Abstract
Voltage-gated sodium channels, which are involved in pain pathways, have emerged as major targets for therapeutic intervention in pain disorders. Nav1.7, the tetrodotoxin-sensitive voltage-gated sodium channel isoform encoded by SCN9A and predominantly expressed in pain-sensing neurons in the dorsal root ganglion, plays a crucial role in nociception. MicroRNAs are highly conserved, small non-coding RNAs. Through binding to the 3′ untranslated region of their target mRNAs, microRNAs induce the cleavage and/or inhibition of protein translation. Based on bioinformatics analysis using TargetScan software, we determined that miR-30b directly targets SCN9A. To investigate the roles of Nav1.7 and miR-30b in neuropathic pain, we examined changes in the expression of Nav1.7 in the dorsal root ganglion by miR-30b over-expression or knockdown in rats with spared nerve injury. Our results demonstrated that the expression of miR-30b and Nav1.7 was down-regulated and up-regulated, respectively, in the dorsal root ganglion of spared nerve injury rats. MiR-30b over-expression in spared nerve injury rats inhibited SCN9A transcription, resulting in pain relief. In addition, miR-30b knockdown significantly increased hypersensitivity to pain in naive rats. We also observed that miR-30b decreased Nav1.7 expression in PC12 cells. Taken together, our results suggest that miR-30b plays an important role in neuropathic pain by regulating Nav1.7 expression. Therefore, miR-30b may be a promising target for the treatment of chronic neuropathic pain.
Keywords: Neuropathic pain, sodium channel, microRNA, analgesia
Introduction
Central or peripheral nervous system damage or dysfunction can cause neuropathic pain, which is characterized by spontaneous pain, hyperalgesia, and allodynia. Because signal transduction and the regulation of pain depend on the activity of ion channels in the afferent fiber and because sodium ion channels in excitable cells are the foundation of the resting and action potential, sodium ion channels are considered a potential target of neuropathic pain therapy.1,2 Nine sodium channel subunits have been identified (Nav1.1–Nav1.9). In particular, Nav1.7 has received intense scrutiny for its contributions to human genetic pain diseases.3
The tetrodotoxin-sensitive voltage-gated sodium channel Nav1.7, with α subunit that is encoded by SCN9A, is mainly expressed in the dorsal root ganglia (DRG) and sympathetic ganglia.4 Loss-of-function mutations in SCN9A can generate a congenital absence of unintentional pain,5–8 and gain-of-function mutations are linked to the pain associated with primary erythromelalgia9,10 and paroxysmal extreme pain disorder.11,12 Therefore, SCN9A is likely to be a crucial target for pain research.
Mature microRNAs (miRNAs) are non-coding RNAs with a length of 19–25 nucleotides that regulate post-transcriptional gene expression. When a miRNA and target mRNA bind completely, the target mRNA is immediately degraded; however, if the two species do not fully bind, the miRNA inhibits the translation of the target mRNA.13 Many studies have confirmed that certain microRNAs are causally involved in the development of neuropathic pain by regulating voltage-gated sodium channel protein expression and neuronal excitability. Sakai et al.14 found that miR-7a is down-regulated in the neurons of injured DRG in neuropathic pain and that the over-expression of miR-7a suppresses the increase in expression of the β2-subunit and normalizes the persistent hyperexcitability of nociceptive neurons. Chen et al.15 confirmed that miR-96 participates in the regulation of neuropathic pain by inhibiting Nav1.3 expression in the DRG of CCI rats. Zhao et al.16 also found that microRNAs regulate the expression of the nociceptor gene in mouse Nav1.8 sensory neurons. However, the mechanism by which the microRNA associates with Nav1.7 has not been reported in neuropathic pain.
We discovered that miR-30b is closely related to SCN9A using TargetScan bioinformatics software analysis. We examined the change in Nav1.7 expression in PC12 cells after over-expressing or knocking down miR-30b. Using the spared nerve injury (SNI) animal model for neuropathic pain, we demonstrated that miR-30b was down-regulated followed by an up-regulation of Nav1.7 α subunits. Furthermore, we demonstrated a novel role for miRNA in the maintenance of pain sensitization, showing that intrathecal applications of miR-30b could repress the up-regulated expression of Nav1.7 α subunits and consequently relieve pain, thereby identifying miR-30b as a potential therapeutic target.
Materials and methods
SCN9A 3′ untranslated region constructs and luciferase assay
The 3′ untranslated region (3’UTR) of SCN9A containing the miR-30b binding site was amplified with the forward primer 5′-GATATCTCGAGAGAAACAAAGTACTAGGGACAAAAAGATT-3′ and the reverse primer 5′-GTATCGCGGCCGCAAAAATGG-3′ from rat genomic DNA samples. The polymerase chain reaction (PCR) products were separated in an agarose gel and extracted, purified, and cloned using the TA cloning Kit (Promega, Madison, WI, USA). The SCN9A 3′UTR was linked to the vector psiCHECK-2 (Promega) containing Renilla and firefly luciferase using the restriction enzymes XhoI and NotI. The resulting construct was verified by sequencing.
HEK293 cells were seeded at 1 × 105 cells per well in 24-well plates. The cells were transfected using Lipofectamine 2000 according to the manufacturer’s suggestions 24 h after plating. In each well, 0.5 µg of psiCHECK-3′UTR vector or Mut-3′UTR vector was cotransfected with 100 pmol miR-30b agomir (GenePharma, Shanghai, China), while 100 pmol negative control was used in each experiment. Three replicates of each sample were evaluated, and each experiment was repeated at least three times. The cells were harvested with passive lysis buffer at 100 µl per well 48 h after transfection. The activity of Renilla luciferase in the cell lysate was measured using the Dual Luciferase assay system (Promega) in a Berthold Centro LB960 luminometer (Berthold Technologies, Germany) and normalized to the activity of firefly luciferase. The miRNA sequences are as follows:
miR-30b agomir: 5′-UGUAAACAUCCUACACUCAGCU-3′
3′-CUGAGUGUAGGAUGUUUACAUU-5′
miR-30b antagomir: 5′-AGCUGAGUGUAGGAUGUUUACA-3′
scramble-miRNA: 5′-ACAUUUGUUCCUACACUCAGCU-3′
3′-UGUAAACAAGGAUGUUUACAUU-5′
mi-RNA negative control: 5′-UUCUCCGAACGUGUCACGUTT-3′
3′-ACGUGACACGUUCGGAGAATT-5′
PC12 cell culture and transient transfections
PC12 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum, 100 kU/L penicillin, and 100 µg/L streptomycin (Gibco-BRL) at 37℃ with 5% CO2. Cells grown in culture flasks to 70%–80% confluence were collected and replated in 24-well plates at approximately 5 × 104 cells/cm2.
Prior to transfection, the cells were pretreated with glutamate (20 nM for 14 h, Sigma, Santa Clara, CA, USA). The transfections with miR-30b agomir/antagomir (GenePharma), scramble-miRNA, or negative control were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). For each sample, the treatments (100 pmol miR-30b agomir, 100 pmol antagomir, 100 pmol scramble-miRNA, 100 pmol negative control) and Lipofectamine reagent (2 μl) were diluted in 50 μl of DMEM lacking serum and antibiotics at room temperature. After 5 min, the diluted treatment sample was mixed with Lipofectamine. After 25 min, the mixture was added to 400 μl of DMEM lacking serum and antibiotics. The final mixture was applied to the cells on per well. After 6 h, the medium was replaced with DMEM containing 10% fetal bovine serum, 100 kU/L penicillin, and 100 µg/L streptomycin, and the cells were harvested at 48–72 h after transfection for mRNA and protein analysis.
