Abstract
Preeclampsia and gestational diabetes mellitus (GDM) are the most common clinical conditions in pregnancy that could result in adverse in utero environments. Fetal exposure to poor environments may raise the long-term risk of postnatal disorders, while epigenetic modifications could be involved. Recent research has implicated involvement of 5-hydroxymethylcytosine (5hmC), a DNA base derived from 5-methylcytosine, via oxidation by ten–eleven translocation (TET) enzymes, in DNA methylation-related plasticity. Here, we show that the TET2 expression and 5hmC abundance are significantly altered in the umbilical veins of GDM and preeclampsia. Genome-wide profiling of 5hmC revealed its specific reduction on intragenic regions from both GDM and preeclampsia compared to healthy controls. Gene Ontology analysis using loci bearing unique GDM- and preeclampsia-specific loss-of-5hmC indicated its impact on several critical biological pathways. Interestingly, the substantial alteration of 5hmC on several transposons and repetitive elements led to their differential expression. The alteration of TET expression, 5hmC levels and 5hmC-mediated transposon activity was further confirmed using established hypoxia cell culture model, which could be rescued by vitamin C, a known activator of TET proteins. Together, these results suggest that adverse pregnancy environments could influence 5hmC-mediated epigenetic profile and contribute to abnormal development of fetal vascular systems that may lead to postnatal diseases.
Introduction
Fetal origins of adult disease (FOAD) is a series of adult-onset disorders, such as cardiovascular diseases, type II diabetes mellitus, obesity and neurological disorders, which are caused by the adverse environments in utero and/or perinatal periods of an individual (1–3). Preeclampsia and gestational diabetes mellitus (GDM) are two of the most common maternal medical problems that could create poor in utero environment to affect fetal development. The offspring exposed to maternal preeclampsia or GDM in utero are associated with the high risk of hypertension, obesity, stroke, type II diabetes, schizophrenia and cancer in late life (4–8). It has been reported that preeclampsia or GDM could cause placental morphological changes as well as alterations in the blood flow patterns in the umbilical vessels; therefore, it is widely accepted that preeclampsia or GDM produces unfavorable in utero environments to the fetus (9–12). However, it is still unclear how those conditions negatively influence the in utero development, and lead to a higher risk of FOAD.
Epigenetic regulations, including DNA methylation, histone modifications and non-coding regulatory RNAs, have been implicated in the pathogenesis of FOAD (13). Epigenetic regulatory mechanisms play important roles in the regulation of gene expression without altering DNA sequence. Unlike genomic alterations, epigenetic modifications are reversible and responsive to the environmental stimuli (14–16). It has been suggested that the early-life environmental conditions could cause epigenetic changes that would persist throughout one’s life span, and could potentially be inherited to the next generation (17–19). Thus, alteration of epigenetic modifications triggered by adverse in utero environments might be ill-matched with fetus’ subsequent extra-uterine environments, and then contribute to an increased risk of FOAD, and ultimately impact on one’s health (20,21).
5-Hydroxymethylcytosine (5hmC) is an oxidative product of 5-methylcytosine (5mC) catalyzed by the ten–eleven translocation (TET) proteins, which belong to 2-oxoglutarate (2OG)- and Fe (II)-dependent enzyme family (22,23). Three paralogs of TET proteins, namely TET1–3, have been defined in mammalian genome, can modulate cytosine modification dynamics in different genomic loci and cellular context (24). 5hmC is not only an intermediate modification during the active DNA demethylation process, but also an independent and critical epigenetic mark for neurodevelopment, aging, ES cell differentiation as well as tumorigenesis (25–28). In addition, 5hmC is much more plastic and dynamic than 5mC in responding to environmental cues (29). Emerging data suggest that aberrant DNA methylation, either hypermethylation or hypomethylation at selective loci in umbilical cord, placental tissues or cord blood, could be due to the exposure to adverse intra-uterus environments (30–33). However, genome-wide analyses of cytosine modification dynamics and their potential effects in response to intra-uterus environmental changes have not been extensively conducted.
In this study, we found that the expression of TET2 was significantly altered in the umbilical veins from preeclampsia and GDM, which led to the concomitant changes of 5hmC levels. Genome-wide profiling of 5hmC revealed differentially hydroxymethylated regions (DhMRs) specifically associated with preeclampsia and GDM. Gene Ontology (GO) analyses indicated that DhMRs were enriched among the genes involved in unique biological pathways for each condition. Intriguingly, 5hmC displayed significant changes at selective transposons in the fetal blood vessels with preeclampsia and led to the alteration of transposon expression. The 5hmC-mediated alteration of transposon expression was further confirmed using the established hypoxia cell culture model and could be rescued by vitamin C, a known activator of TET proteins. Taken together, the results achieved suggest that adverse in utero environments induced by preeclampsia could influence 5hmC-mediated epigenetic profile in the fetal blood vessels. These epigenetic changes likely reflect the similar changes within fetal vascular systems and possibly lead to the predisposition to hypertension or other vascular diseases in later life.
Results
Altered expression of TET2 in the umbilical veins of GDM and preeclampsia
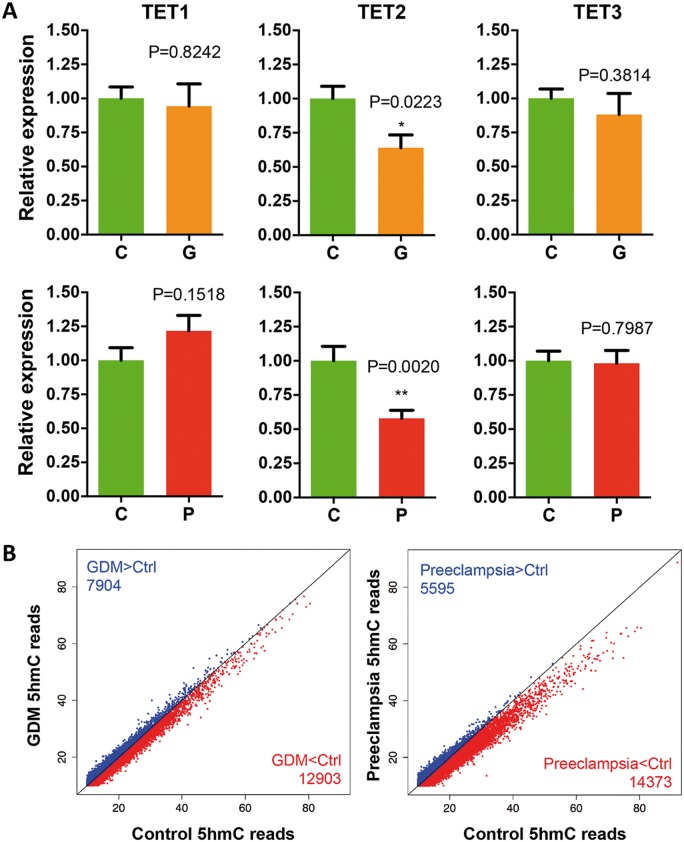
Preeclampsia and GDM are two of the most common medical risks that could cause poor pregnant environments to the fetuses. It has been shown that both preeclampsia and GDM could induce placental and umbilical vessel morphological changes as well as the alterations in umbilical blood flow patterns. To determine the molecular mechanism(s), umbilical cords from healthy, preeclampsia and GDM patients (Table 1 and Supplementary Material, Tables S1–S3) were collected for gene expression analyses and DNA 5hmc study. Given the dynamic nature of 5hmC and role of TET family in 5hmC production, TET1, TET2 and TET3 mRNA in umbilical veins were quantified by real-time quantitative polymerase chain reaction (q-PCR). A significant and specific decrease of TET2 expression was detected in both preeclampsia and GDM, with preeclampsia group in greater extent (P = 0.002) (Fig. 1A). We further confirmed the reduced TET2 protein level in preeclampsia group by western blotting (Supplementary Material, Fig. S1). Since the preeclampsia women tend to have a reduced mean gestation (34), to exclude possibilities that the observed decrease of TET2 expression in human samples was attributed to different development stages, we also examined the expression of TET1–3 using rat umbilical blood vessels during gestational day 18 to day 21 (Supplementary Material, Fig. S2). Furthermore, we compared the expression of TETs between relative shorter or longer gestation within preeclampsia group and found no significant difference (Supplementary Material, Fig. S3). These results exclude the possibility that the observed decrease of TET2 expression in preeclampsia might be due to different developmental stage.
