Abstract
Limb girdle muscular dystrophy 2A is due to loss-of-function mutations in the Calpain 3 (CAPN3) gene. Our previous data suggest that CAPN3 helps to maintain the integrity of the triad complex in skeletal muscle. In Capn3 knock-out mice (C3KO), Ca2+ release and Ca2+/calmodulin kinase II (CaMKII) signaling are attenuated. We hypothesized that calpainopathy may result from a failure to transmit loading-induced Ca2+-mediated signals, necessary to up-regulate expression of muscle adaptation genes. To test this hypothesis, we compared transcriptomes of muscles from wild type (WT) and C3KO mice subjected to endurance exercise. In WT mice, exercise induces a gene signature that includes myofibrillar, mitochondrial and oxidative lipid metabolism genes, necessary for muscle adaptation. C3KO muscles fail to activate the same gene signature. Furthermore, in agreement with the aberrant transcriptional profile, we observe a commensurate functional defect in lipid metabolism whereby C3KO muscles fail to release fatty acids from stored triacylglycerol. In conjunction with the defects in oxidative metabolism, C3KO mice demonstrate reduced exercise endurance. Failure to up-regulate genes in C3KO muscles is due, in part, to decreased levels of PGC1α, a transcriptional co-regulator that orchestrates the muscle adaptation response. Destabilization of PGC1α is attributable to decreased p38 MAPK activation via diminished CaMKII signaling. Thus, we elucidate a pathway downstream of Ca2+-mediated CaMKII activation that is dysfunctional in C3KO mice, leading to reduced transcription of genes involved in muscle adaptation. These studies identify a novel mechanism of muscular dystrophy: a blunted transcriptional response to muscle loading resulting in chronic failure to adapt and remodel.
Introduction
Calpain 3 (CAPN3) belongs to a family of non-lysosomal, Ca2+-dependent cysteine proteinases, all of which share similar proteolytic domains, and are expressed either ubiquitously or tissue specifically (1). CAPN3 is expressed almost exclusively in skeletal muscle. Mutations in CAPN3 cause autosomal recessive limb girdle muscular dystrophy type 2A (LGMD2A) (2), one of the most frequently occurring forms of LGMD, characterized by muscle wasting leading to loss of ambulation about 10 years following clinical onset (3,4). Over 400 different pathogenic LGMD2A mutations have been identified, according to the Leiden Muscular Dystrophy database. The majority of CAPN3 mutations are missense mutations that are spread along the entire length of the gene. Interestingly, not all pathogenic mutations alter proteolytic properties of CAPN3 (5); and it remains unclear why many mutations in CAPN3 lead to LGMD2A.
We previously generated Capn3 knockout mice (C3KO) using a gene trap retroviral vector and demonstrated that these mice replicate several features of LGMD2A including decreased muscle mass and myofiber diameter, abnormal myofiber ultrastructure, mitochondrial dysfunction and low levels of inflammation/regeneration (6,7). Eosinophilic myositis and increased serum creatine kinase levels have been documented in young LGMD2A patients and represent an early and transient feature of LGMD2A (4). LGMD2A muscles usually display low levels of muscle regeneration (<3% on average) (8). Sparse inflammation and regeneration distinguish LGMD2A from other forms of muscular dystrophy. For example, in Duchenne muscular dystrophy and several other LGMDs, abnormalities in the dystrophin-glycoprotein complex cause membrane fragility and repeated cycles of inflammation, degeneration and regeneration that eventually lead to progressive replacement of muscle with fibrotic tissue and fat (9). In contrast, LGMD2A does not involve plasma membrane fragility; rather, calpainopathy stems from insufficiencies in muscle adaptation and growth (6,10,11). The specific role of CAPN3 and its link to deficits in muscle adaptation is still unknown.
One difficulty in discerning the underlying pathomechanism of LGMD2A resides in the lack of insight into the normal biological role of CAPN3. CAPN3 is a protease with no known consensus cleavage site and with numerous potential substrates. Moreover, it is located at several different cellular sites and may possess both proteolytic and non-proteolytic functions (12,13). A major pool of CAPN3 is associated with myofibrils, bound to titin. This pool of CAPN3 may play a role in releasing damaged proteins to facilitate myofibrillar protein turnover (14). It has been shown that CAPN3 mutations that interfere with titin binding are pathogenic, implying that the CAPN3-titin interaction is paramount for CAPN3 stability and function (15); however, not all of the cellular CAPN3 is bound to titin and regulation of myofibrillar turnover is not the only cellular role of CAPN3. A portion of cellular CAPN3 is concentrated at the triad (12), which is the site of Ca2+ release during excitation-contraction coupling. The triad comprised a complex of several proteins including the Ryanodine receptor (RyR), which is a calcium release channel. CAPN3 has been shown to biochemically interact with RyR and to maintain the integrity of the triad complex (12, 13) because loss of CAPN3 leads to decreased RyR concentration in both C3KO and LGMD2A patients, with concomitant reductions in Ca2+ release in C3KO muscle fibers (12,16,17).
Ca2+ transport plays a fundamental role in muscle adaptation by regulating several signaling pathways that control gene expression to allow muscle to adapt to increased functional and metabolic demands (18). We previously demonstrated that one of these signaling pathways, Ca2+-calmodulin kinase II (CaMKII), is blunted in C3KO muscles, along with a concomitant failure to activate the slow myogenic program (16). Activation of the slow myogenic program is only one adaptation induced by exercise. In addition, it is well known that exercise produces global alterations in skeletal muscle metabolism and mitochondrial biogenesis (19). Endurance exercise provokes increases in oxidative phosphorylation and preferential utilization of fatty acids (FAs) as an energy source.
Here, we test the hypothesis that failure of Ca2+ release and CaMKII signaling leads to downstream insufficiencies of transcription of genes involved in muscle adaptation in response to increased contractile activity. We survey the gene expression changes that occur in response to endurance exercise by RNAseq and identify an aberrant gene signature in muscles lacking CAPN3. We focus on three categories of genes that are severely attenuated in C3KO: myofibrillar, mitochondrial and genes involved in lipid metabolism, all of which are necessary for muscle adaptation. These findings have led to the hypothesis that a chronic failure to properly activate gene expression intended to accommodate muscle remodeling is an underlying feature of calpainopathy. This study is the first to suggest that failure to activate genes necessary for muscle adaptation is an underlying mechanism of muscular dystrophy.
Results
RNAseq analysis of transcriptional changes in exercised muscles
We previously demonstrated that both Ca2+ release and CAMKII signaling are blunted in muscles from C3KO mice (16,17). Since CaMKII-mediated signaling is associated with transcriptional regulation, we assessed transcriptional changes in response to endurance exercise, to determine whether blunted CAMK signaling led to consequences on downstream gene expression. Because CaMKII signaling is induced by increased contractile activity, we compared transcriptional profiles of wild type (WT) and C3KO muscles subjected to uphill running exercise. Histological assessment of muscles from exercised mice showed no significant difference in damage or inflammation between the two genotypes (Supplementary Material, Fig. S1). These results eliminate muscle damage and associated inflammation and regeneration as confounding factors for our gene expression analysis.
The total number of genes, differentially expressed in exercised C3KO muscles (compared to sedentary muscles of the same genotype), was lower than the number of genes differentially expressed in WT muscles (344 versus 430, respectively). In addition to making comparisons of total gene expression (i.e. expression of all known mRNA isoforms for a particular gene), we also made comparisons between individual mRNA isoforms (Supplementary Material, Table S1). We did not identify any global changes in splicing spectra as a result of running exercise or as a result of CAPN3 deficiency.
In WT muscles, we observed induction of several groups of genes involved in muscle adaptation such as myofibrillar, mitochondrial and metabolic; however, the same gene signature was not observed in C3KO muscles. The list of genes differentially regulated in C3KO mice after running is presented in Supplementary Material, Table S2.