Animal preparation and induction of peripheral neuropathic pain
Adult male Sprague–Dawley male rats weighing 280–350 g were purchased from the Animal Center of Henan Province. The rats were housed under artificial light from 7:00 a.m. to 7:00 p.m. in individual cages with free access to water and food. The room temperature was maintained at 24℃. The care and use of animals followed the recommendations and guidelines of the National Institutes of Health and was approved by Zhengzhou University Animal Care and Use Committee.
The rats were anesthetized with 2% isoflurane delivered via a nose cone, and an incision was made in the skin on the lateral surface of the thigh directly through the biceps femoris muscle, exposing the sciatic nerve and its three terminal branches: the sural, common peroneal, and tibial nerves. The common peroneal and tibial nerves were tightly ligated with 4.0 silk and sectioned distal to the ligation, removing 2 ± 4 mm of the distal nerve stump. Great care was taken to avoid any contact or stretching of the sural nerve. The muscle and skin were sutured. Sham controls underwent exposure of the sciatic nerve and its branches without the generation of any lesion.
Mechanical stimulus threshold test
The mechanical stimulus threshold was measured with von Frey hairs (North Coast Medical Inc., CA, USA). The animals were placed in wire mesh bottom cages and allowed to acclimatize for 30 min before examination. The plantar surface of the animal’s hind paw was stimulated with von Frey filament. Following application of calibrated von Frey filaments (0.4–15 g) with sufficient force to cause buckling of the filament, the 50% withdrawal threshold was determined according to the up-down method of Chaplan et al.17 in an ascending order of force until a paw withdrawal response were elicited. Each von Frey hair was applied to the paw for 8 s or until a response occurred. Each rat was alternately tested three times with a 5-min intertrial interval.
Intrathecal injections
Under 2% isoflurane anesthesia, the rats were implanted with an intrathecal catheter in the lumbar enlargement (close to the L4–5 segments) for intrathecal drug administration according to the method of Wei-Ping Wu.18 In brief, a longitudinal skin incision was created above vertebrae L5 and L6. The vertebra was accessed through a hole in the muscle made by a 23-gauge needle. The polyethylene catheter (primary erythromelalgia-10, Smiths Medical, England) was pushed into the L5–6 process above the vertebral column and the midline of the vertebra. The catheter was then pushed through the intervertebral space and dura. A sudden movement of tail or the hindlimb indicated dura penetration. The catheter was then pushed gently upward to reach L4 at the lumbar enlargement and fixed into the superficial muscle by suturing a bead made on the catheter at the neck area. The catheter was tunneled under the skin and pulled out at the neck where another skin incision was made and the second bead was fixed into the muscle. The outer end of the catheter was sealed by melting. The both incisions were carefully sutured. After a seven-day recovery, the catheter placement was verified by observing transient hind paw paralysis induced by an intrathecal injection of lidocaine (2%, 5 µl). Those rats exhibiting postoperative neurological deficits (e.g., paralysis) or poor grooming were excluded from the experiments. After peripheral nerve injury, the rats were intrathecally administered the miR30b agomir (20 µM, 10 µl) or scramble miRNA (10 µl as a control) once a day from day 10 (after nerve injury) to day 13. The miR30b antagomir (20 µM, 10 µl) was administered intrathecally in naïve rats four times once daily from seven days after intrathecal catheter implantation. All drugs were injected intrathecally in a 10 μl volume followed by a 10 μl saline flush.
Immunofluoresence
The rats were deeply anesthetized with 2% isoflurane and perfused intracardially with 4% paraformaldehyde in PBS precooled to 4℃. L4–6 DRGs were quickly removed and fixed in 4% paraformaldehyde overnight at 4℃. After dehydration, L4–6 DRGs were sliced into 3.5 µm thick sections. Endogenous peroxidase was inhibited with 3% H2O2 for 15 min. The slices were incubated with 10% normal goat serum to block the non-specific binding of immunoglobulins and then incubated with primary antibodies diluted in blocking solution at 4℃ overnight (mouse anti-Nav1.7 (Abcam, lot number GR51762-1, catalog number ab62758, 1:200), mouse anti-NF200 (Boster, BM0100, 1:100), and rat anti-calcitonin gene-related peptide (CGRP) (Sigma, C8198, 1:100)). Then, sections were washed four times with PBS (0.01M), and incubated with secondary antibodies (Cy3 conjugated donkey anti-mouse antibody (Jackson laboratories 1:400), Alexa 488 conjugated donkey anti-rat antibody (Jackson laboratories 1:400)), diluted in blocking solution at room temperature for 1–2 h, washed again four times with PBS. FIT4 conjugated IB4 (Sigma, L2895) was diluted at 1/200 and incubated together with secondary antibodies. Images were taken with a laser-scanning confocal microscope (Olympus Fluoview FV1000, Japan) and processed with Image J and photoshop CS3 software.
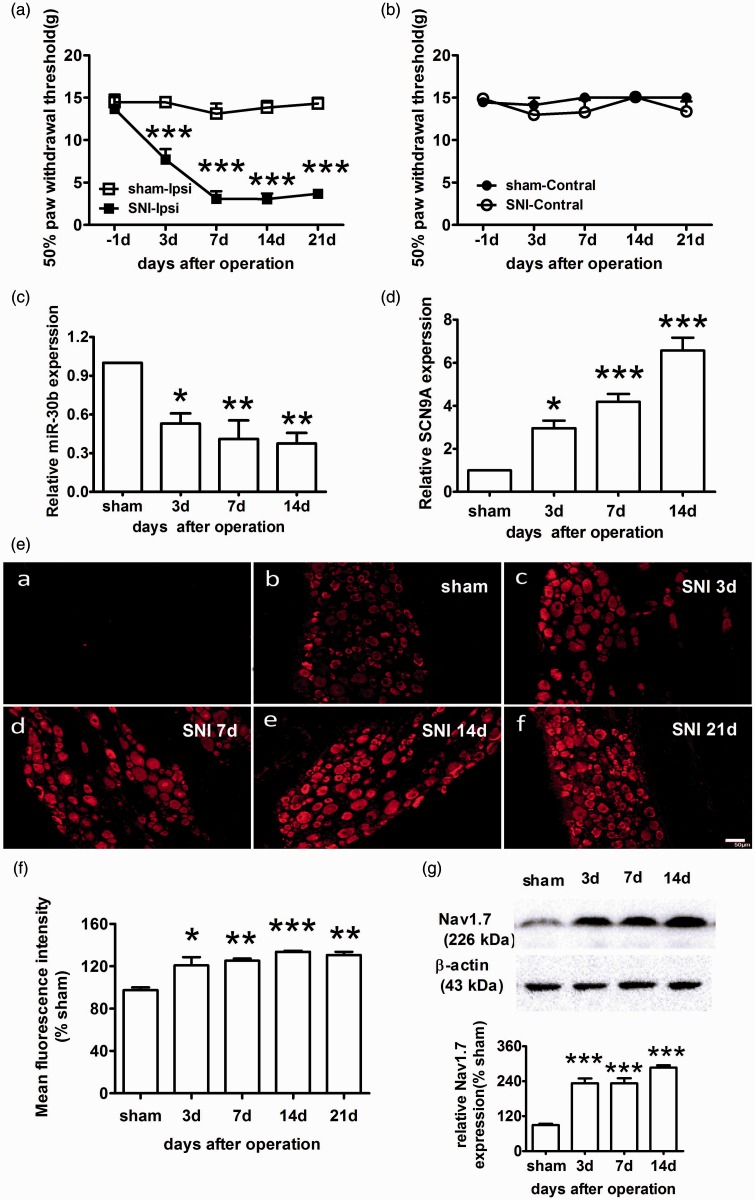
To verify the specificity of Nav1.7 antibody used in the present study, the antibody were preincubated with Nav1.7 protein at the concentration that were fivefold higher than that of the antibody in 4℃ for 1 h, and then the preincubated antibody were used in DRG sections to detect Nav1.7 by immunohistochemistry. As shown in Figure 3, the signal of Nav1.7 detected by the pre-incubated antibody (E-a) was significantly lower than that detected by non-preincubated antibody (E-b).
