Abstract
The favorable biophysical attributes of non-antibody scaffolds make them attractive alternatives to monoclonal antibodies. However, due to the well-known stability-function trade-off, these gains tend to be marginal after functional selection. A notable example is the fibronectin Type III (FN3) domain, FNfn10, which has been previously evolved to bind lysozyme with 1 pM affinity (FNfn10-α-lys), but suffers from poor thermodynamic and kinetic stability. To explore this stability-function compromise further, we grafted the lysozyme-binding loops from FNfn10-α-lys onto our previously engineered, ultra-stable FN3 scaffold, FN3con. The resulting variant (FN3con-α-lys) bound lysozyme with a markedly reduced affinity, but retained high levels of thermal stability. The crystal structure of FNfn10-α-lys in complex with lysozyme revealed unanticipated interactions at the protein–protein interface involving framework residues of FNfn10-α-lys, thus explaining the failure to transfer binding via loop grafting. Utilizing this structural information, we redesigned FN3con-α-lys and restored picomolar binding affinity to lysozyme, while maintaining thermodynamic stability (with a thermal melting temperature 2-fold higher than that of FNfn10-α-lys). FN3con therefore provides an exceptional window of stability to tolerate deleterious mutations, resulting in a substantial advantage for functional design. This study emphasizes the utility of consensus design for the generation of highly stable scaffolds for downstream protein engineering studies.
Keywords: consensus design, loop grafting, protein engineering, stability-function trade-off, X-ray crystallography
Introduction
A major goal of protein engineering is to produce novel proteins that bind to a specified target. Immunoglobulins are a natural scaffold for binding, and antibodies can be generated for virtually any given target (Holliger and Hudson, 2005; Demarest and Glaser, 2008; Beck et al., 2010; Zolot et al., 2013). Despite their success as a rapidly growing class of therapeutics (Walsh, 2014), unmodified immunoglobulins nevertheless are subject to a range of limitations, such as their large size, challenges with over expression, solubility and stability that can limit their applicability (Birch and Racher, 2006; Rouet et al., 2014). To overcome the size and stability limitations of monoclonal antibodies, a large body of work has focused on the engineering of antibody single domains and fragments (Holliger and Hudson, 2005) and on increasing stability through mutation (Chennamsetty et al., 2009; Dudgeon et al., 2012).
An alternative strategy relates to the generation of non-antibody scaffolds, which show great potential in terms of affinity, ease of production, target neutralization and stability for diagnostics, biotechnology and therapeutics (Binz et al., 2005; Stern et al., 2013; Vazquez-Lombardi et al., 2015). Although the stability of parental non-antibody scaffolds is often considerable, this is not always observed for generated binders; indeed a stability-function trade-off is often reported, resulting in suboptimal candidates after functional selection (Schreiber et al., 1994; Shoichet et al., 1995; Beadle and Shoichet, 2002; Nagatani et al., 2007; Tokuriki et al., 2008). This compromise can result in non-antibody binders that are less stable or only marginally better than their monoclonal antibody counterparts (Vazquez-Lombardi et al., 2015).
One particular example of this trade-off is observed in the fibronectin Type III (FN3) domain. The benchmark scaffold from this family is the 10th FN3 repeat from human fibronectin (FNfn10; Koide et al., 1998). This particular domain is chosen because it is the most stable human FN3 repeat, with a midpoint of thermal denaturation (Tm) of 84°C (Koide et al., 1998) and has the capacity to tolerate a number of mutations in three surface-exposed loops (B/C, D/E and F/G which are analogous to the complementary determinant regions of antibodies) (Koide et al., 1998, 2007; Parker et al., 2005; Lipovsek, 2011). Combinatorial libraries have been built into these loops and specific binders selected for several different targets (Lipovsek, 2011). While these engineered FN3 domains have been shown to display very high affinities to their targets, they often exhibit a large reduction in thermodynamic stability, solubility and are prone to aggregation (Parker et al., 2005; Lipovsek, 2011). For example, Wittrup and colleagues evolved FNfn10 to bind lysozyme with high affinity (Hackel et al., 2008). Their resulting clone (DE0.4.1, which we refer to as FNfn10-α-lys herein) bound lysozyme with an affinity of 1 pM, but had a Tm of 51 ± 3°C, which is 33°C lower than wild type FNfn10 (Tm of 84°C) (Hackel et al., 2008).
Although many proteins have been shown to display a stability-function trade-off (Shoichet et al., 1995; Beadle and Shoichet, 2002; Tokuriki et al., 2008; Tokuriki and Tawfik, 2009), it is possible for high stability and functionality to coexist (Serrano et al., 1993; Arnold, 1998; Giver et al., 1998; Arnold et al., 2001; Taverna and Goldstein, 2002; Sánchez et al., 2006). In order to further explore the stability-function trade-off in FN3 domains, we questioned whether the potent binding activity of FNfn10-α-lys could be achieved without a loss in stability. We previously reported the consensus design of a FN3 domain, FN3con, which exhibits an extremely high degree of thermodynamic and kinetic stability (Tm > 100°C, reversible folding and aggregation resistant) (Porebski et al., 2015). In this study, we describe loop grafting from FNfn10-α-lys onto the stable FN3con scaffold (creating FN3con-α-lys). This variant, although destabilized by a relatively small amount, bound lysozyme with a significantly reduced affinity. To investigate the structural reason for reduced affinity, we determined the crystal structure of both FNfn10-α-lys in complex with lysozyme and of FN3con-α-lys alone, thereby enabling the rational redesign of FN3con-α-lys (FN3con-α-lys.v2). Redesign successfully restored binding affinity with a substantially smaller loss in thermodynamic stability, further demonstrating that function and stability are not mutually exclusive. We discuss the implications of our findings for the use of consensus design in the generation of highly stable binding scaffolds that circumvent the stability-function trade-off.