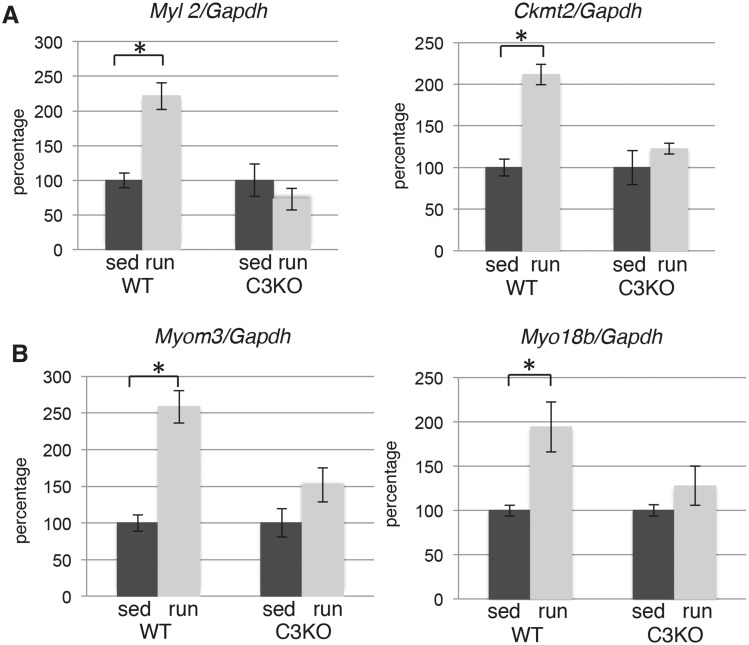
To validate the results of the RNAseq, we performed real-time polymerase chain reaction (PCR) evaluation of the expression levels of several individual genes shown to be significantly different by RNAseq. Figure 1 shows real-time PCR validation of four myofibril-associated genes identified in our current RNAseq analysis. The results confirmed that the ratio of WTrun/WTsed and KOrun/KOsed correlated well with our RNAseq data. Some of the genes that failed to activate in C3KO muscles are genes known to be preferentially expressed in slow-twitch myofibers (e.g. Myl2, Myom3 and Ckmt2), while others Myo18b are not preferential to slow fibers. Thus, these RNAseq data demonstrate that CAPN3-deficient muscles fail to properly induce both slow genes as well as other myofibrillar genes following an exercise stimulus.
Figure 1.
Exercise does not induce proper myofibrillar gene expression in C3KO muscles. RT-PCR validation of gene expression levels for sarcomere-associated proteins (Myl2—regulatory myosin light chain 2, cardiac, slow; Ckmt2—mitochondrial creatine kinase; Myom3—myomesin 3; Myo18b—myosin 18b) in plantaris muscles. In each graph, the expression level of the exercised (run) group is graphed relative to the sedentary (sed) controls; the average expression of each gene in the sedentary control group was set at 100% for each genotype. Asterisks indicate statistical significance by two-tailed Student’s t-test with P < 0.05 considered statistically significant. All groups had n = 4.
Exercise fails to induce genes of lipid metabolism in C3KO muscles
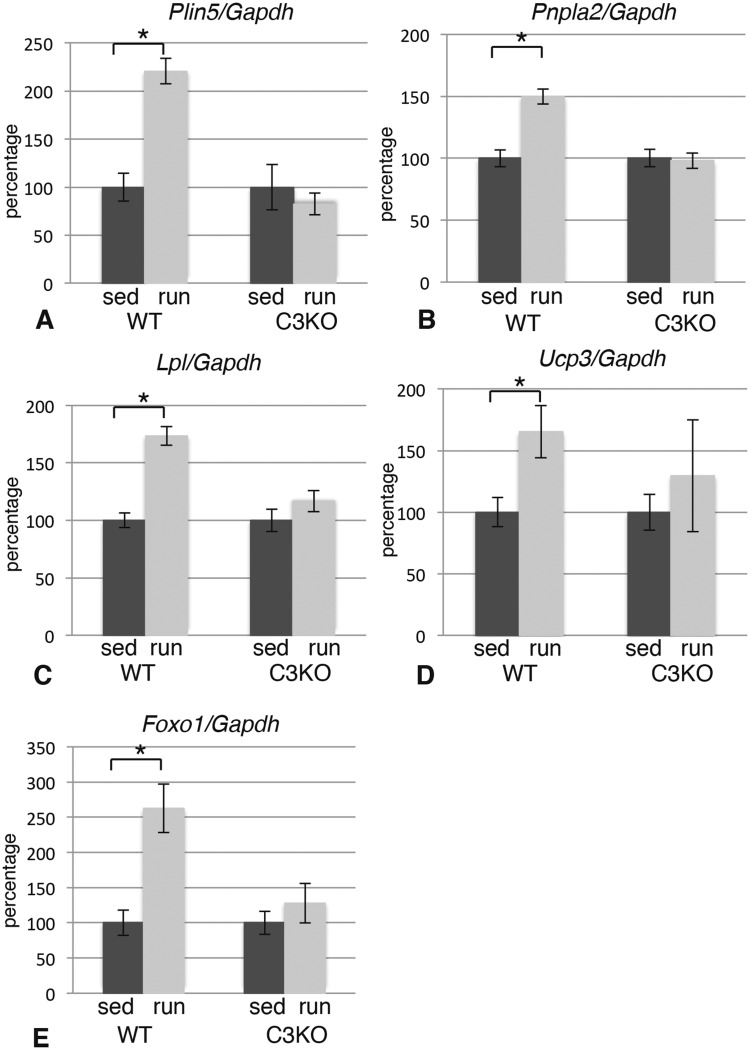
Running exercise prompts increased skeletal muscle utilization of FAs. In our study, we observed that the best defined and most numerous group of genes up-regulated in response to exercise in WT were genes encoding several key mediators of lipid metabolism and oxidative phosphorylation. As shown in Table 1, WT muscles demonstrated a significant increase in genes encoding key proteins/enzymes involved in release of free FA (lipoprotein lipase, Lpl), transport of FA across the sarcolemma (FA translocase, Cd36) and trafficking of FA inside the myofiber (FA binding protein 3, Fabp3) (see schematic in Fig. 5A). These increases were not observed in C3KO muscles following exercise. Several genes encoding proteins associated with lipid droplets (perilipin 3 and 5, Plin3 and Plin5) and enzymes involved in triglyceride metabolism (triglyceride lipase ATGL, Pnpla2; diacylglycerol acyltransferase, Dgat2) were also not properly induced by exercise in C3KO muscles. Furthermore, a large group of mitochondria-associated genes were not up-regulated in C3KO muscles, as compared to WT muscles (Table 1). These RNAseq data were validated by RT-PCR (Fig. 2A–D) confirming that genes involved in lipid metabolism are not appropriately up-regulated in C3KO muscles in response to exercise.
Table 1.
List of lipid metabolism and mitochondrial genes that were not properly induced by 5 days of running exercise in C3KO mice
| Gene | Fold changea, WTrun versus WTsed | Fold changea, KOrun versus KOsed | Protein, function |
|---|---|---|---|
| Cd36 | 1.48 | −1.08 | FA translocase |
| Pnpla2 | 1.54 | 1.01 | Enzyme that catalyzes the first step in the hydrolysis of triglycerides |
| Fabp3 | 1.69 | −1.24 | FA binding protein 3, muscle and heart |
| Plin3 | 1.79 | −1.16 | Perilipin 3, lipid droplets |
| Plin5 | 2.58 | −1.49 | Perilipin 5, lipid droplets |
| Lpl | 1.59 | −1.13 | Lipoprotein lipase, triglyceride metabolism |
| Cfd | 1.93 | −1.33 | Complement factor D (adipsin), lipid metabolism |
| C3 | 2.08 | 1.05 | Complement component 3, control of lipid metabolism |
| Dgat2 | 2.27 | 1.04 | Enzyme, catalyzes the final reaction in the synthesis of triglycerides |
| Cidec | 2.41 | 1.04 | Promotes lipid droplet formation in adipocytes |
| Per2 | 1.87 | 1.13 | Period circadian clock 2, controls lipid metabolism |
| Bdh1 | 2.68 | 1.18 | Short-chain dehydrogenase/reductase |
| Ucp3 | 1.48 | 1.05 | Uncoupling protein 3 (mitochondrial, proton carrier) |
| Nceh1 | 1.70 | 1.01 | Neutral cholesterol ester hydrolase 1 |
| Zbtb16 | 1.90 | 1.05 | Transcription factor, regulates bioenergetics |
| Mid1ip1 | 2.64 | 1.05 | Activates AcetyCoA carboxylase that is involved in FA synthesis |
| Car4 | 1.93 | 1.09 | Carbonic anhydrase 4 |
| Retsat | 1.87 | 1.10 | Retinol saturase, promotes adipogenesis |
| Cfh | 2.09 | 1.17 | Complement factor H, acts as a cofactor for cleavage of C3b |
| Adh1 | 1.83 | 1.15 | Alcohol dehydrogenase ADH1, positive effect on FA oxidation |
| Cyp1a1 | 3.08 | 1.12 | Cytochrome P450 1A, synthesis of cholesterol and other lipids |
| Cyp2e1 | 3.59 | 1.28 | Cytochrome P450 2E1 |
| Cyp2f2 | 2.76 | −1.15 | Cytochrome P450, family 2, subfamily f, polypeptide 2 |
| Slc39a14 | 2.13 | 1.32 | Solute carrier, metal ion transporter |
| Slc7a8 | 2.40 | 1.33 | Solute carrier, cationic amino acid transporter |
| Slc26a10 | 2.46 | 1.13 | Solute Carrier, multifunctional anion exchange |
| Slc45a3 | 2.07 | 1.10 | Solute carrier, Putative sugar transporter |
| Slco3a1 | 1.63 | 1.02 | Solute carrier organic anion transporter family, member 3A |
| Slc25a30 | 2.10 | −1.04 | SLC25 family of mitochondrial carrier proteins |
| Slc25a34 | 1.84 | −1.14 | SLC25 family of mitochondrial carrier proteins |
aFold change was calculated as the higher value always divided by the lower value. If a gene is down-regulated, the value is negative; if a gene is up-regulated, the value is positive.
