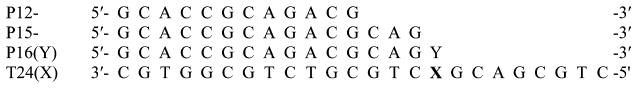Abstract
4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN) are important human carcinogens in tobacco products. They are metabolized to produce a variety 4-(3-pyridyl)-4-oxobutyl (POB) DNA adducts including O2-[4-(3-pyridyl)-4-oxobut-1-yl]thymidine (O2-POB-dT), the most abundant POB adduct in NNK- and NNN-treated rodents. To evaluate the mutagenic properties of O2-POB-dT, we measured the rate of insertion of dNTPs opposite and extension past O2-POB-dT and O2-Me-dT by purified human DNA polymerases η, κ, ι, and yeast polymerase ζ in vitro. Under conditions of polymerase in excess, polymerase η was most effective at the insertion of dNTPs opposite O2-alkyl-dTs. The time courses were biphasic suggesting the formation of inactive DNA-polymerase complexes. The kpol parameter was reduced approximately 100-fold in the presence of the adduct for pol η, κ, and ι. Pol η was the most reactive polymerase for the adducts due to a higher burst amplitude. For all three polymerases, the nucleotide preference was dATP > dTTP ≫ dGTP and dCTP. Yeast pol ζ was most effective in bypassing the adducts; the kcat/Km values were reduced only 3-fold in the presence of the adducts. The identity of the nucleotide opposite the O2-alkyl-dT did not significantly affect the ability of pol ζ to bypass the adducts. The data support a model in which pol η inserts ATP or dTTP opposite O2-POB-dT, and then, pol ζ extends past the adduct.
Graphical Abstract
INTRODUCTION
Lung cancer is the most common cause of cancer death in the US. Cigarette smoking is the main risk factor for lung cancer.1 Tobacco specific nitrosamines (TSNAs) are a significant class of tobacco carcinogens.2–4 N′-Nitrosonornicotine (NNN) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) are the two most potent TSNAs in causing lung tumors in animals.2–4 NNK and NNN are considered to be among the causative agents of lung and oral cavity cancers in individuals who use tobacco products.
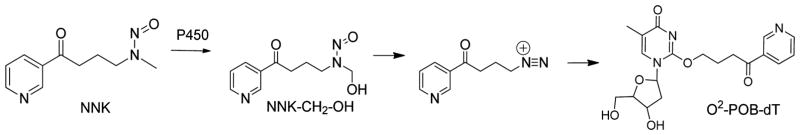
Both NNN and NNK are metabolically activated to alkylating agents. NNK produces both a methyl and a 4-(3-pyridyl)-4-oxobutyl (POB) diazonium ion, while NNN produces the POB-diazonium ion.5 The methylation pathway, with the formation of the mutagenic O6-methyl-2′-deoxyguanosine, is an important contributor to mutagenicity and carcinogenicity of NNK.6,7 The pyridyloxobutylation pathway, although less well studied, also plays a role in carcinogenesis in NNK and NNN. Four POB adducts have been characterized in rodents, in which the POB-group is bound to the N7- and O6-positions of dG and the O2-positions of dC and dT.13,34–36 The chemical toxicology of O6-POB-dG has been the most studied. It is repaired by O6-alkylguanine-DNA alkyltransferase8–10 and is mutagenic in bacteria and human cells causing mostly G to A transitions with some A to T transversions.11,12 DNA polymerase η is the most reactive Y-family polymerase toward O6-POB-dG.13
O2-[4-(3-Pyridyl)-4-oxobut-1-yl]thymidine (O2-POB-dT) may also play a role in carcinogenesis. Its formation is shown in Scheme 1. In long-term studies, in which rats are continuously administered NNK or NNN, O2-POB-dT is the most abundant POB-adduct.14–17 NNK-treated transgenic mice produce approximately equal amounts of mutations at G/C and A/T base pairs.18,19 If the mutagenesis were solely due to the methyl diazonium ion pathway, then the mutagenic spectrum would be dominated by G/C to A/T mutations caused by O6-Me-dG, as observed in mice treated with N,N-dimethyl-N-nitrosamine.20 Therefore, O2-POB-dT is a potential cause of the A/T mutations in NNK-treated rodents.
Scheme 1.
Formation of O2-POB-dT from the Bioactivation of NNK
DNA polymerases are crucial in maintaining genome integrity. Seventeen human DNA polymerases are involved in high fidelity DNA replication, DNA repair, and replication of DNA damage.21–23 Polymerases α, γ, δ, ε, and telomerase are involved in the high fidelity replication of the genome. The Y-family polymerases η, κ, ι, and Rev1 as well as the B-family pol ζ are involved in translesion DNA synthesis (TLS).21,24 Several other polymerases such as λ,25–27 ν,28 θ,29 and PrimPol30 may also be involved in TLS. A current model for replicating past DNA damage is the polymerase switch mechanism.31–35 In this model, the high fidelity polymerase stalls at the DNA damage and is replaced by a TLS polymerase that inserts a dNTP opposite the damage. This polymerase or perhaps a different polymerase then extends past the damage.35 The propensity of a specific DNA polymerase to bypass damage is thought to depend on the relative activity of the polymerase on the DNA substrate, protein–protein interactions, and post-translational modifications. The unique active sites of the polymerases have led to multiple polymerases combining to bypass the damage.35 In many cases, the Y-family polymerase inserts a dNTP opposite the damage and pol ζ extends past the damage a few nucleotides so that the high fidelity polymerase can take over.
The earliest studies on the mutagenic potential of O2-methyl-dT (O2-Me-dT) and O2-ethyl-dT (O2-Et-dT)36–38 were conducted with noneukaryotic polymerases. While O2-Me-dT forms the most stable base pair with dG,36 T7 DNA polymerase and the Klenow fragment of proofreading deficient E. coli DNA polymerase I (Kf(exo-)) preferentially incorporate dATP and dTTP opposite O2-Et-dT.37,38 We previously found that both Kf(exo-) and Sulfolobus solfataricus DNA polymerase IV (Dpo4) bypass both O2-Me-dT and O2-POB-dT with low efficiency and low fidelity, primarily inserting both dATP and dTTP opposite the adduct.39 More recent in vitro replication studies showed that O2-Me-dT is highly blocking for DNA synthesis catalyzed by Kf(exo-), human pol κ, and Saccharomyces cerevisiae DNA polymerase η (yPol η).40 Steady-state kinetic measurements revealed that Kf(exo-) and yPol η preferentially incorporated the correct nucleotide (dAMP) opposite O2-Me-dT.40 However, using a LC-MS/MS to analyze both insertion and extension, hPol κ-mediated the insertion of dG opposite O2-Me-dT. Similar results were obtained with the larger O2-Et-dT, which was bypassed slowly with human DNA polymerases η, κ, and ι, yeast DNA polymerase ζ (yPol ζ), and Kf(exo-) to generate full-length replication products.41
The replication of O2-alkyl-dTs were evaluated in E. coli.42,43 Bypass of O2-Me-dT and O2-POB-dT, in the absence of SOS activation, are low but accurate.42 When SOS is induced, both bypass efficiency and mutagenicity are increased. Pol II, IV, and V are responsible for bypass, with pol V as the most mutagenic. While dA is inserted opposite O2-Me-dT and O2-POB-dT most often, the major mutagenic process is the incorporation of dC opposite the adducts.42 More recently, the bypass of O2-Me-dT and O2-POB-dT were evaluated in mammalian cells.44 Both dATP and dTTP were incorporated opposite the adducts, and pol η, ξ, and REV1 are the primary polymerases involved. To further investigate the mechanisms of mutagenicity of O2-POB-dT in humans, we evaluated the in vitro reactivity of O2-POB-dT in a defined oligodeoxynucleotide substrate with purified human DNA polymerases η, κ, and ι. We found that pol η has the highest reactivity for the incorporation opposite the adducts, incorporating both dATP and dTTP, and that yPol ζ is able to extend the primer past this base pair.
EXPERIMENTAL PROCEDURES
General
[32P]ATP (6000 Ci/mmol) was purchased from PerkinElmer and T4 polynucleotide kinase from USB/Affymetrix. The dNTPs (ultrapure grade) were purchased from GE Healthcare, and the concentrations were determined by UV absorbance.45 yPol ζ was purchased from Enzymax (Lexington, KT).
Oligodeoxynucleotides
Oligodeoxynucleotides containing O2-Me-dT and O2-POB-dT were synthesized, purified, and characterized as described.42,46 The oligodeoxynucleotide containing 6-carboxyfluorescein was purchased from Integrated DNA Technologies (Coralville, Iowa). The sequences of the oligodeoxynucleotides are shown in Chart 1. The concentrations of oligodeoxynucleotides were determined from the absorbance at 260 nm, using the method of Borer47 in which it was assumed that the spectroscopic properties of O2-Me-dT and O2-POB-dT were identical to those of the dT. The primer was 32P-labeled with γ-[32P]ATP and annealed with a 20% excess of the template.48
Chart 1.
Oligodeoxynucleotide Sequences
Polymerase Purification and Concentration
Polymerases κ, η, and ι were purified from Sf9 insect cells as described.49–51 The polymerases are full length proteins with an N-terminal His-tag. The active concentration of the enzymes were determined by evaluating the magnitude of the burst using an undamaged template (data not shown). The polymerases were reacted with 300 nM DNA (P15/T24G) and 100 μM dCTP and fit to eq 2. The amplitude was set as the polymerase concentration.
Polymerase Kinetics
Enzyme reactions were initiated by mixing equal volumes of the DNA (P15/T24)/polymerase solution in buffer with dNTP and Mg2+ in H2O at 37 °C. The final buffer concentration was 40 mM Tris-HCl (pH 8.0), 3 mM DTT, 10 μg/mL BSA, and 2.5% glycerol. The reactions were quenched with equal volumes of STOP solution containing 10% 0.5MNa2EDTA, 90% formamide, 0.025% (w/v) xylene cyanol, and 0.025% (w/v) bromophenol blue. Rapid reactions were performed on a RQF-3 (Kin-Tek Corporation) and quenched with 300 mM EDTA (pH 8).
The time course of the reaction was analyzed by denaturing PAGE, and the radioactivity on the gel was visualized with a Typhoon 9200. The progress of the reaction was quantitated by dividing the total radioactivity in the product band(s) by the radioactivity in the product and reactant bands. Multiple product bands appeared when the incorrect dNTP was added to the reaction.
Polymerase-DNA Binding Experiments
The binding affinity of the DNA to the polymerases was evaluated with fluorescence anisotropy. The DNA consisted of the primer strand (P15) modified on the 5′-end with 6-carboxyfluorescein and the template strand containing dT, O2-Me-dT, or O2-POB-dT. The fluorescence anisotropy was measured with 1 nM DNA and 0–100 nM polymerase at 37 °C. Anisotropy was measured with a Tecan Safire 2 plate reader with half-volume 96 well plates. The excitation and emission wavelengths were 471 and 525 nm, respectively, with an emission bandwidth of 20 nm.
