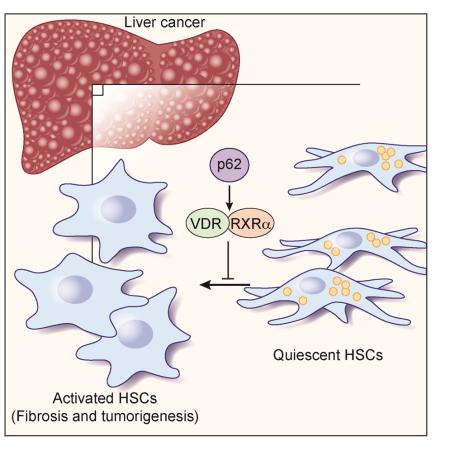
Abstract
Hepatic stellate cells (HSC) play critical roles in liver fibrosis and hepatocellular carcinoma (HCC). Vitamin D receptor (VDR) activation in HSC inhibits liver inflammation and fibrosis. We found that p62/SQSTM1, a protein upregulated in liver parenchymal cells but downregulated in HCC-associated HSC, negatively controls HSC activation. Total body or HSC-specific p62 ablation potentiates HSC and enhances inflammation, fibrosis and HCC progression. p62 directly interacts with VDR and RXR promoting their heterodimerization, which is critical for VDR:RXR target gene recruitment. Loss of p62 in HSC impairs the repression of fibrosis and inflammation by VDR agonists. This demonstrates that p62 is a negative regulator of liver inflammation and fibrosis through its ability to promote VDR signaling in HSC, whose activation supports HCC.
Graphical abstract
INTRODUCTION
Large scale genomic and transcriptomic interrogation of cancer has underscored the complexity and of its genetic landscape. The multitude of genetic alterations makes it difficult to distinguish “driver” from “passenger” mutations. This variability is well exemplified in hepatocellular carcinoma (HCC), in which more than 28,000 different somatic mutations have been identified (Shibata and Aburatani, 2014). This makes the design of therapeutic strategies based on cancer-linked genetic alterations extremely challenging. Microenvironmental inflammation and metabolic stress results in non-genetic vulnerabilities that can be exploited for the design of innovative therapeutics. HCC is the third leading cause of cancer death in the world (Farazi and DePinho, 2006). It develops together with chronic hepatitis, fibrosis, cirrhosis, and stromal activation, which are key mediators of a microenvironment conducive to tumorigenesis (El-Serag, 2011). Whereas infection with hepatitis B and C viruses (HVB and HVC) has been the major HCC risk factor, obesity-induced non-alcoholic fatty liver disease (NAFLD), and its more severe consequence, non-alcoholic steatohepatitis (NASH), has increased lately as critical promoters of HCC (Anstee and Day, 2013; El-Serag, 2011; Toffanin et al., 2010). Epidemiological studies point to obesity and NASH as the leading new risk factors for HCC (Calle and Kaaks, 2004).
Chronic liver injury sets in motion a vicious cycle of hepatocyte cell death, inflammation, and fibrosis that results in cirrhosis and cancer, in which hepatic stellate cells (HSC) play a decisive role (Bataller and Brenner, 2005; Hernandez-Gea and Friedman, 2011). Quiescent HSC store retinoid lipid droplets and express glial fibrillar associated protein (GFAP), synaptophysin, and nerve growth factor receptor p75 (Bataller and Brenner, 2005). In response to injury, HSC differentiate into α smooth muscle actin (αSMA)-expressing myofibroblasts (Bataller and Brenner, 2005). Recent studies suggest the critical role of HSC in generation of pro-inflammatory signals important for HCC development (Lujambio et al., 2013; Yoshimoto et al., 2013). This has since received support from studying pancreatic cancer, which shows that therapeutic targeting of pancreatic stellate cells improves therapeutic responses in pancreatic cancer (Sherman et al., 2014). Therefore, the tumor stroma, and specifically stellate cells and myofibroblasts, emerges as a potential target for therapeutic intervention in cancer. However, the precise molecular mechanisms underlying the activation of myofibroblasts and HSC during NASH and HCC, and their role in creating a microenvironment conducive to cancer remain largely unexplored.
