Abstract
Huntington’s disease (HD) is a devastating illness and at present there is no disease modifying therapy or cure for it; and management of the disease is limited to a few treatment options for amelioration of symptoms. Recently, we showed that the administration of bezafibrate, a pan-PPAR agonist, increases the expression of PGC-1α and mitochondrial biogenesis, and improves phenotype and survival in R6/2 transgenic mouse model of HD. Since the R6/2 mice represent a ‘truncated’ huntingtin (Htt) mouse model of HD, we tested the efficacy of bezafibrate in a ‘full-length’ Htt mouse model, the BACHD mice. Bezafibrate treatment restored the impaired PPARγ, PPARδ, PGC-1α signaling pathway, enhanced mitochondrial biogenesis and improved antioxidant defense in the striatum of BACHD mice. Untreated BACHD mice show robust and progressive motor deficits, as well as late-onset and selective neuropathology in the striatum, which was markedly ameliorated in the BACHD mice treated with bezafibrate. Our data demonstrate the efficacy of bezafibrate in ameliorating both neuropathological features and disease phenotype in BACHD mice, and taken together with our previous studies with the R6/2 mice, highlight the strong therapeutic potential of bezafibrate for treatment of HD.
Introduction
Huntington’s disease (HD) is a devastating neurodegenerative disorder characterized by the triad of progressive psychiatric, movement and cognitive problems. The disease is caused by a dominantly inherited mutation in the huntingtin (Htt) gene, consisting of an abnormal cytosine–adenine–guanine (CAG) repeat expansion, resulting in a polyglutamine expansion in the Htt protein (mutant huntingtin; mHtt) (1). The most striking neuropathological hallmark of this disorder is atrophy of the striatum with preferential loss of GABAergic medium-size spiny neurons (2). Other regions such as cortex, hypothalamus and hippocampus also undergo degeneration in the course of the disease (3,4). Existing evidence strongly supports the case for a defect in energy metabolism as a contributor to the pathogenesis of HD. In HD patients, there is reduced glucose consumption in the brain, especially in basal ganglia, an abnormality which is also present in presymptomatic mutation carriers. Studies of striatal samples from late-stage HD patients show reduced activity for many components of the oxidative phosphorylation (OXPHOS) pathway (5–7).
Htt associates directly with mitochondria, resulting in impaired OXPHOS, abnormal mitochondrial calcium handling and abnormal mitochondria trafficking. Furthermore, mHtt produces impairment of the transcriptional coactivator peroxisome proliferator-activated receptor (PPAR) γ coactivator-1α (PGC-1α) and dysregulation of mitochondrial biogenesis (8–17). In addition to these well-documented effects, other studies provide evidence that astrocytes expressing mHtt may produce adverse metabolic events affecting neurons (18–20).
There is strong evidence supporting a role of impaired PGC-1α expression and downstream signaling changes culminating in mitochondrial dysfunction in HD. Recently, we showed that the administration of the pan-PPAR agonist, bezafibrate, increases the expression of PGC-1α and mitochondrial biogenesis, and thereby results in improved behavior, improved survival and reduced brain, muscle and brown adipose tissue (BAT) pathology in R6/2 transgenic HD mice (21). The R6/2 mice represent a ‘truncated’ Htt mice model of HD with an N-terminal genomic fragment containing exon 1 with ∼130 CAG repeats. Best practice guidelines recommend testing medications for therapeutic effects whenever possible in at least two transgenic HD mouse lines, including at least one full length (22,23). We, therefore, set out to determine whether bezafibrate treatment would induce PGC-1α expression and mitochondrial biogenesis, and show similar beneficial effects in a full-length (FL) Htt mouse model of HD, i.e. the BACHD mice.
Results
Bezafibrate improves phenotype in the BACHD mice
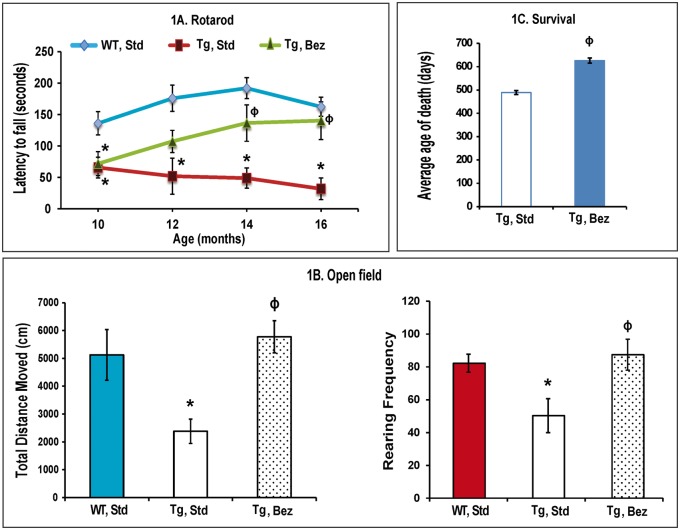
Consistent with previous studies, we noted a robust and progressive phenotype in BACHD mice (24). Motor coordination was assessed by performance on an accelerated rotarod apparatus. Latency to fall was recorded for three trials per day for 3 days monthly assessment and scores were averaged (Fig. 1A). Throughout the trial period, BACHD mice consistently performed poorly on rotarod, with a significantly reduced latency to fall. Bezafibrate-treated mice remained on the rotarod longer than the untreated BACHD mice, indicating better motor coordination in the treated mice (Fig. 1A). To further assess motor function, general activity and exploration, we performed the open field test. BACHD mice were significantly hypoactive, as measured by the total distance covered, and had a significantly reduced rearing frequency as compared with their wild-type (WT) littermates. In the bezafibrate-treated BACHD mice, a significant amelioration of the deficits was seen. The total distance covered and rearing frequency were significantly greater in the treated group than in the BACHD mice on a standard diet (Fig. 1B). In our hands, BACHD mice normally have a life span of an average of 489 days (mean = 489 ± 8.3). Bezafibrate-treated BACHD mice lived 28% longer than the BACHD mice on a standard diet (mean = 626 ± 11.0) (Fig. 1C).
Figure 1.
Bezafibrate improves the phenotype and extends survival in BACHD mice. (A) Assessment of motor coordination in BACHD mice on bezafibrate diet. BACHD mice on a standard diet showed progressive, robust deficits on rotarod, while bezafibrate-treated BACHD mice remained on the rotarod longer and their performance improved progressively. Scores were averaged from three independent experiments. The asterisks and symbols represent the significance levels calculated by one-way ANOVA followed by Tukey–Kramer multiple comparisons test: *P < 0.05, as compared with the WT controls; ϕP < 0.05 when compared with BACHD, standard. (n = 10 for each genotype, bezafibrate or standard diet and bars represent S.E.M.). (B) Measurement of exploratory activity in BACHD mice at 16 months of age. Scores were averaged from three independent experiments. BACHD mice are significantly hypoactive as compared with their WT littermates (*P < 0.05). Bezafibrate significantly restores normal activity and exploration (ϕP < 0.05 when compared with BACHD, standard). One-way ANOVA followed by Tukey–Kramer multiple comparisons test, n=10. (C) Average age of death of BACHD mice on the bezafibrate diet or a standard diet. Unpaired, Student’s two-tailed t-test (ϕP < 0.05 when compared to BACHD, standard; n = 15).
Bezafibrate modulates the PPAR-PGC-1α signaling and antioxidant response pathways
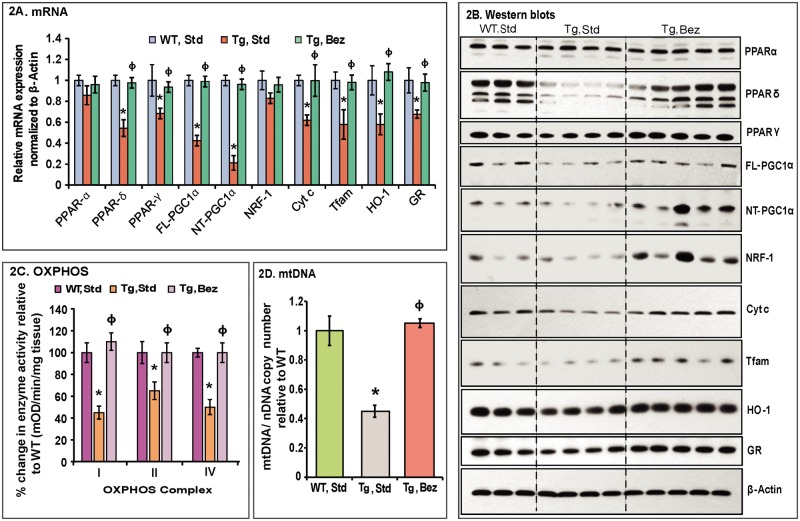
We performed quantitative real-time PCR analysis of cDNA obtained from the striatum of BACHD mice, their WT littermates and BACHD mice treated with bezafibrate. We found that both the PPARγ and PPARδ were significantly downregulated in the BACHD mice on standard diet as compared with their WT littermates. Furthermore, both the FL and N-truncated (NT)-isoforms of PGC-1α as well as the downstream genes Cyt c and Tfam were significantly downregulated in the striatum of BACHD mice compared to their WT littermates, whereas levels of NRF-1 remain unchanged (Fig. 2A). Bezafibrate treatment restored the mRNA expression of PPARs, PGC-1α and downstream targets Cyt c and Tfam in the striatum of BACHD mice, and their levels were not significantly different from those of the WT controls (Fig. 2A). Concomitant with the stimulation of expression of genes involved in mitochondrial energy production, PGC-1α also induces genes responsible for countering reactive oxygen species (ROS) generated as by-products of oxidative metabolism (25–27). In BACHD striatum, we found that genes responsive to ROS, such as, hemoxygenase-1 (HO-1), and glutathione reductase (GR) were significantly downregulated, and bezafibrate restored the levels of these genes to control levels in the BACHD mice (Fig. 2A).