Data Analysis
Data were fitted by nonlinear regression using the program Prism, version 5, for Windows (GraphPad Software, San Diego, California, USA, www.graphpad.com). The Vmax and Km values were determined by fitting the data to eq 1; v0 is the initial rate, E0 the enzyme, and N0 the dNTP concentrations.
| (1) |
The time courses for the insertion enzyme in excess reactions with O2-Me-dT and O2-POB-dT were fitted to eq 2 in which P is the product concentration, A the burst amplitude, k the burst rate constant, kss the steady-state rate constant, and t the time. The dNTP concentration dependence was quantified by fitting the burst equation parameters to the hyperbolic eq 1 in which v0 is the value of the parameter at a specific [dNTP], and Vmax is the maximum value of that parameter. The time courses for the enzyme in excess extension reactions with O2-Me-dT and O2-POB-dT were fitted to eq 3 in which P is the product concentration, A the amplitude, and k the first-order rate constant. The dNTP concentration dependence on the kinetic parameters in eqs 2 and 3 was quantified by fitting the parameters to the hyperbolic eq 1.
| (2) |
| (3) |
The KdDNA was determined by fitting the measured anisotropy (A) to eq 4, in which K is KdDNA, E0 and D0 (set to 1 nM) are the total concentrations of the polymerase and DNA, respectively, and Amax is the increase in anisotropy caused by the polymerase binding to the DNA.
| (4) |
RESULTS
Running Start Incorporation of dNTPs Opposite O2-alkyl-dT
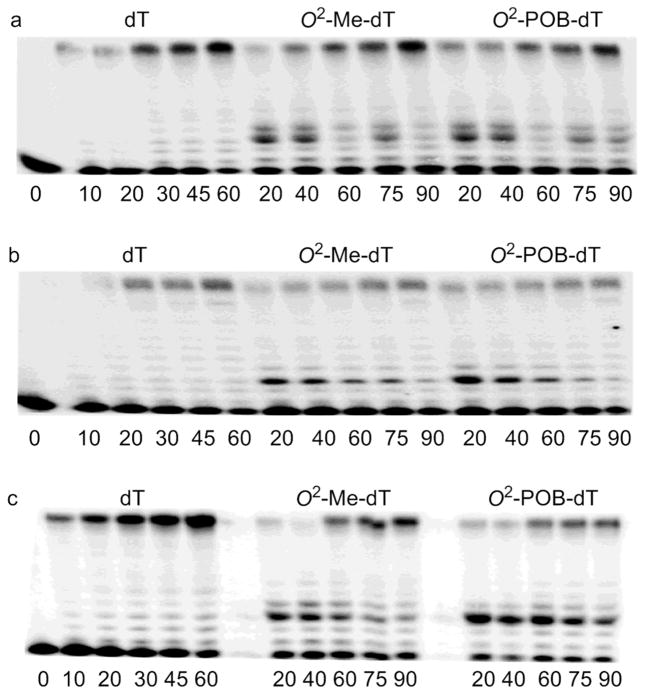
The incorporation of dNTPs into a DNA substrate containing O2-alkyl-dT was examined with the running start analyses as illustrated in Figure 1. The lowest bands represent the 12-mer primer, while the top band is the 24-mer product. The left-most lanes show that templates containing dT are rapidly replicated by all three polymerases. With O2-alkyl-dT in the template, the gels display a band corresponding to a 15-mer, indicating that the polymerases stalled prior to the adduct. A more faint band just above the 15-mer indicates that extension past the adduct is also inefficient.
Figure 1.
PAGE analysis of running start polymerase reactions. DNA substrates (10 nM, P12/T24 with dT, O2-Me-dT, and O2-POB-dT) were reacted with pol κ (a), pol η(b), and pol ι (c) and all four dNTPs for the indicated time (min). The DNA polymerase concentrations were 0.1 nM for dT and 0.5 nM for O2-alkyl-dT substrates. The dNTP concentrations were 5 μM for dT and 50 μM for O2-alkyl-dT substrates.
DNA-Polymerase Binding to Oligodeoxynucleotides Containing O2-alkyl-dT
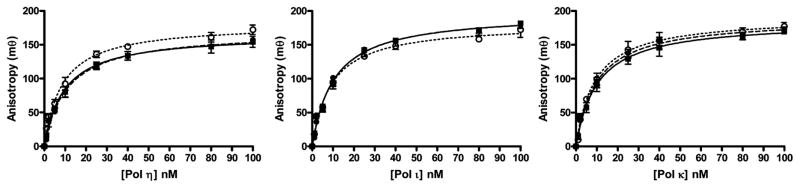
The binding affinities of the DNA substrates to the polymerases were measured by fluorescence anisotropy using a fluorescein-labeled primer strand. The increase in anisotropy was plotted against polymerase concentration and fitted to the quadratic equation as shown in Figure 2. The results, presented in Table 1, indicate that the modification, either methyl or POB, does not affect the affinity of the DNA to the polymerase.
Figure 2.
Polymerase-DNA affinity. Fluorescence anisotropy was determined with 1 nM DNA in which the 5′-terminus of the primer strand was modified with fluorescein with variable concentrations of (A) pol η, (B) pol ι, and (C) pol κ. The DNA contained dT (open circle), O2-Me-dT (black circle), O2–POB-dT (square). The data points are the mean ± standard deviation of three determinations. The lines are the best fit to eq 3.
Steady-State Incorporation of dNTPs Opposite O2-alkyl-dT
The Michaelis–Menten kinetic parameters were determined with polymerase concentration from 0.01 to 0.1 nM, 10 nM DNA, and 50 μM to 1 mM dNTP. The kinetic parameters are presented in Tables 2–4 for the different polymerases. For each enzyme, we also evaluated the correct incorporation of dATP opposite dT and the misincorporation of dTTP opposite dT. The relative kcat/Km values are visualized in Figure 2. Fluorescence anisotropy was determined with 1 nM DNA in which the 5′-terminus of the primer strand was modified with fluorescein with variable concentrations of (A) pol η, (B) pol ι, and (C) pol κ. The DNA contained dT (open circle), O2-Me-dT (black circle), and O2–POB-dT(squrate). The data points are the mean ± standard deviation of three determinations. The lines are the best fit to eq 3.
Table 2.
Steady-State Parameters for the Pol η Catalyzed Insertion Opposite O2-alkyl-dTa
| dNTP | template | kcat (min−1) | Km (μM) | kcat/Km (mM−1 min−1) |
|---|---|---|---|---|
| dATP | dT | 49 ± 2 | 5.3 ± 0.3 | 9230 ± 230 |
| dTTP | dT | 0.140 ± 0.004 | 77.8 ± 5.5 | 1.8 ± 0.1 |
| dATP | O2-Me-dT | 0.068 ± 0.003 | 18.5 ± 2.4 | 3.7 ± 0.4 |
| dCTP | O2-Me-dT | 0.068 ± 0.003 | 130.2 ± 11.4 | 0.52 ± 0.02 |
| dGTP | O2-Me-dT | 0.039 ± 0.001 | 38.4 ± 1.8 | 1.05 ± 0.04 |
| dTTP | O2-Me-dT | 0.119 ± 0.004 | 166 ± 9 | 0.72 ± 0.02 |
| dATP | O2-POB-dT | 0.098 ± 0.004 | 21.7 ± 2.5 | 4.50 ± 0.37 |
| dCTP | O2-POB-dT | 0.141 ± 0.008 | 148.6 ± 16.2 | 0.95 ± 0.05 |
| dGTP | O2-POB-dT | 0.121 ± 0.004 | 126.2 ± 8.0 | 0.96 ± 0.03 |
| dTTP | O2-POB-dT | 0.126 ± 0.005 | 103.3 ± 8.6 | 1.22 ± 0.06 |
Initial rates were conducted with 0.25 to 0.5 nM pol, 5 nM DNA, and 0 to 200 μM dNTP. The values are the mean ± standard deviation of three determinations.
Table 4.
Steady-State Parameters for the Pol κ Catalyzed Insertion Opposite O2-alkyl-dTa
| dNTP | template | kcat (min−1) | Km(μM) | kcat/Km |
|---|---|---|---|---|
| dATP | T | 4.3 ± 0. 2 | 1.4 ± 0.1 | 3200 ± 200 |
| dTTP | T | 0.11 ± 0.01 | 120 ± 20 | 1.0 ± 0.1 |
| dATP | O2-Me-dT | 0.087 ± 0.008 | 35 ± 8 | 2.5 ± 0.3 |
| dCTP | O2-Me-dT | 0.067 ± 0.006 | 160 ± 30 | 0.41 ± 0.03 |
| dGTP | O2-Me-dT | 0.055 ± 0.004 | 64 ± 12 | 0.86 ± 0.10 |
| dTTP | O2-Me-dT | 0.067 ± 0.001 | 34 ± 2 | 1.99 ± 0.07 |
| dATP | O2-POB-dT | 0.047 ± 0.001 | 33 ± 2 | 1.41 ± 0.05 |
| dCTP | O2-POB-dT | 0.022 ± 0.002 | 29 ± 7 | 0.81 ± 0.16 |
| dGTP | O2-POB-dT | 0.13 ± 0.01 | 66 ± 12 | 2.0 ± 0.2 |
| dTTP | O2-POB-dT | 0.14 ± 0.03 | 215 ± 79 | 0.64 ± 0.10 |
Initial rates were conducted with 0.25 to 0.5 nM pol, 5 nM DNA, and 0 to 200 μM dNTP. The values are the mean ± standard deviation of three determinations.
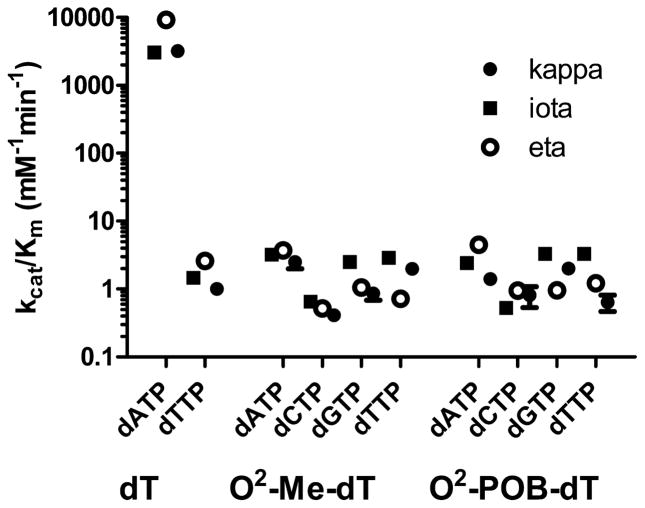
As shown in Figure 3, the incorporation of dATP opposite dT was >1000-fold faster than that of dTTP opposite dT for all three polymerases. The increase in kcat/Km was due to both kcat and Km effects. The kcat/Km values for the incorporation of the dNTPs opposite the O2-alkyl-dTs are similar to the misincorporation kcat/Km values. The selectivity for a particular dNTP is low. The greatest selectivity is for the incorporation of dATP opposite O2-Me-dT by pol η in which the kcat/Km for dATP is 7-, 3.5-, and 5-fold greater than that for dCTP, dGTP, and dTTP, respectively. This selectivity is due to Km differences. A similar selectivity is found with O2-POB-dT in which the kcat/Km for the pol η catalyzed insertion of dATP is about 4-fold greater than that for the other dNTPs. Pol ι is the least selective polymerase with dATP, dGTP, and dTTP having similar kcat/Km values with dCTP having kcat/Km valuees 5-fold less. The kcat/Km values for each enzyme are also very similar. Upon the basis of the steady-state kinetic analysis, we would predict that O2-alkyl-dTs would be bypassed with low fidelity with pol η, κ, and ι.