Figure 3.
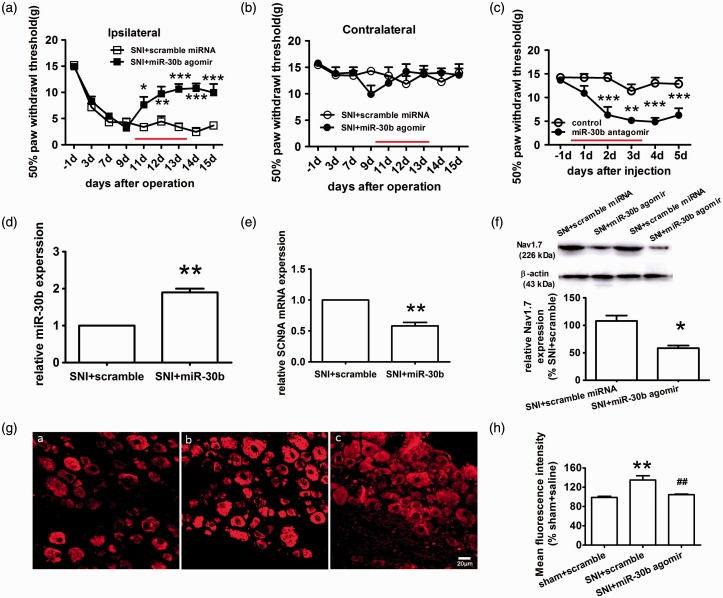
Increased expression of Nav1.7 and decreased expression of miR-30b in the ipsilateral L4–6 DRG of SNI rats. (a, b) The paw withdrawal response to mechanical stimuli was evaluated in the sham and SNI (n = 6). The data are the mean ± SEM. ***P < 0.001 compared with the sham. (a) Responses of ipsilateral paws. (b) Responses of contralateral paws. (c) miR-30b expression levels in the DRG of the sham and SNI ipsilateral sides at 3, 7, and 14 days after the operation (n = 4). *P < 0.05, **P < 0.01 compared with the sham. (d) Expression level of SCN9A mRNA in the DRG (n = 4). *P < 0.05, **P < 0.01, ***P < 0.001 compared with the sham. (e) Representative immunofluorescence images of Nav1.7 expression in the DRG of the sham (b) and SNI ipsilateral sides at 3 (c), 7 (d), 14 (e) and 21 days (f) after the operation. (a) Section of DRG for negative control experiment, in which anti-Nav1.7antibodies was pre-incubated with Nav1.7 protein. Scale bar: 50 µm. (f) Quantification of Nav1.7 labelling intensity in (e) normalized to the sham values. n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the sham. (g) Representative Western blot band and graph showing Nav1.7 expression in the DRG of the SNI ipsilateral sides (n = 4) at different time points after injury. β-actin served as a loading control. ***P < 0.001 compared with the sham.
In situ hybridization with subsequent immunofluorescence
Rats were deeply anaesthetized with 2% isoflurane and perfused transcardially with PBS (pH 7.4) followed by fresh 4% paraformaldehyde in PBS. DRGs were removed and post-fixed in paraformaldehyde overnight, and then cryoprotected in 20% sucrose in PBS overnight at 4℃. Cryostat sections were cut at a thickness of 16 µm. A biotin-anti-digoxigenin-labeled specific detection probe for miR-30b was purchased from Boster (MK10158). Hybridization was performed according to the manufacturer’s instructions. After treatment with pepsin and prehybridization, the sections were hybridized with miR-30b probes (5′—AGCTGAGTGTAGGATGTTTACA—3′) at 42℃ overnight. Slides were rinsed twice with 2 × SSC at 37℃ for 5 min for each wash, and subsequently washed with 0.5 × SSC and 0.2 × SSC at 37℃ for 15 min. Slides were then incubated with blocking solution at 37℃ for 30 min, and then with a mouse anti-digoxigenin antibody at 37℃ for 60 min. In situ hybridization sections were rinsed in PBS, incubated with SABC-FITC/Cy3 at 37℃ for 30 min. Slides were rinsed three times with PBS for 5 min each wash and then incubated with the following primary antibodies overnight at 4℃: mouse anti-Nav1.7 (Abcam, 1:200), mouse anti-NF200 (Boster, 1:100), rat anti-CGRP (Sigma, 1:100), or IB4 conjugated with FITC (Sigma, 1:200) dissolved in PBS-Tween 0.1% at 4℃ overnight. Then, sections were washed four times with PBS (0.01M), and incubated with secondary antibodies (Cy3 conjugated donkey anti-mouse antibody (Jackson Laboratories 1:400), Alexa 488 conjugated donkey anti-rat antibody (Jackson laboratories 1:400)), diluted in blocking solution at room temperature for 1–2 h, washed again four times with PBS. Sections were visualized with confocal microscope (Olympus Fluoview FV1000, Japan).
RNA isolation and quantitative real-time PCR
Total RNA was extracted from the ipsilateral L4–5 DRGs of the SNI/sham rats or PC12 cells using TRIzol according to the manufacturer’s protocol (Invitrogen). Complementary DNA synthesis was performed by reverse transcription using the RevertAid First-strand cDNA Synthesis Kit (Thermo Scientific, USA). Quantitative real-time PCR (qRT-PCR) was performed using the 7500 fast real-time PCR detection system (Applied Biosystems, USA). The Thermo Scientific Maxima SYBR Green qPCR Master Mix kit (Thermo Scientific) was used to detect SCN9A mRNA with the following PCR amplification cycles: initial denaturation, 95℃ for 30 s followed by 40 cycles with denaturation at 95℃ for 5 s and annealing extension at 60℃ for 34 s. The Hairpin-it™ miRNAs qPCR Quantification Kit (GenePharma) was used for miR-30b with the following PCR amplification cycles: initial denaturation at 95℃ for 3 min followed by 40 cycles with denaturation at 95℃ for 12 s and annealing extension at 62℃ for 50 s. A dissociation curve was generated at the end of the 40th cycle to verify that a single product was amplified. β-actin and U6 were used as internal references for SCN9A mRNA and miR-30b, respectively. The relative level of expression was calculated using the comparative (2−ΔCt) method.19 All samples were evaluated in triplicate. The primer sequences were as follows:
U6snRNA-F: 5′-ATTGGAACGATACAGAGAAGATT-3′
U6snRNA-R: 5′-GGAACGCTTCACGAATTTG-3′
miR-30b-F: 5′-CACCAGCCATGTAAACATCCTAC-3′
miR-30b-R: 5′-TATGCTTGTTCTCGTCTCTGTGTC3′
SCN9A-F: 5′-TGGCGTCGTGTCGCTTGT-3′
SCN9A-R: 5′-TGGCCCTTTGCCTGAGAT-3′
GAPDH-F: 5′-TCG GTG TGA ACG GAT TTG GC-3′
GAPDH-R: 5′-CCT TCA GGT GAG CCC CAG C-3′.