Figure 2.
Bezafibrate restores the PPAR–PGC-1α signaling pathway and induces mitochondrial biogenesis in BACHD mice. (A) Relative expression of PPARα, γ, δ, FL and NT-isoforms of PGC-1α, and the downstream target genes, nuclear respiratory factor NRF-1, Cyt c, Tfam, as well as the oxidative stress response genes HO-1 and GR in striatum of BACHD mice on a standard diet or on the bezafibrate diet. The levels of each gene transcript were normalized to that of β-actin and expressed as fold variation relative to the WT mice on a standard diet. The asterisks and symbols represent the significance levels calculated by one-way ANOVA followed by Tukey–Kramer multiple comparisons test: *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls (n = 5 and bars represent S.E.M., each sample run in triplicate). (B) Representative western blots showing protein expression of PPARα, γ, δ, FL- & NT-PGC-1α, NRF-1, Cyt c, Tfam, HO-1 and GR. 20 μg of protein from striatal lysates of BACHD mice and their WT littermates were separated by SDS-PAGE and immunoblotted with the antibodies. β-actin served as a loading control. (C) Mitochondrial respiratory chain enzyme activities were calculated as mean optical density/min/mg of tissue and expressed as percent change with reference to WT controls. Values were averaged from two independent experiments. *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls, n = 5. One-way ANOVA followed by Tukey–Kramer multiple comparisons test. (D) mtDNA copy number was averaged from two separate experiments utilizing Cyt b or 12S rRNA as the reference gene for mtDNA against the nuclear gene β-actin. Values are expressed as mtDNA/nDNA ratio relative to WT controls. *P < 0.001 compared with the WT controls; ϕP < 0.001 compared with BACHD controls, n = 5. One-way ANOVA followed by Tukey–Kramer multiple comparisons test.
Western blot analysis revealed a similar trend for protein levels of PPARγ, PPARδ, PGC-1α, Cyt c, Tfam, HO-1 and GR as their respective mRNA expression levels except NRF-1 (Fig. 2B). Striatum from the BACHD mice on a standard diet showed reduced levels of PPARγ, PPARδ, PGC-1α, NRF-1, Cyt c, Tfam, HO-1 and GR and bezafibrate treatment restored their protein levels (Fig. 2B). In BACHD striatum, the activity of mitochondrial respiratory chain complexes (OXPHOS) and mtDNA copy number is significantly reduced (Fig. 2C and D). Concomitant with the increase in mRNA and protein levels of PGC-1α, we observed increased mitochondrial activity and increased mtDNA copy number in the striatum of BACHD mice treated with bezafibrate (Fig. 2C and D).
Bezafibrate ameliorates neuropathological features in BACHD mice
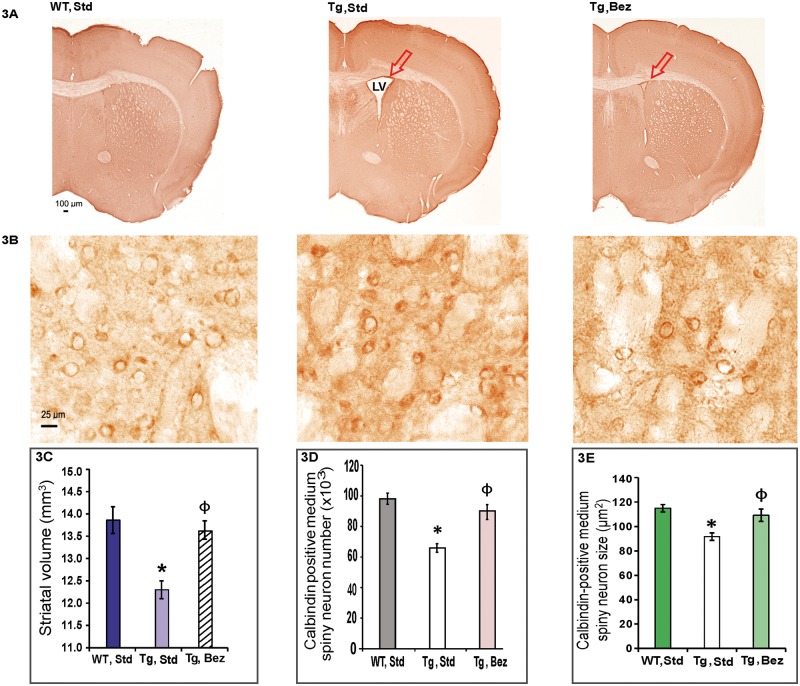
General atrophy of the brain, with losses of projection neurons in the deeper layers of the cortex and calbindin immunoreactive medium spiny neurons (MSNs) in the caudate-putamen are well-known neuropathological features of HD (2). We performed a stereological analysis of calbindin-immunoreactive medium spiny neuronal perikarya in the striatum of 16-month-old BACHD mice. In BACHD mice on a standard diet, enlarged lateral ventricles were consistently present (Fig. 3A) and striatal volume was significantly reduced along with significantly reduced MSN number and size (Fig. 3B–E). Induction of PGC-1α expression by bezafibrate treatment was accompanied by attenuation of these neuropathological features and the MSN size, number and striatal volume in BACHD mice on bezafibrate diet were not significantly different from those in WT littermates (Fig. 3A–E).
Figure 3.
Bezafibrate reduces striatal atrophy in BACHD mice. (A, B) Calbindin immunoreactivity in the striatum of BACHD mice and their WT littermates on bezafibrate or standard diet. Presence of a large lateral ventricle in untreated BACHD mice is demonstrated (A) along with smaller and dark-stained medium spiny neurons at a higher resolution (B). (C–E) Stereological analysis of calbindin-immunoreactive medium spiny neuronal perikarya in the striatum. The decrease in striatal volume (C), MSN number (D) and size (E) is significantly ameliorated by bezafibrate treatment. One-way ANOVA followed by Tukey–Kramer multiple comparisons test: *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls (n = 6 and bars represent S.E.M.).
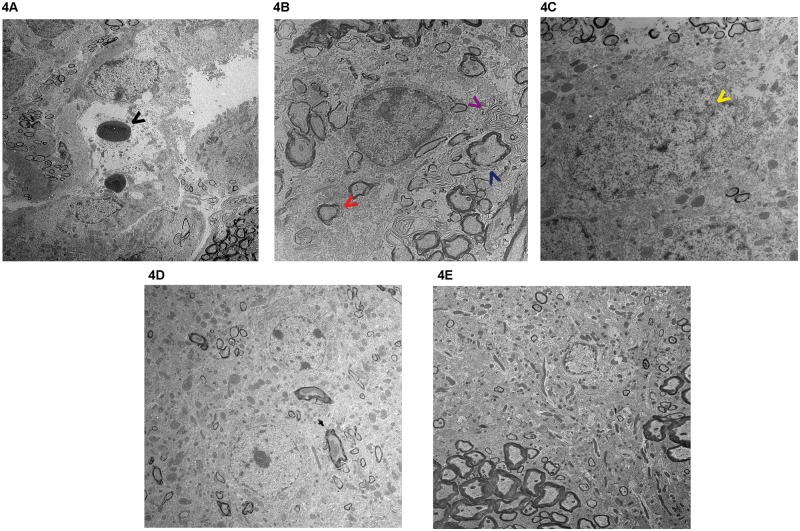
We further examined the striatum of BACHD mice treated with bezafibrate and those on standard diet at the ultrastructural level. In untreated BACHD mice, we observed several apoptotic neurons with condensed cytoplasm and abnormal nuclear shape showing margination and condensation of chromatin (Fig. 4A–C). Enlarged extracellular spaces, cytoplasmic vacuoles and lysosome-like dense bodies were also noted (Fig. 4A–C). There were fewer mitochondria per nm2 area and most of the mitochondria were abnormally shaped. Our observations are consistent with previously reported ultrastructural abnormalities in the brains of HD mice (21,24,28–30). An amelioration of these abnormalities was noted in the bezafibrate-treated BACHD mice. In particular, in the striata from bezafibrate-treated BACHD mice, the cytoplasm of the neurons was preserved, the axonal and dendritic profiles in the neuropil and myelination were relatively intact, and there were an increased number of mitochondria per nm2 area (Fig. 4D and E).