Figure 3.
kcat/Km values ± standard errors for the incorporation of dNTPs opposite dT, O2-Me-dT, and O2–POB-dT catalazed by pol κ (black circle), ι (square), and η (open circle).
Incorporation of dNTPs Opposite O2-alkyl-dT with Polymerase in Excess
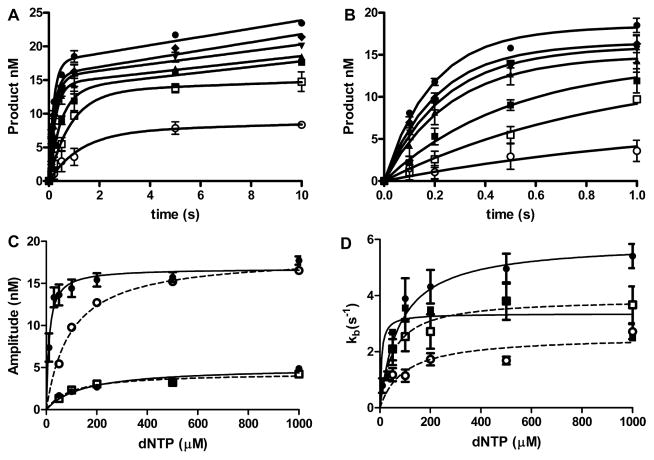
The TLS bypass of DNA adducts does not occur under steady-state kinetic conditions in the cell. The TLS polymerase would be brought to the DNA, perform an insertion reaction, and then hand the DNA off to a subsequent polymerase. This process can be kinetically driven by the relative reactivity of the polymerases or actively driven by protein–protein interactions.31 While kcat/Km values and consequently the selectivity of the polymerase under steady-state conditions is not affected by nonproductive binding complexes, the relative reactivity during single-turnover reaction will be. Consequently, we analyzed the single-nucleotide incorporation reactions with enzyme in excess conditions with polymerase concentrations of 250 nM and a DNA concentration of 25 nM. The DNA was P15/T24 in which the template base was dT, O2-Me-dT, or O2-POB-dT. The time courses for pol η catalyzed incorporation of each dATP opposite O2-POB-dT are shown in Figure 4A, with the early time points in Figure 4B. The data were fit to the burst equation, and the burst amplitude (A), burst rate constant (k), and steady-state rate constant (kss) were determined. Depending on the polymerase and the dNTP, the amplitude, burst rate constant, and/or steady-state rate constants were dependent on the dNTP concentration. If the parameters were dependent on the dNTP concentration, the parameters were fit to the hyperbolic eq 1. Figure 4C and D show the graphs for the amplitude and burst rate constant for pol η with each dNTP. The complete set of time course plots and the dNTP concentration dependence of the kinetic parameters for pol η, ι, and κ are presented in Figures S1–S12. The fitted parameters are presented in Tables 5–7. The parameters in the tables are the Amax, the maximum amplitude, KA, and the dNTP concentration at half maximal amplitude, kpol, the maximum burst rate constant, and KdNTP (app) the apparent dNTP dissociation constant, kssmax, the maximum kss, and Kss, the dNTP concentration at half maximal kssmax. In some cases, the parameter did not exhibit a [dNTP] dependence, in which case the K value was not listed.
Figure 4.
Pol η catalyzed insertion of dNTPs opposite O2–POB-dT. (A) Pol η (250 nM) and O2–POB-dT DNA (25 nM) were reacted with 10 μM (open circle), 30 μM (open square), 50 μM (square), 100 μM (up triangle), 200 μM (down triangle), 500 μM (diamond), and 1000 μM (circle) dATP. (B) Early time points from panel A. The lines are the best fit to the burst equation. The data points are the mean of three experiments ± standard deviation. dNTP concentration dependence on the (C) amplitude and burst rate constant (D) of the pol η catalyzed insertion of dNTPs opposite O2–POB-dT. The error bars are the standard errors. The lines (solid for dATP and dGTP, and dotted for dCTP and dTTP) are the best fit to eq 1 for dATP (black circle), dCTP (open square), dGTP (black square), and dTTP (open square).
Table 5.
Kinetic Parameters for the dNTP Dependence of the Pol κ Catalyzed Insertion Opposite O2-alkyl-dTa
| template | dNTP | amplitude
|
burst rate constant
|
steady-state rate constant
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Amax/[pol] | KA (μM) | Amax/[pol]/KA (mM−1) | kpol (s−1) | Kd dNTP (μM) | kpol/Kd(s−1 mM−1) | kssmax (s−1) | Kss (μM) | ||
| dT | dATP | 0.69 ± 0.03 | 1.9 ± 0.4 | 363 | 155 ± 7 | 7.9 ± 0.8 | 19600 | 0.31 ± 0.03 | 8.0 ± 2.0 |
| O2-Me-dT | dATP | 0.72 ± 0.04 | 22 ± 3 | 33 | 7.4 ± 0.4 | 168 ± 20 | 44 | 0.023 ± 0.003 | 73 ± 41 |
| O2-Me-dT | dCTP | 0.22 ± 0.02 | 100 ± 29 | 2.2 | 3.5 ± 0.28 | 32 ± 15 | 109 | 0.014 ± 0.002 | 130 ± 70 |
| O2-Me-dT | dGTP | 0.25 ± 0.02 | 62 ± 17 | 4 | 3.9 ± 0.23 | 41 ± 12 | 95 | 0.013 ± 0.001 | 41 ± 13 |
| O2-Me-dT | dTTP | 0.72 ± 0.04 | 97 ± 12 | 7.4 | 2.6 ± 0.4 | 96 ± 60 | 27 | 0.020 ± 0.002 | |
| O2-POB-dT | dATP | 0.68 ± 0.04 | 11 ± 2 | 62 | 5.8 ± 0.4 | 78 ± 16 | 74 | 0.026 ± 0.002 | 55 ± 17 |
| O2-POB-dT | dCTP | 0.18 ± 0.02 | 101 ± 29 | 1.8 | 3.89 ± 0.26 | 51 ± 16 | 76 | 0.010 ± 0.002 | 78 ± 45 |
| O2-POB-dT | dGTP | 0.20 ± 0.03 | 149 ± 67 | 1.3 | 3.35 ± 0.45 | 6 ± 15 | 555 | 0.011 ± 0.001 | 37 ± 14 |
| O2-POB-dT | dTTP | 0.72 ± 0.04 | 97 ± 12 | 7.4 | 2.6 ± 0.4 | 96 ± 60 | 27 | 0.020 ± 0.002 | |
Reactions were conducted with 250 nM pol, 25 nM DNA, and 0 to 1000 μM dNTP. The individual time courses were fit to eq 2 to obtain amplitude (A), burst rate constants (kb), and steady-state rate constants (kss) as a function of dNTP concentration. These values were fit to the hyperbolic equation to obtain the parameters in the table. The values are the mean ± standard error of three determinations. If the K values are missing, then the parameter was not dependent on the [dNTP].
Table 7.
Kinetic Parameters for the dNTP Dependence of the Pol ζ Catalyzed Insertion Opposite O2-alkyl-dTa
| template | amplitude
|
burst rate constant
|
steady-state rate constant
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| dNTP | Amax/[pol] | KA (μM) | Amax/[pol]/KA (mM−1) | kpol (s−1) | Kd dNTP (μM) | kpol/Kd (s−1 mM−1) | kssmax (s−1) | Kss (μM) | |
| dT | dATP | 0.3 ± 0.01 | 62 ± 2 | 8 ± 1 | 7750 | 0.86 ± 0.15 | 14 ± 5 | ||
| O2-Me-dT | dATP | 0.49 ± 0.11 | 340 ± 190 | 1.4 | 3.3 ± 0.5 | 0.033 ± 0.005 | 202 ± 84 | ||
| O2-Me-dT | dCTP | 0.18 ± 0.01 | 320 ± 27 | 0.6 | 3.2 ± 0.7 | 0.013 ± 0.003 | 438 ± 215 | ||
| O2-Me-dT | dGTP | 0.14 ± 0.015 | 195 ± 21 | 0.7 | 4.8 ± 0.6 | 210 ± 80 | 22 | 0.010 ± 0.001 | 152 ± 30 |
| O2-Me-dT | dTTP | 0.87 ± 0.47 | 1120 ± 960 | 0.7 | 3.2 ± 0.5 | 0.039 ± 0.006 | 364 ± 139 | ||
| O2-POB-dT | dATP | 0.29 ± 0.03 | 85 ± 37 | 3.4 | 3.3 ± 0.3 | 20 ± 7 | 165 | 0.029 ± 0.006 | 260 ± 150 |
| O2-POB-dT | dCTP | 0.25 ± 0.07 | 860 ± 410 | 0.3 | 2.9 ± 0.4 | 60 ± 40 | 48 | 0.014 ± 0.002 | 490 ± 170 |
| O2-POB-dT | dGTP | 0.80 ± 0.67 | 5200 ± 4800 | 0.2 | 2.3 ± 0.1 | 10 ± 4 | 230 | 0.011 ± 0.002 | 380 ± 200 |
| O2-POB-dT | dTTP | 0.51 ± 0.03 | 350 ± 60 | 1.5 | 2.8 ± 0.3 | 0.027 ± 0.004 | 110. ± 50 | ||
Reactions were conducted with 150 nM pol, 15 nM DNA, and 0 to 1000 μM dNTP. The individual time courses were fit to eq 2 to obtainamplitude (A), burst rate constants (kb), and steady-state rate constants (kss) as a function of dNTP concentration. These values were fit to the hyperbolic equation to obtain the parameters in the table. The values are the mean ± standard error of three determinations. If the K values are missing, then the parameter was not dependent on the [dNTP].