Western blot analysis
The ipsilateral L4–6 DRGs were dissected from each animal. Protein was extracted using tissue protein extraction reagent (Weiao Biotech, Shanghai, China) and separated in 10% sodium dodecyl sulfate polyacrylamide gels, electrophoretically transferred onto polyvinylidene fluoride membranes, incubated with blocking buffer (Tris Buffered Saline with Tween [TBST] containing 5% fat-free dry milk) for 2 h and probed with rabbit anti-Nav1.7 (1:600, Abcam, England, lot number GR51762-1) over two nights at 4℃. The membrane sheet was then incubated with a goat anti-rabbit antibody (1:1000) for 2 h at 37℃, and the staining was visualized using ECL detection reagents followed by exposure with FluorChem ProteinSimple (AlphaImager ProteinSimple, San Jose, USA). The immunoreactive density was analyzed using AlphaView software.
Statistical analyses
The data are presented as the mean ± SEM. GraphPad Prism v5.0 was used for statistical analyses. Normality of data was assessed by the Shapiro–Wilk and Kolmogorov–Smirnov tests, with P < 0.05 accepted as a non-normal distribution. Equality of variance was assessed by F test. Unpaired Student's t-test was used for normally distributed datasets. Comparisons of a non-normally distributed group with any other group were made using the Mann–Whitney U test. Comparisons of more than two groups, all of which were normally distributed, were made using one-way analysis of variance with Tukey post-hoc tests for pairwise comparisons. The data from behavior test were statistically analyzed with two-way analysis of variance. P < 0.05 was considered significant.
Results
SCN9A mRNA is the target of miR-30b
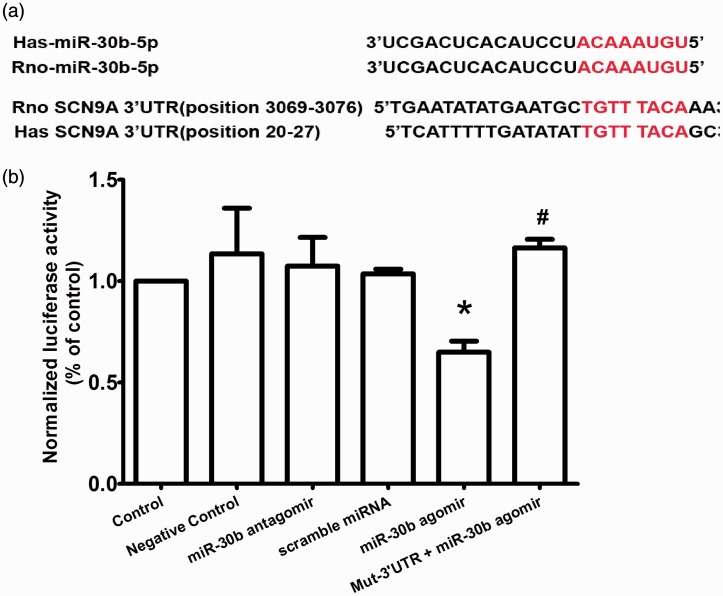
The voltage-gated sodium channel Nav1.7 α subunit is encoded by SCN9A. Using TargetScan algorithms, we found that SCN9A mRNA is a target of several miRNAs, including miR-30b, miR-96, and miR-182. Here, we focused on the relationship between miR-30b and SCN9A. The mature sequences retrieved from miR-base showed that the miR-30b target sequences in the SCN9A 3′UTR were conserved between rats and humans (Figure 1(a)). The TargetScan algorithms showed that the seed sequence of miR-30b (positions 1–8) exactly matched the SCN9A 3′UTR from 3069 to 3076 bp (rat) or from 20 to 27 bp (human) (Figure 1(a)).
Figure 1.
miR-30b directly targets the SCN9A 3′UTR. (a) Sequence alignment of miR-30b and the human and rat SCN9A 3′UTR. The predicted sequences for the pairing of the SCN9A 3′UTR and miR-30b are indicated by red letters. (b) Luciferase activity of luciferase reporter genes bearing SCN9A 3′UTR segments or a mutated (Mut) 3′UTR in HEK293 cells. The data are the mean ± SEM. *indicates P < 0.05 compared with scramble miRNA. # indicates P < 0.05 compared with the miR-30b agomir.
To investigate whether the SCN9A 3′UTR was in fact targeted by miR-30b to negatively regulate Nav1.7 expression, we generated reporter plasmids by fusing the SCN9A mRNA 3′UTR (containing the miR-30b target sites) to the 3′ terminus of a Renilla luciferase coding sequence (abbreviated as 3′UTR). In HEK293 cells, coexpression of the 3′UTR with the miR-30b agomir, which is double-stranded miR-30b, inhibited luciferase expression (Figure 1(b)). Moreover, the miR-30b antagomir, the inhibitor of miR3b, did not influence luciferase expression (Figure 1(b)). These results suggested that miR-30b effectively inhibited translation of the Nav1.7 α subunits. To demonstrate that this inhibition was sequence specific, we fused the mutant SCN9A mRNA 3′UTR to the reporter plasmid (mut-3′UTR) and coexpressed them with the miR-30b agomir in HEK293 cells. As expected, miR-30b had no effect on luciferase expression (Figure 1(b)). To illustrate the specificity of miR-30b targeting, we assessed the binding of the scramble miR-30b sequence (scramble miRNA), which was unable to inhibit luciferase expression (Figure 1(b)).