Our interest in this fundamental biomedical question stems from previous results suggesting a link between the signaling adapter and autophagy substrate p62 and liver cancer (Umemura et al., 2016). p62 is a component of Mallory-Denk Bodies (MDBs) and Hyaline granules (Stumptner et al., 2007), which are protein aggregates that accumulate in the cytoplasm of damaged liver cells in NASH, cirrhosis, and HCC (Stumptner et al., 2007). Importantly, p62 is upregulated not only in liver cancer (Inami et al., 2011), but also in many other epithelial cancers including those of prostate (Valencia et al., 2014), lung (Duran et al., 2008), glioblastoma (Galavotti et al., 2013), and kidney (Li et al., 2013). Such findings strongly suggest that p62-driven pathways could be considered as potential therapeutic targets in NASH and HCC. However, most studies addressing the role of p62 in cancer have focused on its function in transformed epithelial cells (Moscat and Diaz-Meco, 2012), and only very recently the concept of p62 as a tumor suppressor in the cancer stroma has emerged from our studies of prostate cancer (Valencia et al., 2014). The potential role and mechanisms of action of HSC p62 in the context of HCC pathogenesis has not been addressed.
p62 is a scaffold protein that interacts with other signaling intermediates such as the atypical PKCs, Keap-1 and TRAF6, which serve to activate two main stress responsive transcriptional regulators, NRF2 and NF-κB (Moscat and Diaz-Meco, 2011). More recently, p62 has been shown to interact with components of the mTORC1 complex, emerging as a regulator of mTORC1 activation (Duran et al., 2011; Linares et al., 2015; Linares et al., 2013). Interestingly, p62 mRNA expression is induced by oncogenes such as Ras (Duran et al., 2008; Ling et al., 2012), but the protein is subjected to constant degradation via autophagy due to its binding to LC3 (Moscat and Diaz-Meco, 2009). This keeps p62 expression low in non-stressed cells. Upregulation of p62 readily occurs upon disruption of autophagy and is likely to contribute to oncogenic transformation (Moscat and Diaz-Meco, 2012). In this regard, our recent data demonstrate that p62 promotes hepatocyte transformation by impinging into the NRF2 and mTORC1 pathways in several models of HCC (Umemura et al., 2016). These studies were based on disrupting Sqstm1, which encodes p62, in hepatocytes, but its contribution to liver tumorigenesis in other non-parenchymal cell types remains to be determined. Given recent data unveiling the importance of activated myofibroblasts to NASH and tumor promotion, it was of great interest to establish the potential role and mechanisms of action of p62 in HSC during HCC development.
RESULTS
Global p62 loss promotes NASH and HCC
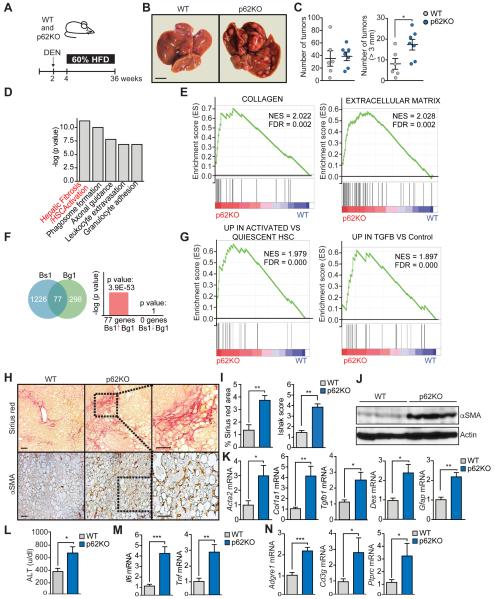
To test whether total body p62 ablation inhibits HCC development in mice, wild type (WT) and total p62 knock out (p62KO) mice were injected at 2 weeks of age with 25 mg/kg of the hepatic carcinogen diethylnitrosamine (DEN) and were next fed with HFD, as a tumor promoter, following a described protocol that recapitulates liver cancer promotion under conditions of obesity (Figure 1A) (Park et al., 2010). Surprisingly, the global loss of p62 did not inhibit HCC and actually enhanced its development (Figure 1B). Although tumor multiplicity was not affected, tumor size was, and the number of large tumors (> 3 mm) was much higher in p62KO than in WT mice (Figure 1C). These results cannot be accounted for by a more severe obesity caused by the loss of p62, since there were no differences in body weight (Figure S1A), fat content (Figure S1B), or levels of steatosis between genotypes (Figures S1C and S1D). Although both genotypes developed adenomas with steatosis and inflammation, only p62KO mice developed HCC (Figure S1E). p62KO tumors displayed higher expression of α fetoprotein (Afp), a common HCC marker (Figure S1F). Histologically, 36% of tumors larger than 3 mm were HCCs, similar in histology to human steatohepatitic HCC (Figure S1G). This was characterized by ballooning cancer cells and inflammatory infiltration as typical in NASH-related HCC (Salomao et al., 2012). In addition, other tumors (6%) displayed a classical thick trabecular pattern, whereas the remaining 64% were either typical or steatohepatitic adenomas (Figure S1G). Consistently, we also found higher numbers of Ki-67 and CK19-positive cells and higher levels of Krt19 transcripts in p62KO livers (Figure S1H).