Results and discussion
Construction of FN3con-α-lys by Loop Grafting
Loop grafting is a common approach in the generation of humanized antibodies and affinity transfer across similar protein scaffolds (Nicaise et al., 2004). Using this approach, we constructed FN3con-α-lys, in which the B/C, D/E and F/G loops from FNfn10-α-lys were grafted onto FN3con. FNfn10-α-lys was purified from insoluble inclusion bodies after recombinant expression in Escherichia coli, with yields of 50 mg/l. Purified FNfn10-α-lys remained soluble for short periods of time (max. of 24 h at 4°C) before visibly precipitating. While similar protein yields were obtained for FN3con-α-lys, this variant expressed as a soluble monomer that, after purification, remained in solution at 10 mg/ml for longer than 90 days at 4°C. These preliminary observations served as an indication of the superior stability and aggregation resistance of the FN3con-α-lys variant.
Biophysical characterization of FNfn10-α-lys and FN3con-α-lys
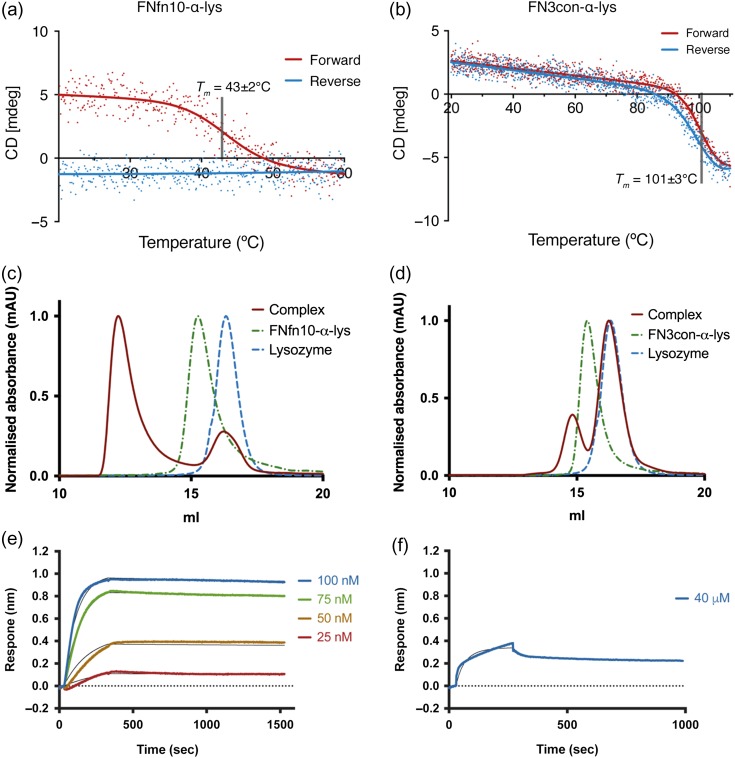
FNfn10-α-lys undergoes irreversible thermal denaturation with a Tm of 43 ± 2°C, as measured by circular dichroism (CD), with complete loss of CD signal and visible precipitate upon cooling (Fig. 1A). This measurement is considerably lower than the previously reported Tm of 51 ± 3°C (Hackel et al., 2008), likely due to the thermal denaturation assay used in the cited study; which assessed binding of yeast-displaying FNfn10-α-lys to lysozyme after heating. In striking contrast, FN3con-α-lys unfolds reversibly with a Tm of 101 ± 3°C (Fig. 1B). Characterization of lysozyme binding by size exclusion chromatography (SEC) and surface plasmon resonance (SPR) shows that FNfn10-α-lys forms a tight complex with lysozyme (Fig. 1C and Supplementary Fig. S1A). However, we observed significant non-specific binding with lysozyme titrations over an immobilized FNfn10-α-lys surface that could not be remedied in SPR by the use of a modified running buffer containing 12 mg/ml CM-Dextran; thus restricting rigorous kinetic evaluation. We therefore employed the use of bio-layer interferometry (BLI) and a reversed surface orientation (see Methods), which resolved the non-specific binding problem. This allowed for a global fit of multiple FNfn10-α-lys concentrations, giving a KD of 2.09 × 10−10 M (Fig. 1E; Supplementary Table S1). Although this is 200-fold weaker than previous reports, the BLI-based kinetics values are not necessarily equivalent to the steady-state equilibrium-based approach previously undertaken by flow cytometry (Hackel et al., 2008). Regardless, the loop-grafted construct FN3con-α-lys binds lysozyme very weakly as shown by SEC, SPR and BLI (Fig. 1D, Supplementary Figs S1B and 1F). A binding curve was barely discernible via BLI when FN3con-α-lys was run at a concentration of 40 μM, with a KD of 7.2 × 10−6 M for the resulting individual curve fit (Fig. 1F; Supplementary Table S1).
Fig. 1.
Biophysical characterization of FNfn10-α-lys and FN3con-α-lys. CD thermal melts of (A) FNfn10-α-lys (Tm of 43 ± 2°C) and (B) FN3con-α-lys (Tm of 101 ± 3°C). SEC, revealing complex formation for (C) FNfn10-α-lys and (D) FN3con-α-lys with lysozyme. Representative BLI sensograms of (E) FNfn10-α-lys and (F) FN3con-α-lys with titrations of the respective FN3 protein against an immobilized HEL surface (fit for each concentration as a thin black line).