Figure 4.
Electron micrographs of BACHD striatum. Electron micrographs showing degenerated neurons in the striatum of BACHD mice (A–C) and its amelioration by Bezafibrate (D–E). (A–C) Apoptotic neurons with condensed cytoplasm and apoptotic vacuoles and dense bodies (black arrow head), loosely packed multi-lamellar bodies (red and purple arrow heads), deficient myelination (blue arrow head), abnormal nuclear shape showing invagination (yellow arrow head), margination and condensation of chromatin and presence of abnormally shaped mitochondria. (D, E) In striata from bezafibrate-treated BACHD mice, the cytoplasm of the neurons is preserved and the axonal and dendritic profiles in the neuropil and myelination are relatively intact. There are an increased number of mitochondria per nm2 area. Magnification A, D, E = 5000× and B and C are at 10 000×.
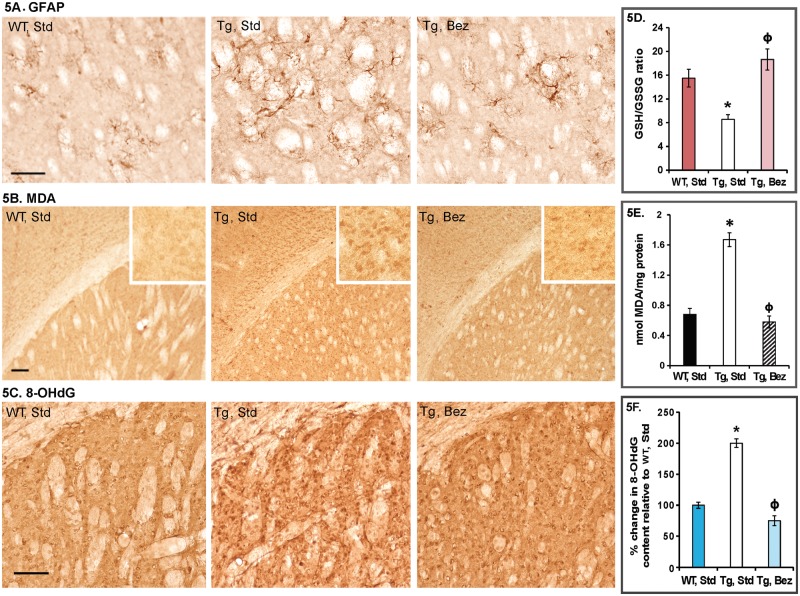
Glial fibrillary acidic protein (GFAP) immunostaining identifies reactive astrogliosis, an early marker of CNS damage in HD (2). Compared with age-matched WT mice striatum, the striatum of BACHD mice showed a remarkable increase in GFAP immunoreactivity, indicated by intense labeling throughout astroglial cell bodies and their fibrous processes and the presence of hypertrophied astrocytes (Fig. 5A). In BACHD mice treated with bezafibrate, only a few sparsely located hypertrophied astrocytes were observed (Fig. 5A).
Figure 5.
Reactive astrogliosis and oxidative stress in BACHD striatum. (A) Photomicrographs showing GFAP immunoreactivity in BACHD striatum. Several GFAP-labeled hypertrophied astrocytes are visible in BACHD mice on standard diet; bezafibrate attenuates reactive astrogliosis in striatum of BACHD mice. (B) Increased MDA immunostaining in striatum of BACHD mice compared with their WT littermates on standard diet. MDA staining is reduced in BACHD mice on bezafibrate diet. Insets show regions at a higher magnification. (C) 8-OHdG immunoreactivity is increased in striatum of BACHD mice on standard diet and bezafibrate markedly reduces it. (D) Reduced and oxidized levels of glutathione were measured using HPLC and are presented as GSH/GSSG ratios. One-way ANOVA followed by Tukey–Kramer multiple comparisons test: *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls (n = 6 and bars represent S.E.M.). (E) MDA measurement by HPLC. One-way ANOVA followed by Tukey–Kramer multiple comparisons test: *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls (n = 6 and bars represent S.E.M.). (F) 8-OHdG measurements by ELISA are presented as percent change relative to the values in WT, standard diet group. One-way ANOVA followed by Tukey–Kramer multiple comparisons test: *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls (n = 6 and bars represent S.E.M.).
Bezafibrate protects against oxidative stress
Increased oxidative stress is implicated in the neurodegeneration and neuronal death in HD (31–33). Levels of malondialdehyde (MDA, a marker for oxidative damage to lipids) are elevated in human HD striatum and cortex as compared with age-matched controls (34). Increased levels of another marker for oxidative stress, 8-hydroxy-2′-deoxyguanosine (8-OHdG), have been reported in the caudate, parietal cortex and peripherally in the serum and leukocytes, in patients diagnosed with HD. Moreover, 8-OHdG concentrations in prodromal HD were shown to increase as a function of proximity to projected disease onset (35). We observed increased immunoreactivity for MDA and 8-OHdG in BACHD striatum, which was ameliorated by the bezafibrate diet (Fig. 5B and C). Furthermore, the reduced to oxidized glutathione ratio, another marker for oxidative stress, was found to be significantly low in BACHD brain by HPLC and bezafibrate reversed this deficit (Fig. 5D). Consonant with the immunohistochemical data, we observed elevated levels of MDA and 8-OHdG in BACHD brains which were significantly reduced with bezafibrate treatment (Fig. 5E and F).
Bezafibrate prevents structural abnormalities in muscle and BAT
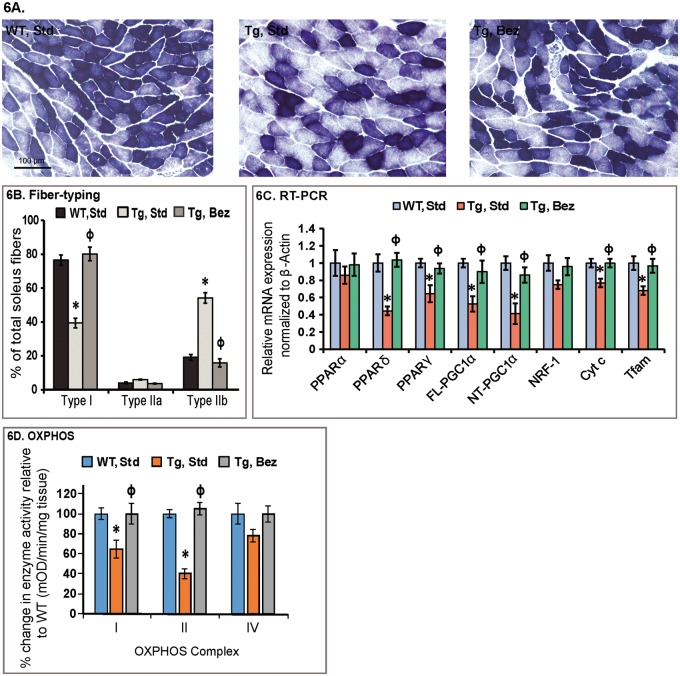
PGC-1α levels are normally high in muscle enriched with type I fibers, such as the soleus muscle, and very low in type II fiber rich muscles such as the extensor digitorum longus and the gastrocnemius (36). We examined the soleus muscle of BACHD mice and their WT littermates for fiber typing using succinate dehydrogenase (SDH) histochemistry (Fig. 6A). There was reduced SDH staining in the soleus of BACHD mice on a standard diet. Quantitation of the SDH histochemistry revealed a significant reduction of type I fibers, and an increase in type II fibers in the soleus muscle of BACHD mice, consistent with the reduced expression of PGC-1α (Fig. 6B and C). A reversal of this fiber-type switching was seen in BACHD mice on the bezafibrate diet, with the type I and II fibers returning back to normal levels seen in WT mice, again following the expression pattern of PGC-1α (Fig. 6B and C). Moreover, the activity of mitochondrial respiratory chain complexes (OXPHOS) is significantly reduced in BACHD soleus muscle compared with that in the WT littermates (Fig. 6D). Concomitant with the increase in mRNA expression of PGC-1α and its downstream targets, we observed increased mitochondrial activity in the soleus of BACHD mice treated with bezafibrate (Fig. 6D).
Figure 6.