Table 1.
Clinical presentations of three groups of patients
| Characteristics | Normal pregnancy | Preeclampsia | GDM | |
|---|---|---|---|---|
| Number of subjects | 36 | 26 | 27 | |
| Maternal age (year) | 28.50 ± 1.97 | 29.12 ± 3.50 | 30.96 ± 3.59 | |
| Gestational age (week) | 39.14 ± 0.78 | 33.31 ± 2.64*** | 37.96 ± 1.17 | |
| Birth weight (g) | 3362.50 ± 249.31 | 2480.58 ± 448.14*** | 3530.37 ± 641.12 | |
| Systolic BP (mmHg) | 118.47 ± 6.00 | 162.43 ± 11.15*** | 122.67 ± 7.65 | |
| Diastolic BP (mmHg) | 74.81 ± 6.63 | 103.43 ± 9.25*** | 76.74 ± 8.19 | |
| Proteinuria (g/24 h) | N/A | 10.49 ± 3.66 | N/A | |
| OGTT-fasting (mmol/l) | N/A | N/A | 5.57 ± 0.87 | |
| OGTT-1 h (mmol/l) | N/A | N/A | 11.17 ± 1.74 | |
| OGTT-2 h (mmol/l) | N/A | N/A | 8.70 ± 1.67 | |
***P<0.001.
Figure 1.
Expression of TET2 and global 5hmC levels were decreased in the umbilical veins of GDM and preeclampsia. (A) Specific decrease of TET2 expression in both GDM and preeclampsia. qRT-PCR indicated a specific and significant decrease of TET2 expression in both GDM and preeclampsia. t-test P-values are indicated. C, control; G, GDM; P; preeclampsia. (B) Global reduction of 5hmC levels in GDM and preeclampsia. Genome-wide normalized 5hmC mapped reads were counted in each 10 kb binned human genome between the two diseases versus their controls. Bins containing less 5hmC reads in each diseases than that of controls are shown in red, whereas bins in blue indicated the opposite. The number of bins is indicated. Bins containing more than 10 5hmC reads are shown.
Genome-wide 5hmC profiling of umbilical veins
Based on the change in TET2 expression, we sought to determine the genome-wide 5hmC distribution using the umbilical cords from control, preeclampsia and GDM patients by chemical labeling and affinity purification, coupled with high-throughput sequencing (35) (Supplementary Material, Table S4). Global 5hmC levels were evaluated first by counting and plotting normalized 5hmC mapped reads in each 10-kb binned human genome between the each disease condition versus controls. Bins with lower 5hmC reads in the diseases than that in the control were shown in red, whereas bins in blue indicated opposite. Interestingly, we found an overall lower number of bins in blue from both plots, indicating a general reduction of global 5hmC in both the diseases compared to controls (Fig. 1B, blue versus red). These results coincided with the reduction of TET2 expression in the diseases (Fig. 1A). Furthermore, there were a larger number of binned genomic regions highlighted in red in the preeclampsia than that in the GDM, suggesting a significant decrease of 5hmC in the preeclampsia (Fig. 1B, 14 343 versus 12 903), which was also consistent with a greater decrease of TET2 expression under that condition (Fig. 1A).
We went on to characterize the 5hmC peaks genome-wide distributions from three conditions, and evaluated their enrichment or depletion on featured genomic regions compared with expected values. 5hmC peaks enriched on intragenic regions, including exons, untranslated regions and promoter (Fig. 2A, log2-fold change from expected > 0) but not intergenic regions, consistent with previous observations that intragenic 5hmC may be involved in modulating gene expression (36). Given the intragenic nature of 5hmC observed from umbilical veins, we investigated the intragenic 5hmC dynamics between control and diseases by plotting average normalized 5hmC reads on known hg19 refseq gene bodies (Fig. 2B, n = 52 291). Reminiscent to bulk 5hmC quantifications in Figure 1B, 5hmC levels in both GDM and preeclampsia were lower on gene bodies than controls, with stronger 5hmC decrease in preeclampsia. Intriguingly, the lower level of 5hmC in diseases appeared to be specific to gene bodies, since less distinction was found in 2 kb upstream and downstream flanking regions (Fig. 2B). These data together indicated an intragenic-associated loss of 5hmC in umbilical cords from both diseases, especially in preeclampsia, implying that aberrant 5hmC alterations may impose impact on transcriptome.
Figure 2.
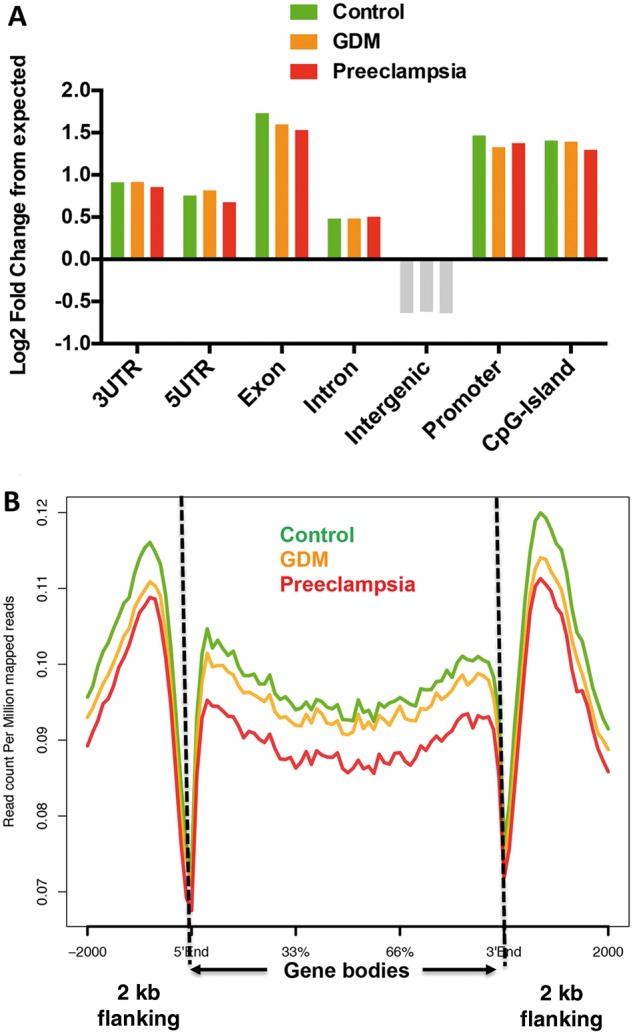
5hmC in umbilical veins preferentially resided in intragenic regions, and reduced on gene bodies under the conditions of GDM and preeclampsia. (A) Umbilical vascular 5hmC preferred intragenic regions. Annotation of umbilical vascular 5hmC peaks from the control, GDM and preeclampsia revealed their enrichment in intragenic features such as exons and promoters. Log2-fold changes over expected values were calculated. Features enriched by 5hmC were indicated by color bars, and depleted were indicated by grey bars. (B) 5hmC specifically reduced on refseq gene bodies in the umbilical veins from GDM and preeclampsia conditions. Normalized 5hmC read densities were calculated as reads per million on known hg 19 refseq gene bodies (n = 52 291), and average reads were plotted in the control and disease conditions using ngsplot. Two kilobase flanking regions upstream and downstream of gene bodies were also included.