Direct loop grafting: structural rationalization of suboptimal binding performance
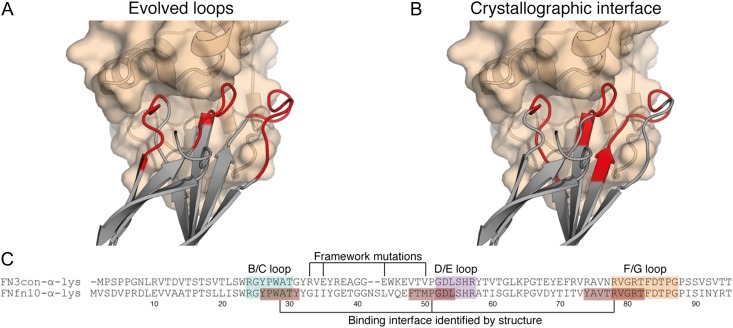
In order to glean structural insight into our biophysical results, we determined the crystal structure of FNfn10-α-lys in complex with lysozyme at 2.54 Å resolution (Table I, Fig. 2 and Supplementary Fig. S2). The asymmetric unit contains two copies of the FN3–lysozyme complex, arranged in a 1:1 binding stoichiometry. Both copies of the complex are highly conserved (root mean square deviation of 0.15 Å between the two complexes over 194 Cα atoms), and clear electron density for both subunits is observed at the protein–protein interface. The interface buries ~930 and ~873 Å2 surface area for FNfn10-α-lys and lysozyme, respectively. The interface area is at the higher end of the scale of antibody–antigen interfaces (Sheriff, 1993) and other FN3-based binders (Ramamurthy et al., 2012). The interaction utilizes all three binding loops of FNfn10-α-lys, which packs well into the lysozyme active site cleft (Fig. 2A and Supplementary Fig. S2). The Sc statistic (Lawrence and Colman, 1993), which is a measure of the binding surface complementarity, is 0.79 (scale from 0.0 to 1.0, with 1.0 being perfect complementarity). This value is greater than the range observed for protease–protease inhibitors (0.71–0.76), oligomeric interfaces (0.70–0.74), antibody–antigen complexes (0.66–0.68) (Lawrence and Colman, 1993) and other FN3 domain–protein complexes (0.64–0.76) (Ramamurthy et al., 2012). Although all three loops play a role in the interface, the structure reveals interactions between the framework residues (those that contribute to β-sheet secondary structure) of FNfn10-α-lys and lysozyme. These residues were not expected to form part of the interaction interface, and were therefore not grafted onto FN3con-α-lys, thus offering a simple explanation of poor affinity transfer upon sequence-based loop grafting (Fig. 2A–C). The binding interface involves 14 hydrogen bonds mediated by 7 residues (P27, A29, Y31, T77, R78, V79 and R81) and 6 salt bridges contributed by two residues (R78 and R81) in FNfn10-a-lys (Supplementary Table S2 and Supplementary Fig. S2). The number of interactions is on the upper end of the scale in comparison to seven other FN3 domain–protein complexes (Ramamurthy et al., 2012), which is consistent with the high affinity of FNfn10-α-lys for lysozyme and size of the interface.
Table I.
Data collection and refinement statisticsa
| Data collection | FNfn10-α-lys complex | FN3con-α-lys |
|---|---|---|
| Wavelength (Å) | 0.9537 | 0.9537 |
| Space group | P212121 | P1211 |
| Unit cell dimensions (Å, °) | a = 54.86, b = 87.73, c = 100.90 | a = 50.22, b = 71.23, c = 126.14, β = 90.4 |
| Resolution (Å) | 2.54 | 2.46 |
| Measured reflections | 111 926 (12 664) | 117 587 (13 558) |
| Unique reflections | 16 719 (1940) | 32 118 (3627) |
| Completeness (%) | 99.0 (96.0) | 98.8 (98.8) |
| Redundancy | 6.7 (6.5) | 3.7 (3.7) |
| Rpim | 0.139 (0.728) | 0.062 (0.949) |
| <I/σI> | 8.90 (2.13) | 11.60 (1.70) |
| Structure refinement | ||
| Reflections | 16 665 (1579) | 32 099 (3181) |
| Protein atoms | 3430 | 4365 |
| Water molecules | 52 | 70 |
| Ligands | 0 | 11 |
| Rwork | 0.2176 | 0.2488 |
| Rfree (5% of data) | 0.2499 | 0.2811 |
| CC1/2 | 0.993 (0.685) | 0.997 (0.408) |
| RMSD bond lengths (Å) | 0.003 | 0.012 |
| RMSD bond angles (º) | 0.56 | 1.53 |
| Mean B-factor (Å2) | 37.52 | 59.13 |
| Protein | 37.75 | 59.23 |
| Solvent | 22.68 | 44.64 |
| Ramachandran | ||
| Favored (%) | 99 | 95 |
| Outliers (%) | 0 | 0.71 |
| MolProbity score | 0.96, 100th percentileb (N = 6642, 2.535 ± 0.25 Å) | 2.21, 89th percentileb (N = 6959, 2.46 ± 0.25 Å) |
| Protein Data Bank (PDB) ID | 5J7C | 5J7K |
aStatistics for the highest-resolution shell are shown in parentheses.
b100th percentile is the best among structures of comparable resolution; 0th percentile is the worst.
Fig. 2.
The FNfn10-α-lys-lysozyme complex reveals a tight binding interface that makes use of framework residues. (A) The loop residues (red) of FNfn10-α-lys (gray) as previously evolved for lysozyme (tan) binding (Hackel et al., 2008). (B) The actual binding interface residues (red) of FNfn10-α-lys (gray) with lysozyme (tan) as determined by crystal structure and the PDBePISA web server (Krissinel and Henrick, 2007). (C) A sequence alignment of FN3con-α-lys with FNfn10-α-lys, highlighting the B/C, D/E and F/G loops (blue, purple and orange) that were previously evolved for lysozyme binding (Hackel et al., 2008), the actual residues involved in the binding interface (red) and positions of the FNfn10-α-lys framework mutations previously introduced (Hackel et al., 2008).
A sequence alignment between FNfn10-α-lys and FN3con-α-lys (Fig. 2C) revealed nine framework positions that were not grafted (Supplementary Table S2), that may be involved in recognition of the lysozyme epitope. In order to accurately discriminate the framework differences between FNfn10-α-lys and FN3con-α-lys, we determined the crystal structure of unbound FN3con-α-lys (Table I). There are eight molecules within the asymmetric unit, revealing prevalent interactions of the C-terminal His tag with strands from adjacent molecules (Supplementary Fig. S3). Fragmented electron density for the N-terminus and parts of the C-terminal His tag is observed in all eight copies of the molecule, and density for the B/C and D/E loops is variable between the molecules, however, density for the F/G loop is not observed in any monomer. By using all eight molecules and homology modeling (for the FG loop only), it was possible to generate a complete model of FN3con-α-lys (Supplementary Fig. S3).