Bezafibrate rescues PPAR–PGC-1α signaling pathway and structural abnormalities in soleus muscle. (A) Histochemical staining for SDH in soleus muscle sections from WT and BACHD mice with or without bezafibrate treatment. (B) Fiber typing of soleus muscle. Decreased proportion of mitochondria enriched oxidative type I fibers can be seen in soleus from BACHD mice as compared with their WT littermates. An enrichment of type I fibers and a decrease in glycolytic type IIb fibers can be seen in soleus muscle from BACHD mice on bezafibrate diet. *P < 0.05 as compared with WT controls; ϕP < 0.001 compared with BACHD controls (n = 6 in each group). (C) Relative expression of PPARα, γ, δ, FL and NT-isoforms of PGC-1α, and the downstream target genes, nuclear respiratory factor NRF-1, Cyt c and Tfam in soleus muscle of BACHD mice on a standard diet or on the bezafibrate diet. The levels of each gene transcript were normalized to that of β-actin and expressed as fold variation relative to the WT mice on a standard diet. The asterisks and symbols represent the significance levels calculated by one-way ANOVA followed by Tukey–Kramer multiple comparisons test: *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls (n = 5 and bars represent S.E.M., each sample run in triplicate). (D) Mitochondrial respiratory chain enzyme activities were calculated as mean optical density/min/mg of tissue and expressed as percent change with reference to WT controls. Values were averaged from two independent experiments. *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls, n = 6. One-way ANOVA followed by Tukey–Kramer multiple comparisons test.
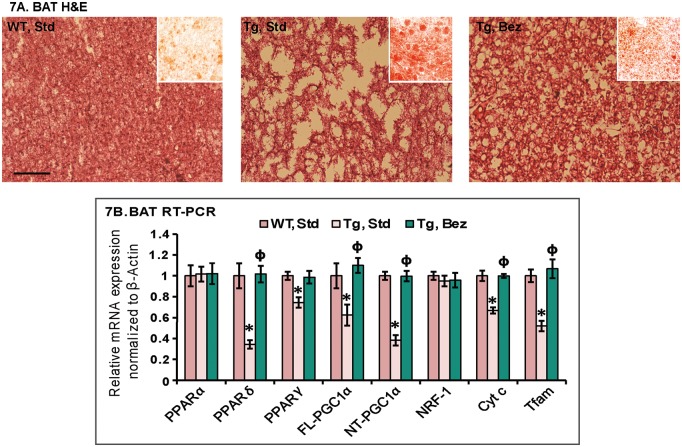
BAT is the principal tissue that mediates adaptive thermogenesis, a phenomenon which is defective in HD transgenic mice (10,17). BAT is distinguished from white fat by its high degree of vascularization and high mitochondrial density (37). PGC-1α is highly expressed in BAT and is a key mediator of adaptive thermogenesis by activating uncoupling protein 1 (38). As seen with the other HD mice models, the Hematoxylin and Eosin staining of BAT from the BACHD mice showed marked reductions in cell density and nuclei numbers and increased vacuolization when compared with WT mice (Fig. 7A). The white-fat like appearance of BAT was due to accumulation of neutral lipids as revealed by Oil red O staining (Fig. 7A, inset). Bezafibrate reduced vacuolization in the BAT of the BACHD mice as compared with BACHD mice fed a standard diet, and reduced Oil red O staining was also observed along with an increased expression of PPARγ, PPARδ, PGC-1α and its downstream mitochondrial biogenesis genes (Fig. 7A and B).
Figure 7.
Bezafibrate rescues structural abnormalities in BAT. (A) BATs of BACHD mice and their WT littermates stained with hematoxylin-and-eosin showing increased apparent vacuolization in the BACHD mice. Oil red O staining (red staining, inset) revealed abundant accumulation of larger lipid droplets in the BACHD mice as compared with WT mice. Bezafibrate reduces the accumulation of lipids and apparent vacuolization in the BACHD mice. (B) Relative expression of PPARα, γ, δ, FL and NT-isoforms of PGC-1α, and the downstream target genes, NRF-1, Cyt c and Tfam normalized to β-actin in BAT of BACHD mice on a standard or on the bezafibrate diet. One-way ANOVA followed by Tukey–Kramer multiple comparisons test: *P < 0.05 compared with the WT controls; ϕP < 0.05 compared with BACHD controls (n = 5 and bars represent S.E.M., each sample run in triplicate).
Discussion
Mitochondrial dysfunction and oxidative stress are intertwined in HD and there is substantial evidence for an impaired function of PGC-1α, the master co-regulator of mitochondrial biogenesis and antioxidant defense (9,10,12–14,17,39,40). It follows, therefore, that an upregulation of this critical transcriptional coactivator sitting at the crossroads of impaired mitochondrial function, increased oxidative damage and neurodegeneration would be beneficial in HD. We showed that upregulation of PGC-1α activity can be achieved by activating PPARs. PPAR agonists increase OXPHOS capacity in mouse and human cells, and enhance mitochondrial biogenesis; PPARγ agonists such as rosiglitazone, thiazolidinedione and pioglitazone exert neuroprotective effects in vitro and in vivo (41–45).
We utilized the pan-PPAR-agonist, bezafibrate, which was earlier shown to be effective in increasing life span and delaying the onset of symptoms in a mouse model of mitochondrial myopathy (46). Bezafibrate also improved mitochondrial function in the central nervous system of a mouse model of mitochondrial encephalopathy (47). Stimulation of the PPAR–PGC-1α axis by bezafibrate produces wide-spread beneficial effects in brain and peripheral tissues in the R6/2 transgenic mouse model of HD (21). According to consensus best practice guidelines, it is recommended that potential neuroprotective therapeutics should be tested whenever possible in at least two transgenic HD mouse lines, including at least one FL HD transgenic mouse model (22,23). We, therefore, performed studies to investigate effects of bezafibrate in the BACHD FL Htt mouse model.
We found that the impairment of PGC-1α pathway can be reversed in the striatum, soleus muscle and BAT of BACHD mice by bezafibrate. PGC-1α co-activates nuclear transcription factor NRF-1 increasing its activity and thereby activating downstream genes involved in mitochondrial biogenesis such as Cyt c and Tfam, increased activity of mitochondrial respiratory chain complexes and increased mtDNA copy number. We found wide-spread beneficial effects of bezafibrate on behavior, survival and histopathological features in the striatum, soleus muscle and BAT of BACHD mice. BACHD mice show robust and progressive deficits in motor coordination and exploratory activity and bezafibrate rescued the phenotype. The increase in survival observed with the bezafibrate diet (28%) is comparable to the highest range of percent increases in survival seen in other therapeutic trials in transgenic mouse models of HD (48). We showed that administration of creatine or triterpenoid compounds to N171-82Q mice increased survival by 19 or 21.9%, respectively, and that administration of coenzyme Q10 with remacemide or administration of mithramycin increased survival by 31.8 or by 29.1% in R6/2 mice, respectively (21,49–52). Bezafibrate, therefore, improves the behavioral phenotype and survival of BACHD mice in a comparable range to the best therapeutic interventions thus far tested. Compounds previously demonstrated to be beneficial in various animal models of HD have not met with much success in clinical trials so far, limiting the extent of predictive response to experimental treatments. In general, it is difficult to mimic the heterogeneity of the human population, comorbidities that may be present, and to fully recapitulate the course of human disease in the relatively short lifespan of animals. There are some successes, however, which encourage preclinical studies. One such example can be noted in pridopidine, a dopamine stabilizer, which shows therapeutic benefit in preclinical models of HD as well as in clinical trials (53). Bezafibrate, with its wide range of beneficial effects in both the R6/2 as well as BACHD models, two of the best animal models of HD that mimic juvenile and adult-onset disease, respectively, is a promising agent for clinical trials since it is already being used in patients to treat hyperlipidemia.
We found that the improved behavioral phenotype, increased survival and the induction of the PGC-1α signaling pathway was accompanied by reduced neuropathological features in striatum of BACHD mice treated with bezafibrate. Moderate increases in PGC-1α activity/expression are neuroprotective especially in the disease models where there is a deficiency to begin with (13,21,54–56). Administration of a lentiviral vector expressing PGC-1α into the striatum of R6/2 mice increased the mean neuronal volume of medium spiny neurons (12). Overexpression of PGC-1α was shown to enhance the mitochondrial membrane potential and to reduce mitochondrial toxicity in in vitro models of HD (17). A cross between mice inducibly overexpressing PGC-1α with a transgenic mouse model of HD revealed that PGC-1α overexpression virtually eradicates aggregates of mHtt protein in the brains of the HD mice by switching on the expression of the transcription factor EB, a key regulatory transcription factor that activates genes in the autophagy-lysosome pathway of protein turnover (57).