Figure 1. p62 total ablation promotes DEN-induced hepatocarcinogenesis and fibrosis.
(A) Schematic diagram of DEN-HFD induced HCC model. Two week-old mice were i.p. injected with DEN (25 mg/kg) and two weeks later were fed 60% fat diet for 32 weeks. (B) Representative images of livers from WT (n=6) and p62KO (n=7) treated as in (A). Scale bar, 1 cm. (C) Total number of tumors and number of tumors bigger than 3 mm in WT and p62KO livers. (D) Top canonical pathways from Ingenuity analysis (IPA) of upregulated genes in p62KO as compared to WT livers (n=3). (E) GSEA plot of enrichment in “Collagen” and “Extracellular Matrix” signatures in p62KO liver tumors (n=3) using C5 MSigDB database. (F) NextBio analysis of gene overlap between genes up-regulated in p62KO versus WT (n=3) livers (Bs1, Bioset1) with Extracellular Matrix Geneset (Bg1, Biogroup1). (G) GSEA plot of enrichment of “UP IN ACTIVATED VS QUIESCENT HSC” and “UP IN TGFB VS Control” genesets associated with gene expression in p62KO liver tumors (n=3) using C2 MSigDB database. (H) Sirius red and αSMA staining of WT and p62KO livers. Scale bars, 100 μm. (I) Sirius red positive area and Ishak score of WT and p62KO livers. (J) Immunoblot analysis of αSMA and Actin in WT and p62KO livers. (K) qPCR analysis of mRNA of fibrosis markers in WT and p62KO livers. (L) ALT levels in serum of WT and p62KO mice. (M) qPCR analysis of mRNA of inflammation markers. (N) qPCR analysis of mRNA of immune cell infiltration. Results are presented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001. See also Figure S1.
We next carried out a genome wide transcriptomic analysis by RNAseq of livers of mice of both genotypes. Gene Set Enrichment Analysis (GSEA) using the curated gene set compilation C2 (MSigDb C2) showed several “Liver Cancer” and “Cancer Proliferation” related gene sets among the TOP10 most positively enriched in p62KO condition (NES>2.5) (Figure S1I), in agreement with the increased tumorigenic phenotype of p62KO livers. “Core Pathway Analysis” (IPA) of differentially up-regulated genes (p.adj<0.05) in p62KO showed enrichment for “Hepatic Fibrosis/Hepatic Stellate Cell Activation” canonical pathway (Figure 1D) with significant up-regulation of collagen and genes encoding soluble factors such as TGFβ and PDGF (Figure S1J). TGFβ1 appeared as the most significantly up-regulated upstream regulator (p=6.56E-31). Moreover, Gene Ontology gene set compilation C5 (MSigDb C5) showed enrichment for “COLLAGEN” and “EXTRACELLULAR MATRIX” gene sets (Figures 1E and S1I). Using NextBio software to compute GO canonical pathways we identified an enrichment of differentially expressed genes in the “extracellular matrix” category (Figure 1F). GSEA using recently developed human stellate cells (HSC) signatures (Ding et al., 2013) showed a positive enrichment of upregulated genes in p62KO for genes upregulated in activated HSC vs. quiescent, and in HSC stimulated by TGFβ treatment vs. control (Figure 1G). These results establish that at a gene transcriptional level there is a more aggressive phenotype in p62KO that correlates with increased HSC activation.