Identification of SCN9A as a target of miR-30b in PC12 cells by miR-30b antagomir transfection
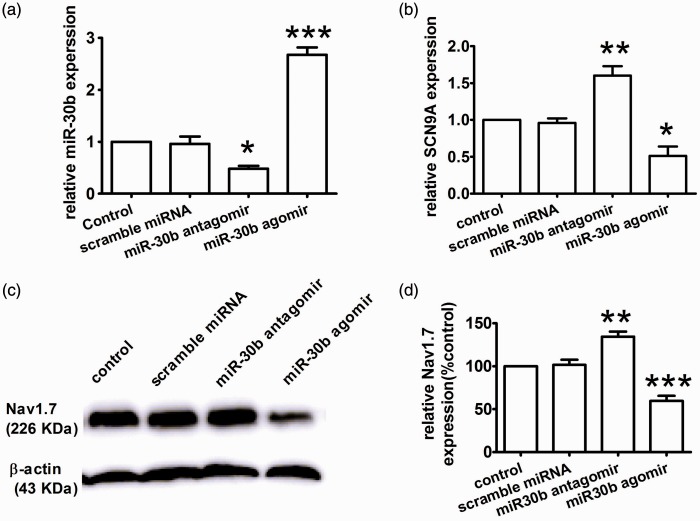
To determine whether endogenous miR-30b could regulate the translation of SCN9A, we over-expressed and knocked down endogenous miR-30b by infecting PC12 cells with miR-30b agomir and antagomir, respectively. We measured the expression of SCN9A mRNA and miR-30b by qRT-PCR. Compared with the control, miR-30b agomir transfection induced a significant up-regulation of miR-30b expression (Figure 2(a)) and down-regulation of SCN9A (Figure 2(b)). Conversely, miR-30b expression decreased after miR-30b antagomir transfection (Figure 2(a)). Concomitantly, miR-30b knockdown increased SCN9A mRNA expression (Figure 2(b)). Accordingly, in PC12 cells, Nav1.7 expression changed together with the change in miR-30b (Figure 2(c)). As shown in Figure 2(c), compared with control non-transfected conditions, over-expression of miR-30b induced a significant decrease in Nav1.7 expression (Figure 2(c) and (d)). In contrast, knockdown of miR-30b with the miR-30b antagomir caused increase in Nav1.7 expression (Figure 2(c) and (d)), supporting a role for endogenous miR-30b in limiting Nav1.7 expression in PC12 cells. These results indicate that the inhibition of Nav1.7 translation by miR-30b may rely on mRNA decay. Thus, we demonstrated that the ability of miR-30b to silence Nav1.7 expression occurs via binding to the SCN9A 3′UTR and repressing mRNA transcription at the molecular level.
Figure 2.
Nav1.7 down-regulation by miR-30b over-expression in PC12 cells. (a and b) The relative expression levels of miR-30b (a) and SCN9A mRNA (b) were determined by qRT-PCR in PC12 cells treated with the miR-30b agomir/antagomir or scramble miRNA. *P < 0.05, **P < 0.01, ***P < 0.001 versus the control. (c and d) Expression level of Nav1.7 protein in PC12 cells. **P < 0.01, ***P < 0.001 versus the control.
Increases in Nav1.7 expression inversely correlate with the down-regulation of miR-30b in the DRG of SNI rats
We chose the SNI animal model for neuropathic pain. SNI produced a significant reduction of the paw withdrawal threshold (PWT) to mechanical stimulation of the ipsilateral hind paw (Figure 3(a)) but had no effect on the contralateral side (Figure 3(b)). At baseline, the PWT was approximately 15 g, while the post-SNI PWT gradually decreased to a minimum of 3.0 ± 0.6 g on day 14 after SNI (P < 0.001).
To evaluate the expression and localization of Nav1.7 in the DRG of SNI rats, we performed immunofluorescence microscopy using antibodies against Nav1.7 (red fluorescence). Nav1.7 immunoreactivity was predominantly detected in the cytoplasm of neurons in rat DRGs (Figure 3(e)). Compared with the sham group, nerve injury induced a significant up-regulation of Nav1.7 expression in rat DRGs (Figure 3(e) and (f)), which was consistent with the data obtained in the behavior test. A time course analysis by Western blotting showed that Nav1.7 protein was significantly increased at three days after the operation and reached a maximum level at day 14 (Figure 3(g)). Then, we used qRT-PCR to evaluate the expression of SCN9A mRNA and miR30b. SCN9A mRNA expression increased and miR-30b expression gradually decreased, both showing a maximum on day 14 after SNI (Figure 3(d) and (c)). Therefore, the increase in SCN9A mRNA and protein expression inversely correlated with miR-30b expression in the DRGs of SNI rats. This phenomenon appeared to be due to transcriptional regulation because SCN9A mRNA expression changed after the operation.
Nav1.7 and miR-30b colocalize in normal rat DRG neurons
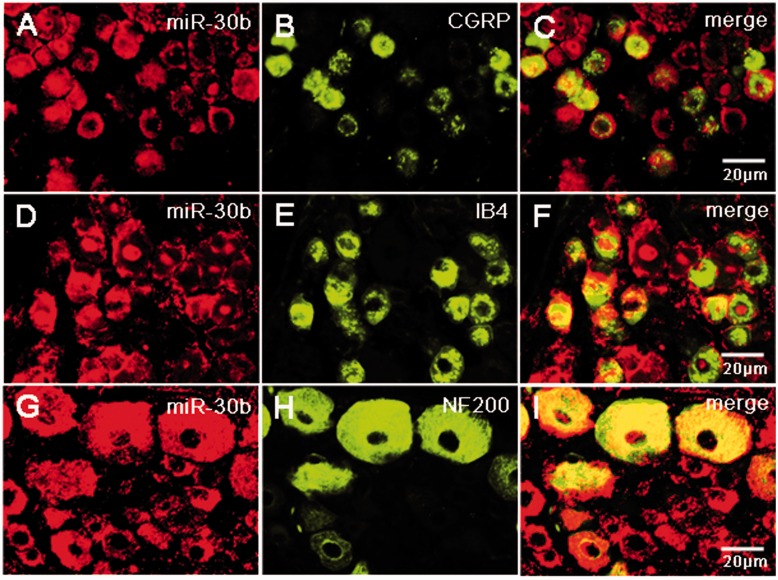
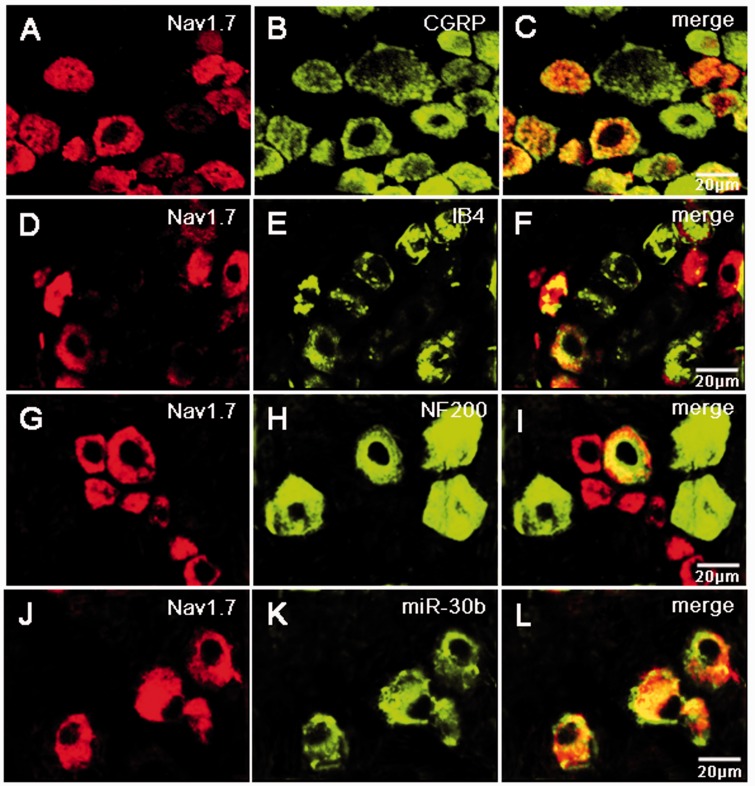
To define the cellular localization of miR-30b and Nav1.7 in the DRG, we performed in situ hybridization using antisense probe for miR-30b and coimmunostainings. In situ hybridization combined with immunostaining showed that miR-30b was colocalized with either CGRP, a marker for peptidergic neurons (Figure 4(a) to (c)), isolectin B4 (IB4), a marker for a fraction of small, non-myelinated nociceptive neurons (Figure 4(d) to (f)), or neurofilament-200 (NF200), a marker for large, myelinated non-nociceptive neurons (Figure 4(g) to (i)). In line with a previous study using Nav1.7 antibody,20–22 Nav1.7 is widely expressed in DRG all size neurons, but it predominantly expressed in the small neurons (Figure 5(a) to (i)). Figure 5(j) to (l) shows that miR-30b-labeled neurons in the DRG also express Nav1.7 protein. Therefore, Nav1.7 and miR-30b are both preferentially expressed in nociceptive neurons, indicating that they might contribute to nociceptive processing in the PNS. On the other hand, Nav1.7 protein is localized in the same cells in which miR30b is expressed (Figure 5(j) to (l)), thus indicating their possible interaction.