In addition to the stimulation of PPAR–PGC-1α axis, the beneficial effects of bezafibrate have been associated with its ability to reduce inflammation and stimulate lipid metabolism. We recently showed that bezafibrate is neuroprotective in the P301S transgenic mice, a mouse model of tauopathy, where it activated mitochondrial biogenesis and reduced inflammatory markers via PPAR mediated inhibition of inducible nitric oxide synthase and cyclooxygenase 2 (58). Earlier, in an early-onset partial COX-deficiency model (Surf1-KO mice), it was shown that bezafibrate induced expression of PPARα and PPARβ/δ, but also caused weight loss and hepatomegaly (59). It increased genes involved in fatty acid oxidation but did not increase mtDNA content and mitochondrial respiratory chain activities in Surf1-KO mice. In a late-onset mitochondrial myopathy mice, bezafibrate delayed the accumulation of COX-negative fibers and mtDNA deletions (60). However, bezafibrate did not induce mitochondrial biogenesis in this model and produced marked hepatomegaly. The authors suggested that the adverse effects of hepatomegaly may mask beneficial effects of bezafibrate in these mice. We also observed slight increase in liver weight, which is a known effect of PPAR stimulation in mice, but not in primates. Consistent with our previous studies with the R6/2 mice, we, however, did observe an increase in PGC-1α, NRF1, Cyt c, Tfam and numbers of mitochondria. This shows that background strain differences, treatment regimen and origin and severity of mitochondrial dysfunction alter the efficacy of bezafibrate in producing increases in PGC-1α and downstream genes, since in both our work and that of Wenz et al. (46,47,61), there were increases in PGC-1α.
Previous studies showed that the expression of mHtt in primary oligodendrocytes results in decreased expression of PGC-1α, and decreased expression of myelin basic protein and deficient myelination were found in the R6/2 and BACHD mouse models of HD; and a decrease in myelin basic protein and deficient postnatal myelination occurs in the striatum of PGC-1α knockout mice (29). In accordance with earlier studies, we observed ultrastructural abnormalities in the striatum of BACHD mice, including the presence of pyknotic nuclei, margination and condensation of cytoplasm, apoptotic bodies, deficient myelination, and abnormally shaped mitochondria, all features ameliorated by bezafibrate in the treated mice. Bezafibrate restored the mitochondrial density in striatum of BACHD mice. Astrocytes are closely associated with neurons and play critical roles in neuronal sustenance providing not only a structural frame work, but also functional roles in homeostasis, regulation of blood flow, regulation of synapse function, recycling neurotransmitter precursors, etc. Destruction of neurons results in a change of molecular expression and morphology of the astrocytes in the vicinity, resulting in reactive astrocytosis or astrogliosis. Astrogliosis is consistently observed in the brains of HD transgenic mice, including the BACHD mice (present study) and there were fewer hypertrophied astrocytes in the bezafibrate-treated BACHD mice brain. Overexpression of PGC-1α was previously shown to markedly increase the resistance of human astrocytes against oxidative stress, increase mitochondrial antioxidant capacity and completely abolish oxidative stress-induced intracellular ROS production (62).
Studies in both HD patients and experimental models of HD support a role for oxidative stress and ensuing mitochondrial dysfunction in mediating the neuronal degeneration in HD (31–33,63–66). There is increased mtDNA damage in STHdhQ111/Q111 striatal cells and in human HD striata and skin fibroblasts, which was ascribed to mHtt-associated oxidative stress (67). PGC-1α plays a role in the suppression of oxidative stress by directly regulating the activity/expression of antioxidant enzymes and together with PPARs, PGC-1α induces HO-1 (25,27). In concert with the increase in PGC-1α expression, we observed that the oxidative stress response genes such as HO-1 and GR also increase in the striatum of BACHD mice treated with bezafibrate. We show that the markers of oxidative stress, such as MDA and 8-OHdG, are significantly increased in BACHD mice and GSH/GSSG ratio is significantly reduced in brain. Bezafibrate reverses these anomalies and restores the antioxidant capacity in BACHD striatum.
PGC-1α plays a critical role in mitochondrial biogenesis in muscle, and in influencing whether muscle contains slow-twitch fatigue resistant oxidative fibers (type I and IIa fibers, rich in mitochondria, use OXPHOS to generate ATP) or fast-twitch fatiguable glycolytic fibers (type IIx and IIb fibers, containing fewer mitochondria, use glycolysis to generate ATP) (36). In PGC-1α knockout mice, there is a shift from type I and type IIa toward type IIx and IIb muscle fibers which is accompanied by myopathy and exercise intolerance (68). Impaired generation of ATP in muscle and a myopathy occurs in gene-positive individuals at risk for HD, HD patients and HD transgenic mice (69–71). We observed reduced PGC-1α activity in muscle of transgenic mouse models of HD, and in myoblasts and muscle biopsies from HD patients; and fiber-type switching in the NLS-N171-82Q and R6/2 transgenic mouse models of HD (9,21). We observed a significant increase of type IIb fibers and a significant reduction of type I fibers in soleus muscle of BACHD mice, consistent with reduced PGC-1α levels and reduced mitochondrial activity. The rescue of PGC-1α signaling pathway by bezafibrate treatment in BACHD mice was associated with reversal of the fiber-type switching back to normal and restoration of OXPHOS enzyme activity.
BAT is a highly specialized tissue consisting of lipid droplets surrounded by numerous mitochondria. PGC-1α is rapidly induced in response to cold exposure and regulates key components of adaptive thermogenesis including the uncoupling of respiration via uncoupling proteins, resulting in heat production in BAT. This process is impaired in HD transgenic mice and they develop significant hypothermia at both baseline and following cold exposure (10,17). In brown fat adipocytes, there are reduced ATP/ADP ratios and mitochondrial numbers, similar to the findings in PGC-1α knock-out mice (72,73). In line with the previous findings, we observed that in BAT of the BACHD mice, there is a marked vacuolization, which is due to accumulation of neutral lipids. Bezafibrate reduced the vacuolization and Oil red O staining in the BAT of BACHD mice, indicating amelioration of neutral lipid accumulation.
In conclusion, we show that stimulation of PPAR–PGC-1α axis by bezafibrate is neuroprotective, produces beneficial effects in peripheral tissues and results in improved phenotype and survival of the BACHD mice. Bezafibrate is an attractive agent for clinical studies since it has been used in patients for more than 25 years, and it is well tolerated with few side effects. It is therefore particularly attractive for clinical trials in neurodegenerative diseases such as HD. Our study showing beneficial effects of bezafibrate in the BACHD mice, combined with our earlier studies using bezafibrate in R6/2 mice, provides compelling evidence that bezafibrate may prove to be an effective neuroprotective agent for treatment of HD.
Materials and Methods
Reagents
Bezafibrate and other chemicals were purchased from Sigma (St. Louis, MO). Perchloric acid (69–72%) was purchased from J.T. Baker (Phillipsburg, NJ). Anti-calbindin and anti-8-OHdG antibodies were from Chemicon (Temecula, CA); anti-MDA-modified protein was a gift from Dr. Craig Thomas, and anti-GFAP was from Dako, Denmark. The sequences of all the primers used in this study have been published elsewhere and/or are available on request (14,21).
Animals and treatment
BACHD founder mice expressing expanded human Htt with 97 mixed CAA–CAG repeats were kindly provided by Dr. X. William Yang (University of California, Los Angeles, CA). These mice were bred with FVB/NJ mice (Jackson Laboratory, Bar Harbor) to generate male and female BACHD and WT littermates. All experiments were conducted within National Institutes of Health guidelines for animal research and were approved by the Weill Cornell Medical College Animal Care and Use Committee. The animals were kept on a 12-h light/dark cycle, with food and water available ad libitum. Mice were fed standard diet containing 0.5% bezafibrate or standard diet (Purina-Mills, Richmond, IN), starting at 10 months up to 16 months of age.
Behavioral tests
Experimenters were blind to the genotype during all testing, at least until the appearance of a robust phenotype in the mutants. We utilized a behavioral testing battery consisting of: rotarod and open field. On the rotarod (Economex, Columbus Instruments, Columbus, OH), mice were tested over three consecutive days, in three 5 min trials, with an accelerating speed (from 0 to 40 RPM in 5 min) separated by a 45 min inter-trial interval. The latency to fall from the rod was recorded. Exploratory behavior was recorded in the open-field (45 cm × 45 cm; height: 20 cm), for 10 min per day using a video tracking system (Ethovision 3.0, Noldus Technology, Attleborough, MA) and averaged over 3 days.