More detailed analysis of p62KO livers confirmed liver bridging fibrosis and collagen deposition, as determined by Sirius Red staining and concomitant increase in cells positive for αSMA, a marker of stellate activation to myofibroblasts (Bataller and Brenner, 2005) (Figure 1H). Quantification of fibrosis was performed by Sirius Red area analysis and histological fibrotic scoring in a blind fashion by a pathologist (Figure 1I). These results correlated with increased αSMA protein (Figure 1J) and mRNA levels of Acta2, Col1a1, and Tgfb1 as well as the HSC markers Des and Gfap (Figure 1K). p62KO livers exhibited higher serum alanine aminotransferase (ALT), reflecting hepatocytic damage (Figure 1L), and liver inflammation as determined by increased expression of Il6 and Tnf mRNAs (Figure 1M), as well as of markers of macrophage (Adgre1) and lymphocyte infiltration (Cd3g and Ptprc; Figure 1N). These are characteristic features of NASH (Toffanin et al., 2010). Collectively these results show that global loss of p62 in DEN-HFD-fed mice results in a NASH-like microenvironment. These results, unveiled an unexpected possible non-cell autonomous tumor suppressor function of p62 probably exerted through inhibition of HSC activation.
Role of p62 in fibrosis and myofibroblast/HSC cell activation
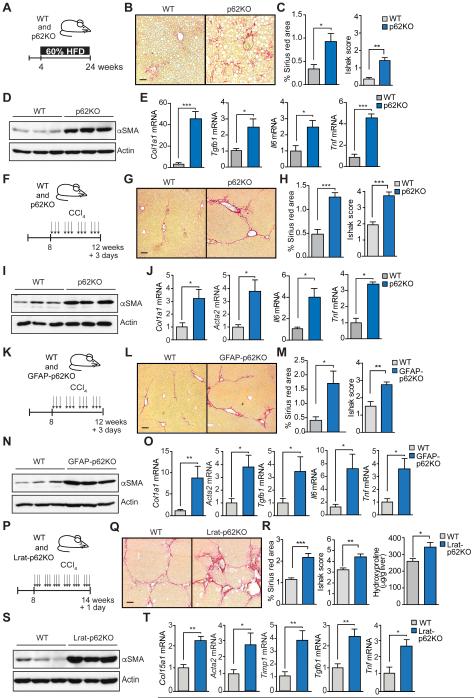
The molecular mechanisms controlling a NASH-like phenotype were also observed in p62 total KO mice kept on HFD without DEN injection (Figures 2A-2E). p62KO mice on regular diet did not show fibrosis (Figures S2A and S2B), although they have increased Acta2 and Col1a1 (Figure S2C). Therefore, p62 deletion predisposes mice for fibrosis that is further promoted by HFD. These results are of significance since activated HSC were proposed to play a critical role in not only fibrosis but also in promotion of HCC in the context of steatohepatitis and chronic liver injury (Lujambio et al., 2013). Therefore, the negative role of p62 in HSC activation may account for its ability to attenuate HFD-promoted HCC development. To further explore the role of p62 in HSC, we subjected mice to chronic carbon tetrachloride (CCl4) challenge (Ding et al., 2013). p62KO and WT mice were repeatedly injected with CCl4 for 4 weeks and livers were analyzed 3 days after the final CCl4 injection (Figure 2F). p62KO mice displayed higher collagen deposition (Figures 2G and 2H), and increased αSMA levels (Figure 2I), as well as of Col1a1 and Acta2, and inflammatory cytokine expression (Figure 2J). To exclude that our data may be specific to the CCl4 model, we confirmed our results in a second well-established model of toxic liver fibrosis driven by HSC activation, induced by thioacetamide (TAA) treatment. Livers from TAA-treated p62KO mice displayed an enhanced robust fibrogenic response as compared to WT controls similar to the CCl4 model (Figures S2D-2G). To test whether p62 in hepatocytes plays any role in CCl4-induced fibrosis, we crossed Sqstm1fl/fl mice with Alb-Cre mice (from now on termed L-p62KO) and subjected WT and L-p62KO mice to the same CCl4 regime. L-p62KO mice exhibited a fibrotic phenotype identical to that of WT mice (Figures S2H-S2K). This demonstrates that cells other than hepatocytes are responsible for enhancement of liver fibrosis upon total body p62 deficiency.
Figure 2. Selective p62 deficiency in hepatic stellate cells increases fibrosis upon liver injury.