Identification of DhMRs in GDM and preeclampsia
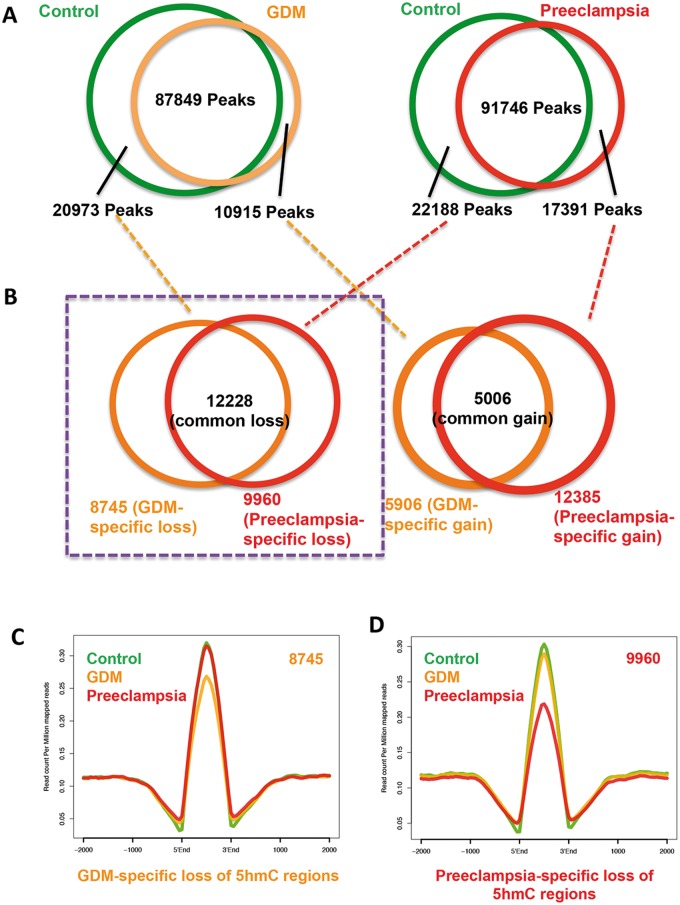
Since we observe global 5hmC alteration in umbilical veins from GDM and preeclampsia, we went on to identify the gain- or loss-of-5hmC regions (DhMRs) in both diseases compared to their controls. 20 973 and 22 188 5hmC-containing genomic regions in controls but loss in either GDM or preeclampsia were obtained. Meanwhile, 10 915 and 17 391 regions acquired 5hmC in either disease were also collected (Fig. 3A and Supplementary Material, Table S5). In an effort to identify unique DhMRs associating with particular disease, DhMRs from Figure 3A were further analyzed. As a result, 8745 GDM- and 9960 preeclampsia-specific loss-of-5hmC were readily obtained, respectively (Fig. 3B). On the other hand, 5906 GDM- and 12 385 preeclampsia-specific gain-of-5hmC were also identified. Since global and intragenic-associated 5hmC were consistently decreased, we focused on these loss-of-5hmC regions uniquely associated with each disease and further investigate their biological roles (Fig. 3B, purple box highlight). Plotting of 5hmC normalized reads from the control, GDM and preeclampsia on those regions confirmed their specific loss of 5hmC in GDM (Fig. 3C) or preeclampsia (Fig. 3D). Thus, unique DhMRs associated with GDM and preeclampsia were obtained.
Figure 3.
Identification of disease-specific DhMRs. (A) Common and differential peaks between diseases and controls were identified and indicated. In total, 20 973 and 22 188 5hmC-containing genomic regions associated with controls but lost in either GDM or preeclampsia were obtained. Meanwhile, 10 915 and 17 391 regions acquired 5hmC in each disease were also collected. (B) Identification of unique 5hmC peaks associated to the diseases. Unique gain- and loss-of 5hmC regions associated with individual disease were obtained by further overlapping disease-differential peaks identified in A. 8745 GDM- and 9960 preeclampsia-specific loss-of-5hmC were readily obtained, respectively. Meanwhile, 5906 GDM- and 12 385 preeclampsia-specific gain-of-5hmC were also found. Loss-of-5hmC regions uniquely associated with each disease were highlighted by purple box for further study. (C and D) Validation of unique loss-of-5hmC peaks associated with GDM and preeclampsia. Normalized 5hmC read densities from the control and diseases were calculated and their average reads per millions were plotted on unique loss-of-5hmC regions associated with GDM (C) and preeclampsia (D). Specific reduction of 5hmC in the particular condition was observed.