Figure 4.
Subpopulation distribution of miR-30b-containing neurons in DRG of naive rats. In situ hybridization of miR-30b and immunofluorescence staining of CGRP (a–c), IB4 (d–f), and NF200 (g–i) show that miR-30b was colocalized with nociceptive neuronal and non-nociceptive neurons marker. n = 3, Scale bars: 20 µm.
Figure 5.
Subpopulation distribution of Nav1.7 protein-containing neurons and double labeling of miR-30b with Nav1.7 protein in DRG of naive rats. Neurons were double labeled with Nav1.7 and CGRP (a–c), IB4 (d–f), or NF200 (g–i). Representative examples showing that most miR-30b labeled neurons in the DRG express Nav1.7 protein (j–l). n = 3 rats. Scale bar: 20 µm.
Nav1.7 is responsible for miR-30b-mediated analgesia of neuropathic pain
To assess the direct contribution of miR-30b to neuropathic pain, we performed daily intrathecal injections of the miR-30b agomir in SNI rats for four days and measured the sensitivity to mechanical stimulation. At day 10 after SNI, when neuropathic pain was stably established, the miR-30b agomir was administered once daily for four days. Beginning at day 2 after drug administration, mechanical allodynia induced by SNI was attenuated by the intrathecal administration of the miR-30b agomir but not the scrambled miRNA (Figure 6(a)). Because over-expression of miR-30b in SNI rats relieves pain, the inhibition of miR-30b in normal rats may induce pain. To investigate this possibility, we performed four daily intrathecal injections of miR-30b antagomir in naïve animals and measured their sensitivity to mechanical stimulation. Compared with control animals that received saline, miR-30b antagomir-injected rats demonstrated a moderate but significant hypersensitivity to pain (P < 0.05; Figure 6(c)), which suggested that endogenous miR-30b may function as a basal antinociceptive control.
Figure 6.
Nav1.7 down-regulation by miR-30b over-expression relieves neuropathic pain. (a, b) The paw withdrawal response to mechanical stimuli was evaluated on the ipsilateral side (a) and contralateral side (b). The scramble miRNA or miR-30b agomir was delivered intrathecally once daily for four days, as indicated by red lines. The data represent the mean ± S.E.M, n = 6. *P < 0.05, **P < 0.01, ***P < 0.001 versus the scrambled miRNA group. (c) The sensitivity to pain of naïve animals subjected to four daily intrathecal injections of miR-30b antagomir was measured by mechanical stimulation. The data are expressed as the mean ± S.E.M. n = 5. *P < 0.05, **P < 0.01, ***P < 0.001 compared with the control. (d, e) The relative expression of miR-30b (d) and SCN9A (e) was determined by qRT-PCR in the DRG of SNI rats that were intrathecally injected with miR-30b agomir. **P < 0.01 versus the SNI+scramble group. (f) Nav1.7 protein expression in the DRG from the SNI side at 14 days after SNI (n = 5). * P < 0.05 versus the SNI+scramble miRNA group. (g) Representative immunofluorescence images of Nav1.7 expression in the DRG of sham rats injected with scramble miRNA (i) and SNI rats injected with scramble miRNA (ii), or miR-30b agomir (iii). Scale bar: 20 µm. (h) Quantification of Nav1.7 labeling intensity in (g) normalized to the sham + scramble values. n = 3. **P < 0.01 compared with the sham + scramble. ##P < 0.01 compared with the SNI + scramble.
To examine the potential causal role of miR-30b down-regulation in the up-regulation of Nav1.7, SNI rats received four daily intrathecal applications of miR-30b agomir or scramble miR-30b sequence as a control. As determined by qRT-PCR, miR-30b expression was markedly increased above basal levels in response to intrathecal applications of the miR-30b agomir (Figure 6(d)). As expected, the miR-30b injections induced SCN9A mRNA expression decrease (Figure 6(e)). The immunofluorescence staining results showed that miR-30b over-expression led to a significant reduction in Nav1.7 expression (Figure 6(g)-iii, (h)). In contrast, the scramble miR-30b sequence (Figure 6(g)-i,ii, (h)) had no effect on Nav1.7 expression. In accordance with the immunofluorescence data, Nav1.7 protein expression, as determined by Western blot analysis, was suppressed in the DRG following miR-30b over-expression (Figure 6(f)). Thus, the down-regulation of miR-30b results in an up-regulation of Nav1.7 mRNA and protein expression that, at least in part, causes tactile allodynia, although miR-30b regulates the expression of multiple genes.23–26
Discussion
The voltage-gated sodium channel Nav1.7 plays a vital role in initiating action potentials in response to the depolarization of sensory neurons by noxious stimuli.5,27 Nav1.7 participates in many different pain states. The expression of Nav1.7 increases in nociceptive neurons during the development of inflammatory hyperalgesia, while the knockdown or ablation of Nav1.7 expression relieves inflammatory pain and hyperalgesia.28–29 In rodent models of painful diabetic neuropathy, an increase in Nav1.7 expression in DRG neurons correlates with pain-related behaviors.30–32 Similarly, Nav1.7 is also involved in nerve injury-induced neuropathic pain.33–35 The results showed that Nav1.7 mRNA expression is up-regulated in DRGs after peripheral inflammation induced by carrageenan or complete Freund’s adjuvant28,36–37 and is decreased in certain animal models of neuropathic pain.38,39 However, in the present study, we assessed the time course of Nav1.7 expression in rat DRGs after SNI and found that the mRNA level of SCN9A increased in L4–5 DRGs after SNI (Figure 3(d)). This result seemed to contrast with the significant down-regulation of SCN9A in the SNL experiment and suggested that different types of lesion may have differing effects depending on the isoform. Furthermore, this up-regulation of SCN9A also contrasted with the previous study on the mice SNI model,38 further supporting differences between mice and rats. In accordance with the qRT-PCR data, Nav1.7 protein expression was up-regulated beginning at day 3 after SNI (Figure 3(e) to (g)). These data confirmed the involvement of Nav1.7 in nerve injury-induced neuropathic pain. On the other hand, these data may indicate that epigenetic modifications are involved in this phenomenon.