(A) Schematic diagram of HFD protocol. Four week-old WT (n=5) and p62KO (n=5) mice were fed 60% fat diet during 20 weeks. (B) Sirius red staining of livers of WT and p62KO mice. (C) Sirius red positive area and Ishak score of WT and p62KO livers. (D) Immunoblot analysis of αSMA and Actin in WT and p62KO livers. (E) qPCR analysis of indicated mRNA. (F) Schematic diagram of CCl4-induced fibrosis protocol. Eight week-old mice were i.p. injected with 0.5 ml/kg body weight CCl4 (1:50 v/v in corn oil) three times per week during 4 weeks. Mice were sacrificed 3 days after the last injection (n=6, per genotype). (G) Sirius red staining of livers of WT and p62KO mice (n=6). (H) Sirius red positive area and Ishak score. (I) Immunoblot analysis of αSMA and Actin in WT and p62KO livers. (J) qPCR analysis of indicated mRNA in livers of mice described in (F). (K) Scheme of CCl4-induced fibrosis model as in (F) in WT (n=5) and GFAP-p62KO (n=9) mice. (L) Sirius red staining of livers of WT and GFAP-p62KO mice. (M) Sirius red positive area and Ishak score of WT and GFAP-p62KO livers. (N) Immunoblot analysis of αSMA and Actin in WT and GFAP-p62KO livers. (O) qPCR analysis of indicated mRNA in livers from the mice described in (K). (P) Scheme of CCl4-induced fibrosis protocol. Eight week-old WT (n=6) and Lrat-p62KO (n=5) mice were i.p. injected with 0.5 ml/kg body weight CCl4 (1:10 v/v in corn oil) three times per week during 6 weeks. Mice were sacrificed 1 day after the last injection. (Q) Sirius red staining of livers of WT and Lrat-p62KO mice. (R) Sirius red positive area, Ishak score, and hydroxyproline content of livers of WT and Lrat-p62KO mice. (S) Immunoblot analysis of αSMA and Actin in WT and Lrat-p62KO livers. (T) qPCR analysis of indicated mRNA of livers from the mice described in (P). Scale bars, 100 μm. Results are presented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001. See also Figure S2.
To test whether p62 functions in HSC in vivo, we generated GFAP-p62KO mice by crossing Sqstm1fl/fl mice with GFAP-Cre mice, a previously described model for selective deletion of HSC (Kisseleva and Brenner, 2008; Lujambio et al., 2013). Livers from untreated GFAP-p62KO mice did not display fibrosis, although basal levels of Col1a1 were increased (Figures S2L-2N). Exposure of GFAP-p62KO mice to chronic CCl4 treatment resulted in a fibrotic phenotype identical to that of p62KO mice, including increased Sirius red staining (Figures 2L and 2M), increased αSMA (Figure 2N), and elevated Col1a1, Acta2, Tgfb1, Il6 and Tnf mRNA (Figures 2O). p62 was expressed not only in hepatocytes but also in HSC, as demonstrated by the co-localization of p62 with Desmin in WT but not in GFAP-p62KO mice (Figure S2O). It should be noted that Cre-LoxP-induced recombination in GFAP-Cre mice was reported to result in three outcomes: (1) recombination only in HSC (Kisseleva and Brenner, 2008; Krizhanovsky et al., 2008; Lujambio et al., 2013); (2) recombination only in cholangiocytes (Mederacke et al., 2013); (3) recombination in both HSC and cholangiocytes (Yang et al., 2008). We measured the p62 mRNA level in HSC purified from GFAP-p62KO mice and found that they expressed no more p62 mRNA than HSC from p62 total KO mice (Figure S2P). However, immunofluorescence staining showed that GFAP-p62KO mice exhibited p62 deficiency in both HSC (Figure S2O) and cholangiocytes (Figure S2Q), which could confound our interpretation of the role of p62 in HSC during fibrosis. Analysis of p62 expression in L-p62KO mice revealed that p62 was similarly deleted in not only hepatocytes (as expected) but also cholangiocytes (Figure S2R). This is in agreement with the specifications of the Jackson laboratory that the Alb-Cre line also results in gene inactivation in cholangiocytes (Jax Stock# 003574). Since the L-p62KO mice did not phenocopy GFAP-p62KO or total p62KO mice in terms of fibrosis and inflammation upon CCl4 (Figures S2H-S2K), we concluded that p62 mainly acts to prevent liver fibrosis in HSC and not in cholangiocytes. To further confirm the role of p62 in HSCs, we used a recently reported Cre-driven HSC-specific mouse model (Lrat-Cre) (Mederacke et al., 2013). We crossed this mouse line with Sqstm1fl/fl mice to generate Lrat-p62KO mice. After confirming the selective deletion of Sqstm1 in the HSC population (Figure S2S), we subjected these mice to the CCl4 protocol but using a more aggressive regime, consisting of a higher CCl4 dose and longer treatment period (Figure 2P), and analyzing the livers one day after the final CCl4 injection in this case. Following this strategy we seek to analyze the effects of selective loss of p62 in HSCs under conditions where fibrosis is more extensive/mature and is not subjected to resolution. p62 inactivation in HSCs in this model also led to increased fibrotic response, pathology score, hepatic hydroxyproline content, and increased expression of αSMA and other fibrogenic and inflammatory genes (Figures 2Q to 2T).