Aberrant 5hmC changes in GDM and preeclampsia affect key biological pathways and potential hypoxia-inducible factor 1 binding affinity
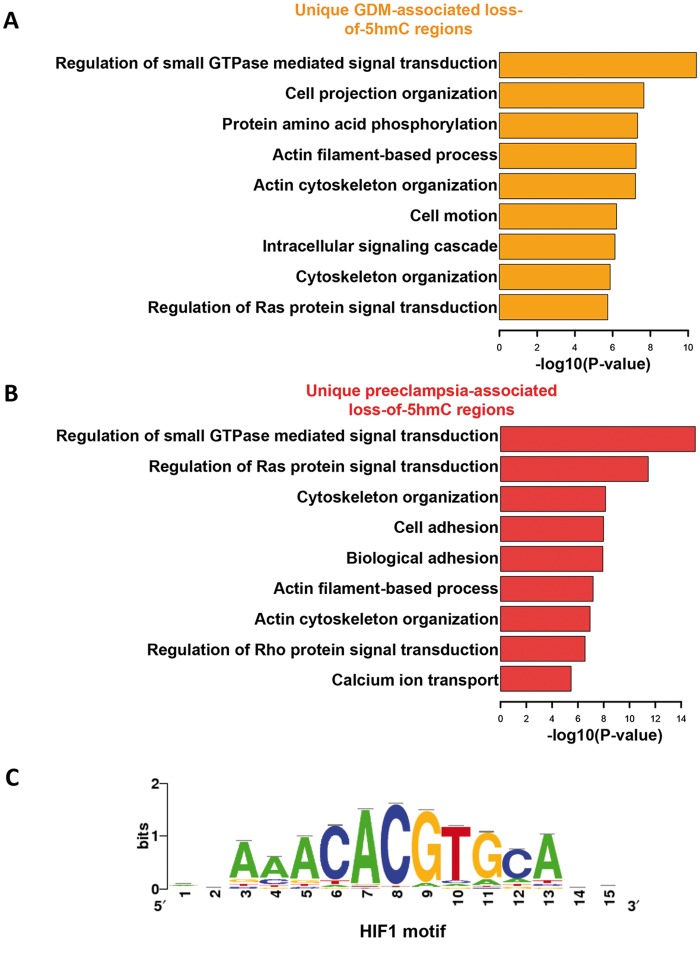
In order to understand the biological roles underlying dynamic 5hmC changes in the umbilical veins from GDM and preeclampsia, we annotated their unique loss-of-5hmC regions to hg19 human refseq genes, and further subjected these genes to GO pathway analysis (37). Interestingly, several biological processes, such as regulation of small GTPase-mediated signal transduction, actin filament-based process and cytoskeleton organization, were affected by loss-of-5hmC in the umbilical vein of both GDM and preeclampsia (Fig. 4A and B). These common pathways may reflect the clinical features of these diseases, such as umbilical vessel morphological changes as well as the alterations in the blood flow patterns in the umbilical vessels. On the contrary, GO analysis also revealed unique pathways associated with either GDM or preeclampsia. For instance, the pathways related to cell adhesion and calcium ion transport were uniquely featured in preeclampsia, suggesting a disease-specific alteration of these biological processes (Fig. 4B). To further understand how the 5hmC alteration at these loci could lead to the change of gene expression, we performed quantitative real time PCR (qRT-PCR) on the selective genes that could potentially contribute to GDM or preeclampsia using the RNA isolated from the umbilical cords from control, preeclampsia and GDM patients, and observed a significant reduction of FKBP5 and NOV in preeclampsia (Supplementary Material, Fig. S4). Indeed previous works have found the decreased expression of NOV associated with preeclampsia (38).
Figure 4.
Characterization of unique loss-of-5hmC regions associated with GDM and preeclampsia. (A and B) GO analysis revealed loss-of-5hmC may impact many critical biological processes. The Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis were fed by the gene loci annotated with unique loss-of-5hmC regions associated with GDM (A) and preeclampsia (B) from Figure 3C and 3D. Significance of these processes was indicated by the rank of −log10 P-value. (C) Motif search using unique loss-of-5hmC regions associated with GDM and preeclampsia suggested potential HIF1 binding alteration to these 5hmC dynamic regions. Motif search were performed by Hypergeometric Optimization of Motif EnRichment (HOMER).
It has been well documented that many transcription factor genome-wide occupancy was sensitive to DNA covalent modification dynamics (39). We employed motif search algorithms to comprehensively predict the potential transcription factor binding on unique 5hmC dynamic loci in GDM and preeclampsia. Remarkably, hypoxia-inducible factor 1 (HIF1), a critical transcription factor responsible for mastering transcriptome in response to normoxia and hypoxia (40), can bind to the common motif from both GDM and preeclamsia unique loss-of-5hmC regions (Fig. 4C). Fetal hypoxia is an important issue accompanied with intrauterine growth retardation or restriction, which is common during preeclampsia pregnancies (12). These data taken together suggested loss-of-5hmC in GDM and preeclamsia could not only impact on the transcripts that associated with critical biological processes, but also may alter critical transcription factor binding dynamics, such as HIF1, which might contribute to the disease onset.
Aberrant 5hmC changes in GDM and preeclampsia resulted in the alteration of transposon and repetitive element expression
It has been recently suggested that aberrant DNA methylation can affect transposon and repetitive element expression, such as long interspersed nuclear element (LINE) and arthrobacter luteus (Alu) elements in short interspersed nuclear elements (SINEs), in hypoxia condition (41). We therefore investigated the 5hmC distributions and dynamics on these elements by aligning sequencing reads to human repeat masker genomic index. Comparing to controls, altered 5hmC levels were found in LINE, SINE and other repetitive elements in GDM or preeclampsia (Fig. 5A). We hypothesized the altered 5hmC could cause aberrant expression of these repetitive elements. qRT-PCR analyses revealed that both Alu and LINE elements showed significant reduced expression in preeclampsia (Fig. 5B). These data sustain the notion that altered 5hmC can also impact site-specific transposon and repetitive element expression, possibly associated with hypoxia condition.
Figure 5.
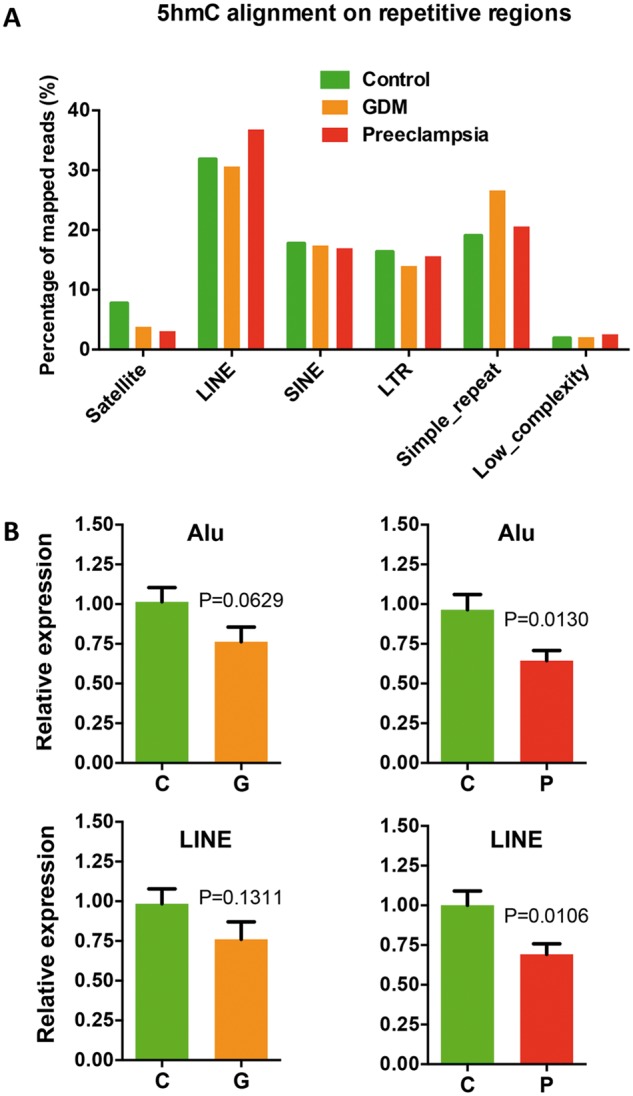
Aberrant 5hmC distributions in GDM and preeclampsia resulted in the alteration of transposon and repetitive element expression. (A) Aberrant 5hmC distributions in GDM and preeclampsia were observed on several repetitive elements in human genome. Unique mapped reads were aligned to the RepeatMasker track of NCBI37v1/mm9 using Bowtie—q—best parameters, allowing no more than two mismatches across the entire 38 bp read. Aligned reads were assigned to repeat class annotations defined by RepeatMasker. 5hmC reads were counted in each repetitive class, and then divided by total aligned reads by Bowtie to achieve their percentage. (B) The expression of LINE and Alu decreased in GDM and preeclampsia. qRT-PCR were performed to determine the expression of LINE and Alu in GDM and preeclampsia compared to controls. t-test P-values were indicated.