Real-time PCR
Total RNA was isolated from liquid nitrogen snap frozen tissues using Trizol reagent, according to manufacturer’s instructions (Invitrogen, Life Technologies, Carlsbad, CA). Genomic DNA was removed using RNase free DNase (Ambion, Life Technologies, CA) in RNA pellets re-suspended in DEPC-treated water (Ambion, Life Technologies). Total RNA purity and integrity was confirmed and equal amounts of RNA were reverse transcribed using the cDNA Synthesis Kit (Invitrogen, Life Technologies). Real-time RT-PCR was performed using the ABI prism 7900 HT sequence detection system (Applied Biosystems, Foster City, CA). Expression of the gene β-actin served as a control to normalize values. Relative expression was calculated using the 2−(ΔΔCt) method. For mtDNA copy number quantification method described by Chen et al. (74) was used. Essentially, DNA was extracted from striatum samples using the Qiagen kit (Qiagen, CA) and used for amplification of the mitochondrial gene 12S rRNA and Cyt b and normalized against nuclear gene β-actin (two actin gene copies).
Western blot
Striatum tissue from BACHD mice and their littermates were homogenized in tissue extraction buffer containing 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 0.5% NP-40, 0.5% deoxycholate supplemented with protease and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO) and protein concentration was determined using the BCA protein assay (Pierce, ThermoFisher Scientific, Waltham, MA). Equal amounts of protein were run on a 4–12% Tris–glycine gel (Invitrogen, Life Technologies). The gel was blotted on polyvinylidene fluoride membrane (Bio-Rad) or nitrocellulose (Bio-Rad). After transfer, membranes were blocked for 1 h at room temperature or 4°C overnight, depending on the protein being detected, in Tris–buffered saline/Tween-20 (TBST) (50 mM Tris–HCl, 150 mM NaCl, pH 7.4, 0.1% Tween-20) containing 5% non-fat dried milk. Primary antibodies used were either PPARα, γ, δ, NRF-1, HO-1, GR (all from Abcam, Cambridge, MA), PGC-1α (Calbiochem/EMD Millipore, Temecula, CA), Cyt c (BD Biosciences, San Jose, CA), Tfam or β-actin (both from Sigma-Aldrich). Membranes were then washed three times with TBST and incubated for 1 h with HRP-conjugated secondary antibody and the immunoreactive proteins detected using a chemiluminescent substrate (Pierce, ThermoFisher Scientific).
Subcellular fractionation
Mitochondrial and cytosolic fractions of the striatum and muscle were isolated with the Mitochondria Isolation Kit for Tissue (Abcam) per manufacturer’s instruction. In brief, striata were washed with washing buffer, homogenized with a Kontes Microtube Pellet Pestle Rod with motor in isolation buffer, centrifuged at 1000g for 10 min., the pellet was saved (nuclear enriched fraction) and supernatant was centrifuged again at 12 000g for 15 min. The supernatants (cytosolic fraction) were saved, the pellets (mitochondrial fraction) washed with Isolation buffer twice and re-suspended with isolation buffer with complete protein inhibitor cocktail (Roche Applied Bioscience, Indianapolis, IN). Mitochondrial respiratory chain complex activities were measured using enzyme activity microassay kits (Abcam).
Histological analysis
Mice intended for neuropathologic analysis were deeply anesthetized by intraperitoneal injection of sodium pentobarbital and perfused with 0.9% sodium chloride followed by 4% paraformaldehyde. Tissues were harvested, sectioned and processed for immunohistochemistry as described previously (9,10,18,21,52,75).
Transmission electron microscopy
Transmission electron microscopy was performed using previously published methods (9), except that for striatum the post-fixation was performed in 1% osmium tetroxide in 0.1 M buffer for 60 min at room temperature.
Measurement of oxidative stress markers
The HPLC determination of MDA was carried out by method introduced by Agarwal and Chase (76) with slight modifications. We used HPLC system coupled with Waters 474 scanning fluorescence detector, Waters 717 plus autosampler with cooled platform and Perkin Elmer Binary pump. The system was controlled by ESA501 software (ESA, Inc Chelmsford, MA). The mobile phase contained acetonitrile and 50 mM potassium monobasic phosphate pH 6.8 at a v/v ratio 4:6, pumped out in an isocratic mode at the rate 1.0 ml/min. Fluorescence detector was set at 515 nm excitation, 553 nm emission, ×1000 gain, 3 s digital filter and attitude 2. The metabolite separation was performed on Phenomenex, C18, 150×3 mm, 5 μm analytical column housed at 30°C. Under these conditions, MDA was eluted at 2.22 min. Stock solutions were prepared using distilled deionized water unless otherwise indicated. The MDA standard was prepared with 40% ethanol solution. Butylated hydroxytoluene (BHT) solution was prepared in 95% ethanol to a final concentration of 0.05% BHT. 2-Thiobarbituric acid (TBA) was dissolved in water on a stirring hot-plate at 50–55°C to a final concentration 42 mM. Tissue samples were homogenized in 40% ethanol. 50 µl of BHT solution, 400 µl of 0.44 M phosphoric acid solution and 100 µl of TBA solution were added to 50 µl of the sample homogenate or 50 µl of the MDA standard. The tubes were capped with screw cap tightly, vortexed and heated on a dry bath for 1 h at 100°C. After heat derivatization, samples were immediately placed on an ice-cold water bath for 5 min. 250 µl of n-butanol was subsequently added into each vial for extraction of the MDA–TBA complex. Tubes were then vortexed for 5 min and centrifuged at 14 000g for 3 min, separating the two phases. From each sample 100 µl aliquots were transferred from the n-butanol layer into HPLC vials for analysis. The concentration of MDA in a sample was calculated using standard curve equation as nmol per mg of protein.
HPLC determination of GSH and GSSG was carried out as described previously (77) with minor modifications. We used HPLC system coupled with 8-channel CoulArray electrochemical detector (ESA Inc), Shimadzu pump, and Waters 717-plus autosampler with cooled platform. The mobile phase consisted of 50 mM LiH2PO4, 1.0 mM 1-octanesulfonic acid and 1.5% (v/v) methanol, pumped out in an isocratic mode at 0.8 ml/min. Metabolite separation was performed on C18, 80×4.6 mm, 3 μm analytical column (ESA Inc) equipped with Phenomenex Security guard column (cartridge C18, 4×2 mm). The potentials for an electrochemical detector were set up at the range 460–880 µV with increments of 60 µV. Under such conditions GSH and GSSG were eluted at 1.42 and 2.42 min. Samples for the GSH/GSSG analysis were prepared by addition of 300 µl 0.1 M PCA followed by brief (6 s) sonication, vortex and centrifugation at 14 000g for 20 min (4°C). Supernatant was transferred into fresh tube and centrifuged again at the same conditions. Clear supernatant (100 µl) was placed into HPLC vial for analysis, with an injection volume of 20 µl. Concentrations of GSH and GSSG were calculated as nmol per mg protein and are expressed as GSH/GSSG ratio.
8-OHdG levels in brain were measured using commercial assay kit according to the manufacturer’s protocol (Abcam).
Statistical analysis
All analysis was performed using GraphPad Prism software. Data are represented as mean ± SEM. Analysis of significance was performed using Unpaired, Student’s two-tailed t-test or one-way ANOVA followed by Tukey–Kramer multiple comparisons test as indicated. Values were considered significant at P < 0.05.
Acknowledgements
The authors would like to thank Mr. Thomas Hennessey and Mr. Daniel Ho for their technical assistance during preliminary studies.
Conflict of Interest statement. None declared.
Funding
This work was supported by National Institutes of Health grant [P01AG14930]. A.J. and M.F.B. are supported by National Institutes of Health/National Institute of Neurological Disorders & Stroke grant [1R01 NS086746-01A1] and a gift from ‘Sons of HD’.