p62 negatively regulates the pro-tumorigenic functions of HSC
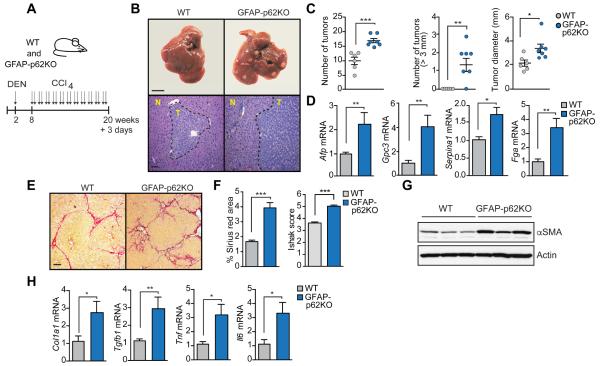
GFAP-p62KO mice were subjected to HCC induction through a protocol combining DEN with CCl4 injections to create a fibrogenic microenvironment (Lujambio et al., 2013) (Figure 3A). HCC induction was more efficient in GFAP-p62KO mice relatively to identically treated WT controls (Figures 3B and 3C). GFAP-p62KO livers also expressed higher HCC markers, including Afp, Gpc3, Serpina1 and Fga (Figure 3D), indicating a more aggressive phenotype, and exhibited more fibrosis and inflammation (Figures 3E-3H). Collectively these results establish that p62 acts as a repressor of HSC function, and a non-cell-autonomous tumor suppressor in HCC.
Figure 3. Selective p62 deficiency in hepatic stellate cells promotes hepatocarcinogenesis.
(A) Schematic diagram of DEN-CCl4-induced HCC model. Two week-old mice were i.p. injected with DEN (25 mg/kg) and 6 weeks later injected with CCl4 (2 ml/kg) twice per week for 12 weeks. (B) Gross morphology (top) and H&E staining (bottom) of livers of WT (n=6) and GFAP-p62KO (n=7) mice described in (A). N, normal liver tissue. T, liver tumor area. Scale bar, 1 cm (top) and 100 μm (bottom). (C) Total number of tumors, number of tumors bigger than 3 mm and maximal tumor diameters. (D) qPCR analysis of mRNA of HCC markers in WT and GFAP-p62KO livers described in (A). (E) Sirius red staining of livers of WT and GFAP-p62KO mice. Scale bar, 100 μm. (F) Sirius red positive area and Ishak score of WT and GFAP-p62KO livers. (G) Immunoblot analysis of αSMA and Actin in WT and GFAP-p62KO livers. (H) qPCR analysis of indicated mRNA. Results are presented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001.