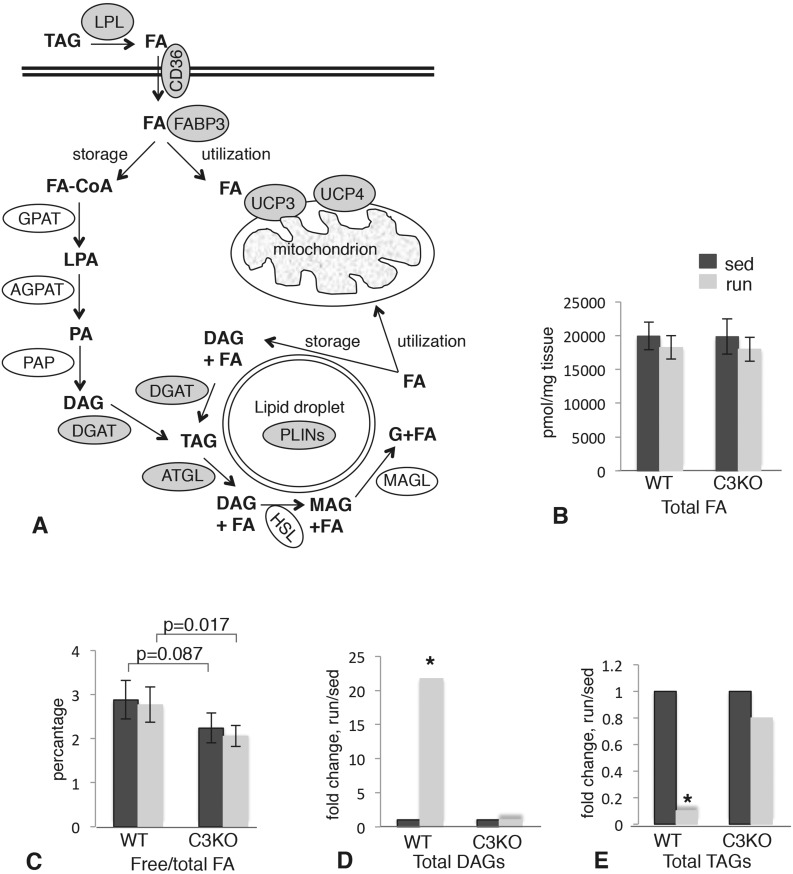
Figure 5.
Mass spectrometric analysis of FA metabolites in WT and C3KO muscles. (A) Diagram summarizing lipid metabolic pathways in skeletal muscle. All of the genes that were induced by exercise in WT but not in C3KO muscles are shown in gray ovals. LPL, lipoprotein lipase; FABP3, FA binding protein 3; GPAT, glycerol-3-phosphate acyltransferase; AGPAT, acyl-CoA:acylglycerol-3-phosphate acyltransferase; PAP, phosphatidic acid phosphohydrolase; DGAT, diacylglycerol acyltransferase; ATGL, adipose triglyceride lipase; HSL, hormone-sensitive lipase; MAGL, monoacylglycerol lipase; PLINs, perilipins; UCP3 and 4, uncoupling proteins 3 and 4; DAG, diacylglycerol; MAG, monoacylglycerol; G, glycerol; FA-CoA, fatty acyl-CoA; LPA, lysophosphatidic acid; PA, phosphatidic acid. (B) Total FA content in sedentary and run muscles; (C) pool of free FA in in sedentary and run muscles; (D) total DAGs in sedentary and run muscles; (E) total TAGS in sedentary and run muscles. Asterisk indicates statistical significance by t-test (P < 0.05). All groups had n = 5.
Figure 2.
Exercise does not properly induce expression of lipid metabolism genes in C3KO muscles. RT-PCR validation of the expression levels of genes encoding proteins involved in lipid metabolism and mitochondrial oxidation (Plin5—perilipin 5; Pnpla2—diacylglycerol acyltransferase, ATGL; Lpl—lipoprotein lipase; Ucp3—uncoupling protein 3; Foxo1—transcription factor FOXO1) in plantaris muscles. In each graph, the expression level of the exercised (run) group is graphed relative to the sedentary (sed) control; the average expression of each gene in the sedentary control group was set at 100% for each genotype. Asterisk indicates statistical significance by two-tailed Student’s t-test with P < 0.05 considered statistically significant. All groups had n = 4.
Next, we examined expression of FOXO transcription factor genes, since both FOXO1 and FOXO3 play important roles in regulation of energy metabolism in skeletal muscles. FOXO1 shifts fuel utilization toward lipid instead of glucose under stress conditions such as fasting or exercise (20). Our RNA seq analysis showed that both Foxo genes were up-regulated in WT muscles after 5 days of running exercise. RT-PCR evaluation confirmed that Foxo1 increased by 2.5–3-fold in WT muscles; however, this change was not observed in C3KO muscles (Fig. 2E). Thus, several metabolic adaptations to exercise were blunted in C3KO muscles compared to WT muscles.
We also compared WT and C3KO transcriptomes under sedentary conditions to determine whether levels of these metabolic genes might be elevated in C3KO muscles at rest. We identified 13 down-regulated and 21 up-regulated genes related to metabolism/mitochondria in C3KO compared to WT muscles (Supplementary Material, Table S3); however, the baseline levels of lipid metabolism genes induced by exercise in WT muscles (Table 1) were not elevated in sedentary C3KO muscles compared to sedentary WT muscles. Thus, the failure of C3KO to increase the gene signature in response to an exercise stimulus is not related to elevated gene expression at baseline.
Identification of molecular pathways affected by the absence of CAPN3
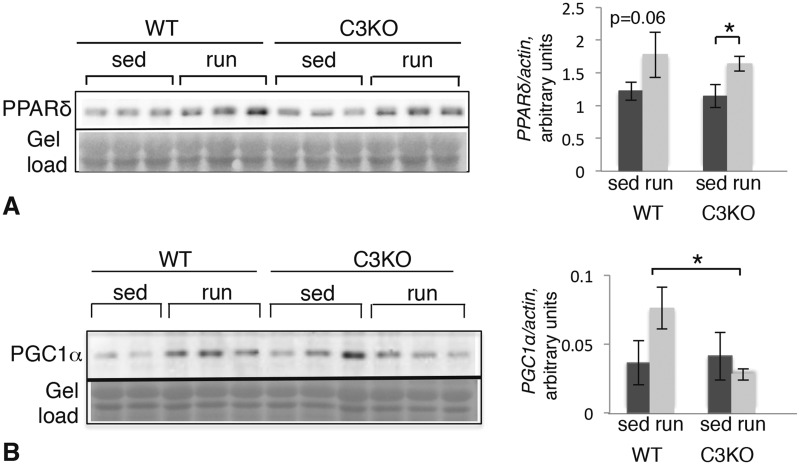
Many genes involved in lipid metabolism are controlled by peroxisome proliferator-activated receptor (PPAR) transcription factors (21). We tested whether a deficiency of PPAR expression might underlie the defective induction of lipid metabolism genes in C3KO muscles. As demonstrated by RNAseq, expression of Ppar genes did not change significantly with exercise. Pparδ is the most highly expressed Ppar gene in skeletal muscle (Supplementary Material, Fig. S2A). Although running did not increase PPARδ mRNA levels, it increased PPARδ protein in both WT and C3KO muscles (Fig. 3A), suggesting that misregulation of PPARδ cannot explain the reduction in PPARδ target genes in C3KO muscles.