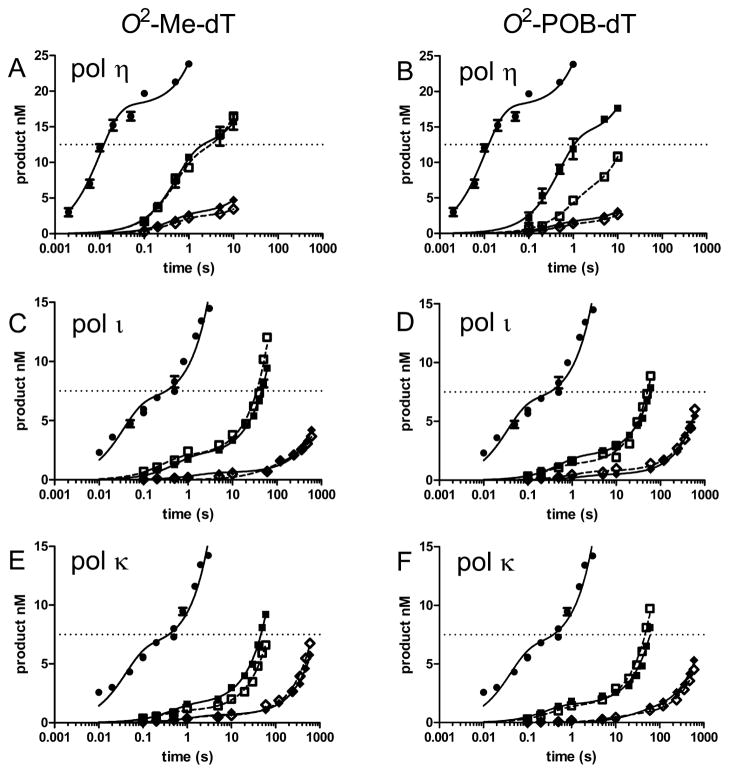
The relative reactivity of the polymerase/DNA/dNTP pairings are shown in the time course plots in Figure 5 in which the dNTP concentration was 50 μM. This dNTP concentration is shown because cellular dNTP concentrations typically range from 5 to 50 μM.52 The left panels have O2-Me-dT, and the right panels have O2-POB-dT as the template. Each panel also shows the correct incorporation of dATP opposite dT (black cricle). Several general observations can be made. First, the rate of incorporation of dATP opposite dT is 100–1000-fold faster than incorporation opposite the O2-alkyl-dTs. Second, pol η is the most reactive polymerase toward the alkylated substrates. Third, the incorporation of dATP and dTTP are faster than the incorporation of dCTP and dGTP opposite the O2-alkyl-dTs. These observations are discussed further below.
Figure 5.
Comparisons of the relative reactivity of polymerases with DNA and dNTP. The pol and O2-alkyl-dT DNA pairing in each panel is as follows: (A) pol η and O2-Me-dT; (B) pol η and O2-POB-dT; (C) pol ι and O2-Me-dT; (D) pol ι and O2-POB-dT; (E) pol ι and O2-Me-dT; (F) pol ι and O2-POB-dT. Each panel shows the insertion of 25 μM dATP opposite dT (black circle). The polymerase (250 nM) and DNA (25 nM) were reacted with 50 μM dATP (black square), dCTP (open diamond), dGTP (black diamond), dTTP (open square); dT, O2-Me-dT, or O2-POB-dT. The solid lines are the best fit to the burst equation with the purine dNTP represented by the solid line and the pyrimidine dNTP represented by the dashed line.
O2-alkyl-dT Are Poorer Substrates than dT
The decreased reactivity of the incorporation of dATP opposite the alkylated substrates is due to decreased kpol and increased KddNTP and KA values. This is evident in Tables 5–7, in which the kpol values for dATP decreases from 160, 72, and 62 s−1 to less than 10 s−1 with O2-alkyl-dT for all three enzymes. For pol η, the KddNTP(app) increased ~10-fold, but for ι and κ, the KddNTP(app) did not appreciably change. The Amax for pol η remained at ~0.7 for both dT and the O2-alkyl-dTs, while the KA and KddNTP increased 5–10-fold for the O2-alkyl-dTs.
Pol η Is More Reactive than Pol ι or κ with Both O2-Me-dT and O2-POB-dT
In Figure 5, the increased reactivity of pol η is evident by comparing the half-lives of the reactions. The half-lives of the pol η-catalyzed incorporation of dATP is ~1 s, while that for pol ι and κ are ~30 s. The increased reactivity of pol η is due to an increased burst amplitude for pol η relative to pols ι and κ. These conclusions are evident in Tables 5–7 in which the Amax is the highest and the KA is the lowest for pol η when compared with those of pols ι and κ. For example, for dATP, the Amax values are 0.72 and 0.68, and KA values are 25 and 14 μMfor O2-Me-dT and O2-POB-dT, respectively. The Amax values drop to between 0.24 and 0.49, and the KA values rise to 90–340 μMfor pols ι and κ. In contrast, the kpol and KddNTP(app) parameters are very similar for each polymerase. For example, for pol η and O2-alkyl-dT, the kpol ranges from 5.8 to 7.3 s−1, and very similar values are observed for pol ι (3–3.7 s−1) and pol κ (3 s−1). The KddNTP(app) values are also very similar: η (67–162 μM), ι (8–43 μM), and κ (8–11 μM).
dATP and dTTP Are the Most Reactive Nucleotides
The increased reactivity of dATP and dTTP over dCTP and dGTP is due primarily to the burst amplitudes. Examination of Table 5 for pol η shows that the Amax/[pol] for dATP and dTTP is ~0.7, while that for dCTP and dGTP is ~0.2. In contrast, the KA, kpol, and KddNTP(app) parameters are quite similar for each dNTP.
Extension Past O2-alkyl-dT
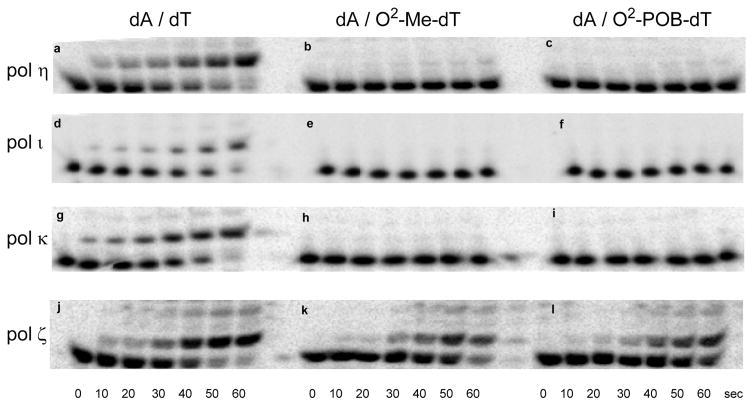
We examined the ability of human DNA pols η, κ, and ι, and yeast pol ζ to extend past X/Y base pairs using the P16/T24 DNA substrates. Figure 6 shows polyacrylamide gels for the extension past dA/dT (left), dA/O2-Me-dT (middle), and dA/O2-POB-dT (right) by pols η, κ, ι, and ζ. In each panel, the lower band is the 16-mer starting material, and the upper bands are product bands. The polymerase concentrations were set so that the reactivity with the dA/dT DNA substrate was similar for each polymerase. As can be seen in the left panels, the polymerases react with the DNA almost to completion within 1 min. Also evident is the robust activity of yeast pol ζ (lower panels), while the human Y-family polymerases show little if any activity. These experiments were duplicated for pol ζ with dC, dG, and dT as the base pair partner for the O2-alkyl-dTs. Plots showing the quantification of these data are shown in the Supporting Information. These experiments indicate that pol ζ catalyzes the extension past the dN/O2-alkyl-dT more efficiently than the Y-family polymerases and that the extension is not dependent on the base pair partner or the alkyl chain.
Figure 6.
Relative bypass of O2-alkyl-dT by human (a–c) pol η, (d–f) pol κ, (g–i) pol ι, and (j–l) yeast pol ζ. The concentration of the next incoming dNTP was 50 μM, and the concentration of the DNA containing a dA/dT (left), dA/O2-Me-dT (middle), and dA/O2-POB-dT (left) base pair was 3 nM. The concentration of pol η was 0.05 nM, pol κ and ι were 0.1 nM, and pol ζ was 6 μg/μL.
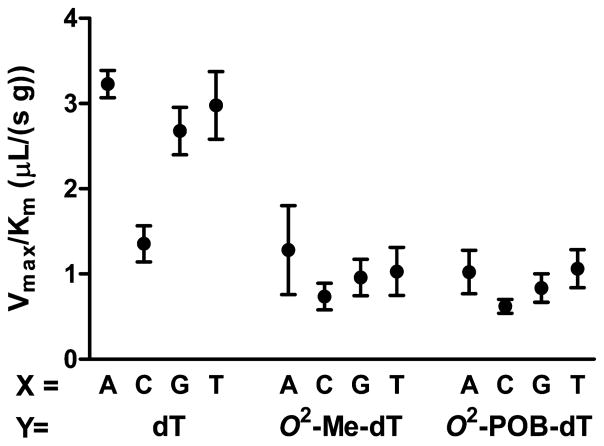
To quantitate the relative reactivity of pol ζ in extending undamaged versus NNK-damaged base pairs, we performed steady-state kinetic analysis. The initial rate kinetics of the bypass of dN/O2-alkyl-dT were performed at various concentrations of the next correct dNTP. The data were fit to eq 1, and the resulting Michaelis–Menten parameters are presented in Table 8. The Vmax/Km parameters are summarized in Figure 7. Several observations are made. First, the presence of mispairs in the terminal base pair (X/Y) does not inhibit the insertion of the correct dNTP. This result is consistent with previous reports that pol ζ is effective at extending mispairs.53 Second, the presence of the O2-alkyl-dT only inhibits replication by a factor of 3. Third, the identity of the alkyl chain, being methyl or POB, does not affect the Vmax/Km parameter. Finally, the base pair opposite the O2-alkyl-dT does not affect the rate of extension.
Table 8.
Steady-State Kinetic Parameters for Pol ζ Catalyzed the Extension Past O2-alkyl-dTa
| Y | Z | dNTP | X | Vmax nmol s−1 g−1 | Km μM | Vmax/Km μ L/(s g) |
|---|---|---|---|---|---|---|
| A | T | A | dT | 0.0674 ± 0.0067 | 20.9 ± 2.4 | 3.22 ± 0.16 |
| A | T | C | dT | 0.0472 ± 0.0047 | 35.7 ± 8.2 | 1.35 ± 0.21 |
| A | T | G | dT | 0.0674 ± 0.0040 | 25.2 ± 1.3 | 2.68 ± 0.28 |
| A | T | T | dT | 0.0678 ± 0.0090 | 23.4 ± 6.5 | 2.98 ± 0.40 |
| T | A | A | O2-Me-dT | 0.0421 ± 0.0078 | 38 ± 18 | 1.28 ± 0.52 |
| T | A | C | O2-Me-dT | 0.0411 ± 0.011 | 60 ± 31 | 0.73 ± 0.16 |
| T | A | G | O2-Me-dT | 0.0424 ± 0.00035 | 46 ± 14 | 0.96 ± 0.21 |
| T | A | T | O2-Me-dT | 0.0444 ± 0.0103 | 48 ± 26 | 1.03 ± 0.28 |
| T | A | A | O2-POB-dT | 0.0431 ± 0.0075 | 44 ± 16 | 1.02 ± 0.25 |
| T | A | C | O2-POB-dT | 0.0401 ± 0.0048 | 66 ± 16 | 0.62 ± 0.08 |
| T | A | G | O2-POB-dT | 0.0416 ± 0.0013 | 51 ± 11 | 0.83 ± 0.17 |
| T | A | T | O2-POB-dT | 0.0371 ± 0.0038 | 36 ± 11 | 1.06 ± 0.24 |
Data are expressed as the mean ± SD of three independent experiments. The DNA substrate was P16/T24 as in Chart 1.