Recent studies have shown that gene expression is controlled by epigenetic modifications,40–42 which include DNA methylation, covalent histone modifications (e.g., acetylation and methylation), and non-coding RNAs (e.g., miRNAs and long non-coding RNAs). Accumulating evidence indicates that peripheral noxious stimulation changes epigenetic modifications and that these changes may be related to the induction of pain hypersensitivity under chronic pain conditions.43–47 Because Nav1.7 expression changes at the mRNA and protein levels, understanding the regulation of Nav1.7 by epigenetic modifications is important for unraveling the molecular mechanism underlying the neuropathic pain modulation. This information may help us to identify new therapeutic methods to relieve pain.
MicroRNAs exist extensively in vivo and participate in many physiological and pathological processes in which they alter and modulate the expression of proteins.13 Here, using TargetScan algorithms, we found that SCN9A mRNA is a target of several miRNAs, including miR-30b, miR-96, and miR-182. Aldrich et al.48 found that miR-96, miR-182, and miR-183 are highly enriched in the DRG of adult rats and reduced in ipsilateral DRG neurons in the L5 SNL model of chronic neuropathic pain. However, what about miR-30b? Does it target the SCN9A gene, and is it involved pain? In the present study, our preliminary experiments found that miR-30b expression was decreased in the L4–5 DRG of rats with SNI (Figure 3(c)). Then, we constructed a dual luciferase reporter vector with the 3′UTR of SCN9A containing the miR-30b binding site inserted and found that miR-30b targeted SCN9A 3′UTR and inhibited the expression of luciferase after the cotransfection of miR-30b and dual luciferase reporter vector (Figure 1(b)). In contrast, there was no effect on luciferase expression after the cotransfection of miR-30b and a mutated dual luciferase reporter vector (Figure 1(b)).
It is known that miRNAs silence genes either by initiating the cleavage of their respective target mRNA after complete binding to their target sequences or by inhibiting gene translation after partial binding to their target sequences. We found that both Nav1.7 protein and mRNA changed in both rat groups after SNI (Figure 6) and in PC12 cells (Figure 2) treated with the miR-30b agomir. Our data indicate that miR-30b can inhibit Nav1.7 expression by preventing SCN9A transcription in DRGs. However, the precise mechanism of how miR-30b controls Nav1.7 expression needs further studies in the future.
In addition, over-expression of miR-30b not only efficiently inhibited the up-regulation of Nav1.7 caused by SNI but also relieved mechanical allodynia (Figure 6(a)). Conversely, naive animals demonstrated a moderate but significant hypersensitivity to mechanical stimulation following intrathecal administration of the miR-30b antagomir (Figure 6(c)), suggesting the occurrence of basal antinociceptive control by endogenous miR-30b. Therefore, miR-30b is involved in pain perception and has become novel target for analgesic therapies that function by regulating the expression of SCN9A, which is the relevant pain-related gene. Since a single miRNA can act on multiple target genes and one gene can be simultaneously regulated by multiple miRNA molecules, miR-30b could take part in pain by other target genes. In fact, we discovered that miR-30b is not only related to SCN9A but also related to other genes such as SCN3A and SCN8A which are associated with pain using TargetScan bioinformatics software analysis. We are investigating the relationship between them in pain. Yeomans et al.29 found Nav1.7 participates in inflammatory pain. Whether miR-30b take part in inflammatory pain by regulating Nav1.7 is unknown and it needs more experiments to prove.
In conclusion, we demonstrated that the expression of Nav1.7 and miR-30b was changed in rat DRGs after SNI. Furthermore, the down-regulation of miR-30b expression in naive animals induced a hypersensitivity to mechanical stimulation. In contrast, miR-30b repressed the up-regulation of Nav1.7 both in vivo and in vitro and subsequently relieved the pain sensitization in SNI rats. Our study is the first to report that miR-30b is involved in the regulation of neuropathic pain by interacting with Nav1.7. Thus, the results of our study identify miR-30b as a novel potential therapeutic target in neuropathic pain.
Authors’ contributions
WDZ conceived the project, supervised all experiments, and wrote manuscript. JPS and JC designed the project, researched data, and wrote manuscript. JNW, SSX, and XHR researched data and reviewed/edited manuscript. ML, ZHL, and QZZ reviewed/edited manuscript. All authors read and approved the final manuscript. JPS and JC contributed equally to this study.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (No. 81471144), the Outstanding Young talent Research Fund of Zhengzhou University (No. 1421328058), and the Normal Project of Henan Province Education Department (No. 14A310012).
References
- 1.Wang W, Gu J, Li YQ, et al. Are voltage-gated sodium channels on the dorsal root ganglion involved in the development of neuropathic pain? Mol Pain 2011; 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waxman SG, Zamponi GW. Regulating excitability of peripheral afferents: emerging ion channel targets. Nat Neurosci 2014; 17: 153–163. [DOI] [PubMed] [Google Scholar]
- 3.Drenth JP, Waxman SG. Mutations in sodium-channel gene SCN9A cause a spectrum of human genetic pain disorders. J Clin Invest 2007; 117: 3603–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toledo-Aral JJ, Moss BL, He ZJ, et al. Identification of PN1, a predominant voltage-dependent sodium channel expressed principally in peripheral neurons. Proc Natl Acad Sci U S A 1997; 94: 1527–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox JJ, Reimann F, Nicholas AK, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature 2006; 444: 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox JJ, Sheynin J, Shorer Z, et al. Congenital insensitivity to pain: novel SCN9A missense and in-frame deletion mutations. Hum Mutat 2010; 31: E1670–E1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg YP, MacFarlane J, MacDonald ML, et al. Loss-of-function mutations in the Nav1.7 gene underlie congenital indifference to pain in multiple human populations. Clin Genet 2007; 71: 311–319. [DOI] [PubMed] [Google Scholar]
- 8.Nilsen KB, Nicholas AK, Woods CG, et al. Two novel SCN9A mutations causing insensitivity to pain. Pain 2009; 143: 155–158. [DOI] [PubMed] [Google Scholar]
- 9.Cregg R, Laguda B, Werdehausen R, et al. Novel mutations mapping to the fourth sodium channel domain of Nav1.7 result in variable clinical manifestations of primary erythromelalgia. Neuromolecular Med 2013; 15: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberhardt M, Nakajima J, Klinger AB, et al. Inherited pain: sodium channel Nav1.7 A1632T mutation causes erythromelalgia due to a shift of fast inactivation. J Biol Chem 2014; 289: 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dib-Hajj SD, Estacion M, Jarecki BW, et al. Paroxysmal extreme pain disorder M1627K mutation in human Nav1.7 renders DRG neurons hyperexcitable. Mol Pain 2008; 4: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estacion M, Dib-Hajj SD, Benke PJ, et al. NaV1.7 gain-of-function mutations as a continuum: A1632E displays physiological changes associated with erythromelalgia and paroxysmal extreme pain disorder mutations and produces symptoms of both disorders. J Neurosci 2008; 28: 11079–11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 14.Sakai A, Saitow F, Miyake N, et al. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain 2013; 136: 2738–2750. [DOI] [PubMed] [Google Scholar]