Figure 3.
Expression of PPARδ and PGC1α in WT and C3KO muscle after 5 days of running exercise. Western blotting of whole muscle extracts from sedentary (sed) and exercised (run) animals. (A) Blot probed with a PPARδ-specific antibody. Quantification of the blot by densitometry is shown to the right. (B) Blot probed with a PGC1α-specific antibody. Quantification by densitometry is shown to the right. All vertical bars indicate standard error. Asterisk indicates statistical significance by two-tailed t-test with P < 0.05 considered significant.
PPARδ is activated by free FA and this transcription factor co-operates with PGC1α to induce transcription of target genes. In fact, PGC1α works in conjunction with several transcription factors (e.g. PPARs and MEF2), to coordinate the transcriptional response to increased muscle loading (19). As shown in Supplementary Material, Figure S2B, expression of the Ppargc1 gene, which encodes PGC1α, is increased with exercise in both WT and C3KO mice; however, western blotting revealed that PGC1α protein only increased in WT muscles but not C3KO muscles (Fig. 3B). Thus, failure to activate or maintain proper levels of PGC1α protein may underlie the reduction in expression of PPARδ-responsive genes in C3KO.
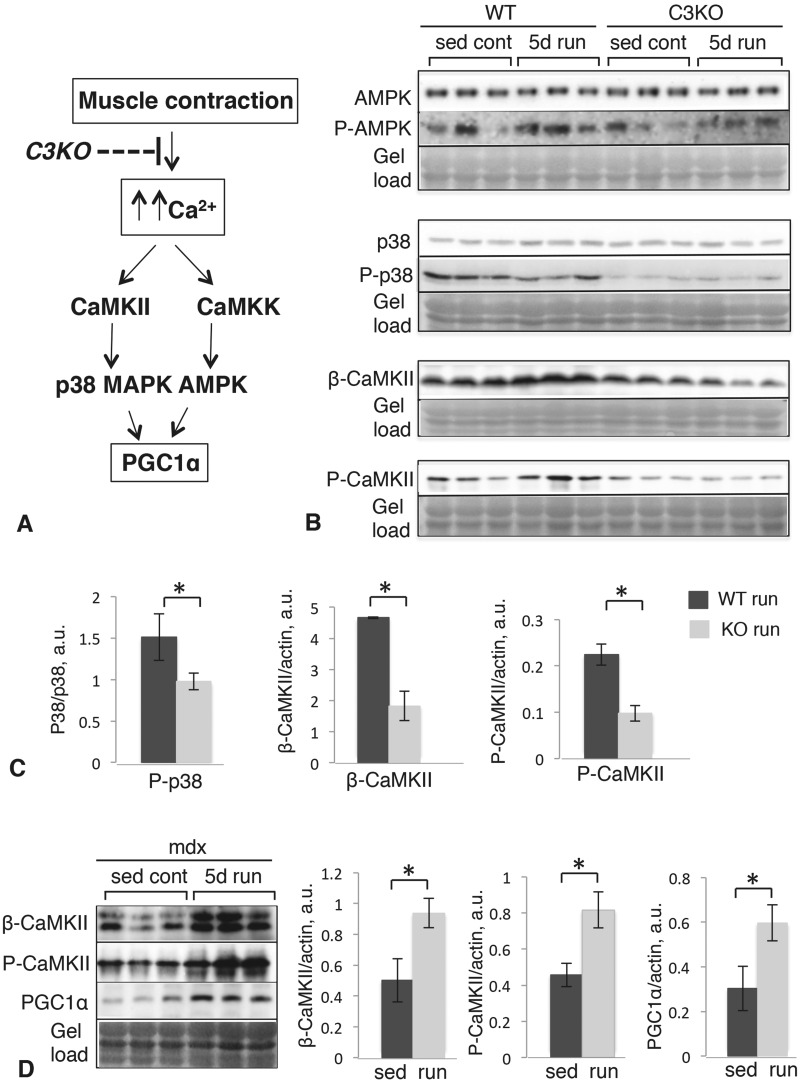
To gain additional insight into why PGC1α protein levels are not elevated in exercised C3KO muscles, we examined upstream regulatory kinases of PGC1α. PGC1α activity and stability are mediated by phosphorylation; and two protein kinases, p38 MAPK and AMP-activated kinase (AMPK), have been implicated in its phosphorylation (22,23). The pathways controlling PGC1α in skeletal muscle are shown in Figure 4A (adapted from (24)). Both p38 and AMPK are themselves activated by phosphorylation, so we assessed the ratio of their phosphorylated (activated) forms relative to the total amount of each kinase in each mouse genotype. Western blotting (Fig. 4B and C) revealed that phospho-AMPK and total AMPK levels were not different between WT and C3KO muscles; however, the ratio of phospho-p38/total p38 was significantly lower in C3KO muscles. Calcium calmodulin kinase signaling (CaMKII) is a pathway implicated in activation of p38 MAPK during muscle contraction. We previously demonstrated attenuated CaMKII signaling in C3KO muscles (16). In this study, we observed a similar reduction in CaMKII and P-CaMKII between WT and C3KO muscles, suggesting that reductions in CaMKII likely underlie the reductions in p38 phosphorylation (Fig. 4B and C). These data support the hypothesis that destabilization of PGC1α in C3KO is attributable to decreased activation of p38 MAPK, via diminished CaMKII signaling.
Figure 4.
Western blot analysis of the upstream components of the pathways controlling PGC1α levels in WT and C3KO muscles. (A) Schematic representation of the molecular pathways controlling PGC1α. (B) Western blots of whole plantaris muscle extracts stained with antibodies to total AMPK and phospho-AMPK (top), total p38 and phospho-p38 (middle), β-CaMKII and phospho-CaMKII (bottom). (C) Quantification of the corresponding blots by densitometry; asterisk indicates statistical significance by t-test (P < 0.05). (D) Western blots of whole plantaris muscle extracts of sedentary and exercised mdx muscles stained with antibodies to β-CaMKII (top), phospho-CaMKII (middle) and PGC1a (bottom). Quantification of the corresponding blots by densitometry is shown to the right; asterisk indicates statistical significance by t-test (P < 0.05). Vertical bars represent standard error.
To validate the specificity of our results for calpainopathy, we also tested this pathway in one of the best-characterized mouse models of muscular dystrophy, the mdx mouse, which carries a mutation in the Dmd gene and is a model of Duchenne muscular dystrophy. For these experiments, mdx mice were subjected to the same running protocol used in our study, and then their muscles were examined for the expression of β-CaMKII, P-CaMKII and PGC1α by western blotting. The results demonstrated that all three of these proteins showed proper induction by exercise in mdx muscles, in contrast to C3KO muscles. These results are shown in Figure 4D. Thus, these data suggest that the inability to up-regulate β-CaMKII, P-CaMKII and PGC1α in response to exercise are specific features of calpainopathy and are not a generalized feature of dystrophic muscles.
Functional validation of RNAseq data by lipidomics
To investigate whether the aberrant gene expression of lipid metabolism genes observed in C3KO translates to functional changes in lipid composition, we performed a comprehensive lipidomic analysis by mass spectrometry (MS). Figure 5A shows a schematic overview of the proteins involved in skeletal muscle lipid metabolism (25). Since FAs circulate in a conjugated form, they need to be released by lipoprotein lipases and transported into the muscle fiber, where they can be directed to the mitochondria for immediate utilization or converted to triacylglycerol (TAG) for storage in lipid droplets. The process is reversible and TAGs can be broken down to release free FA when needed.
Lipidomic analysis did not reveal any differences in the total FA content between WT and C3KO muscles in sedentary versus running conditions (Fig. 5B); however, the pool of free FA was diminished in C3KO muscles (Fig. 5C). The data are consistent with the observed reduction in expression of the genes encoding lipases (such as LPL and ATGL) and those involved in the transport of free FA across the sarcolemma (e.g. FA translocase CD36).