Figure 7.
Pol ζ catalyzed extension of O2-alkyl-dT base pairs. The relative Vmax/Km values for dA, dC, dG, and dT as at the primer terminus (X) with dT, O2-Me-dT, and O2-POB-dT as base pair partner (Y). The steady-state kinetics were performed with 30 nM DNA (P16/T24) and 1 μg/μL pol ζ. The data points are the mean ± SD of three independent determinations.
Since pol ζ is effective in extending dN/O2-POB-dT base pairs, we briefly examined if the enzyme was capable of inserting a nucleotide opposite the adducts. As is shown in Figure S18, the enzyme is very inefficient at inserting dATP opposite O2-alkyl-dT.
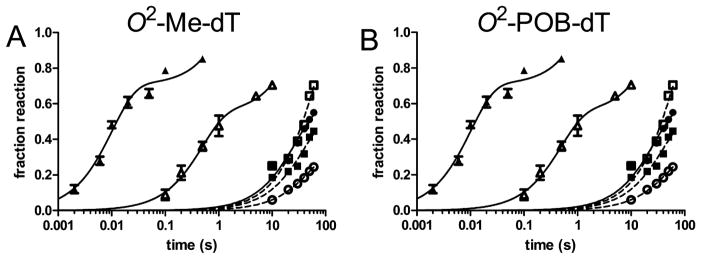
Extension Past O2-alkyl-dT with Polymerase in Excess
As discussed above, the steady-state kinetic parameters may not accurately reflect the activity of the Y-family polymerases during the incorporation of a single nucleotide at the replication fork. We therefore examined the extension past the adducts with enzyme in excess. We found that extension past the adducts was slower than the insertion opposite the adducts for the three Y-family polymerases studied. The reactions with 3 nM DNA, 30 nM pol η, and 50 μM dNTP are shown in Figure 8. The black triangles show the rapid insertion of dATP opposite dT, and the open triangles show the slower insertion of dATP opposite O2-alkyl-dT. The extension past the dN/O2-POB-dT base pairs are represented by the solid and open circles and squares. As is evident, the pol η catalyzed extension is much slower than the insertion reaction. The extension past the adducts by pols ι and κ are shown in Figure S19.
Figure 8.
Insertion and extension reactivity of pol η past O2-Me-dT (A) and O2-POB-dT (B). Pol η (30 nM) and DNA (3 nM) were reacted with 50 μM dNTP. Each panel shows the insertion of dATP opposite dT (triangle) and O2-alkyl-dT (open triangle) and the extension past dA (square), dC (open circle), dG (black circle), and dT (open square) opposite O2-alkyl-dT. The solid lines are the best fit to the burst equation, and the dashed lines are fit to a first-order equation.
DISCUSSION
NNK is a potent human lung carcinogen.4 It is unique among lung carcinogens in that irrespective of the route of administration, rodents given NNK develop lung tumors.5 Understanding the molecular mechanisms underlying its mutagenicity may shed some light on its remarkable organ-specificity. NNK is a bifunctional alkylating agent, producing both methyl and POB-DNA adducts. While similar sites on the DNA are alkylated, the relative proportions are different. In particular, O2-POB-dT is the most abundant POB-DNA adduct in rats chronically treated with NNK or NNN.14,54–56 In contrast, the corresponding O2-Me-dT is a minor adduct from the corresponding methylating agent. Rodents given NNK have increased levels of mutations at AT base pairs when compared with rodents given a methylating agent,19,20 supporting the role of NNK in mutagenesis at AT base pairs.
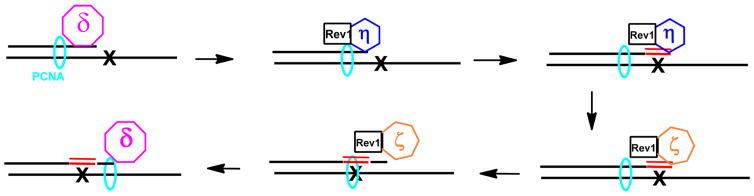
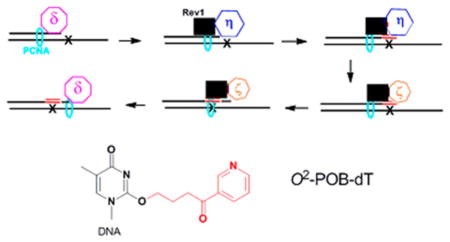
Recently, we found that siRNA knockdown of pol η, ζ, and Rev1 impacted the bypass of O2-Me-dT and O2-POB-dT in human cells.44 These results, along with known reactivities of the enzymes, led to the hypothesis illustrated in Scheme 2 in which pol η is involved in the insertion of dNTPs opposite the adducts, pol ζ is involved in the extension past the adduct, while Rev1 is a structural protein. To test the hypotheses that catalytic activities of pols η and ζ are critical to bypass and mutagenesis of O2-POB-dT, we evaluated the in vitro kinetics of insertion of dNTPs opposite and bypass of O2-POB-dT by pols η, ι, κ, and ζ.
Scheme 2.
Model for the Bypass of O2-alkyll-dT
We initially examined the steady-state kinetics for the incorporation of each dNTP opposite both O2-Me-dT and O2-POB-dT. The kinetic parameters in Tables 2–4 show that pols η, κ, and ι are equally poor at insertion opposite the adducts. This result is inconsistent with a role for pol η in bypass. However, TLS polymerases do not operate under steady-state kinetics in vivo. A current model of TLS bypass is the handoff model in which polymerases are recruited for insertion opposite the adduct, while another polymerase can perform the extension past the adduct.21,34 In this model, a polymerase acts once, and consequently, steady-state kinetics do not accurately describe the reactivity.
Under conditions of polymerase in excess, the time courses for the insertion of dNTPs opposite the adducts are biphasic with a rapid burst followed by a slower reaction. As illustrated in Figure 5, we show that pol η is more effective than pols ι and κ in the insertion of dNTPs opposite O2-POB-dT. All three enzyme are more effective than Kf(exo-) and Dpo4.39 The biphasic kinetic behavior under conditions with polymerase in excess can be explained by two mechanisms: (1) formation of nonproductive enzyme–substrate complexes39,57–61 and (2) a rate limiting step after rapid phosphodiester bond formation.62,63 The formation of nonproductive complexes has been observed in DNA damage bypass.39,57–61 In addition, pol κ exhibits this behavior during the formation of a correct base pair.64
The substrate specificity of polymerases have been reported by their relative kcat/Km and presteady-state kpol/KddNTP values.65,66 However, this relationship does not hold in this system. For example, as shown in Table 5, the kpol/Kd value for pol η-catalyzed incorporation of dGTP (555 mM−1 s−1) is greater than that for dATP (74 mM−1 s−1). However, it is clearly evident in Figure 5B that the incorporation of dATP (solid square) is faster than the incorporation of dGTP (solid diamond). Therefore, the relative reactivity of the substrates is not simply the kpol/Kd ratios but must take into account the amplitude and the steady-state parameters.
Pol κ and its E. coli analogue pol IV accurately bypass N2-dG adducts of various sizes with good efficiency.51,67–70 For example, human pol κ bypasses the very bulky N2-benzopyrene adduct, while the prokaryotic analogue (pol IV/dinB) bypasses the less bulky N2-furfuryl-dG and N2-(1-carboxyethyl)-2′-dG adducts. The crystal structure of pol κ displays an open area on the minor groove of DNA.71 Thus, one can envision that pol κ could bypass O2-POB-dT by binding the POB-group in the minor groove pocket of pol κ, while dGTP would bind to dT in a Watson–Crick-like conformation in Figure 9d. However, we found that neither O2-POB-dT nor the smaller O2-Me-dT is a good substrate for pol κ. This result agrees with Andersen et al., who reported similar findings for O2-Me-dT and O2-Et-dT.40,41
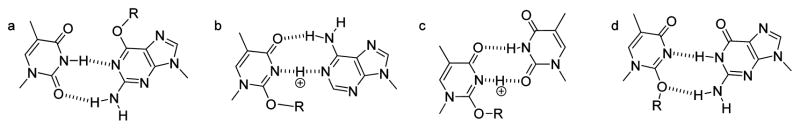
Figure 9.
Potential base pair structures.
The ability of pol κ to accommodate N2-alkyl-dG DNA damage is not a strictly steric issue. For example, the replacement of Phe13 by the smaller valine eliminates the ability of pol IV to bypass N2-furfuryl-dG.69 This result appears to suggest that van der Waals interactions play a role in pol κ’s substrate specificity. In addition, functional/kinetic studies show that pol κ requires Watson–Crick hydrogen bonds for rapid catalysis.72 Adifference between N2-dG adducts and O2-POB-dT is that O2-POB-dT cannot form Watson–Crick hydrogen bonds with the incoming dNTP. While O2-POB-dT has a bulky adduct in the minor groove, perhaps the inability of O2-POB-dT to form Watson–Crick base pairs makes this a poor substrate for pol κ. This rationale was proposed to explain why pol IV is not involved in the bypass of O2-alkyl-dT adducts in E. coli.73 In this article, we extend this rationale to human pol κ.
Pol ι flips template purines from the anti- to the syn-configuration thereby displaying the Hoogsteen hydrogen bonding face to the incoming dNTP.74–77 This property allows for the replication of adducts in which the Watson–Crick hydrogen bonding face is blocked.78 In addition, O6-Et-dG and N2-Et-dG adducts are bypassed in this manner.79,80 However, pol ι is inefficient at bypassing more bulky N2-dG adducts.50 Template pyrimidines react with pol ι in the anti-conformation. Kinetic evidence indicates that dGTP/dC is replicated with Watson–Crick base pairs, while dGTP/dT occurs via wobble base pairing.81 Upon the basis of these properties, there is no reason to expect that pol ι would be efficient at bypassing O2-POB-dT.