References
- 1.HDCRG (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 2.Vonsattel J.P., DiFiglia M. (1998) Huntington disease. J. Neuropathol. Exp. Neurol., 57, 369–384. [DOI] [PubMed] [Google Scholar]
- 3.Rosas H.D., Koroshetz W.J., Chen Y.I., Skeuse C., Vangel M., Cudkowicz M.E., Caplan K., Marek K., Seidman L.J., Makris N, et al. (2003) Evidence for more widespread cerebral pathology in early HD: an MRI-based morphometric analysis. Neurology, 60, 1615–1620. [DOI] [PubMed] [Google Scholar]
- 4.Waldvogel H.J., Dragunow M., Faull R.L. (2015) Disrupted vasculature and blood-brain barrier in Huntington disease. Ann. Neurol., 78, 158–159. [DOI] [PubMed] [Google Scholar]
- 5.Beal M.F. (2005) Mitochondria take center stage in aging and neurodegeneration. Ann. Neurol., 58, 495–505. [DOI] [PubMed] [Google Scholar]
- 6.Chandra A., Johri A., Beal M.F. (2014) Prospects for neuroprotective therapies in prodromal Huntington's disease. Mov. Disord, 29, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johri A., Beal M.F. (2012) Mitochondrial dysfunction in neurodegenerative diseases. J. Pharmacol. Exp. Ther., 342, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossy-Wetzel E., Petrilli A., Knott A.B. (2008) Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci., 31, 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaturvedi R.K., Adhihetty P., Shukla S., Hennessy T., Calingasan N., Yang L., Starkov A., Kiaei M., Cannella M., Sassone J, et al. (2009) Impaired PGC-1alpha function in muscle in Huntington's disease. Hum. Mol. Genet., 18, 3048–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaturvedi R.K., Calingasan N.Y., Yang L., Hennessey T., Johri A., Beal M.F. (2010) Impairment of PGC-1alpha expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington's disease following hronic energy deprivation. Hum. Mol. Genet., 19, 3190–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa V., Giacomello M., Hudec R., Lopreiato R., Ermak G., Lim D., Malorni W., Davies K.J., Carafoli E., Scorrano L. (2010) Mitochondrial fission and cristae disruption increase the response of cell models of Huntington's disease to apoptotic stimuli. EMBO Mol. Med., 2, 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. (2006) Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell, 127, 59–69. [DOI] [PubMed] [Google Scholar]
- 13.Johri A., Chandra A., Beal M.F. (2013) PGC-1alpha, mitochondrial dysfunction, and Huntington's disease. Free Radic. Biol. Med., 62, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johri A., Starkov A.A., Chandra A., Hennessey T., Sharma A., Orobello S., Squitieri F., Yang L., Beal M.F. (2011) Truncated peroxisome proliferator-activated receptor-gamma coactivator 1alpha splice variant is severely altered in Huntington's disease. Neurodegener. Dis., 8, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirendeb U., Reddy A.P., Manczak M., Calkins M.J., Mao P., Tagle D.A., Reddy P.H. (2011) Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington's disease: implications for selective neuronal damage. Hum. Mol. Genet., 20, 1438–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R, et al. (2011) Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med., 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weydt P., Pineda V.V., Torrence A.E., Libby R.T., Satterfield T.F., Lazarowski E.R., Gilbert M.L., Morton G.J., Bammler T.K., Strand A.D, et al. (2006) Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab., 4, 349–362. [DOI] [PubMed] [Google Scholar]
- 18.Boussicault L., Herard A.S., Calingasan N., Petit F., Malgorn C., Merienne N., Jan C., Gaillard M.C., Lerchundi R., Barros L.F, et al. (2014) Impaired brain energy metabolism in the BACHD mouse model of Huntington's disease: critical role of astrocyte-neuron interactions. J. Cereb. Blood Flow Metab., 34, 1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford J., Shin J.Y., Roberts M., Wang C.E., Sheng G., Li S., Li X.J. (2010) Mutant huntingtin in glial cells exacerbates neurological symptoms of Huntington disease mice. J. Biol. Chem., 285, 10653–10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee W., Reyes R.C., Gottipati M.K., Lewis K., Lesort M., Parpura V., Gray M. (2013) Enhanced Ca(2+)-dependent glutamate release from astrocytes of the BACHD Huntington's disease mouse model. Neurobiol. Dis., 58, 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johri A., Calingasan N.Y., Hennessey T.M., Sharma A., Yang L., Wille E., Chandra A., Beal M.F. (2012) Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington's disease. Hum. Mol. Genet., 21, 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menalled L., Brunner D. (2014) Animal models of Huntington's disease for translation to the clinic: best practices. Mov. Disord., 29, 1375–1390. [DOI] [PubMed] [Google Scholar]
- 23.Menalled L., Lutz C., Ramboz S., Brunner D., Lager B., Noble S., Park L., Howland D. (2014) A Field guide to working with mouse models of Huntington’s disease. Available from: http://www.psychogenics.com/newsletter/A_field_Guide_to_Working_with_Mouse_Models_of_Huntington_Disease/index.html, last accessed April 05, 2016
- 24.Gray M., Shirasaki D.I., Cepeda C., Andre V.M., Wilburn B., Lu X.H., Tao J., Yamazaki I., Li S.H., Sun Y.E, et al. (2008) Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci., 28, 6182–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali F., Ali N.S., Bauer A., Boyle J.J., Hamdulay S.S., Haskard D.O., Randi A.M., Mason J.C. (2010) PPARdelta and PGC1alpha act cooperatively to induce haem oxygenase-1 and enhance vascular endothelial cell resistance to stress. Cardiovasc. Res., 85, 701–710. [DOI] [PubMed] [Google Scholar]
- 26.Lin J., Handschin C., Spiegelman B.M. (2005) Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab., 1, 361–370. [DOI] [PubMed] [Google Scholar]
- 27.St-Pierre J., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W, et al. (2006) Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell, 127, 397–408. [DOI] [PubMed] [Google Scholar]
- 28.Davies S.W., Scherzinger E. (1997) Nuclear inclusions in Huntington's disease. Trends Cell Biol., 7, 422. [DOI] [PubMed] [Google Scholar]
- 29.Xiang Z., Valenza M., Cui L., Leoni V., Jeong H.K., Brilli E., Zhang J., Peng Q., Duan W., Reeves S.A, et al. (2011) Peroxisome-proliferator-activated receptor gamma coactivator 1 alpha contributes to dysmyelination in experimental models of Huntington's disease. J. Neurosci., 31, 9544–9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Z.X., Li S.H., Evans J., Pillarisetti A., Li H., Li X.J. (2003) Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease. J. Neurosci., 23, 2193–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Browne S.E., Beal M.F. (2006) Oxidative damage in Huntington's disease pathogenesis. Antioxid. Redox Signal., 8, 2061–2073. [DOI] [PubMed] [Google Scholar]
- 32.Johri A., Beal M.F. (2012) Antioxidants in Huntington's disease. Biochim. Biophys. Acta, 1822, 664–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stack E.C., Matson W.R., Ferrante R.J. (2008) Evidence of oxidant damage in Huntington's disease: translational strategies using antioxidants. Ann. N. Y. Acad. Sci., 1147, 79–92. [DOI] [PubMed] [Google Scholar]
- 34.Browne S.E., Bowling A.C., MacGarvey U., Baik M.J., Berger S.C., Muqit M.M., Bird E.D., Beal M.F. (1997) Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann. Neurol., 41, 646–653. [DOI] [PubMed] [Google Scholar]
- 35.Long J.D., Matson W.R., Juhl A.R., Leavitt B.R., Paulsen J.S., Investigators, P.-H. and Coordinators of the Huntington Study, G. (2012) 8OHdG as a marker for Huntington disease progression. Neurobiol. Dis, 46, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N, et al. (2002) Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature, 418, 797–801. [DOI] [PubMed] [Google Scholar]
- 37.Cannon B., Nedergaard J. (2004) Brown adipose tissue: function and physiological significance. Physiol. Rev., 84, 277–359. [DOI] [PubMed] [Google Scholar]
- 38.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell, 92, 829–839. [DOI] [PubMed] [Google Scholar]
- 39.McConoughey S.J., Basso M., Niatsetskaya Z.V., Sleiman S.F., Smirnova N.A., Langley B.C., Mahishi L., Cooper A.J., Antonyak M.A., Cerione R.A, et al. (2010) Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol. Med., 2, 349–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weydt P., Soyal S.M., Gellera C., Didonato S., Weidinger C., Oberkofler H., Landwehrmeyer G.B., Patsch W. (2009) The gene coding for PGC-1alpha modifies age at onset in Huntington's disease. Mol. Neurodegener., 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiang M.C., Chen C.M., Lee M.R., Chen H.W., Chen H.M., Wu Y.S., Hung C.H., Kang J.J., Chang C.P., Chang C, et al. (2010) Modulation of energy deficiency in Huntington's disease via activation of the peroxisome proliferator-activated receptor gamma. Hum. Mol. Genet., 19, 4043–4058. [DOI] [PubMed] [Google Scholar]