Vitamin C treatment restored the TET expression, 5hmC levels and repetitive elements expression in HUVECs
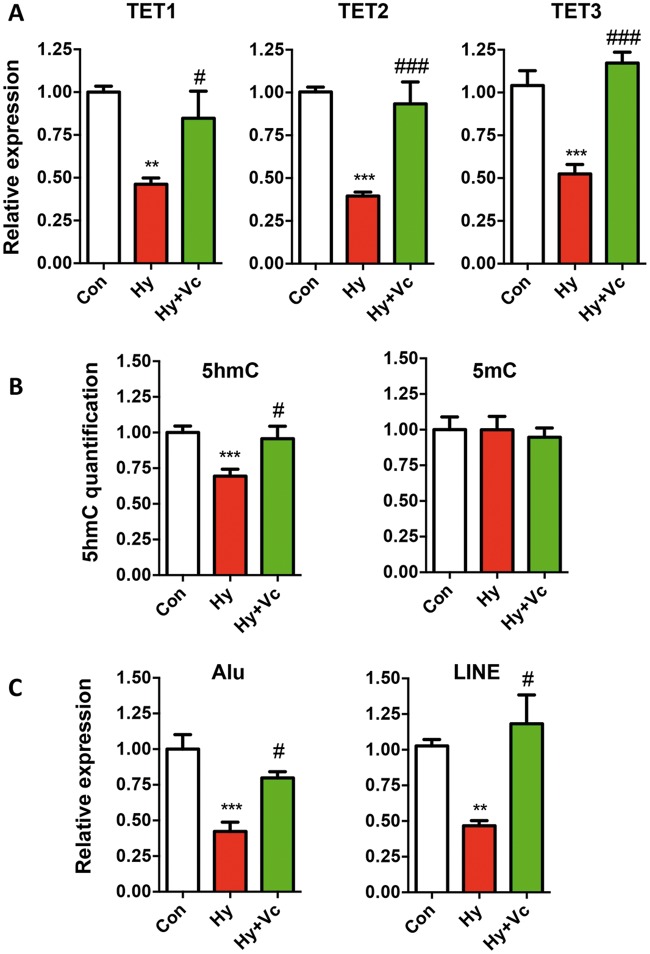
HUVECs are the endothelial cells (ECs) isolated from the veins of umbilical cord, which consists two main cellular components: ECs and smooth muscle cells, and have been shown to be subject to DNA methylation regulation (42). In order to draw a definite link between 5hmC alteration and hypoxia under the condition of GDM and preeclampsia, we treated HUVECs under hypoxia condition with vitamin C, one of the most important co-factors of TETs (43). We then tested the expression of TET proteins and global 5hmC changes. Hypoxia caused significant decrease of all three TET proteins in HUVECs, and exposure of vitamin C resulted in a complete restoration of the expression comparable to controls (Fig. 6A). In addition, 5hmC not 5mC displayed the same dynamics with TET proteins in HUVECs with vitamin C treatment (Fig. 6B). These data provided strong evidence that hypoxia served as a critical factor in GDM and preeclampsia to down-regulated TET2 and 5hmC in umbilical cords. Since reduced repetitive element expression was observed in GDM and preeclampsia, we tested their expression in HUVEC cells as well. Intriguingly, the expression of Alu and LINE positively correlated with TET and 5hmC dynamics, suggesting 5hmC as a central epigenetic modulator in regulating their expression under hypoxia condition (Fig. 6C).
Figure 6.
Vitamin C treatment restored the TET expression, 5hmC levels and repetitive elements expression in HUVECs. (A) Decreased TET protein expressions by hypoxia were restored by vitamin C treatment. qRT-PCR was performed to determine the expression levels of TET proteins in the control (Con), hypoxia (Hy) and Hypoxia with vitamin C treatment (Hy+Vc). t-test P-values were indicated. * indicated statistical analysis between Hy and Con, and #: Hy versus Hy+Vc. */#, P < 0.05; **/##, P < 0.01; ***/###, P < 0.001. (B) Decreased 5hmC but not 5mC levels by hypoxia was restored by vitamin C treatment. 5hmC and 5mC dot blots were performed by specific antibodies. (C) The expression of Alu and LINE were restored by vitamin C. qRT-PCR was performed to determine the expression levels of Alu and LINE.
Discussion
Over the years it has been shown that a number of common adult diseases could be linked to adverse intrauterine environments, which has been defined as FOAD (1). Barker (44) proposed that a fetus reprograms his/her systems to adapt adverse intrauterine environmental stress, which increases the risk of chronic diseases in adulthood. Glucocortioid exposure, hypothalamic pituitary axis change, insulin resistance, epigenetic programming and mitochondrial damage have been shown potentially involved in that process, however the molecular mechanism(s) underlying FOAD remains unclear (45). Here, we demonstrate that the DNA modification, 5hmC, could be involved in the process in the context of preeclampsia or GDM. We found that the expression of TET2 and 5hmC abundance significantly altered in the umbilical blood vessels of preeclampsia and GDM. Genome-wide profiling of 5hmC revealed DhMRs associated with preeclampsia and GDM. In particular, 5hmC significantly changed in selective transposons and led to the alteration of transposon activity. The 5hmC-mediated alteration of transposon activity was further confirmed using the established hypoxia cell model and could be rescued by vitamin C, a known activator of TET proteins. Together our results suggest that adverse in utero environments induced by GDM and preeclampsia could influence 5hmC-mediated epigenetic profile in fetal blood vessels, which may contribute to FOAD.
It has been demonstrated that TETs requires oxygen for their enzymatic activities. Since fetal hypoxia and intrauterine growth retardation are commonly associated with preeclampsia and GDM (10,12), it is likely that the changes of 5hmC abundance and TET2 in human umbilical cord from preeclampsia could be caused by hypoxia. To test this hypothesis, using hypoxia culture assay of human umbilical vein ECs, we found that along with the significant decrease of all three TETs, 5hmC levels were reduced. This strongly suggested that hypoxia could be one of critical adverse factors in pregnancy under condition of preeclampsia and GDM, to influence TET enzyme expression directly; therefore, result in decreased 5hmC across the genome of umbilical blood vessels during fetal stage.
GO analyses of the loss-of-5hmC DhMRs indicate the enrichment of the genes involved in the regulation of small GTPase-mediated signal transduction as well as the pathways involved in cell morphology in both GDM and preeclampsia, which suggests a potential role of small GTPase in the response to the adverse in utero environments. Our gene expression analyses also suggest that the loss of 5hmC could potentially lead to the reduction of gene expression, such as FKBP5 and NOV. Systematic examination of the correlation between the loss of 5hmC and gene expression alteration is certainly needed in the future.
