Am. Chem. Soc., 136, 5407–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.A., Lu Y.Y., Van Deventer J.A. and Tirrell D.A. (2010) Curr. Opin. Chem. Biol., 14, 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junutula J.R., Bhakta S., Raab H., Ervin K.E., Eigenbrot C., Vandlen R., Scheller R.H. and Lowman H.B. (2008) J. Immunol. Methods, 332, 41–52. [DOI] [PubMed] [Google Scholar]
- Klis F.M., de Jong M., Brul S. and de Groot P.W. (2007) Yeast, 24, 253–258. [DOI] [PubMed] [Google Scholar]
- Kuzmin A., Poloukhtine A., Wolfert M.A. and Popik V. (2010) Bioconjug. Chem., 21, 2076–2085. [DOI] [PubMed] [Google Scholar]
- Kwon O.H., Yoo T.H., Othon C.M., Van Deventer J.A., Tirrell D.A. and Zewail A.H. (2010) Proc. Natl. Acad. Sci. U S A, 107, 17101–17106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.H., Madabhushi S.R., Mann J., Neelamegham S. and Park S. (2010) Biotechnol. Bioeng., 106, 27–41. [DOI] [PubMed] [Google Scholar]
- Link A.J., Vink M.K.S. and Tirrell D.A. (2004) J. Am. Chem. Soc., 126, 10598–10602. [DOI] [PubMed] [Google Scholar]
- Lipovsek D., Lippow S.M., Hackel B.J., Gregson M.W., Cheng P., Kapila A. and Wittrup K.D. (2007) J. Mol. Biol., 368, 1024–1041. [DOI] [PubMed] [Google Scholar]
- Liu C.C., Mack A.V., Tsao M.L., Mills J.H., Lee H.S., Choe H., Farzan M., Schultz P.G. and Smider V.V. (2008) Proc. Natl. Acad. Sci. U S A, 105, 17688–17693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Jafari M.R. and Derda R. (2012) ACS Chem. Biol., 7, 123–138. [DOI] [PubMed] [Google Scholar]
- Ng S., Jafari M.R., Matochko W.L. and Derda R. (2012) ACS Chem. Biol., 7, 1482–1487. [DOI] [PubMed] [Google Scholar]
- Ng S., Lin E., Kitov P.I., et al. (2015) J. Am. Chem. Soc., 137, 5248–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue P., Ling J., Wang Y.S. and Soll D. (2013) Nat. Chem. Biol., 9, 594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y., Morimoto J., Murakami H. and Suga H. (2008) J. Am. Chem. Soc., 130, 7232–7234. [DOI] [PubMed] [Google Scholar]
- Shen B.Q., Xu K., Liu L., et al. (2012) Nat. Biotechnol., 30, 184–189. [DOI] [PubMed] [Google Scholar]
- Sun S.B., Schultz P.G. and Kim C.H. (2014) Chembiochem, 15, 1721–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanrikulu I.C., Schmitt E., Mechulam Y., Goddard W.A. III and Tirrell D.A. (2009) Proc. Natl. Acad. Sci. U S A, 106, 15285–15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxlmayr M.W., Hasenhindl C., Hackl M., Stadlmayr G., Rybka J.D., Borth N., Grillari J., Ruker F. and Obinger C. (2012) J. Mol. Biol., 423, 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxlmayr M.W., Lobner E., Antes B., et al. (2013) Protein Eng. Des. Sel., 26, 255–265. [DOI] [PubMed] [Google Scholar]
- Van Deventer J.A., Kelly R.L., Rajan S., Wittrup K.D. and Sidhu S.S. (2015) Protein Eng. Des. Sel., 28, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deventer J.A. and Wittrup K.D. (2014) Methods Mol. Biol., 1131, 151–181. [DOI] [PubMed] [Google Scholar]
- Van Deventer J.A., Yuet K.P., Yoo T.H. and Tirrell D.A. (2014) Chembiochem, 15, 1777–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanAntwerp J.J. and Wittrup K.D. (2000) Biotechnol. Prog., 16, 31–37. [DOI] [PubMed] [Google Scholar]
- Wang Q. and Wang L. (2008) J. Am. Chem. Soc., 130, 6066–6067. [DOI] [PubMed] [Google Scholar]
- Wiltschi B., Wenger W., Nehring S. and Budisa N. (2008) Yeast, 25, 775–786. [DOI] [PubMed] [Google Scholar]
- Ye S., Zaitseva E., Caltabiano G., Schertler G.F., Sakmar T.P., Deupi X. and Vogel R. (2010) Nature, 464, 1386–1389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.