Free FAs represent a very small fraction of total FAs, as they are quickly consumed for energy production and do not accumulate in the cell. Thus, the change in free FA itself is not the best indicator of compromised lipid metabolism. However, we also observed significant differences in the TAG/DAG pools in C3KO compared to WT muscles (Fig. 5D and E), and these differences correlated well with the RNAseq data. Exercise induces rapid hydrolysis of TAG to DAG to release FA for immediate utilization; this step is catalyzed by ATGL (adipose TAG lipase). As we showed in Fig. 2, expression of the Pnpla2 gene, encoding ATGL, was induced by exercise in WT but not in C3KO muscles. In agreement with this observation, exercise induced a 20-fold increase in total DAGs and an approximately 10-fold decrease in total TAGs (Fig. 5D and E). No significant changes in TAG/DAG pools were observed in C3KO muscles after running exercise. Thus, these data support the hypothesis that failure to induce lipid metabolism genes in C3KO muscles leads to aberrant lipid metabolism in response to exercise.
Compensatory up-regulation of glycolytic enzymes in C3KO muscles
To examine whether the observed abnormalities in lipid metabolism in C3KO muscles were associated with a compensatory switch to glycolytic metabolism, we assayed the activities of several glycolytic enzymes. Two muscles were used: the diaphragm, which is one of the few mouse muscles containing both slow and fast twitch muscle fibers, and the tibialis anterior, which is primarily made up of fast-twitch muscle fibers. This analysis revealed higher glycogen phosphorylase activity in both of these muscles from C3KO mice (Fig. 6A). This enzyme catalyzes the release of glucose-1-phosphate from glycogen that is the rate-limiting step of glycogen breakdown. Furthermore, activities of the glycolytic enzymes, lactate dehydrogenase and aldolase, were also higher in C3KO muscles (Fig. 6B and C). These data suggest that the glycolytic pathway is up-regulated in C3KO muscles to compensate for defects in lipid metabolism.
Figure 6.
Activities of glycolytic enzymes are increased in C3KO muscles. Activities of glycogen phosphorylase (A), lactate dehydrogenase (B) and aldolase (C) were measured in tibialis anterior (TA) and diaphragm muscles from WT and C3KO mice. All enzyme activities are expressed as units per min per gram of wet muscle weight. Asterisk indicates statistical significance by two-tailed Student’s t-test with P < 0.05 considered statistically significant. All groups had n = 5.
C3KO mice demonstrate decreased exercise endurance compared with WT mice
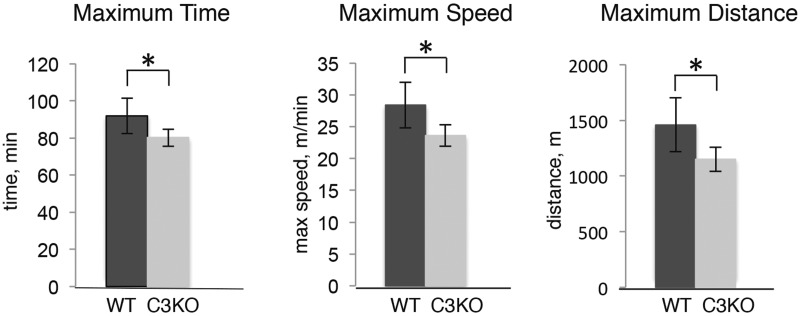
To investigate whether the impaired lipid metabolism resulted in commensurate functional deficits, the endurance of C3KO mice was evaluated in a run to exhaustion test. Mice were subjected to a running protocol that lasted approximately 90 min (60 min at low speed followed by gradual increase in speed every 5 min until a mouse stops from exhaustion). As shown in Figure 7, C3KO mice were unable to run for the same amount of time as WT mice or achieve the same maximum speed or distance as WT mice. Thus, endurance of C3KO muscles was decreased in accordance with the gene expression and metabolic changes we observed in their muscles.
Figure 7.
C3KO mice show decreased exercise performance. Results of the run to exhaustion test, performed as described in Materials and Methods section. All mice were age-matched males (n = 5 for each genotype). Asterisk indicates statistical significance by t-test (P < 0.05). Vertical bars represent standard errors.
Discussion
The studies reported here extend upon our previous findings of blunted Ca2+ release and CaMKII signaling in C3KO muscles. Those prior findings led us to hypothesize that reductions in Ca2+-mediated signaling might result in an impairment of CAPN3-deficient muscles to sense changes in loading and to subsequently respond with appropriate changes in gene expression necessary for adaptation (16). Here we test this hypothesis by performing an unbiased comparison of gene expression profiles of muscles isolated from C3KO and WT mice that had been subjected to an exercise protocol. For these studies, we used RNAseq because it offers several advantages over gene arrays: it has enhanced sensitivity, larger dynamic range and allows for highly accurate and quantitative comparisons independent of transcript abundance (26). Moreover, by challenging muscles with an exercise stimulus, gene expression changes could be examined within a genotype (WTsed versus WTrun, C3KOsed versus C3KOrun). Using this approach, we identified more than 400 differentially expressed genes that were induced by exercise in plantaris muscles of WT mice. By comparison, only 15 genes and ESTs were identified by microarray analysis in similar studies by other laboratories (27). In our investigation, RNAseq revealed that C3KO mice showed significant alterations in their transcriptional response: they failed to activate expression of genes related to lipid metabolism, energy production and sarcomere maintenance.
In general, the specific type of adaptation induced in skeletal muscle depends on the type of exercise training to which it is subjected. For example, resistance exercise (high intensity, short duration) stimulates fast twitch muscle fibers and glycolytic metabolism, while endurance exercise (long duration, low intensity, such as treadmill running at moderate speed) induces slow twitch muscle phenotype and oxidative metabolism (28). Our data revealed blunted activation of genes of the slow phenotype and those involved in oxidative metabolism. Analysis of global changes in FA metabolites by MS showed significantly decreased levels of free FA in C3KO muscles compared to WT muscles. Furthermore, analysis of total TAGs and DAGs revealed impaired lipid metabolism in C3KO muscles. These results are in agreement with our RNAseq data, demonstrating an inability for C3KO muscle to activate expression of genes involved in lipid metabolism (Table 1) in response to exercise.
The lipidomic analyses conducted in this investigation also showed an abnormal accumulation of total DAGs and a decrease in total TAGs in sedentary C3KO muscles compared to sedentary WT controls (Supplementary Material, Table S4). Interestingly, these changes did not correlate with differences in the expression of key genes involved in lipid metabolism under sedentary condition. As shown in Supplementary Material, Table S5, none of the genes normally up-regulated upon exercise (a condition that leads to an increase in total DAGs and a decrease in total TAGs) were notably elevated in sedentary C3KO muscles. Thus, excessive accumulation of DAGs in sedentary C3KO muscles is not likely caused by abnormal induction of lipid metabolism genes. Alternatively, elevated DAG content in C3KO muscles could be explained by mitochondrial dysfunction. In this scenario, accumulation of lipid substrates might occur if the mitochondria are unable to utilize substrates for energy production (29). Our prior studies showed that C3KO mitochondria have ultra-structural and functional abnormalities (7). Thus, it is possible that DAG accumulation in the sedentary condition may result from impaired mitochondrial function and an inability to efficiently utilize lipids for ATP production. Taken together, data presented in this article and in our prior studies strongly suggest that C3KO muscles experience deficits in oxidative metabolism. It is noteworthy that Siciliano et al. (30) recently reported a significantly reduced PCr recovery rate in LGMD2A at the end of the exercise in comparison with LGMD2B subjects and healthy controls, suggesting that altered oxidative metabolism is a common feature of C3KO and LGMD2A muscles.
There are several molecular sensors that detect skeletal muscle energy requirements (31). One of them, AMPK, senses the ratio of AMP/ATP, which normally increases in actively contracting muscle. The other metabolite that accumulates during exercise, and which may act as a messenger between muscle contractile activity and gene transcription, is free FA (21). Free FA serves as a ligand for PPARδ, a transcription factor that regulates many lipid metabolism genes. Although we did not find any evidence of decreased expression or activation of AMPK or PPARδ in exercised C3KO muscles, we observed a decrease in PGC1α, which is a co-activator for PPARs and several other transcription factors and nuclear receptors including MEF2, ERRα, HRFs and FOXO (19,28). Even a single bout of exercise induces transcription of PGC1α (32), emphasizing its important role in coordinating the adaptation response. PGC1α is also regulated by post-translational modifications including phosphorylation, acetylation, methylation and ubiquitination (22,23,33). p38 MAPK is a kinase important for stabilizing PGC1α by phosphorylation (23) and we found that levels of both activated p38 and PGC1α were decreased in C3KO muscles. Taken together with our previous studies demonstrating reduced CaMKII signaling (16), these data suggest that the molecular pathway involving CaMKII-mediated activation of p38 MAPK and phosphorylation of PGC1α is defective in C3KO muscles and is likely responsible for deregulation of genes driving muscle adaptation.