Further sequence analyses of the DhMRs also revealed an enrichment of the HIF1 binding motif among both GDM and preeclampsia unique loss-of-5hmC regions. It has been shown that HIF1 is a critical transcription factor responsible for mastering transcriptome in response to normoxia and hypoxia (40). Given that it has been demonstrated that individual transcription factor could be sensitive to DNA covalent modification dynamics, we further examined the binding of HIF1 in HUVECs under normoxia and hypoxia at the loci with the loss of 5hmC modification in both GDM and preeclampsia (46). Intriguingly, HIF1 binding significantly reduced under hypoxia condition at the loci with the loss of 5hmC specifically in preeclampsia condition (Supplementary Material, Fig. S5). Thus the loss-of-5hmC in umbilical veins with GDM and preeclampsia conditions could not only impact on the transcripts that associated with critical biological processes, but also might alter critical transcription factor binding dynamics such as HIF1 that may contributed to the disease onset in fetal origins.
Repeat elements such as SINEs and LINEs together comprised around 35% of the genome (47,48). DNA methylation within these repetitive elements are thought to play a critical role in genomic defense and structural integrity by silencing expression of these transposon elements, thereby limiting chromosomal rearrangement and translocation events (48). The present study showed interesting finding that reduced 5hmC and decreased expression of Alu and LINE were associated with the exposure to adverse in utero environments with GDM or preeclampsia. The modulation of Alu and LINE activity by 5hmC was further evidenced in cell culture experiments. Importantly, vitamin C treatment could induce the TETs expression and increase 5hmC abundance, and therefore increased the expression of Alu and LINE. It was reported that the reduced H3K9me3 was concomitant with increased 5hmC and TET1 binding (49). Meanwhile, increases in H3K9me3 have been reported in a number of cell lines exposed to hypoxia with the subsequent repression of a number of different genes (50). Therefore, the reduced expression of Alu and LINE under conditions of preeclampsia or hypoxia associated with decreased 5hmC in fetal blood vessels could be caused by the increased H3K9me3 under hypoxic conditions, which deserves future investigation.
Previous works have demonstrated that ascorbate, the dominant form of vitamin C, serves as a direct cofactor for TETs that catalyze the oxidation of 5mC into 5hmC. Addition of vitamin C to mouse ES cells promoted TET activity, and led to an increase in 5hmC followed by DNA demethylation (43). In this study, we found that vitamin C could rescue the decreased TETs and 5hmC as well as LINE and Alu under the hypoxia condition in HUVECs. One of the features in epigenetic modification is reversible. Our finding suggested that vitamin C was useful to reverse the decreased 5hmC following the exposure to adverse in utero environments. Therefore, vitamin C could work as a potential treatment tool in preventing adverse outcomes caused by abnormal pregnancy. It would be interesting to further investigate the long-term effect of prenatal vitamin C supplementation on adverse maternal or fetal/perinatal outcomes as well as FOAD in general (51–53).
In summary, the present study provides the first link between 5hmC alteration and the genomic DNA in human blood vessels during the fetal stage with two of the most common clinical conditions: preeclampsia and GDM. Our results showed that the altered 5hmC is one of the direct consequences in response to an adverse intrauterine environment like hypoxia under the disease conditions (while 5hmC was reduced in the fetal blood vessels from both the diseases, greater decrease was noted in preeclampsia), which could potentially increase the offspring’s sensitivity to postnatal challenges after birth and predispose individuals to FOAD (54). In addition, umbilical arteries and veins are only blood vessels can be used for research from healthy human in groups, the present study was the first to use them in determination of certain epigenetic changes (5hmC) that can be linked to early development of FOAD, there are sufficient reasons this good example will be followed by numerous studies using human subjects.
Materials and Methods
Samples collection
This study was approved by Soochow University Institutional Ethic Board. Pregnant women were recruited from local hospitals. Written consent was obtained from all participates. Data on gestational age at birth, birth weight, blood pressure and oral glucose tolerance test (OGTT) results were summarized in Table 1 and Supplementary Material, Table S1–S3. Umbilical veins next to fetuses (∼5 cm) and cord blood were collected from three different pregnancy groups: normal pregnancy without any abnormal symptom (Control), pregnancy with preeclampsia and pregnancy with GDM. Preeclampsia diagnosis is defined as an increase in blood pressure (>140/90 mmHg) and accompanied by proteinuria (≥0.3 g/24 h or ≥1+ on dipstick). A 75 g OGTT was conducted at 24–28 weeks of pregnancy. GDM was diagnosed when glucose levels exceeds the thresholds: 5.1 mm at fasting; 10.0 mm at 1 h or 8.5 mm at 2 h.
Immuno-dot blot assay
Genomic DNA from HUVECs was extracted by using the phenol–chloroform methods and quantified by Nanodrop 2000 (Thermo, USA). After denatured in 1 M NaOH for 2 h, 100–300 ng genomic DNAs were spotted onto a charged nylon-based membrane at room temperature for 30 min and then baked at 75°C for 1 h. The membrane was blocked with 5% non-fat milk for 1 h. After washing in phosphate buffered saline (PBS) three times, the blot was incubated in rabbit polyclonal anti-5hmC antibody (Active Motif; 1:10 000) or mouse monoclonal anti-5mc antibody (Millipore; 1:2000) at 4°C overnight. On the following day, the blot was washed in PBS three times incubated with peroxidase-conjugated anti-rabbit IgG secondary antibody for 1 h. The signal was visualized by using ECL-Plus system (Amersham Pharmacia Biotech).
5hmC-specific chemical labeling, affinity purification and sequencing
Genomic DNA of four samples (C1–4, P1–4 and G1–4) from each human group was used for 5hmC genome-wide profiling. 5hmC enrichment was performed using a previously described procedure with an improved selective chemical labeling method (28). DNA libraries were generated following the Illumina protocol for ‘Preparing Samples for ChIP Sequencing of DNA’ (Part# 111257047 Rev. A) using 25–50 ng of input genomic DNA or 5hmC-captured DNA to initiate the protocol.
Bioinformatic data analyses
Processing of sequencing data was performed as previously described (55). Briefly, FASTQ sequence files from biological replicates were concatenated and aligned to the Mus musculus reference genome (NCBI37v1/mm9) using Bowtie 0.12.6, keeping only unique non-duplicate genomic matches with no more than two mismatches within the first 25 bp. Unique, non-duplicate reads from non-enriched input genomic DNA of ischemic brain regions and each 5hmC-enriched sequence set were counted in 100-, 1000-, and 10 000-bp bins genome-wide and subsequently normalized to the total number of non-duplicate reads in millions. Input-normalized values were then subtracted from 5hmC-enriched values per bin to generate normalized 5hmC signals. To determine genomic regions with altered 5hmC profiles, we identified true 5hmC-enriched regions or ‘peaks’ using the Model-based Analysis of ChIP-Seq algorithm. GO analyses were performed as previously described using the DAVID Bioinformatics Resources 6.7 Functional Annotation Tool. Gene sets were identified by joining subsets of DhMRs with RefSeq tables obtained from the UCSC genome browser tables.