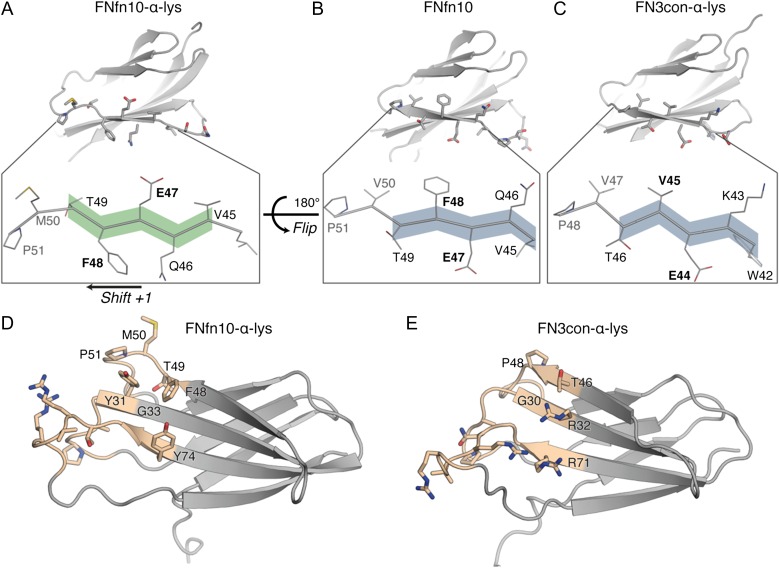
A structural alignment between FNfn10-α-lys and FN3con-α-lys reveals two features that are consistent with the poor affinity transfer. First, a conformational change is present in strand D of FNfn10-α-lys that results in a 180° flip and register shift (Fig. 3A), which is not present in FNfn10 (Fig. 3B) or FN3con-α-lys (Fig. 3C). Although this conformational change may be the result of induced fit on binding with lysozyme, we could not predict this event from sequence alone. Crystallization attempts of FNfn10-α-lys alone were unsuccessful, preventing an analysis of structural adaptations upon lysozyme binding. We speculate that the relatively low stability of FNfn10-α-lys may have contributed to crystallization difficulties. The flip and shift alters the physiochemical properties of the binding surface (paratope) on strand D (Fig. 3D), moving from polar and charged residues in FNfn10 and FN3con-α-lys to hydrophobic and uncharged residues in FNfn10-α-lys (Fig. 3A–C). The flip and shift is particularly important as M50 from FNfn10-α-lys packs between two tryptophan residues (W62 and W63) from lysozyme (Supplementary Figs S2B and S4). As the FN3con-α-lys structure does not display this conformational change, the position of P48 in FN3con-α-lys may impose a steric clash with W62 from lysozyme when modeled into the same binding site, thereby preventing tight complex formation (Supplementary Fig. S4).
Fig. 3.
Structural comparison between FNfn10-α-lys and FN3con-α-lys reveals framework incompatibilities that likely prevent tight complex formation. A conformational change is observed between FNfn10-α-lys (A) and FNfn10 (B) resulting in a 180° flip and +1 register shift of strand D that is also lacking in the unbound FN3con-α-lys crystal structure (C). Differences in framework residues of the lysozyme-binding interface (tan region) between FNfn10-α-lys (D) and FN3con-α-lys (E) highlight the potential for cavity formation due to the lack of Y31 (G30 in FN3con-α-lys) and steric clashes as a result of R32 and R71 in FN3con-α-lys. These characteristics may impact the formation of a tight binding interface.
Furthermore, when the entire framework-binding region of FNfn10-α-lys is compared to FN3con-α-lys, it becomes clear that the physiochemical incompatibilities with lysozyme binding extend beyond strand D. In particular, Y31 in FNfn10-α-lys plays a pivotal role in packing and forming hydrogen bonds with D48, S50 and N59 from lysozyme (Fig. 3D, Supplementary Figs S2A and S4). The analogous residue in FN3con-α-lys is G30, which not only results in a loss of three hydrogen bonds, but may also leave a large unfilled cavity in the binding interface (Fig. 3E, Supplementary Figs S2A and S4). In close proximity to G30 is R32 and R71, which belong to a stability enhancing electrostatic mesh on the surface of FN3con (Porebski et al., 2015) that were retained in the grafting process. As these are long and charged residues, we therefore hypothesize that R32 and R71 in FN3con-α-lys (G33 and Y74 in FNfn10-α-lys) may impose steric clashes with lysozyme, as suggested by modeling of the complex, thereby further restricting tight complex formation (Fig. 3D and E, Supplementary Fig. S4).