p62 expression is reduced in HSC of human HCC patients
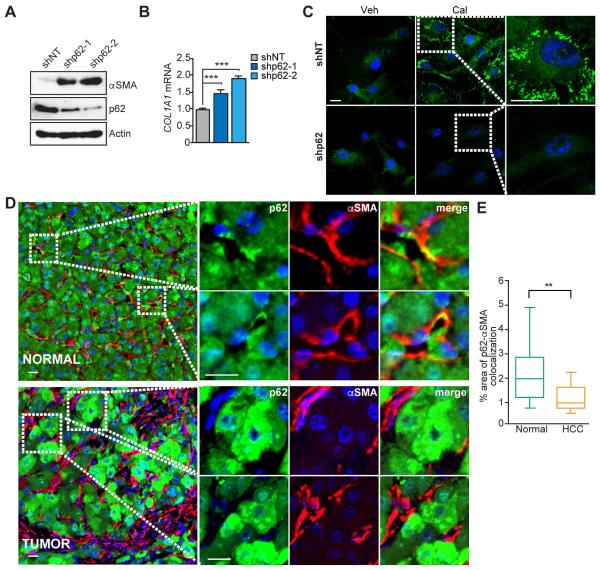
To examine the relevance of the above finding to human biology, we knocked down p62 in immortalized human HSC and found increased αSMA protein and COL1A1 mRNA expression (Figures 4A and 4B). Recent studies suggest that reversion of activated HSC into a quiescent-like phenotype could be a major mechanism underlying reversal of liver fibrosis (Kisseleva et al., 2012). Exposure to the low-calcemic vitamin D analogue calcipotriol (Cal) induced lipid droplet formation, a critical hallmark of quiescent stellate cells, in human pancreatic stellate cells (PSCs) (Sherman et al., 2014). To test if p62 is involved in this process, we treated human HSC, either shNT or shp62, with Cal and stained them with BODIPY. Whereas Cal induced a robust accumulation of lipid droplets in shNT cells, this effect was completely blocked in shp62 cells (Figure 4C). Analysis of human HCC specimens showed that expression of p62 was significantly lower in tumoral HSC than in normal tissue HSC, as measured by double staining for p62 and αSMA (Figures 4D and 4E). Consistently, exposure of human HSC to conditioned media from a human liver cancer cell line (HepG2) promoted p62 downregulation at protein and mRNA levels, concomitant with increased expression of ACTA2 (Figures S3A to S3C). p62 loss in HSC observed in human HCC samples was also confirmed in mouse liver tumors from DEN-HFD treated mice (Figure S3D). Analysis of public dataset showed that Sqstm1 mRNA levels were downregulated in primary mouse HSCs upon activation in cell culture and in HSCs isolated from mice with bile duct–ligation or after CCl4 injections (Figures S3E). Although p62 was downregulated in the liver tumor stroma, its levels in the tumor hepatocyte were highly upregulated (Figures 4D and S3D).
Figure 4. p62 is lost in HSC in human HCC.
(A) Levels of p62, αSMA and Actin in immortalized human HSC expressing non-targeting shRNA (shNT) or one of two different shRNAs for p62 (shp62-1 and shp62-2) via lentiviral infection were analyzed by immunoblot. (B) qPCR analysis of mRNA of COL1A1 in human HSC either shNT or shp62. Results are presented as mean ± SEM. (C) BODIPY 493/503 staining for neutral lipids in shNT or shp62 human HSC treated with vehicle (DMSO) or 100 nM Cal for 48 hr. Scale bars, 20 μm. (D) Representative images of double immunofluorescence of p62 (green) and αSMA (red) in normal liver (NORMAL) and HCC samples (TUMOR). Scale bars, 25 μm (NORMAL) and 10 μm (TUMOR). (E) Box-and-whisker plots showing median (horizontal line), interquartile range (box) and 10th-90th percentiles (whiskers) of the % colocalization area between p62 and αSMA (n=14 per group). **p<0.01, ***p<0.001. See also Figure S3.