Pol η evolved to accurately bypass cis-syn cyclobutane pyrimidine dimers. Inactivation of the POLH gene leads to the XPV form of xeroderma pigmentosum in which individuals are predisposed to skin cancer.82–84 Pol η is also proficient at bypassing other adducts such as N-(deoxyguanosin-8-yl)-1-aminopyrene,61 O6-alkyl-dG,13 and 8-oxo-dG.85–88 Synthesis opposite the bulky N2-dG adducts is slow,67 although pol η may play a role in the mutagenic bypass of N2-BP-dG.68,89,90 These studies do not provide any mechanistic rationale for the ability of pol η to insert dTTP and dATP opposite O2-alkyl-dT. Our in vitro kinetic data do agree with the mammalian cell culture studies in which pol η is crucial to the bypass.44
The mechanism underlying the pol η catalyzed insertion of dTTP and dATP opposite O2-alkyl-dT is not clear. The ability to form Watson–Crick-like structures has been proposed to explain the mutagenic potential of DNA adducts. This type of simplistic structural analysis worked well with O6-Me-dG in which the Watson–Crick-like structure (Figure 9a) initially proposed in 197691 explains the mutagenic incorporation of dTTP.92–94 However, an attempt to predict the base pairing of O2-alkyl-dT is not as satisfying. Figure 9b and c illustrates the potential hydrogen-bonding interactions between dA and dT with O2-alkyl-dT. While the dA/O2-alkyl-dT structure requires protonation, the alkyl group can also make a hydrophobic interaction with the 2-position of adenine stabilizing the structure.95 Figure 9c displays a base-pair structure between dT and O2-alkyl-dT that is similar to the wobble structure that was observed in an oligodeoxynucleotide duplex containing a dT/dT mispair.96 Figure 9b and c does not exhibit any chemical interactions that would explain the preference for pol η to preferentially insert dATP and dTTP opposite O2-alkyl-dT more often than dGTP. Figure 9d shows a potential Watson–Crick-like structure between dG and O2-alkyl-dT. This structure is very similar to that in Figure 9a, except that the alkyl group is in the minor groove. Perhaps this structure explains the mutagenic incorporation of dGTP opposite O2alkyl-dT by pol V in E. coli.42,73 However, the preference for the incorporation of dT and dA opposite O2-alkyl-dG in humans cells is not evident. Structural and functional studies must be undertaken to elucidate the interactions that control the identity of the nucleotide inserted opposite O2-alkyl-dTs.
While pol η is the most efficient Y-family polymerase to incorporate dNTPs opposite O2-alkyl-dG, neither it nor pol ι or κ were effective at further extending the primer. We did find that yeast pol ζ is able to extend past the O2-alkyl-dT adducts. Pol ζ is a B-family polymerase that does not insert dNTPs opposite DNA damage but readily extends past mismatches,97,98 abasic sites,99 γ-hydroxy-1,N2-propano-dG,100 8-oxo-dG,101 O6-Me-dG,101 thymine glycol,102 and mismatched N2-BP-dG/T.103,104 The polymerase we employed was the heterodimer between yeast Rev3 and Rev7. Human Rev3 is twice as large as the yeast protein.105,106 Human and yeast Rev7 are homologous, and the activity of the catalytic subunit, Rev3, is increased 10-fold by Rev 7.106 In vivo, the likely active species is a tetramer among Rev3, Rev7, and the pol δ subunits, pol31 and pol32.107–110 We found that yeast pol ζ readily extends dN/O2-alkyl-dT base pairs, with kcat/Km values one-third that of undamaged DNA. There is no sequence specificity with respect to the base pair partner.
Our model for the bypass of O2-POB-dT is illustrated in Scheme 1. A replicative polymerase, such as pol δ, synthesizes up to the adduct. We found that the high fidelity polymerase Kf(exo-) can synthesize up to the adduct, but have very little activity at the insertion of a dNTP opposite the adduct.39 The replicative polymerase stalls at the adduct and is replaced by a Y-family polymerase. Pol η has higher activity toward O2-POB-dT than κ or ι, and based upon relative kinetics, it will be the polymerase that inserts the correct dA or an incorrect dT opposite the adduct.40,41 Pol η, as well as ι and κ, has low activity extending past O2-POB-dT. Following insertion, pol η will be replaced by pol ζ, which can extend the primer past the adduct. Subsequently, a high fidelity polymerase can continue DNA synthesis. The activity of pol ζ does not depend on the identity of the nucleotide that is inserted opposite the adduct. In our model, we have Rev1 acting as a scaffold protein. The involvement of Rev1 is supported from cell studies using siRNA to deplete polymerases. 44 Rev1 is highly selective for inserting C opposite normal and adducted template G because it uses arginine as a template for dCTP binding.111,112
Supplementary Material
Table 1.
Dissociation Constants for Normal and Damaged DNA to Polymerasesa
| X | pol η (nM) | pol κ (nM) | pol ι (nM) |
|---|---|---|---|
| dT | 7.9 ± 0.7 | 8.3 ± 0.7 | 8.4 ± 0.7 |
| O2-Me-dT | 10.1 ± 0.7 | 9.1 ± 0.5 | 9.7 ± 0.5 |
| O2-POB-dT | 8.9 ± 0.8 | 9.4 ± 0.9 | 9.7 ± 0.8 |
Calculated value with standard error. The fluorescence anisotropy was determined with 1 nM DNA with 0–100 nM polymerase in the appropriate buffer. The experiment was performed three times, and the data were fitted to the quadratic equation.
Table 3.
Steady-State Parameters for the Pol ι Catalyzed Insertion Opposite O2-alkyl-dTa
| dNTP | template | kcat (min−1) | Km (μM) | kcat/Km (mM−1 min−1) |
|---|---|---|---|---|
| dATP | dT | 8.0 ± 0.6 | 2.6 ± 0.4 | 3070 ± 270 |
| dTTP | dT | 0.103 ± 0.003 | 70 ± 4 | 1.46 ± 0.06 |
| dATP | O2-Me-dT | 0.076 ± 0.002 | 24 ± 1 | 3.2 ± 0.1 |
| dCTP | O2-Me-dT | 0.061 ± 0.004 | 93 ± 14 | 0.65 ± 0.06 |
| dGTP | O2-Me-dT | 0.114 ± 0.002 | 46 ± 2 | 2.5 ± 0.1 |
| dTTP | O2-Me-dT | 0.121 ± 0.005 | 42 ± 5 | 2.9 ± 0.2 |
| dATP | O2-POB-dT | 0.086 ± 0.003 | 35 ± 3 | 2.4 ± 0.1 |
| dCTP | O2-POB-dT | 0.059 ± 0.003 | 112 ± 13 | 0.53 ± 0.03 |
| dGTP | O2-POB-dT | 0.094 ± 0.003 | 28 ± 3 | 3.3 ± 0.2 |
| dTTP | O2-POB-dT | 0.121 ± 0.001 | 37 ± 1 | 3.27 ± 0.08 |
Initial rates were conducted with 0.25 to 0.5 nM pol, 5 nM DNA, and 0 to 200 μM dNTP. The values are the mean ± standard deviation of three determinations.
Table 6.
Kinetic Parameters for the dNTP Dependence of the Pol ι Catalyzed Insertion Opposite O2-alkyl-dTa
| template | dNTP | amplitude
|
burst rate constant
|
steady-state rate constant
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Amax/[pol] | KA (μM) | Amax/[pol]/KA (mM−1) | kpol (s−1) | Kd dNTP (μM) | kpol/Kd (s−1 mM−1) | kssmax (s−1) | Kss (μM) | ||
| dT | dATP | 3.0 ± 0.4 | 22 ± 6 | 0.25 ± 0.03 | 10 ± 5 | ||||
| O2-Me-dT | dATP | 0.420 ± 0.047 | 190 ± 60 | 8.9 | 3.3 ± 0.3 | 0.027 ± 0.267 | 78 ± 37 | ||
| O2-Me-dT | dCTP | 0.180 ± 0.020 | 601 ± 130 | 0.3 | 2.3 ± 0.6 | 0.013 ± 0.003 | 250 ± 160 | ||
| O2-Me-dT | dGTP | 0.093 ± 0.007 | 213 ± 56 | 0.4 | 5.5 ± 1.1 | 217 ± 121 | 25 | 0.017 ± 0.006 | 592 ± 426 |
| O2-Me-dT | dTTP | 0.380 ± 0.033 | 104 ± 34 | 3.6 | 3.6 ± 0.2 | 0.023 ± 0.003 | 110 ± 40 | ||
| O2-POB-dT | dATP | 0.313 ± 0.020 | 90 ± 20 | 3.5 | 3.7 ± 0.2 | 43 ± 12 | 86 | 0.017 ± 0.001 | 80 ± 20 |
| O2-POB-dT | dCTP | 0.187 ± 0.013 | 300 ± 53 | 0.6 | 2.53 ± 0.05 | 27 ± 3 | 94 | 0.004 ± 0.001 | 8 ± 15 |
| O2-POB-dT | dGTP | 0.147 ± 0.013 | 421 ± 105 | 0.3 | 2.42 ± 0.24 | 45 ± 22 | 54 | 0.007 ± 0.001 | 186 ± 63 |
| O2-POB-dT | dTTP | 0.307 ± 0.033 | 82 ± 31 | 3.7 | 3.7 ± 0.3 | 42 ± 18 | 88 | 0.024 ± 0.004 | 170 ± 90 |
Reactions were conducted with 150 nM pol, 15 nM DNA, and 0 to 1000 μM dNTP. The individual time courses were fit to eq 2 to obtain amplitude (A), burst rate constants (kb), and steady-state rate constants (kss) as a function of dNTP concentration. These values were fit to the hyperbolic equation to obtain the parameters in the table. The values are the mean ± standard error of three determinations. If the K values are missing, then the parameter was not dependent on the [dNTP].
Acknowledgments
The oligodeoxynucleotide synthesis and MS analysis were performed in the Macromolecular Core facility at the PSU College of medicine. Core Facility services and instruments used in this project were funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds.
Funding
This project was funded under an NIH grant (ES021762).
ABBREVIATIONS
- 7-POB-dG
7-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine
- Dpo4
Sulfolobus solfataricus DNA polymerase IV
- Kf(exo-)
Klenow fragment of proofreading deficient E. coli DNA polymerase I
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNN
N′-nitrosonornicotine
- O2-Me-dT
O2-methylthymidine
- O2-POB-dC
O2-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxycytidine
- O2-POB-dT
O2-[4-(3-pyridyl)-4-oxobut-1-yl]-thymidine
- O6-POB-dG
O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine
- POB
4-(3-pyridyl)-4-oxobut-1-yl
- pol
polymerase
- yPol
Saccharomyces cerevisiae DNA polymerase
Footnotes
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions. The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.5b00468.