Rational redesign of FN3con-α-lys restores binding
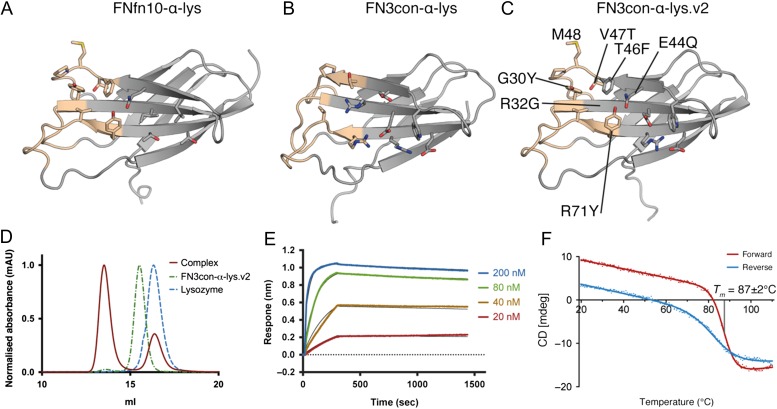
Given our high-resolution structural insights for the FNfn10-α-lys-lysozyme interface and direct comparison with a structure of the low-affinity FN3con-α-lys graft, we sought to rationally redesign FN3con-α-lys. Through redesign, we aimed to restore binding by addressing the conformational change seen in strand D and the physiochemical incompatibilities of framework residues in the lysozyme-binding interface (Fig. 4A and B). We have previously shown FN3con to be extremely rigid due to an optimized hydrophobic core (Porebski et al., 2015). For this reason, we did not expect to be able to easily engineer strand D to mimic the conformation seen in the FNfn10-α-lys structure (Fig. 3B and C). Instead, we decided to retain the hydrophobic core residues of FN3con and simply mutate the surface residues of strand D to match that of FNfn10-α-lys. This involved the mutation of E44Q, T46F, V47T and the insertion of a methionine residue between V47T and P48 (Fig. 4C). To mimic the remaining FNfn10-α-lys paratope, we made three more mutations (G30Y, R32G and R71Y), which predominantly removed charged residues from FN3con's stability enhancing electrostatic mesh (Porebski et al., 2015). We did so with the hypothesis that this will restore packing and remove the predicted steric clashes, thereby allowing for tighter complex formation. In total, seven mutations (Supplementary Table S3) were made to FN3con-α-lys, producing the variant FN3con-α-lys.v2 (Fig. 4C and Supplementary Data S1). FN3con-α-lys.v2 expressed as a soluble monomer to ~50 mg/l of culture and was visibly soluble when concentrated to 10 mg/ml for over 30 days.
Fig. 4.
Framework residues in the lysozyme-binding interface of FN3con-α-lys were redesigned by alignment of FNfn10-α-lys and the FN3con-α-lys crystal structures. Redesign restored binding at the cost of thermodynamic stability. (A) The crystal structure of FNfn10-α-lys showing the paratope surface residues (tan) and surrounds; (B) the composite crystal structure of FN3con-α-lys showing the paratope surface residues (tan) and surrounds; (C) a homology model of the redesigned FN3con-α-lys.v2 based on FNfn10-α-lys showing the redesigned binding interface residues (tan); (D) SEC complex formation shift of FN3con-α-lys.v2; (E) representative BLI sensograms of FN3con-α-lys.v2 titrations against an immobilized HEL surface (fit for each concentration as a thin black line); (F) variable temperature CD melt of FN3con-α-lys.v2 showing a Tm of 87 ± 2°C and incomplete reversible folding.
Assessment of binding to lysozyme was performed by SEC, SPR (Fig. 4D and Supplementary Fig. S1C) and BLI revealing a substantially improved KD (4.37 × 10−10 M) that is remarkably similar to FNfn10-α-lys (Figs 4E and 1E; Supplementary Table S1). As lysozyme binding was successfully restored, these results support our hypothesis that steric hindrance and the presence of a cavity were limiting tight complex formation in the FN3con-α-lys graft. Subsequent characterization of the thermodynamic stability of FN3con-α-lys.v2 revealed a Tm of 87 ± 2°C (Fig. 4F). Although thermal denaturation is not completely reversible, as demonstrated by the small loss of CD signal on cooling (Fig. 4F), the remaining and exceptionally high Tm of 87 ± 2°C positions FN3con-α-lys.v2 well above the Tm of both FNfn10-α-lys (43 ± 2°C) and FNfn10 (84°C). Crystallization of FN3con-α-lys.v2 alone and in complex with lysozyme is currently in progress and may allow structural rationalization of the redesign, as well as further insights. Together, these results highlight the remarkable capacity of consensus design to generate highly stable and mutationally tolerant scaffolds. As restoration of lysozyme binding required the degeneration of known stability enhancing features in FN3con, future directed evolution studies could be designed with this in mind, thereby retaining a greater degree of thermodynamic stability and favorable biophysical properties for the same level of function.
Conclusions
Non-antibody scaffolds are attractive alternatives to monoclonal antibodies, but experience stability-function trade-offs after selection, and are thus only marginally more stable than their antibody counterparts. This study sought to circumvent the stability-function trade-off in the FN3 domain. Crystallographic structure determination provided key structural insight into the binding interface between FNfn10-α-lys and lysozyme, which allowed for successful transfer of binding affinity onto the FN3con scaffold by a combination of loop grafting and rational design. Biophysical characterization subsequently showed FN3con to exhibit a smaller loss in thermodynamic stability after the engineering of function. These results therefore highlight that the effect of loop mutagenesis and stability-function trade-off is not equivalent across homologous proteins, with the FN3con scaffold imparting a greater resistance to destabilizing loop sequences (ΔTm of 41°C between FNfn10 and FNfn10-α-lys and ΔTm of ~20°C between FN3con and FN3con-α-lys.v2). This study underlines the utility of consensus design for the generation of highly stable and mutationally-tolerant scaffolds that may be suited to further protein engineering and directed evolution studies.
Methods
Loop grafting and homology modeling
Loop grafting was performed in PyMol v. 1.8.0.6 using structural alignments of FN3con (PDB: 4U3H) and an FNfn10-α-lys homology model based on FNfn10 (PDB: 1FNF). Loop boundaries were identified and grafting was conducted on the FN3con sequence. A homology model of FN3con-α-lys was generated based on FN3con (PDB: 4U3H) using Modeller v. 9.12 (Eswar et al., 2007). In each instance, 50 models were built and the lowest DOPE (Discrete Optimized Protein Energy) scoring model was selected for further analysis. Modeller was also used to complete the FN3con-α-lys crystal structure, and generate a model of FN3con-α-lys.v2.