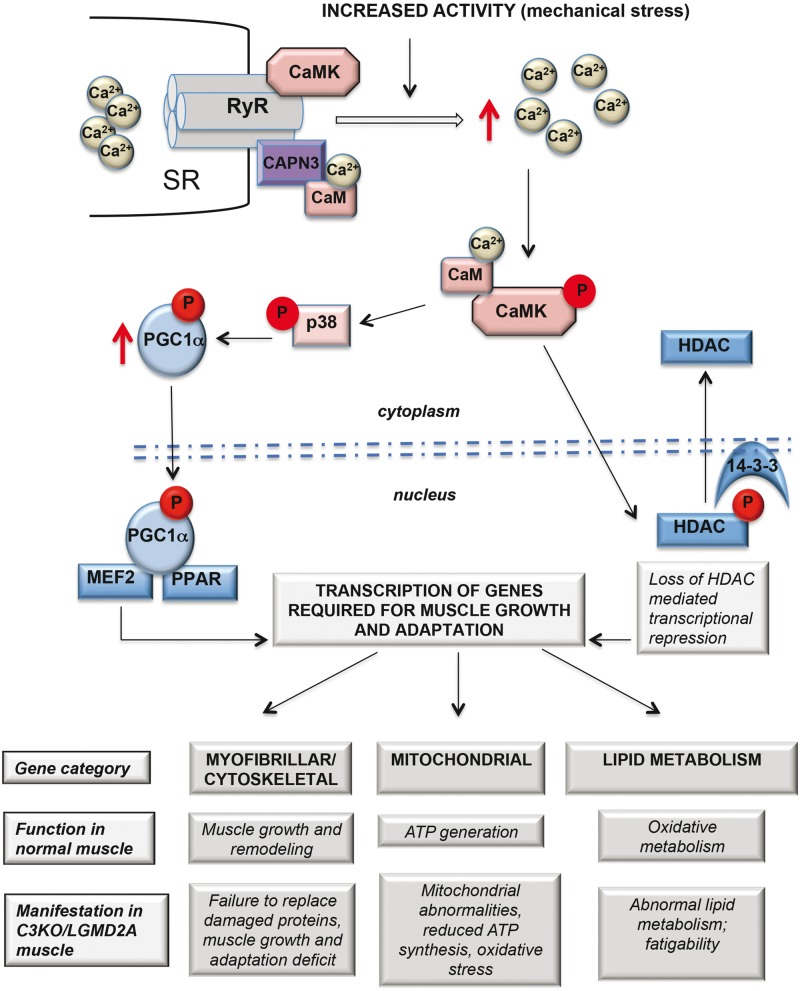
Figure 8 summarizes our current hypothesis about the relationship between CAPN3, Ca2+-mediated signaling via CaMKII and downstream targets. We believe that CAPN3 preserves the integrity of the triad complex and may serve as a sensor of muscle loading, transmitting downstream signals via CaMKII to induce transcription of genes in response to increased contractile activity. In our previous article (16), we demonstrated that expression of slow myofiber-associated genes (controlled by Mef2) is attenuated in C3KO muscle; current research identifies another branch of the pathway that affects expression of lipid metabolism genes corroborating our hypothesis of blunted CaMKII signaling in C3KO muscle. Even though it is understandable how altered CaMKII signaling might lead to the observed transcriptional changes and to pathogenic outcomes, there is a gap in our understating of the exact role for CAPN3 in this process, especially regarding CAPN3’s proteolytic function. Interestingly, similar to CaMKII, CAPN3 itself is activated by Ca2+-calmodulin (34) but so far none of the triad-associated proteins that we have examined appears to behave as a CAPN3 substrate (i.e. accumulated in the muscle deficient of CAPN3 proteolytic activity). Future studies will be focused on identifying CAPN3 substrates and binding partners that are present at the triad to provide insights into its role in these proposed activities.
Figure 8.
Proposed hypothesis of regulation of muscle adaptation genes mediates by Ca2+-CaMKII-mediated signaling. According to the hypothesis, CAPN3 (shown in purple) is associated with the muscle triad through its interaction with RyR1. In WT mice, muscle activity leads to increased Ca2+ release through RyR and concomitant activation of CaMKII signaling, which acts (1) to activate p38 MAPK, which stabilizes the transcriptional co-activator PGC1α, leading to increased transcription of muscle adaptation genes controlled by MEF2, PPAR and, possibly, other transcription factors and (2) to phosphorylate transcriptional inhibitor HDAC, leading to its relocation from the nucleus to the cytoplasm, thus alleviating transcriptional repression. As was demonstrated here and in our previous studies (12, 16), in the absence of CAPN3 the levels of RyR and the amplitude of Ca2+ release are reduced leading to decreased activation of CaMKII. As a result, both branches of downstream events are blunted leading to an inability to up-regulate transcription of genes, necessary for muscle adaptation. Possible physiological and pathogenic consequences of impairments in the proposed pathways are presented below the schematic.
In conclusion, the most significant finding of this study is that CAPN3-deficient muscles fail to up-regulate a set of genes associated with muscle adaptation. These data are in agreement with our hypothesis that blunted Ca2+-CaMKII mediated signaling in C3KO muscles hinders the ability of CAPN3-deficient muscles to adapt to changes in muscle loading. The data may relate to several of our prior findings in the C3KO and those of others in LGMD2A patients such as mitochondrial abnormalities (7,35), growth failure (10), sarcomere disorganization (6,36), abnormal energy production (7), etc. Thus, the finding of blunted CaMKII signaling and aberrant transcription in response to muscle loading may provide a unifying explanation for many phenotypic observations in the literature that were previously disjointed. A chronic failure to adapt and remodel in LGMD2A may ultimately lead to a muscle that cannot bear the loads placed on it, leading to myofiber degeneration and eventual muscle wasting. These findings are the first to suggest failure to activate genes necessary for muscle adaptation as a mechanism of muscular dystrophy.
Materials and Methods
Animals
All experimental protocols and the use of animals were in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals and approved by the UCLA Institutional Animal Care and Use Committee. C57 BL/6 mice were obtained from the Jackson Laboratories. C3KO mice were previously described (6) and they are congenic to C57/BL6.
Treadmill exercise
Male mice (n = 6 for each genotype) were accustomed to the treadmill (Columbus Instruments) with a 15 min run once per day at 10 m/min for 3 days. For the next 5 days, mice were subjected to forced running exercise performed on a 10% positive incline at 10 m/min for the first 20 min followed by 40 min at 15 m/min. At the end of the experiment, mice were sacrificed and soleus, plantaris and medial gastrocnemius muscles were collected for analyses. Age-matching males (n = 6 for each genotype) that were not subjected to exercise were used as sedentary control.
For the run to exhaustion experiment, mice were run at 10 m/min for the first 10 min (0–10 min), at 12 m/min during 10–30 min and 14 m/min during 30–60 min. After 60 min, the speed was increased by 2 m/min every 5 min until a mouse was unable to perform. All mice used for these experiments were males (n = 6 per group for each genotype), aged 5–6 months.
RNAseq
RNA was isolated from the plantaris muscle using RNeasy Fibrous Tissue Mini Kit (Qiagen) according to the manufacturer’s recommendations. RNA sequencing libraries were constructed with the TruSeq RNA Sample Prep Kits v.2 (Illumina). Eight RNA samples were indexed with different adapters and pooled for paired-end 100 bp sequencing on two lanes of Illumina HiSeq2000. RNAseq reads were aligned with TopHat v.2.0.2 (37) to the mouse genome version mm9. The average TopHat alignment rate was 72.54%, resulting in an average of 29.6 million reads per sample. Transcripts were assessed by Cufflinks (v.2.0.0) (38), using a GTF file based on Ensembl Mus_musculus_NCBI37. Differentially expressed genes were found by Cuffdiff, with significant genes satisfying a threshold of FDR multiple testing correction value less than 0.05.