Kinetic plots for the incorporation opposite and extension past dT, O2-Me-dT, and O2-POB-dT by pol η, ζ, ι, and κ are presented (PDF)
References
- 1.American Cancer Society. Cancer Facts and Figures, 2014. American Cancer Society; Vienna, VA: 2014. [Google Scholar]
- 2.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 4.IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. World Health Organization, International Agency for Research on Cancer; Lyon, France: 2004. Tobacco Smoke and Involuntary Smoking; pp. 53–119. [PMC free article] [PubMed] [Google Scholar]
- 5.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 6.Peterson LA, Mathew R, Murphy SE, Trushin N, Hecht SS. In vivo and in vitro persistence of pyridyloxobutyl DNA adducts from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 1991;12:2069–2072. doi: 10.1093/carcin/12.11.2069. [DOI] [PubMed] [Google Scholar]
- 7.Peterson LA, Hecht SS. O6 -Methylguanine is a critical determinant of 4-(methylnitrosamino)-1-(3-pyridyl)-1-buta-none tumorigenesis in A/J mouse lung. Cancer Res. 1991;51:5557–5564. [PubMed] [Google Scholar]
- 8.Liu XK, Spratt TE, Murphy SE, Peterson LA. Pyridyloxobutylation of guanine residues by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem Res Toxicol. 1996;9:949–953. doi: 10.1021/tx960067t. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Spratt TE, Liu XK, Hecht SS, Pegg AE, Peterson LA. Pyridyloxobutyl adduct O6-[4-oxo-4-(3-pyridyl)-butyl]guanine, is present in 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone-treated DNA and is a substrate for O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 1997;10:562–567. doi: 10.1021/tx9602067. [DOI] [PubMed] [Google Scholar]
- 10.Mijal RS, Thomson NM, Fleischer NL, Pauly GT, Moschel RC, Kanugula S, Fang Q, Pegg AE, Peterson LA. The repair of the tobacco specific nitrosamine derived adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine by O6 -alkylguanine-DNA alkyltransferase variants. Chem Res Toxicol. 2004;17:424–434. doi: 10.1021/tx0342417. [DOI] [PubMed] [Google Scholar]
- 11.Pauly GT, Peterson LA, Moschel RC. Mutagenesis by O6-[4-oxo-4-(3-pyridyl)butyl]guanine in Escherichia coli and human cells. Chem Res Toxicol. 2002;15:165–169. doi: 10.1021/tx0101245. [DOI] [PubMed] [Google Scholar]
- 12.Mijal RS, Loktionova NA, Vu CC, Pegg AE, Peterson LA. O6-Pyridyloxobutylguanine adducts contribute to the mutagenic properties of pyridyloxobutylating agents. Chem Res Toxicol. 2005;18:1619–1625. doi: 10.1021/tx050139t. [DOI] [PubMed] [Google Scholar]
- 13.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J Biol Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 14.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high performance liquid chromatographyelectrospray ionization-tandem mass spectrometry. Chem Res Toxicol. 2006;19:674–682. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Upadhyaya P, Lindgren BR, Hecht SS. Comparative levels of O6-methylguanine, pyridyloxobutyl-, and pyridylhydroxybutyl-DNA adducts in lung and liver of rats treated chronically with the tobacco-specific carcinogen 4-(methylnitrosami-no)-1-(3-pyridyl)-1-butanone. Drug Metab Dispos. 2009;37:1147–1151. doi: 10.1124/dmd.109.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Analysis of pyridyloxobutyl DNA adducts in F344 rats chronically treated with (R)- and (S)-N′-nitrosonornicotine. Chem Res Toxicol. 2007;20:246–256. doi: 10.1021/tx060208j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto K, Ohsawa Ki, Kimura M. Mutations induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in the lacZ and cII genes of Muta(TM) Mouse. Mutat Res, Genet Toxicol Environ Mutagen. 2004;560:119–131. doi: 10.1016/j.mrgentox.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Sandercock LE, Hahn JN, Li L, Luchman HA, Giesbrecht JL, Peterson LA, Jirik FR. Mgmt deficiency alters the in vivo mutational spectrum of tissues exposed to the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) Carcinogenesis. 2008;29:866–874. doi: 10.1093/carcin/bgn030. [DOI] [PubMed] [Google Scholar]
- 20.Shane BS, Smith-Dunn DL, de Boer JG, Glickman BW, Cunningham ML. Mutant frequencies and mutation spectra of dimethylnitrosamine (DMN) at the lacI and cII loci in the livers of Big Blue transgenic mice. Mutat Res, Fundam Mol Mech Mutagen. 2000;452:197–210. doi: 10.1016/s0027-5107(00)00081-6. [DOI] [PubMed] [Google Scholar]
- 21.Yang W. An overview of Y-family DNA polymerases and a case Study of human DNA polymerase η. Biochemistry. 2014;53:2793–2803. doi: 10.1021/bi500019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shcherbakova PV, Bebenek K, Kunkel TA. Functions of Eukaryotic DNA Polymerases. Sci Aging Knowl Environ. 2003;2003:re3. doi: 10.1126/sageke.2003.8.re3. [DOI] [PubMed] [Google Scholar]
- 24.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 25.Skosareva LV, Lebedeva NA, Rechkunova NI, Kolbanovskiy A, Geacintov NE, Lavrik OI. Human DNA polymerase lambda catalyzes lesion bypass across benzo[a]-pyrene-derived DNA adduct during base excision repair. DNA Repair. 2012;11:367–373. doi: 10.1016/j.dnarep.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Maga G, Villani G, Ramadan K, Shevelev I, Tanguy Le Gac N, Blanco L, Blanca G, Spadari S, Hubscher U. Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem. 2002;277:48434–48440. doi: 10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 27.Maga G, Crespan E, Markkanen E, Imhof R, Furrer A, Villani G, Hubscher U, van Loon B. DNA polymerase delta-interacting protein 2 is a processivity factor for DNA polymerase lambda during 8-oxo-7,8-dihydroguanine bypass. Proc Natl Acad Sci U S A. 2013;110:18850–18855. doi: 10.1073/pnas.1308760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takata K-i, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) Is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 29.Yousefzadeh MJ, Wood RD. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair. 2013;12:1–9. doi: 10.1016/j.dnarep.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, Taylor E, Stevanovic I, Green AJ, Stracker TH, Lindsay HD, Doherty AJ. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell. 2013;52:566–573. doi: 10.1016/j.molcel.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T. The Y-Family of DNA Polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 33.Yang W, Woodgate R. What a difference a decade makes: Insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livneh Z, ZO, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 36.Xu YZ, Swann PF. Oligodeoxynucleotides containing O2-alkylthymine: Synthesis and characterization. Tetrahedron Lett. 1994;35:303–306. [Google Scholar]
- 37.Grevatt PC, Solomon JJ, Bhanot OS. In vitro mispairing specificity of O2-ethylthymine. Biochemistry. 1992;31:4181–4188. doi: 10.1021/bi00132a005. [DOI] [PubMed] [Google Scholar]
- 38.Bhanot OS, Grevatt PC, Donahue JM, Gabrielides CN, Solomon JJ. In vitro DNA replication implicates O2− ethyldeoxythymidine in transversion mutagenesis by ethylating agents. Nucleic Acids Res. 1992;20:587–594. doi: 10.1093/nar/20.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gowda ASP, Krishnegowda G, Suo Z, Amin S, Spratt TE. Low Fidelity Bypass of O2-(3-Pyridyl)-4-oxobutylthymine, the Most Persistent Bulky Adduct Produced by the Tobacco Specific Nitrosamine 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone by Model DNA Polymerases. Chem Res Toxicol. 2012;25:1195–1202. doi: 10.1021/tx200483g. [DOI] [PubMed] [Google Scholar]
- 40.Andersen N, Wang J, Wang P, Jiang Y, Wang Y. In-vitro replication studies on O2-methylthymidine and O4-methyl-thymidine. Chem Res Toxicol. 2012;25:2523–2531. doi: 10.1021/tx300325q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersen N, Wang P, Wang Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem Res Toxicol. 2013;26:1730–1738. doi: 10.1021/tx4002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jasti VP, Spratt TE, Basu AK. Tobacco-specific nitrosamine-derived O2-alkylthymidines are potent mutagenic lesions in SOS-induced. Chem Res Toxicol. 2011;24:1833–1835. doi: 10.1021/tx200435d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai Q, Wang P, Wang Y. Cytotoxic and mutagenic properties of regioisomeric O2-, N3- and O4-ethylthymidines in bacterial cells. Carcinogenesis. 2014;35:2002–2006. doi: 10.1093/carcin/bgu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weerasooriya S, Jasti VP, Bose A, Spratt TE, Basu AK. Roles of translesion synthesis DNA polymerases in the potent mutagenicity of tobacco-specific nitrosamine-derived O-alkylthymidines in human cells. DNA Repair. 2015;35:63–70. doi: 10.1016/j.dnarep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunn DB, Hall RH. In: Handbook of Biochemistry and Molecular Biology. Fasman GD, editor. CRC Press; Boca Raton, FL: 1986. pp. 65–215. [Google Scholar]
- 46.Krishnegowda G, Sharma AK, Krzeminski J, Gowda ASP, Lin JM, Desai D, Spratt TE, Amin S. Facile syntheses of O2-[4-(3-pyridyl-4-oxobut-1-yl]thymidine, the major adduct formed by tobacco specific nitrosamine 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in vivo, and Its site-specifically adducted oligodeox-ynucleotides. Chem Res Toxicol. 2011;24:960–967. doi: 10.1021/tx200127j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borer P. Optical Properties of Nucleic Acids, Absorption, and Circular Dichroism Spectra. In: Fasman GD, editor. Handbook of Biochemistry and Molecular Biology. CRC Press; Boca Raton, FL: 1977. p. 589. [Google Scholar]
- 48.Meyer AS, Blandino M, Spratt TE. E. coli DNA polymerase I (Klenow fragment) uses a hydrogen bonding fork from Arg668 to the primer terminus and incoming deoxynucleotide triphosphate to catalyze DNA replication. J Biol Chem. 2004;279:33043–33046. doi: 10.1074/jbc.C400232200. [DOI] [PubMed] [Google Scholar]
- 49.Choi JY, Guengerich FP. Adduct Size Limits Efficient and Error-free Bypass Across Bulky N2-Guanine DNA Lesions by Human DNA Polymerase eta. J Mol Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 50.Choi JY, Guengerich FP. Kinetic Evidence for Inefficient and Error-prone Bypass across Bulky N2-Guanine DNA Adducts by Human DNA Polymerase iota. J Biol Chem. 2006;281:12315–12324. doi: 10.1074/jbc.M600112200. [DOI] [PubMed] [Google Scholar]