Hypoxia cell culture
HUVECs were purchased (ATCC) and cultured in medium DMEM supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in the condition with 5.0% CO2 and saturated humidity. After grow up to 80% coverage, HUVECs are incubated with 1.0% O2 for hypoxia or normoxia, respectively. After cultured for 48 h, cells were harvested. DNA and RNA were extracted from cells for experiments. All the experiments were repeated for three times.
Quantitative real time PCR
Total RNA was extracted from umbilical blood vessels and cord blood with TaKaRa MiniBEST Universal RNA Extraction Kit or RNAiso Reagent (TaKaRa, Japan). RNA was reverse transcribed with PrimeScript™ II by using 1st Strand cDNA Synthesis Kit (TaKaRa, Japan). The reference sequence of studied genes was acquired from the UCSC Genome Browser. The information of primers was shown in Supplementary Material, Table S6. The q-PCR was performed on a Bio-Rad MyiQ2 Thermal Cycler QPCR machine (Bio-Rad, USA) with a SYBR® Premix Ex Taq™ mix (TaKaRa, Japan). Data were normalized against beta-actin or glyceraldehyde-3-phosphate dehydrogenase as internal controls and calibrated with a normal control cDNA. The relative expression ratio was calculated with the 2−ΔΔCt method. For human umbilical veins study, we checked at least 20 samples for each group.
Vitamin C treatment
HUVECs were cultured with l-ascorbic acid (A5960, Sigma) in a concentration range from 0.05 to 1 mm under hypoxia (1.0% O2) condition for 48 h. TET1–3 mRNA expression was evaluated by q-PCR to determine the best working concentration of l-ascorbic acid. The HUVECs then were treated with 0.5 mm l-ascorbic acid under the hypoxia condition for 48 h. Expression of TETs, Alu and LINE was checked by q-PCR. 5hmC levels were determined by dot blot assay. Both untreated HUVECs under normoxia or hypoxia were used as controls. Each group consisted of three wells and was repeated three times.
Rat cord experiments
Sprague-Dawley rats were obtained from Soochow University Experimental Animal Center. All procedures and protocols were approved by the Institutional Animal Care and Use Committee and followed the Guidelines for the Care and Use of Laboratory Animals. When vaginal plugs were observed in female rats, the day was designated embryo day 1 (E1). At E18, E19, E20 and E21, the pregnant rats were anesthetized intraperitoneally with 4% chloral hydrate (0.8 ml/100 g), fetuses were removed through surgical hysterotomy. The umbilical veins from the entire litter embryos were collected and frozen in liquid nitrogen, and then stored in a −80 °C freezer for molecular experiments.
Statistical analysis
GraphPad Prism software was used for the statistical analyses. Either t-test or one-way ANalysis Of VAriance was performed to evaluate differences between the groups. All results were expressed in mean ± SEM. P < 0.05 was considered statistically significant differences.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We would like to thank all the family members for their participation in this study.
Conflict of interest Statement. None declared.
Funding
This study was in part supported by grants from Ministry of Science and Technology of China (2013CB945400, 2012CB947600 and 2013BAI04B05), National Natural Science Foundation of (China 81320108006, 81570960 and 81401244), and National Institutes of Health (NS079625 and HG006699). SM was a trainee in the International Collaborative Genetics Research Training Program (NIH grant D43 TW06176 to Jabs EW).
References
- 1.Barker D.J. (1990) The fetal and infant origins of adult disease. Br. Med. J., 301, 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker D.J., Osmond C., Golding J., Kuh D., Wadsworth M.E. (1989) Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. Br. Med. J., 298, 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker D.J., Winter P.D., Osmond C., Margetts B., Simmonds S.J. (1989) Weight in infancy and death from ischaemic heart disease. Lancet, 2, 577–580. [DOI] [PubMed] [Google Scholar]
- 4.Calkins K., Devaskar S.U. (2011) Fetal origins of adult disease. Curr. Probl. Pediatr. Adolesc. Health Care, 41, 158–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Page K.A., Romero A., Buchanan T.A., Xiang A.H. (2014) Gestational diabetes mellitus, maternal obesity, and adiposity in offspring. J. Pediatr, 164, 807–810., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holder T., Giannini C., Santoro N., Pierpont B., Shaw M., Duran E., Caprio S., Weiss R. (2014) A low disposition index in adolescent offspring of mothers with gestational diabetes: a risk marker for the development of impaired glucose tolerance in youth. Diabetologia, 57, 2413–2420. [DOI] [PubMed] [Google Scholar]
- 7.Kajantie E., Eriksson J.G., Osmond C., Thornburg K., Barker D.J. (2009) Pre-eclampsia is associated with increased risk of stroke in the adult offspring: the Helsinki birth cohort study. Stroke, 40, 1176–1180. [DOI] [PubMed] [Google Scholar]
- 8.Palmsten K., Buka S.L., Michels K.B. (2010) Maternal pregnancy-related hypertension and risk for hypertension in offspring later in life. Obstet. Gynecol., 116, 858–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daskalakis G., Marinopoulos S., Krielesi V., Papapanagiotou A., Papantoniou N., Mesogitis S., Antsaklis A. (2008) Placental pathology in women with gestational diabetes. Acta Obstet. Gynecol. Scand., 87, 403–407. [DOI] [PubMed] [Google Scholar]
- 10.Koech A., Ndungu B., Gichangi P. (2008) Structural changes in umbilical vessels in pregnancy induced hypertension. Placenta, 29, 210–214. [DOI] [PubMed] [Google Scholar]
- 11.Cetin A., Kukner A., Ozturk F. (2002) Ultrastructure of human umbilical vessels in pre-eclampsia. J. Matern. Fetal. Neonatal. Med., 12, 178–184., [DOI] [PubMed] [Google Scholar]
- 12.Inan S., Sanci M., Can D., Vatansever S., Oztekin O., Tinar S. (2002) Comparative morphological differences between umbilical cords from chronic hypertensive and preeclamptic pregnancies. Acta Med. Okayama, 56, 177–186. [DOI] [PubMed] [Google Scholar]
- 13.Gao Q., Tang J., Chen J., Jiang L., Zhu X., Xu Z. (2014) Epigenetic code and potential epigenetic-based therapies against chronic diseases in developmental origins. Drug Discov. Today, 19, 1744–1750. [DOI] [PubMed] [Google Scholar]
- 14.Cutfield W.S., Hofman P.L., Mitchell M., Morison I.M. (2007) Could epigenetics play a role in the developmental origins of health and disease? Pediatr. Res., 61, 68r–75r. [DOI] [PubMed] [Google Scholar]
- 15.Raychaudhuri N., Raychaudhuri S., Thamotharan M., Devaskar S.U. (2008) Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J. Biol. Chem., 283, 13611–13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons R.A. (2007) Developmental origins of beta-cell failure in type 2 diabetes: the role of epigenetic mechanisms. Pediatr. Res., 61, 64r–67r. [DOI] [PubMed] [Google Scholar]
- 17.Heijmans B.T., Tobi E.W., Stein A.D., Putter H., Blauw G.J., Susser E.S., Slagboom P.E., Lumey L.H. (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl Acad. Sci. U. S. A., 105, 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehan V.K., Liu J., Naeem E., Tian J., Sakurai R., Kwong K., Akbari O., Torday J.S. (2012) Perinatal nicotine exposure induces asthma in second generation offspring. BMC Med., 10, 129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leslie F.M. (2013) Multigenerational epigenetic effects of nicotine on lung function. BMC Med., 11, 27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanaka-Gantenbein C. (2010) Fetal origins of adult diabetes. Ann. N. Y. Acad. Sci., 1205, 99–105. [DOI] [PubMed] [Google Scholar]