Real-time PCR
cDNA was generated using iScript Reverse Transcriptase Supermix (Bio-Rad) and was used for real-time PCR with ITaq Universal SYBR Green Supermix (BioRad) according to manufacturer’s instructions. All real-time PCR reactions were run in CFX Connect Real-Time PCR System (Bio-Rad). Primers for real-time PCR were selected to span intron–exon junctions (when possible) and were first tested in regular PCR amplification to ensure the production of a single band in each case. The primer pairs are presented in Table 2.
Table 2.
Primers used for real-time PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Myl2 | AGTTCAAGGAAGCCTTCACAATC | ATTGGACCTGGAGCCTCTTTGAT |
| Ckmt2 | ATAGGCAGAAGGTATCTGCTGATG | GTGTCATCTTGTTTCGGAGTTTGG |
| Actc1 | CCCCTAGCACGCCTACAGAACC | TGTCCCATACCCACCATGACAC |
| Myom3 | CGGCAAGTACCGTATCACCA | ATCTCCGAGTCAAAGCCAGC |
| Myo18 | TGAGGTCGTCATGGAAAGGC | CCAGACTTTCTGTGCCTCGT |
| Plin5 | CAAGTCGGAGAAGCTGGTGG | GACCCCAGACGCACAAAGTA |
| Pnpla2 | CTCACATCTACGGAGCCTCG | CCAGGTTGAAGGAGGGATGC |
| Lpl | TCGTCATCGAGAGGATCCGA | TGTTTGTCCAGTGTCAGCCA |
| Ucp3 | CGGACCTCCTCACTTTTCCC | TCTGTGCGCACCATAGTCAG |
| Foxo1 | GCCGCCACATTCAACAGGC | GAGCCTCCGAGTTCGTGCC |
Western blot analysis
For western blot analysis, plantaris muscles were homogenized in reducing sample buffer (80 mm Tris–HCl [pH 6.8], 0.1 M dithiothreitol, 2% sodium dodecyl sulphate and 10% glycerol) and Halt phosphatase and protease inhibitor cocktail (Thermo Scientific) using a Dounce homogenizer. An equal amount of total protein for each sample was loaded on sodium dodecyl sulphate-PAAG followed by transfer to nitrocellulose membrane. Ponceau red was used to verify the transfer onto the nitrocellulose. The following primary antibodies were used for western blotting: anti-AMPK, anti-phospho (T172)—AMPK, anti-p38 and anti-phospho (T180/Y182) p38 (Cell Signaling), anti-β-CaMKII (Life Technologies), phospho-CaMKII (Thermo Scientific), anti-PGC1α (Santa Cruz) and anti-PPARδ (Aviva Systems Biology). Secondary antibodies conjugated with HRP were from Sigma-Aldrich. Specific signals were developed using ChemiGlow chemiluminescent substrate for HRP (Protein Simple). Images of the blots were acquired using FluorChem FC2 Imager (Alpha Innotech). Quantitative analysis was performed using ImageJ software.
Lipidomics
For glycerolipid analysis, medial gastrocnemius muscles (∼50 mg) were homogenized in 1 ml of 10% methanol/PBS and neutral lipids were extracted from each sample using 50 μl of homogenate by a modified Bligh and Dyer extraction method substituting dichloromethane for chloroform, as described (39). Glycerolipid analysis was performed by liquid chromatography and MS as described previously with some modifications (40). An aliquot (10 μl) of sample was used for UPLC separation (Acuity UPLC, Waters, Milford, MA) using a CSH C18 column (Waters, Milford, MA). The lipids were eluted by linear gradient chromatography consisting of 100% buffer A to 100% buffer B within 3.5 min (buffer A: water/acetonitrile = 60/40 includes 10 mm ammonium acetate; buffer B: isopropanol/methanol/dichloromethane = 60/20/20 includes 10 mm ammonium acetate). Mass spectrometric analysis ESI MS was carried out using a QTRAP 6500 MS system (Sciex, Redwood Shores, CA). Instrument parameters were optimized by infusion of a mixture of standard glycerolipids. For data analysis, the data from neutral loss experiments were processed with the Lipid Profiler software (Sciex, Redwood Shores, CA) to identify and quantify detected DAG and TAG molecular species. All data were normalized to internal standard and tissue weight.
Free FAs were extracted and analyzed by gas chromatography MS essentially as described previously (40,41). For the analysis of total, free and matrix bound FA composition, a total lipid extraction was carried out first, as described for the glycerolipids, followed by a saponification step. To minimize oxidation, 50 μg of butylated hydroxytoluene was added. For saponification, the lipids were dried under argon, resuspended in 500 μl of methanol:15% KOH (1:1) and incubated at 37 °C for 30 min followed by acidification with 0.005 N HCl. The lipid hydrolates were then extracted following the procedure for the free FAs. The FA esters were analyzed by gas chromatography and MS on an Agilent 6890N gas chromatograph equipped with an Agilent 5973 mass selective detector (Agilent, Santa Clara, CA). FA quantitation was achieved by the stable isotope dilution method.
Muscle glycolytic enzyme activities
Frozen muscles (diaphragm or tibialis anterior) were homogenized in 10 volumes/weight of buffer (50 Tris–HCl, 0.15 M KCl, 1 mm ethylenediaminetetraacetic acid, 0.2 mm DTT, pH 7.5) and homogenates were centrifuged at 10 000g for 10 min at 4 °C. The supernatants were kept on ice until analyzed. For measuring total phosphorylase activity (a + b) (EC 2.4.1.1), 1% glycogen was added to the homogenates to activate phosphorylase b according to (42). After 5 min incubation at room temperature, concentration of NADPH was assessed by spectrophotometry at 340 nm as described previously (43) with some modifications. The following assay buffer was used (50 mm Imidazole, 10 mm MgCl2, 2 mm ethylenediaminetetraacetic acid, pH 7.0 containing 0.01% bovine serum albumin). Lactate dehydrogenase (EC 1.1.1.27) activity was measured at 25 °C as described (44) except that the assay was performed in 0.1 M Potassium Phosphate buffer, pH 7.4. Aldolase (EC 4.1.2.13) activity was measured at 25 °C in 0.1 M Tris–HCl, pH 7.4 by assessing NADH concentration as described (38). All enzyme activities are expressed as units per min per gram of wet muscle weight.
Statistics
Statistical analysis of all data was carried out by two-tailed Student’s t-test. Differences were considered statistically significant if the P-value was less than 0.05.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
RNAseq was performed in the Center for DMD Bioinformatics and Genomics Core (Director: S.F. Nelson) and Mouse Phenotyping was carried out in the Center for DMD Muscle Phenotyping and Imaging Core (Director: M.J. Spencer).
Conflict of Interest statement. None declared.
Funding
National Institute of Arthritis, Musculoskeletal and Skin Diseases (NIAMS-P30AR057230 to M.J.S., RO1 AR048177 to M.J.S., U54AR052646); the Coalition to Cure Calpain 3; the Muscular Dystrophy Association; My Directives; the Crystal Ball Fund. National Institute of Health (UCSD-UCLA DRC NIH P30DK063491); UCLA Department of Medicine and the Iris Cantor Women’s Health Foundation.