Protein expression and purification
Genes encoding FNfn10-α-lys, FN3con-α-lys and FN3con-α-lys.v2 were chemically synthesized and provided in a pD444-CH (C-terminal 6× His tag, ampicillin resistance) vector by DNA2.0. The resulting plasmids were transformed into competent C41 E. coli cells for expression. A single colony from each transformation was picked and grown overnight at 37°C in 100 ml of 2×YT (16.0 g/l tryptone, 10.0 g/l yeast extract, 5.0 g/l NaCl) media containing 100 μg/ml of ampicillin. These cultures were then used to seed 1 l of 2× YT media. Cultures were induced at an OD600 of 0.9 with IPTG (0.5 mM final concentration), and grown for a further 4 h at 37°C. The cells were harvested by centrifugation. FN3con-α-lys and FN3con-α-lys.v2 had their cell pellets resuspended in 5 ml/g of native lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) and were lysed by sonication. Cell debris was removed by centrifugation and recombinant protein was isolated from the supernatant by nickel affinity chromatography using loose NiNTA resin (Sigma). Protein eluted from NiNTA resin was filtered and then loaded onto a size exclusion column (Superdex 75 16/60, GE Healthcare) equilibrated in either phosphate buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM PO4 3−, pH 7.4) for biophysical characterization or tris buffered saline (TBS) (50 mM Tris, 200 mM NaCl, pH 7.4) for protein crystallography. Protein concentration was determined by Nanodrop ND-1000 (ThermoFisher) and protein was stored at 4°C until use (biophysical characterization) or used immediately (protein crystallography).
Refolding and purification of FNfn10-α-lys
FNfn10-α-lys expressed insolubly under the same conditions as FN3con-α-lys and FN3con-α-lys.v2 (above) to ~50 mg/ml. The culture was harvested and resuspended in 5 ml/g of native lysis buffer and lysed by sonication. The supernatant was discarded and the insoluble fraction resolublized in denaturing buffer (8 M urea, 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0). Remaining insoluble material was cleared by centrifugation and filtration with a 0.8-μm syringe filter (Merck-Millipore). Urea solubilized FNfn10-α-lys was dialyzed against 4 L of TBS, 0.5 M Arginine, pH 8.0. The dialyzed material was filtered through a 0.2-μm syringe filter and concentrated in a 3000 molecular weight cut-off (MWCO) filter (Merck-Millipore). The concentrate was then separated by SEC (Superdex 75 16/60, GE Healthcare) in 1× TBS, pH 7.4 for crystallography, 1× TBS, 10% glycerol, 0.5M L-arginine, 1 mM 2-(2-aminoethyl)isothiourea dihydrobromide (AET-Sigma Aldrich), pH 8.0 for BLI and 1× PBS for CD thermal melts and SPR.
Expression and purification of the HyHEL10 Anti-Lysozyme FAb
Constructs for the light and heavy chains of anti-lysozyme monoclonal HyHEL10 were designed in a fragment antibody (FAb) format based on the previously reported sequences (Padlan et al., 1989). A C-terminal His tag was added to the heavy chain construct. The constructs were both cloned into the pCEP4 vector and co-transfected at a 2:1 light to heavy chain ratio in HEK293 using the Expi293 expression system (ThermoFisher Scientific). FAb was purified from culture via the heavy chain His tag using HisTrap FF crude columns based on the manufacturer's instructions (GE Healthcare). Purified FAb was biotinylated at a 5:1 molar ratio using EZ-Link NHS-PEG4 biotinylation reagent based on the manufacturer's instructions (ThermoFisher Scientific).
CD thermal melts
Thermal stability of purified FNfn10-α-lys, FN3con-α-lys and FN3con-α-lys.v2 was measured by CD. CD measurements were performed using a Jasco 815 spectropolarimeter with 0.2 mg/ml protein in PBS in a 1 mm path length quartz cuvette. Thermal denaturation was measured by observing signal changes at 222 nm during heating at a rate of 1°C/min. The melting temperature (Tm) was obtained by fitting to a sigmoidal dose–response (variable slope) equation.
SPR binding measurements
The binding profiles of FNfn10-α-lys, FN3con-α-lys and FN3con-α-lys.v2 were measured using SPR (BIAcore T-100, GE Healthcare). FN3 domains were captured (90 μl at 5 μl/min) on a NiNTA sensor chip (GE Healthcare) at a concentration of 2.5 μg/ml in HBS-EP+ (10 mM HEPES, 200 mM NaCl, 3 mM EDTA, 0.05% (v/v) Tween 20, pH 7.4). A serial 2-fold dilution series from 125 to 0.936 nM of lysozyme (Sigma Aldrich) in HBS-EP+ with 12 mg/ml CM-Dextran was injected at a flow rate of 50 μl/min. The NiNTA sensor chip was regenerated using regeneration buffer (HBS-EP+ with 350 mM EDTA) and the NiNTA chip was recharged with NiCl2 (2.5 mM in MiliQ H2O) before each new cycle. Binding curve analysis was performed in triplicate, and signals for FNfn10-α-lys, FN3con-α-lys and FN3con-α-lys.v2 were background subtracted from a flow channel containing wild type FN3con.
BLI binding measurements
The binding affinities of FNfn10-α-lys, FN3con-α-lys and FN3con-α-lys.v2 were measured using BLI (BLItz, ForteBio). Streptavidin biosensors were rehydrated and blocked for 1 h at RT in PBS containing 0.1% w/v bovine serium albumin. Runs were performed with all proteins diluted in 20 mM Tris 200 mM NaCl, pH 7.4. Sensors were first subjected to ‘on-line’ loading with a 50 μg/ml solution of biotinylated HyHEL10 anti-Lysozyme FAb. Sensors were incubated with 1 μM Hen Egg Lysozyme (Sigma) until a saturation of binding to HyHEL10 has been reported. After a 30 s baseline, association curves were obtained for up to 300 s with various concentrations of anti-HEL FN3 proteins. Dissociation curves were then obtained for up to 1200 s by transferring the sensors back to tubes containing buffer only. Runs were referenced against a run where no FN3 protein was added to the buffer for the association step. Global fits for the affinity of each FN3 protein were then obtained using the BLItz Pro 1.2.1.3 software (ForteBio).