- 21.Gluckman P.D., Hanson M.A., Spencer H.G., Bateson P. (2005) Environmental influences during development and their later consequences for health and disease: implications for the interpretation of empirical studies. Proc. Biol. Sci., 272, 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science, 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pastor W.A., Aravind L., Rao A. (2013) TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol., 14, 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W. (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature, 473, 398–402. [DOI] [PubMed] [Google Scholar]
- 26.Kraus T.F., Guibourt V., Kretzschmar H.A. (2015) 5-Hydroxymethylcytosine, the “Sixth Base”, during brain development and ageing. J. Neural Transm., 122, 1035–1043. [DOI] [PubMed] [Google Scholar]
- 27.Kriaucionis S., Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science, 324, 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I. et al. (2011) 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci., 14, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irier H., Street R.C., Dave R., Lin L., Cai C., Davis T.H., Yao B., Cheng Y., Jin P. (2014) Environmental enrichment modulates 5-hydroxymethylcytosine dynamics in hippocampus. Genomics, 104, 376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomura Y., Lambertini L., Rialdi A., Lee M., Mystal E.Y., Grabie M., Manaster I., Huynh N., Finik J., Davey M. et al. (2014) Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod. Sci., 21, 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J., Zhang A., Fang M., Fang R., Ge J., Jiang Y., Zhang H., Han C., Ye X., Yu D. et al. (2013) Methylation levels at IGF2 and GNAS DMRs in infants born to preeclamptic pregnancies. BMC Genom., 14, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D., Zhang A., Fang M., Fang R., Ge J., Jiang Y., Zhang H., Han C., Ye X., Huang H. et al. (2014) Increased methylation at differentially methylated region of GNAS in infants born to gestational diabetes. BMC Med. Genet., 15, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Y., Zhang X., Li Q., Xu J., Zhou X., Wang T., Xing Q., Liu Y., Wang L., He L. et al. (2013) Promoter hypomethylation of TIMP3 is associated with pre-eclampsia in a Chinese population. Mol. Hum. Reprod., 19, 153–159. [DOI] [PubMed] [Google Scholar]
- 34.Lal A.K., Gao W., Hibbard J.U. (2013) Eclampsia: Maternal and neonatal outcomes. Pregnancy Hypertens., 3, 186–190. [DOI] [PubMed] [Google Scholar]
- 35.Yao B., Lin L., Street R.C., Zalewski Z.A., Galloway J.N., Wu H., Nelson D.L., Jin P. (2014) Genome-wide alteration of 5-hydroxymethylcytosine in a mouse model of fragile X-associated tremor/ataxia syndrome. Hum. Mol. Genet., 23, 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y., Chavez L., Chang X., Wang X., Pastor W.A., Kang J., Zepeda-Martinez J.A., Pape U.J., Jacobsen S.E., Peters B. et al. (2014) Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl Acad. Sci. U. S. A, 111, 1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang da W., Sherman B.T., Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 38.Gellhaus A., Schmidt M., Dunk C., Lye S.J., Winterhager E. (2007) The circulating proangiogenic factors CYR61 (CCN1) and NOV (CCN3) are significantly decreased in placentae and sera of preeclamptic patients. Reprod. Sci., 14, 46–52. [DOI] [PubMed] [Google Scholar]
- 39.Tsankov A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I., Gnirke A., Meissner A. (2015) Transcription factor binding dynamics during human ES cell differentiation. Nature, 518, 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G.L., Jiang B.H., Rue E.A., Semenza G.L. (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. U. S. A., 92, 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pal A., Srivastava T., Sharma M.K., Mehndiratta M., Das P., Sinha S., Chattopadhyay P. (2010) Aberrant methylation and associated transcriptional mobilization of Alu elements contributes to genomic instability in hypoxia. J. Cell Mol. Med., 14, 2646–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krause B.J., Costello P.M., Munoz-Urrutia E., Lillycrop K.A., Hanson M.A., Casanello P. (2013) Role of DNA methyltransferase 1 on the altered eNOS expression in human umbilical endothelium from intrauterine growth restricted fetuses. Epigenetics, 8, 944–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martinez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A. et al. (2013) Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature, 500, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barker D.J. (1998) In utero programming of chronic disease. Clin. Sci., 95, 115–128. [PubMed] [Google Scholar]
- 45.Brenseke B., Prater M.R., Bahamonde J., Gutierrez J.C. (2013) Current thoughts on maternal nutrition and fetal programming of the metabolic syndrome. J. Pregnancy, 2013, 368461.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mimura I., Nangaku M., Kanki Y., Tsutsumi S., Inoue T., Kohro T., Yamamoto S., Fujita T., Shimamura T., Suehiro J. et al. (2012) Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol. Cell Biol., 32, 3018–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kazazian H.H., Jr. (2004) Mobile elements: drivers of genome evolution. Science, 303, 1626–1632. [DOI] [PubMed] [Google Scholar]
- 48.Wilson A.S., Power B.E., Molloy P.L. (2007) DNA hypomethylation and human diseases. Biochim. Biophys. Acta, 1775, 138–162. [DOI] [PubMed] [Google Scholar]
- 49.Leung D., Du T., Wagner U., Xie W., Lee A.Y., Goyal P., Li Y., Szulwach K.E., Jin P., Lorincz M.C. et al. (2014) Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase Setdb1. Proc. Natl Acad. Sci. U. S. A., 111, 6690–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olcina M.M., Leszczynska K.B., Senra J.M., Isa N.F., Harada H., Hammond E.M. (2016) H3K9me3 facilitates hypoxia-induced p53-dependent apoptosis through repression of APAK. Oncogene, 35, 793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conde-Agudelo A., Romero R., Kusanovic J.P., Hassan S.S. (2011) Supplementation with vitamins C and E during pregnancy for the prevention of preeclampsia and other adverse maternal and perinatal outcomes: a systematic review and metaanalysis. Am. J. Obstet. Gynecol., 204, 503 e1–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poston L., Briley A.L., Seed P.T., Kelly F.J., Shennan A.H. (2006) Vitamin C and vitamin E in pregnant women at risk for pre-eclampsia (VIP trial): randomised placebo-controlled trial. Lancet, 367, 1145–1154. [DOI] [PubMed] [Google Scholar]
- 53.Abramovici a., Gandley R., Clifton R., Leveno K., Myatt L., Wapner R., Thorp J., Jr., Mercer B., Peaceman A., Samuels P. et al. (2015) Prenatal vitamin C and E supplementation in smokers is associated with reduced placental abruption and preterm birth: a secondary analysis. Bjog, 122, 1740–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Debnath M., Venkatasubramanian G., Berk M. (2015) Fetal programming of schizophrenia: select mechanisms. Neurosci. Biobehav. Rev., 49, 90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen L., Shao N., Liu X., Nestler E. (2014) ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics, 15, 284.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.