References
- 1.Ono Y., Sorimachi H. (2012) Calpains: an elaborate proteolytic system. Biochim. Biophys. Acta, 1824, 224–236. [DOI] [PubMed] [Google Scholar]
- 2.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C. et al. (1995) Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell, 81, 27–40. [DOI] [PubMed] [Google Scholar]
- 3.Kramerova I., Beckmann J.S., Spencer M.J. (2007) Molecular and cellular basis of calpainopathy (limb girdle muscular dystrophy type 2A). Biochim. Biophys. Acta, 1772, 128–144. [DOI] [PubMed] [Google Scholar]
- 4.Fanin M., Angelini C. (2015) Protein and genetic diagnosis of limb girdle muscular dystrophy type 2A: the yield and the pitfalls. Muscle Nerve, 52, 163–173. [DOI] [PubMed] [Google Scholar]
- 5.Milic A., Daniele N., Lochmuller H., Mora M., Comi G.P., Moggio M., Noulet F., Walter M.C., Morandi L., Poupiot J. et al. (2007) A third of LGMD2A biopsies have normal calpain 3 proteolytic activity as determined by an in vitro assay. Neuromuscul. Disord., 17, 148–156. [DOI] [PubMed] [Google Scholar]
- 6.Kramerova I., Kudryashova E., Tidball J.G., Spencer M.J. (2004) Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet., 13, 1373–1388. [DOI] [PubMed] [Google Scholar]
- 7.Kramerova I., Kudryashova E., Wu B., Germain S., Vandenborne K., Romain N., Haller R.G., Verity M.A., Spencer M.J. (2009) Mitochondrial abnormalities, energy deficit and oxidative stress are features of calpain 3 deficiency in skeletal muscle. Hum. Mol. Genet., 18, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanin M., Nardetto L., Nascimbeni A.C., Tasca E., Spinazzi M., Padoan R., Angelini C. (2007) Correlations between clinical severity, genotype and muscle pathology in limb girdle muscular dystrophy type 2A. J. Med. Genet., 44, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg A.S., Puig M., Nagaraju K., Hoffman E.P., Villalta S.A., Rao V.A., Wakefield L.M., Woodcock J. (2015) Immune-mediated pathology in Duchenne muscular dystrophy. Sci. Transl. Med., 7, 299rv294.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramerova I., Kudryashova E., Venkatraman G., Spencer M.J. (2005) Calpain 3 participates in sarcomere remodeling by acting upstream of the ubiquitin-proteasome pathway. Hum. Mol. Genet., 14, 2125–2134. [DOI] [PubMed] [Google Scholar]
- 11.Mellgren R.L., Miyake K., Kramerova I., Spencer M.J., Bourg N., Bartoli M., Richard I., Greer P.A., McNeil P.L. (2009) Calcium-dependent plasma membrane repair requires m- or mu-calpain, but not calpain-3, the proteasome, or caspases. Biochim. Biophys. Acta, 1793, 1886–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramerova I., Kudryashova E., Wu B., Ottenheijm C., Granzier H., Spencer M.J. (2008) Novel role of calpain-3 in the triad-associated protein complex regulating calcium release in skeletal muscle. Hum. Mol. Genet., 17, 3271–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ojima K., Ono Y., Ottenheijm C., Hata S., Suzuki H., Granzier H., Sorimachi H. (2011) Non-proteolytic functions of calpain-3 in sarcoplasmic reticulum in skeletal muscles. J. Mol. Biol., 407, 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beckmann J.S., Spencer M. (2008) Calpain 3, the “gatekeeper” of proper sarcomere assembly, turnover and maintenance. Neuromuscul. Disord., 18, 913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ermolova N., Kudryashova E., DiFranco M., Vergara J., Kramerova I., Spencer M.J. (2011) Pathogenity of some limb girdle muscular dystrophy mutations can result from reduced anchorage to myofibrils and altered stability of calpain 3. Hum. Mol. Genet., 20, 3331–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramerova I., Kudryashova E., Ermolova N., Saenz A., Jaka O., Lopez de Munain A., Spencer M.J. (2012) Impaired calcium calmodulin kinase signaling and muscle adaptation response in the absence of calpain 3. Hum. Mol. Genet., 21, 3193–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Franco M.K.I., Vergara J., Spencer M.J. (2016) Attenuated Ca2+ release in a mouse model of limb girdle muscular dystrophy 2A. Skelet. Muscle, 6, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gehlert S., Bloch W., Suhr F. (2015) Ca2+-dependent regulations and signaling in skeletal muscle: from electro-mechanical coupling to adaptation. Int. J. Mol. Sci., 16, 1066–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olesen J., Kiilerich K., Pilegaard H. (2010) PGC-1alpha-mediated adaptations in skeletal muscle. Pflugers Arch. Eur. J. Physiol., 460, 153–162. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez A.M., Candau R.B., Bernardi H. (2014) FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci.: CMLS, 71, 1657–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura M.T., Yudell B.E., Loor J.J. (2014) Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res., 53, 124–144. [DOI] [PubMed] [Google Scholar]
- 22.Jager S., Handschin C., St-Pierre J., Spiegelman B.M. (2007) AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl Acad. Sci. U. S. A., 104, 12017–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puigserver P., Rhee J., Lin J., Wu Z., Yoon J.C., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M. (2001) Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol. Cell, 8, 971–982. [DOI] [PubMed] [Google Scholar]
- 24.Lira V.A., Benton C.R., Yan Z., Bonen A. (2010) PGC-1alpha regulation by exercise training and its influences on muscle function and insulin sensitivity. Am. J. Physiol. Endocrinol. Metab., 299, E145–E161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watt M.J., Hoy A.J. (2012) Lipid metabolism in skeletal muscle: generation of adaptive and maladaptive intracellular signals for cellular function. Am. J. Physiol. Endocrinol. Metab., 302, E1315–E1328. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z., Gerstein M., Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet., 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H., Gallardo T., Olson E.N., Williams R.S., Shohet R.V. (2003) Transcriptional analysis of mouse skeletal myofiber diversity and adaptation to endurance exercise. J. Muscle Res. Cell Motil., 24, 587–592. [DOI] [PubMed] [Google Scholar]
- 28.Ferraro E., Giammarioli A.M., Chiandotto S., Spoletini I., Rosano G. (2014) Exercise-induced skeletal muscle remodeling and metabolic adaptation: redox signaling and role of autophagy. Antioxid. Redox Signal, 21, 154–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen K.F., Dufour S., Befroy D., Garcia R., Shulman G.I. (2004) Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med., 350, 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siciliano G., Simoncini C., Giannotti S., Zampa V., Angelini C., Ricci G. (2015) Muscle exercise in limb girdle muscular dystrophies: pitfall and advantages. Acta Myol., 34, 3–8. [PMC free article] [PubMed] [Google Scholar]
- 31.Gundersen K. (2011) Excitation-transcription coupling in skeletal muscle: the molecular pathways of exercise. Biol. Rev. Camb. Philos. Soc., 86, 564–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baar K., Wende A.R., Jones T.E., Marison M., Nolte L.A., Chen M., Kelly D.P., Holloszy J.O. (2002) Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J., 16, 1879–1886. [DOI] [PubMed] [Google Scholar]
- 33.Dominy J.E., Jr., Lee Y., Gerhart-Hines Z., Puigserver P. (2010) Nutrient-dependent regulation of PGC-1alpha's acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim. Biophys. Acta, 1804, 1676–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ermolova N., Kramerova I., Spencer M.J. (2015) Autolytic activation of calpain 3 proteinase is facilitated by calmodulin protein. The Journal of biological chemistry, 290, 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chae J., Minami N., Jin Y., Nakagawa M., Murayama K., Igarashi F., Nonaka I. (2001) Calpain 3 gene mutations: genetic and clinico-pathologic findings in limb-girdle muscular dystrophy. Neuromuscul. Dis., 11, 547–555. [DOI] [PubMed] [Google Scholar]
- 36.Kawai H., Akaike M., Kunishige M., Inui T., Adachi K., Kimura C., Kawajiri M., Nishida Y., Endo I., Kashiwagi S. et al. (1998) Clinical, pathological, and genetic features of limb-girdle muscular dystrophy type 2A with new calpain 3 gene mutations in seven patients from three Japanese families. Muscle Nerve, 21, 1493–1501. [DOI] [PubMed] [Google Scholar]
- 37.Trapnell C., Salzberg S.L. (2009) How to map billions of short reads onto genomes. Nat. Biotechnol., 27, 455–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol., 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bligh E.G., Dyer W.J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol., 37, 911–917. [DOI] [PubMed] [Google Scholar]
- 40.Quehenberger O., Armando A.M., Brown A.H., Milne S.B., Myers D.S., Merrill A.H., Bandyopadhyay S., Jones K.N., Kelly S., Shaner R.L. et al. (2010) Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res., 51, 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quehenberger O., Armando A.M., Dennis E.A. (2011) High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys Acta, 1811, 648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krebs E.G., Fischer E.H. (1955) Phosphorylase activity of skeletal muscle extracts. J. Biol. Chem., 216, 113–120. [PubMed] [Google Scholar]
- 43.Cori G.T., Illingworth B., Keller P.J. (1955) Muscle phosphorylase. Method Enzymol., 1, 200–205. [Google Scholar]
- 44.Passonneau J.V, Lowry O.H. (1993). Enzymatic Analysis: A Practical Guide. Humana Press, New Jersey, pp. 235–281. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.