- 51.Choi JY, Angel KC, Guengerich FP. Translesion Synthesis across Bulky N2-Alkyl Guanine DNA Adducts by Human DNA Polymerase kappa. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 52.Traut T. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 53.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 54.Upadhyaya P, Kalscheuer S, Hochalter JB, Villalta PW, Hecht SS. Quantitation of Pyridylhydroxybutyl-DNA Adducts in Liver and Lung of F-344 Rats Treated with 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone and Enantiomers of Its Metabolite 4-(Methyl-nitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2008;21:1468–1476. doi: 10.1021/tx8001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S, Wang M, Villalta PW, Lindgren BR, Upadhyaya P, Lao Y, Hecht SS. Analysis of Pyridyloxobutyl and Pyridylhydroxybutyl DNA Adducts in Extrahepatic Tissues of F344 Rats Treated Chronically with 4-(Methylnitrosamino)-1-(3-pyridyl)-1-buta-none and Enantiomers of 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2009;22:926–936. doi: 10.1021/tx900015d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balbo S, Johnson CS, Kovi RC, James-Yi SA, O’Sullivan MG, Wang M, Le CT, Khariwala SS, Upadhyaya P, Hecht SS. Carcinogenicity and DNA adduct formation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol in F-344 rats. Carcinogenesis. 2014;35:2798–2806. doi: 10.1093/carcin/bgu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furge LL, Guengerich FP. Explanation of pre-steady-state kinetics and decreased burst amplitude of HIV-1 reverse transcriptase at sites of modified DNA bases with an additional, non-productive enzyme-DNA-nucleotide complex. Biochemistry. 1999;38:4818–4825. doi: 10.1021/bi982163u. [DOI] [PubMed] [Google Scholar]
- 58.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo - and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 59.Woodside AM, Guengerich FP. Misincorporation and stalling at O6-methylguanine and O6-benzylguanine: evidence for inactive polymerase complexes. Biochemistry. 2002;41:1039–1050. doi: 10.1021/bi011496f. [DOI] [PubMed] [Google Scholar]
- 60.Woodside AM, Guengerich FP. Effect of the O6-substituent on misincorporation kinetics catalyzed by DNA polymerases at O6-methylguanine and O6-benzylguanine. Biochemistry. 2002;41:1027–1038. doi: 10.1021/bi011495n. [DOI] [PubMed] [Google Scholar]
- 61.Sherrer SM, Sanman LE, Xia CX, Bolin ER, Malik CK, Efthimiopoulos G, Basu AK, Suo Z. Kinetic Analysis of the Bypass of a Bulky DNA Lesion Catalyzed by Human Y-Family DNA Polymerases. Chem Res Toxicol. 2012;25:730–740. doi: 10.1021/tx200531y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 63.Gowda AS, Moldovan GL, Spratt TE. Human DNA Polymerase nu Catalyzes Correct and Incorrect DNA Synthesis with High Catalytic Efficiency. J Biol Chem. 2015;290:16292–16303. doi: 10.1074/jbc.M115.653287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlson KD, Johnson RE, Prakash L, Prakash S, Washington MT. Human DNA polymerase kappa forms nonproductive complexes with matched primer termini but not with mismatched primer termini. Proc Natl Acad Sci U S A. 2006;103:15776–15781. doi: 10.1073/pnas.0605785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fersht A. Structure and Mechanism in Protein Science. W.H. Freeman and Co; New York: 1999. [Google Scholar]
- 66.Bertram JG, Oertell K, Petruska J, Goodman MF. DNA Polymerase Fidelity: Comparing Direct Competition of Right and Wrong dNTP Substrates with Steady State and Pre-Steady State Kinetics. Biochemistry. 2010;49:20–28. doi: 10.1021/bi901653g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 68.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J Biol Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 69.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 70.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2’-deoxyguano-sine by DinB DNA polymerase in vitro and in vivo. Proc Natl Acad Sci U S A. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA Polymerase [kappa] Encircles DNA: Implications for Mismatch Extension and Lesion Bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 72.Wolfle WT, Washington MT, Kool ET, Spratt TE, Helquist SA, Prakash L, Prakash S. Evidence for a Watson-Crick Hydrogen Bonding Requirement in DNA Synthesis by Human DNA Polymerase kappa. Mol Cell Biol. 2005;25:7137–7143. doi: 10.1128/MCB.25.16.7137-7143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhai Q, Wang P, Cai Q, Wang Y. Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 2014;42:10529–10537. doi: 10.1093/nar/gku748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson RE, Haracska L, Prakash L, Prakash S. Role of hoogsteen edge hydrogen bonding at template purines in nucleotide incorporation by human DNA polymerase iota. Mol Cell Biol. 2006;26:6435–6441. doi: 10.1128/MCB.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson RE, Prakash L, Prakash S. Biochemical evidence for the requirement of Hoogsteen base pairing for replication by human DNA polymerase iota. Proc Natl Acad Sci U S A. 2005;102:10466–10471. doi: 10.1073/pnas.0503859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Washington MT, Johnson RE, Prakash L, Prakash S. Human DNA Polymerase ι Utilizes Different Nucleotide Incorporation Mechanisms Dependent upon the Template Base. Mol Cell Biol. 2004;24:936–943. doi: 10.1128/MCB.24.2.936-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 78.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat Struct Mol Biol. 2006;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- 79.Pence MG, Choi JY, Egli M, Guengerich FP. Structural Basis for Proficient Incorporation of dTTP Opposite O6-Methylguanine by Human DNA Polymerase ι. J Biol Chem. 2010;285:40666–40672. doi: 10.1074/jbc.M110.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pence MG, Blans P, Zink CN, Hollis T, Fishbein JC, Perrino FW. Lesion bypass of N2-ethylguanine by human DNA polymerase iota. J Biol Chem. 2009;284:1732–1740. doi: 10.1074/jbc.M807296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi JY, Lim S, Eoff RL, Guengerich FP. Kinetic analysis of base-pairing preference for nucleotide incorporation opposite template pyrimidines by human DNA polymerase iota. J Mol Biol. 2009;389:264–274. doi: 10.1016/j.jmb.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci U S A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 84.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 85.Carlson KD, Washington MT. Mechanism of Efficient and Accurate Nucleotide Incorporation Opposite 7,8-Dihydro-8-Oxoguanine by Saccharomyces cerevisiae DNA Polymerase η. Mol Cell Biol. 2005;25:2169–2176. doi: 10.1128/MCB.25.6.2169-2176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patra A, Zhang Q, Lei L, Su Y, Egli M, Guengerich FP. Structural and Kinetic Analysis of Nucleoside Triphosphate Incorporation Opposite an Abasic Site by Human Translesion DNA Polymerase η. J Biol Chem. 2015;290:8028–8038. doi: 10.1074/jbc.M115.637561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee DH, Pfeifer GP. Translesion synthesis of 7,8-dihydro-8-oxo-2’-deoxyguanosine by DNA polymerase eta in vivo. Mutat Res, Fundam Mol Mech Mutagen. 2008;641:19–26. doi: 10.1016/j.mrfmmm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Avkin S, Livneh Z. Efficiency, specificity and DNA polymerase-dependence of translesion replication across the oxidative DNA lesion 8-oxoguanine in human cells. Mutat Res, Fundam Mol Mech Mutagen. 2002;510:81–90. doi: 10.1016/s0027-5107(02)00254-3. [DOI] [PubMed] [Google Scholar]
- 89.Xie Z, Braithwaite E, Guo D, Zhao B, Geacintov NE, Wang Z. Mutagenesis of benzo[a]pyrene diol epoxide in yeast: requirement for DNA polymerase zeta and involvement of DNA polymerase eta. Biochemistry. 2003;42:11253–11262. doi: 10.1021/bi0346704. [DOI] [PubMed] [Google Scholar]
- 90.Klarer AC, Stallons LJ, Burke TJ, Skaggs RL, McGregor WG. DNA polymerase eta participates in the mutagenic bypass of adducts induced by benzo[a]pyrene diol epoxide in mammalian cells. PLoS One. 2012;7:e39596. doi: 10.1371/journal.pone.0039596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O6-methyl-guanine lesions. Proc Natl Acad Sci U S A. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eoff RL, Angel KC, Egli M, Guengerich FP. Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase Dpo4. J Biol Chem. 2007;282:13573–13584. doi: 10.1074/jbc.M700656200. [DOI] [PubMed] [Google Scholar]
- 94.Spratt TE, Levy DE. Structure of the hydrogen bonding complex of O6-methylguanine with cytosine and thymine during DNA replication. Nucleic Acids Res. 1997;25:3354–3361. doi: 10.1093/nar/25.16.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loechler EL. Molecular modeling studies of O2-alkylthymines and O4-alkylthymines in DNA: Structures that may be pertinent to the incorporation of the corresponding dAlkTTP into DNA by DNA polymerases in vitro. Mutat Res, Fundam Mol Mech Mutagen. 1990;233:39–44. doi: 10.1016/0027-5107(90)90149-x. [DOI] [PubMed] [Google Scholar]
- 96.Gervais V, Cognet JA, Le Bret M, Sowers LC, Fazakerley GV. Solution structure of two mismatches A.A and T.T in the K-ras gene context by nuclear magnetic resonance and molecular dynamics. Eur J Biochem. 1995;228:279–290. [PubMed] [Google Scholar]
- 97.Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 98.Lemontt JF. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PMJ, Prakash S, Prakash L. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and Error-Free Replication past a Minor-Groove N2-Guanine Adduct by the Sequential Action of Yeast Rev1 and DNA Polymerase ζ. Mol Cell Biol. 2004;24:6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haracska L, Prakash S, Prakash L. Yeast DNA Polymerase ζ Is an Efficient Extender of Primer Ends Opposite from 7,8-Dihydro-8-Oxoguanine and O6-Methylguanine. Mol Cell Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase zeta (ζ) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao B, Wang J, Geacintov NE, Wang Z. Pol eta, Pol zeta and Rev1 together are required for G to T transversion mutations induced by the (+)- and (–)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells. Nucleic Acids Res. 2006;34:417–425. doi: 10.1093/nar/gkj446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hashimoto K, Cho Y, Yang IY, Akagi J-i, Ohashi E, Tateishi S, de Wind N, Hanaoka F, Ohmori H, Moriya M. The Vital Role of Polymerase ζ and REV1 in Mutagenic, but Not Correct, DNA Synthesis across Benzo[a]pyrene-dG and Recruitment of Polymerase ζ by REV1 to Replication-stalled Site. J Biol Chem. 2012;287:9613–9622. doi: 10.1074/jbc.M111.331728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gibbs PEM, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc Natl Acad Sci U S A. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murakumo Y, Roth T, Ishii H, Rasio D, Numata S-i, Croce CM, Fishel R. A Human REV7 Homolog That Interacts with the Polymerase ζ Catalytic Subunit hREV3 and the Spindle Assembly Checkpoint Protein hMAD2. J Biol Chem. 2000;275:4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- 107.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci U S A. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA Polymerase δ and ζ Switch by Sharing Accessory Subunits of DNA Polymerase δ. J Biol Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee YS, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi JY, Guengerich FP. Kinetic Analysis of Translesion Synthesis Opposite Bulky N2- and O6-Alkylguanine DNA Adducts by Human DNA Polymerase REV1. J Biol Chem. 2008;283:23645–23655. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.