- 15.Chen HP, Zhou W, Kang LM, et al. Intrathecal miR-96 inhibits Nav1.3 expression and alleviates neuropathic pain in rat following chronic construction injury. Neurochem Res 2014; 39: 76–83. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, Lee MC, Momin A, et al. Small RNAs control sodium channel expression, nociceptor excitability, and pain thresholds. J Neurosci 2010; 30: 10860–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaplan SR, Bach FW, Pogrel JW, et al. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods 1994; 53: 55–63. [DOI] [PubMed] [Google Scholar]
- 18.Wu WP, Xu XJ, Hao JX. Chronic lumbar catheterization of the spinal subarachnoid space in mice. J Neurosci Methods 2004; 133: 65–69. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 20.Ho C, O'Leary ME. Single-cell analysis of sodium channel expression in dorsal root ganglion neurons. Mol Cell Neurosci 2011; 46: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Black JA, Dib-Hajj S, McNabola K, et al. Spinal sensory neurons express multiple sodium channel alpha-subunit mRNAs. Brain Res Mol Brain Res 1996; 43: 117–131. [DOI] [PubMed] [Google Scholar]
- 22.Fukuoka T, Kobayashi K, Yamanaka H, et al. Comparative study of the distribution of the alpha-subunits of voltage-gated sodium channels in normal and axotomized rat dorsal root ganglion neurons. J Comp Neurol 2008; 510: 188–206. [DOI] [PubMed] [Google Scholar]
- 23.Agrawal R, Tran U, Wessely O. The miR-30 miRNA family regulates Xenopus pronephros development and targets the transcription factor Xlim1/Lhx1. Development 2009; 136: 3927–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez I, Cazalla D, Almstead LL, et al. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc Natl Acad Sci U S A 2011; 108: 522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song PP, Hu Y, Liu CM, et al. Embryonic ectoderm development protein is regulated by microRNAs in human neural tube defects. Am J Obstet Gynecol 2011; 204: 544, e9–544.e17. [DOI] [PubMed] [Google Scholar]
- 26.Tang B, Li N, Gu J, et al. Compromised autophagy by MIR30B benefits the intracellular survival of Helicobacter pylori. Autophagy 2012; 8: 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dib-Hajj SD, Yang Y, Waxman SG. Genetics and molecular pathophysiology of Na(v)1.7-related pain syndromes. Adv Genet 2008; 63: 85–110. [DOI] [PubMed] [Google Scholar]
- 28.Minett MS, Nassar MA, Clark AK, et al. Distinct Nav1.7-dependent pain sensations require different sets of sensory and sympathetic neurons. Nat Commun 2012; 3: 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeomans DC, Levinson SR, Peters MC, et al. Decrease in inflammatory hyperalgesia by herpes vector-mediated knockdown of Nav1.7 sodium channels in primary afferents. Hum Gene Ther 2005; 16: 271–277. [DOI] [PubMed] [Google Scholar]
- 30.Chattopadhyay M, Mata M, Fink DJ. Continuous delta-opioid receptor activation reduces neuronal voltage-gated sodium channel (NaV1.7) levels through activation of protein kinase C in painful diabetic neuropathy. J Neurosci 2008; 28: 6652–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chattopadhyay M, Zhou Z, Hao S, et al. Reduction of voltage gated sodium channel protein in DRG by vector mediated miRNA reduces pain in rats with painful diabetic neuropathy. Mol Pain 2012; 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong S, Morrow TJ, Paulson PE, et al. Early painful diabetic neuropathy is associated with differential changes in tetrodotoxin-sensitive and -resistant sodium channels in dorsal root ganglion neurons in the rat. J Biol Chem 2004; 279: 29341–29350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dabby R, Sadeh M, Gilad R, et al. Chronic non-paroxysmal neuropathic pain – Novel phenotype of mutation in the sodium channel SCN9A gene. J Neurol Sci 2011; 301: 90–92. [DOI] [PubMed] [Google Scholar]
- 34.Lee JH, Park CK, Chen G, et al. A monoclonal antibody that targets a NaV1.7 channel voltage sensor for pain and itch relief. Cell 2014; 157: 1393–404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.McGowan E, Hoyt SB, Li X, et al. A peripherally acting Na(v)1.7 sodium channel blocker reverses hyperalgesia and allodynia on rat models of inflammatory and neuropathic pain. Anesth Analg 2009; 109: 951–958. [DOI] [PubMed] [Google Scholar]
- 36.Black JA, Liu S, Tanaka M, et al. Changes in the expression of tetrodotoxin-sensitive sodium channels within dorsal root ganglia neurons in inflammatory pain. Pain 2004; 108: 237–247. [DOI] [PubMed] [Google Scholar]
- 37.Strickland IT, Martindale JC, Woodhams PL, et al. Changes in the expression of NaV1.7, NaV1.8 and NaV1.9 in a distinct population of dorsal root ganglia innervating the rat knee joint in a model of chronic inflammatory joint pain. Eur J Pain 2008; 12: 564–572. [DOI] [PubMed] [Google Scholar]
- 38.Laedermann CJ, Pertin M, Suter MR, et al. Voltage-gated sodium channel expression in mouse DRG after SNI leads to re-evaluation of projections of injured fibers. Mol Pain 2014; 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakor DK, Lin A, Matsuka Y, et al. Increased peripheral nerve excitability and local NaV1.8 mRNA up-regulation in painful neuropathy. Mol Pain 2009; 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olzscha H, Sheikh S, La Thangue NB. Deacetylation of chromatin and gene expression regulation: a new target for epigenetic therapy. Crit Rev Oncog 2015; 20: 1–17. [DOI] [PubMed] [Google Scholar]
- 41.Saha A, Jha HC, Upadhyay SK, et al. Epigenetic silencing of tumor suppressor genes during in vitro Epstein-Barr virus infection. Proc Natl Acad Sci U S A 2015; 112: E5199–E5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidoli S, Cheng L, Jensen ON. Proteomics in chromatin biology and epigenetics: elucidation of post-translational modifications of histone proteins by mass spectrometry. J Proteomics 2012; 75: 3419–3433. [DOI] [PubMed] [Google Scholar]
- 43.Lutz BM, Bekker A, Tao YX. Noncoding RNAs: new players in chronic pain. Anesthesiology 2014; 121: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauck M, Van de Ven T, Shaw AD. Epigenetics of chronic pain after thoracic surgery. Curr Opin Anaesthesiol 2014; 27: 1–5. [DOI] [PubMed] [Google Scholar]
- 45.Seo S, Grzenda A, Lomberk G, et al. Epigenetics: a promising paradigm for better understanding and managing pain. J Pain 2013; 14: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone LS, Szyf M. The emerging field of pain epigenetics. Pain 2013; 154: 1–2. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Stefano GB, Kream RM. Epigenetic modification of DRG neuronal gene expression subsequent to nerve injury: etiological contribution to complex regional pain syndromes (Part II). Med Sci Monit 2014; 20: 1188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aldrich BT, Frakes EP, Kasuya J, et al. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience 2009; 164: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]