Crystallization, X-ray data collection, structure determination and refinement
All crystals were grown using the hanging drop vapor diffusion method, with 1:1 (v/v) ratio of protein to mother liquor (500 μl well volume). For the FNfn10-α-lys-lysozyme complex, purified FNfn10-α-lys at a concentration of 0.5 mg/ml was mixed in a roughly 1:1 molar ratio with hen egg-white lysozyme (HEL) (Sigma Aldrich) for a total volume of 5 ml, incubated for 30 min at room temperature, then purified by SEC (Superdex 75 16/60) in 20 mM Tris, 200 mM NaCl, pH 7.4. Peak fractions corresponding to the size of an FNfn10-α-lys-lysozyme complex were concentrated to 8.6 mg/ml. Long thin needle like crystals formed in 10% polyethylene glycol (PEG) 6000, 0.1 M Bicine, pH 8.8 within 4 h. A single crystal was extracted and cryoprotected in 20% ethylene glycol, 12% PEG 6000, 0.1 M Bicine, pH 8.8 prior to collection.
For the unbound FN3con-α-lys loop graft, purified protein was concentrated to 29 mg/ml. Large plates were formed in 5% glycerol, 10% 2-propanol, 0.2 M zinc acetate, 0.1 M sodium cacodylate, pH 6.0 within 2 days. Crystals were dehydrated by stepwise equilibration of the crystallization drops over wells with progressively increasing concentration of glycerol and decreasing concentration of 2-propanol, with 24 h between transfers. The final reservoir solution contained 15% glycerol, 0.2 M zinc acetate, 0.1 M sodium cacodylate pH 6.0. Crystals were subjected to a final soak in 20% glycerol, 0.2 M zinc acetate, 0.1 M sodium cacodylate pH 6.0 for 20 min prior to data collection.
Data for both crystals were collected at 100 K at the Australian Synchrotron micro crystallography MX2 beamline. FNfn10-α-lys-lysozyme complex crystals diffracted to 2.54 Å resolution, and given the small size of these crystals, radiation damage became a significant problem. This was mitigated by collecting 4× 45° wedges along the crystal and later merging the wedges together. Crystals for the unbound FN3con-α-lys diffracted to 2.46 Å. Diffraction images were processed using iMosflm (Battye et al., 2011) and Aimless from the CCP4 suite (Winn et al., 2011). Each data set was initially processed in P1 and Laue group determination was achieved using Pointless within Aimless (Winn et al., 2011). Data sets were reintegrated, scaled and merged in their respective space group and 5% of each data set was flagged for calculation of Rfree, with neither a sigma nor a low-resolution cutoff applied to any data set. A summary of statistics is provided in Table I.
Structure determination proceeded using molecular replacement and the program PHASER (McCoy et al., 2007). A search model for the FNfn10-α-lys-lysozyme complex was constructed from the crystal structure of FNfn10 (PDB:1FNF) by removing solvent molecules and loops that lack homology, and from the crystal structure of HEL (PDB:4Z98) that had solvent molecules, acetate ion and hydrogen atoms removed. PHASER identified two complexes in the asymmetric unit, for a total of two HEL and two FNfn10-α-lys molecules. A single clear peak for both the rotation and translation functions was evident and the molecules packed well within the asymmetric unit. Model building was conducted using COOT (Emsley and Cowtan, 2004) and refined using Buster (Bricogne et al., 2011) and Phenix (Adams et al., 2010).
The FN3con-α-lys crystal was initially identified to be of the space group P22121, with pseudo-translational non-crystallographic symmetry (NCS). A molecular replacement search model for FN3con-α-lys was constructed from the crystal structure of FN3con (PDB:4U3H) by removing solvent molecules and loops that lack homology. PHASER identified four molecules in the asymmetric unit, however, electron density failed to align well with two of the four molecules, and refinement stalled with an Rfree ~0.4. We found that by lowering the symmetry of the space group to P21 (P1211) the crystal also exhibited twinning (twinned fraction of 0.48), which was not detected in P1 or P22121 (twin law of h, –k, –l). With these corrections, molecular replacement was repeated with PHASER, identifying eight molecules in the asymmetric unit (two tetramers). Initial electron density maps were significantly improved, with all eight chains fitting well. Model building was conducted in COOT (Emsley and Cowtan, 2004) and refinement using Buster (Bricogne et al., 2011) and Phenix (Adams et al., 2010).
Structural Analysis
Structural analysis of FNfn10-α-lys-lysozyme complex used chains A (lysozyme) and C (FNfn10-α-lys) in the asymmetric unit. Interface analysis was performed using the PDBePISA web server (Krissinel and Henrick, 2007). Shape complementarity calculations were performed using the Sc program (Lawrence and Colman, 1993) from the CCP4 software suite (Winn et al., 2011). Electrostatic surfaces in Supplementary Fig. S1 were produced by APBS (Baker et al., 2001) and all structures were visualized with PyMol v. 1.8.0.6 (DeLano, 2002).
Supplementary Material
Acknowledgements
We thank Harry Powell, Herman Schreuder, Mirko Velic, Blake Riley, Remy Robert, Andrew Ellisdon and the CCP4BB for helpful discussions and advice during the course of this research. We thank the Australian Synchrotron for beam-time and technical assistance. This work was funded by the Australian Research Council (DP150101371), and the Multi-modal Australian ScienceS Imaging and Visualisation Environment (MASSIVE) (www.massive.org.au). We acknowledge the Monash Macromolecular Crystallization Facility. A.M.B. and D.C. hold fellowships from the National Health and Medical Research Council (1022688, 1050146).
Author contribution
B.T.P. and A.M.B. designed the study. B.T.P. performed loop grafting and design. B.T.P., D.E.H., P.S. and M.R.H. performed protein expression, refolding and purification. B.T.P. performed the CD thermal melts and biophysical characterization. B.T.P., N.D. and S.M. performed the crystallography, data collection and structure determination. B.T.P. P.J.C. and P.S. performed the affinity characterization experiments. B.T.P. generated figures. B.T.P., D.E.H., M.R.H., R.V.L., D.C. and A.M.B. wrote the paper.