- 42.Jin J., Albertz J., Guo Z., Peng Q., Rudow G., Troncoso J.C., Ross C.A., Duan W. (2013) Neuroprotective effects of PPAR-gamma agonist rosiglitazone in N171-82Q mouse model of Huntington's disease. J. Neurochem., 125, 410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Napolitano M., Costa L., Palermo R., Giovenco A., Vacca A., Gulino A. (2011) Protective effect of pioglitazone, a PPARgamma ligand, in a 3 nitropropionic acid model of Huntington's disease. Brain Res. Bull., 85, 231–237. [DOI] [PubMed] [Google Scholar]
- 44.Quintanilla R.A., Jin Y.N., Fuenzalida K., Bronfman M., Johnson G.V. (2008) Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in the pathogenesis of Huntington disease. J. Biol. Chem., 283, 25628–25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morato L., Galino J., Ruiz M., Calingasan N.Y., Starkov A.A., Dumont M., Naudi A., Martinez J.J., Aubourg P., Portero-Otin M, et al. (2013) Pioglitazone halts axonal degeneration in a mouse model of X-linked adrenoleukodystrophy. Brain, 136, 2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenz T., Diaz F., Spiegelman B.M., Moraes C.T. (2008) Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab., 8, 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Noe N., Dillon L., Lellek V., Diaz F., Hida A., Moraes C.T., Wenz T. (2013) Bezafibrate improves mitochondrial function in the CNS of a mouse model of mitochondrial encephalopathy. Mitochondrion, 13, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hersch S.M., Ferrante R.J. (2004) Translating therapies for Huntington's disease from genetic animal models to clinical trials. NeuroRx, 1, 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andreassen O.A., Dedeoglu A., Ferrante R.J., Jenkins B.G., Ferrante K.L., Thomas M., Friedlich A., Browne S.E., Schilling G., Borchelt D.R, et al. (2001) Creatine increase survival and delays motor symptoms in a transgenic animal model of Huntington's disease. Neurobiol. Dis., 8, 479–491. [DOI] [PubMed] [Google Scholar]
- 50.Beal M.F., Ferrante R.J. (2004) Experimental therapeutics in transgenic mouse models of Huntington's disease. Nat. Rev. Neurosci., 5, 373–384. [DOI] [PubMed] [Google Scholar]
- 51.Ferrante R.J., Ryu H., Kubilus J.K., D'Mello S., Sugars K.L., Lee J., Lu P., Smith K., Browne S., Beal M.F, et al. (2004) Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington's disease. J. Neurosci, 24, 10335–10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stack C., Ho D., Wille E., Calingasan N.Y., Williams C., Liby K., Sporn M., Dumont M., Beal M.F. (2010) Triterpenoids CDDO-ethyl amide and CDDO-trifluoroethyl amide improve the behavioral phenotype and brain pathology in a transgenic mouse model of Huntington's disease. Free Radic. Biol. Med., 49, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Squitieri F., de Yebenes J.G. (2015) Profile of pridopidine and its potential in the treatment of Huntington disease: the evidence to date. Drug Des. Devel. Ther., 9, 5827–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komen J.C., Thorburn D.R. (2014) Turn up the power—pharmacological activation of mitochondrial biogenesis in mouse models. Br. J. Pharmacol., 171, 1818–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang H., Ward W.F., Jang Y.C., Bhattacharya A., Bokov A.F., Li Y., Jernigan A., Richardson A., Van Remmen H. (2011) PGC-1alpha protects neurons and alters disease progression in an amyotrophic lateral sclerosis mouse model. Muscle Nerve, 44, 947–956. [DOI] [PubMed] [Google Scholar]
- 56.Mudo G., Makela J., Di Liberto V., Tselykh T.V., Olivieri M., Piepponen P., Eriksson O., Malkia A., Bonomo A., Kairisalo M, et al. (2012) Transgenic expression and activation of PGC-1alpha protect dopaminergic neurons in the MPTP mouse model of Parkinson's disease. Cell. Mol. Life Sci., 69, 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsunemi T., Ashe T.D., Morrison B.E., Soriano K.R., Au J., Roque R.A., Lazarowski E.R., Damian V.A., Masliah E., La Spada A.R. (2012) PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med., 4, 142ra197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dumont M., Stack C., Elipenahli C., Jainuddin S., Gerges M., Starkova N., Calingasan N.Y., Yang L., Tampellini D., Starkov A.A, et al. (2012) Bezafibrate administration improves behavioral deficits and tau pathology in P301S mice. Hum. Mol. Genet., 21, 5091–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Viscomi C., Bottani E., Civiletto G., Cerutti R., Moggio M., Fagiolari G., Schon E.A., Lamperti C., Zeviani M. (2011) In vivo correction of COX deficiency by activation of the AMPK/PGC-1alpha axis. Cell Metab., 14, 80–90., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yatsuga S., Suomalainen A. (2012) Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum. Mol. Genet., 21, 526–535. [DOI] [PubMed] [Google Scholar]
- 61.Wenz T., Wang X., Marini M., Moraes C.T. (2011) A metabolic shift induced by a PPAR panagonist markedly reduces the effects of pathogenic mitochondrial tRNA mutations. J. Cell. Mol. Med., 15, 2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nijland P.G., Witte M.E., van het Hof B., van der Pol S., Bauer J., Lassmann H., van der Valk P., de Vries H.E., van Horssen J. (2014) Astroglial PGC-1alpha increases mitochondrial antioxidant capacity and suppresses inflammation: implications for multiple sclerosis. Acta Neuropathol. Commun., 2, 170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C.M., Wu Y.R., Cheng M.L., Liu J.L., Lee Y.M., Lee P.W., Soong B.W., Chiu D.T. (2007) Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington's disease patients. Biochem. Biophys. Res. Commun., 359, 335–340. [DOI] [PubMed] [Google Scholar]
- 64.Klepac N., Relja M., Klepac R., Hecimovic S., Babic T., Trkulja V. (2007) Oxidative stress parameters in plasma of Huntington's disease patients, asymptomatic Huntington's disease gene carriers and healthy subjects: a cross-sectional study. J. Neurol., 254, 1676–1683. [DOI] [PubMed] [Google Scholar]
- 65.Sorolla M.A., Reverter-Branchat G., Tamarit J., Ferrer I., Ros J., Cabiscol E. (2008) Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic. Biol. Med., 45, 667–678. [DOI] [PubMed] [Google Scholar]
- 66.Sorolla M.A., Rodriguez-Colman M.J., Tamarit J., Ortega Z., Lucas J.J., Ferrer I., Ros J., Cabiscol E. (2010) Protein oxidation in Huntington disease affects energy production and vitamin B6 metabolism. Free Radic. Biol. Med., 49, 612–621. [DOI] [PubMed] [Google Scholar]
- 67.Siddiqui A., Rivera-Sanchez S., Castro Mdel R., Acevedo-Torres K., Rane A., Torres-Ramos C.A., Nicholls D.G., Andersen J.K., Ayala-Torres S. (2012) Mitochondrial DNA damage is associated with reduced mitochondrial bioenergetics in Huntington's disease. Free Radic. Biol. Med., 53, 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Handschin C., Chin S., Li P., Liu F., Maratos-Flier E., Lebrasseur N.K., Yan Z., Spiegelman B.M. (2007) Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J. Biol. Chem., 282, 30014–30021. [DOI] [PubMed] [Google Scholar]
- 69.Gizatullina Z.Z., Lindenberg K.S., Harjes P., Chen Y., Kosinski C.M., Landwehrmeyer B.G., Ludolph A.C., Striggow F., Zierz S., Gellerich F.N. (2006) Low stability of Huntington muscle mitochondria against Ca2+ in R6/2 mice. Ann. Neurol., 59, 407–411. [DOI] [PubMed] [Google Scholar]
- 70.Kosinski C.M., Schlangen C., Gellerich F.N., Gizatullina Z., Deschauer M., Schiefer J., Young A.B., Landwehrmeyer G.B., Toyka K.V., Sellhaus B, et al. (2007) Myopathy as a first symptom of Huntington's disease in a Marathon runner. Mov. Disord., 22, 1637–1640. [DOI] [PubMed] [Google Scholar]
- 71.Turner C., Cooper J.M., Schapira A.H. (2007) Clinical correlates of mitochondrial function in Huntington's disease muscle. Mov. Disord., 22, 1715–1721. [DOI] [PubMed] [Google Scholar]
- 72.Leone T.C., Lehman J.J., Finck B.N., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C, et al. (2005) PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol., 3, e101.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin J., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M, et al. (2004) Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell, 119, 121–135. [DOI] [PubMed] [Google Scholar]
- 74.Chen M., Wu L., Wu F., Wittert G.A., Norman R.J., Robker R.L., Heilbronn L.K. (2014) Impaired glucose metabolism in response to high fat diet in female mice conceived by in vitro fertilization (IVF) or ovarian stimulation alone. PLoS One, 9, e113155.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stack C., Jainuddin S., Elipenahli C., Gerges M., Starkova N., Starkov A.A., Jove M., Portero-Otin M., Launay N., Pujol A, et al. (2014) Methylene blue upregulates Nrf2/ARE genes and prevents tau-related neurotoxicity. Hum. Mol. Genet., 23, 3716–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agarwal R., Chase S.D. (2002) Rapid, fluorimetric-liquid chromatographic determination of malondialdehyde in biological samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci., 775, 121–126. [DOI] [PubMed] [Google Scholar]
- 77.Yang L., Calingasan N.Y., Thomas B., Chaturvedi R.K., Kiaei M., Wille E.J., Liby K.T., Williams C., Royce D., Risingsong R, et al. (2009) Neuroprotective effects of the triterpenoid, CDDO methyl amide, a potent inducer of Nrf2-mediated transcription. PLoS One, 4, e5757.. [DOI] [PMC free article] [PubMed] [Google Scholar]