Supplementary data
References
- Adams P.D., Afonine P.V., Bunkóczi G. et al. (2010) Acta Crystallogr. D Biol. Crystallogr., 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F.H. (1998) Proc. Natl. Acad. Sci., 95, 2035–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold F.H., Wintrode P.L., Miyazaki K. and Gershenson A. (2001) Trends Biochem. Sci., 26, 100–106. [DOI] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J. and McCammon J.A. (2001) Proc. Natl. Acad. Sci., 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye T. G. G., Kontogiannis L., Johnson O., Powell H.R. and Leslie A. G. W. (2011) Acta Crystallogr. D Biol. Crystallogr., 67, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle B.M. and Shoichet B.K. (2002) J. Mol. Biol., 321, 285–296. [DOI] [PubMed] [Google Scholar]
- Beck A., Wurch T., Bailly C. and Corvaia N. (2010) Nat. Rev. Immunol., 10, 345–352. [DOI] [PubMed] [Google Scholar]
- Binz H.K., Amstutz P. and Plückthun A. (2005) Nat. Biotechnol., 23, 1257–1268. [DOI] [PubMed] [Google Scholar]
- Birch J.R. and Racher A.J. (2006) Adv. Drug Deliv. Rev., 58, 671–685. [DOI] [PubMed] [Google Scholar]
- Bricogne G., Blanc E., Brandl M., Flensburg C. and Keller P. (2011) BUSTER v. 2.10.2 Global Phasing Ltd.
- Chennamsetty N., Voynov V., Kayser V., Helk B. and Trout B.L. (2009) Proc. Natl. Acad. Sci. U.S.A., 106, 11937–11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. (2002) The PyMOL molecular graphics system.
- Demarest S.J. and Glaser S.M. (2008) Curr. Opin. Drug Discov. Dev., 11, 675–687. [PubMed] [Google Scholar]
- Dudgeon K., Rouet R., Kokmeijer I., Schofield P., Stolp J., Langley D., Stock D. and Christ D. (2012) Proc. Natl. Acad. Sci. U.S.A., 109, 10879–10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. and Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr., 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M.Y., Pieper U. and Sali A. (2007) Curr. Protoc. Protein Sci. Chapter 2, Unit 2.9. [DOI] [PubMed] [Google Scholar]
- Giver L., Gershenson A., Freskgard P.O. and Arnold F.H. (1998) Proc. Natl. Acad. Sci., 95, 12809–12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel B.J., Kapila A. and Dane Wittrup K. (2008) J. Mol. Biol., 381, 1238–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliger P. and Hudson P.J. (2005) Nat. Biotechnol., 23, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Koide A., Bailey C.W., Huang X. and Koide S. (1998) J. Mol. Biol., 284, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Koide A., Gilbreth R.N., Esaki K., Tereshko V. and Koide S. (2007) Proc. Natl. Acad. Sci., 104, 6632–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E. and Henrick K. (2007) J. Mol. Biol., 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Lawrence M.C. and Colman P.M. (1993) J. Mol. Biol., 234, 946–950. [DOI] [PubMed] [Google Scholar]
- Lipovsek D. (2011) Protein Eng. Des. Sel., 24, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C. and Read R.J. (2007) J. Appl Crystallogr, 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani R.A., Gonzalez A., Shoichet B.K., Brinen L.S. and Babbitt P.C. (2007) Biochemistry, 46, 6688–6695. [DOI] [PubMed] [Google Scholar]
- Nicaise M., Valerio-Lepiniec M., Minard P. and Desmadril M. (2004) Protein Sci., 13, 1882–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padlan E.A., Silverton E.W., Sheriff S., Cohen G.H., Smith-Gill S.J. and Davies D.R. (1989) Proc. Natl. Acad. Sci., 86, 5938–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.H., Chen Y., Danehy F. et al. (2005) Protein Eng. Des. Sel., 18, 435–444. [DOI] [PubMed] [Google Scholar]
- Porebski B.T., Nickson A.A., Hoke D.E., Hunter M.R., Zhu L., McGowan S., Webb G.I. and Buckle A.M. (2015) Protein Eng. Des. Sel., 28, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V., Krystek S.R., Bush A. et al. (2012) Structure, 20, 259–269. [DOI] [PubMed] [Google Scholar]
- Rouet R., Lowe D. and Christ D. (2014) FEBS Lett., 588, 269–277. [DOI] [PubMed] [Google Scholar]
- Sánchez I.E., Tejero J., Gómez-Moreno C., Medina M. and Serrano L. (2006) J. Mol. Biol., 363, 422–432. [DOI] [PubMed] [Google Scholar]
- Schreiber G., Buckle A.M. and Fersht A.R. (1994) Structure, 2, 945–951. [DOI] [PubMed] [Google Scholar]
- Serrano L., Day A.G. and Fersht A.R. (1993) J. Mol. Biol., 233, 305–312. [DOI] [PubMed] [Google Scholar]
- Sheriff S. (1993) Immunol. Methods, 3, 191–196. [Google Scholar]
- Shoichet B.K., Baase W.A., Kuroki R. and Matthews B.W. (1995) Proc. Natl. Acad. Sci., 92, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern L.A., Case B.A. and Hackel B.J. (2013) Curr. Opin. Chem. Eng., 2, 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna D.M. and Goldstein R.A. (2002) Proteins, 46, 105–109. [DOI] [PubMed] [Google Scholar]
- Tokuriki N. and Tawfik D.S. (2009) Curr. Opin. Struct. Biol., 19, 596–604. [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Stricher F., Serrano L. and Tawfik D.S. (2008) PLoS Comput. Biol., 4, e1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Lombardi R., Phan T.G., Zimmermann C., Lowe D., Jermutus L. and Christ D. (2015) Drug Discov. Today, 20, 1271–1283. [DOI] [PubMed] [Google Scholar]
- Walsh G. (2014) Nat. Biotechnol., 32, 992–1000. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D. et al. (2011) Acta Crystallogr. D Biol. Crystallogr., 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolot R.S., Basu S. and Million R.P. (2013) Nat. Rev. Drug Discov., 12, 259–260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.