p62 is critical for vitamin D receptor-induced inactivation of HSC
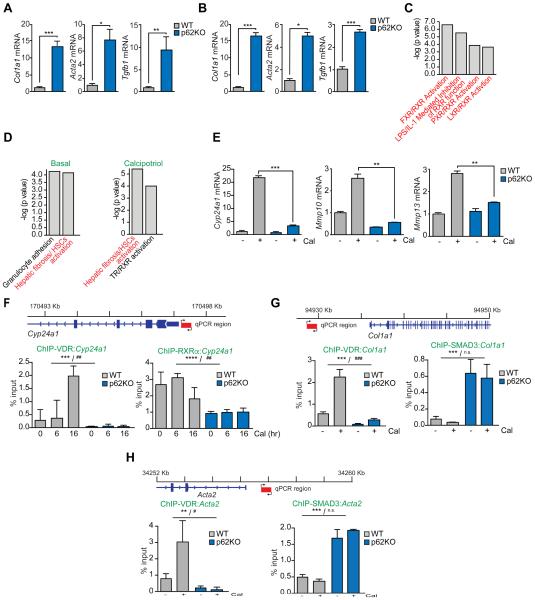
To characterize the potential role of p62 as a cell autonomous repressor of HSC activity, we isolated HSC from WT and p62KO mice. p62 deficiency enhanced the basal levels of Col1a1, Acta2, and Tgfb1 in primary quiescent HSC (Figure 5A). To facilitate further mechanistic studies, HSC of both genotypes were immortalized. Activated immortalized p62KO HSC also displayed a similar increased activation phenotype (Figure 5B). These results demonstrate that p62 inactivation in HSC largely recapitulates the pro-fibrogenic HSC activation effect observed in mouse livers. IPA analysis of the significantly downregulated genes in the transcriptomic RNAseq experiment described in Figure 1D, identified a striking presence of nuclear receptors (NRs)-related pathways, including RXR, FXR and LXR (Figure 5C), suggesting that p62 deficiency might interfere with the HSC suppressive activity of vitamin D. To test this, we treated WT and p62KO HSC with Cal and performed RNAseq transcriptomics. IPA analysis of differentially expressed genes showed that p62-deficient HSC displayed an upregulated profile of “Hepatic Stellate Cell Activation” under basal conditions and, even more markedly, after Cal stimulation (Figure 5D). Un-supervised clustering of these genes not only validated the observation of VDR inability to revert HSC activation but also showed a boost of the majority of pro-fibrotic genes in p62KO HSC (Figure S4A).
Figure 5. p62 is required for hepatic stellate activation and VDR function.
(A and B) qPCR analysis of mRNA of HSC activation markers in primary quiescent (A) or immortalized activated (B) WT and p62KO HSC. (C) Nuclear-receptor-related pathways from Ingenuity analysis (IPA) of downregulated genes in liver tumors of total p62KO and WT mice described in Figure 1A. (D) Top canonical pathways from Ingenuity analysis (IPA) of differentially expressed genes between p62KO and WT HSC in basal conditions and upon calcipotriol (Cal) treatment. (E) qPCR analysis of mRNA of Cyp24a1, Mmp10, and Mmp13 in WT and p62KO HSC. (F) Primer design and ChIP-qPCR analysis of Cyp24a1 promoter occupancy of VDR and RXRα. (G) Primer design and ChIP-qPCR analysis of Col1a1 promoter occupancy of VDR and SMAD3. (H) Primer design and ChIP-qPCR analysis of Acta2 promoter occupancy of VDR and SMAD3. Results are presented as mean ± SEM. p values were calculated using a two-way ANOVA; **p<0.01, ***p<0.001 for all comparisons of p62KO versus WT; #p<0.05, ##p<0.01, ###p<0.001, n.s. non-significant, for all comparisons of Cal versus vehicle. See also Figure S4.
Consistent with the notion that VDR effects are impaired in p62KO HSC, transcripts that were robustly induced in Cal-treated WT HSC were blunted in p62KO HSC (Figure S4B). This was confirmed by qPCR analysis of three of these VDR targets (Figure 5E). Similar results were obtained when HSC were stimulated with Vit1,25(OH)2D3 (Figure S4C). In contrast, p62 deletion did not modulate ligand-induced target genes or the profibrotic genes in response to an agonist of LXR, another nuclear receptor partner of RXR (Figure S4D). No defects in VDR or RXRα expression were found in p62KO HSC (Figure S4E). We next determined whether the loss of p62 impaired the ability of VDR to interact with the Cyp24a1 promoter by chromatin immunoprecipitation (ChIP). Basal and Cal-induced recruit of the VDR, and the associated RXRα, to the Cyp24a1 promoter region was dramatically reduced in p62KO HSC relative to VDR and RXRα promoter recruitment in WT cells (Figure 5F and Figure S4F). These results indicate that p62 plays a role in interaction of VDR with its target promoter. Interestingly, whereas Cal promoted the recruitment of VDR to the Col1a1 and Acta2 promoters in WT HSC, this was not observed in p62KO HSC (Figures 5G, 5H and S4F). These results lend support to our hypothesis that the inability of VDR to interact with its targets genes underlies the hyperactivated phenotype of p62-deficient HSC. In this regard, it was suggested that VDR prevents activation of pro-fibrotic genes by blocking the occupancy of their promoters by SMAD3 (Ding et al., 2013; Ito et al., 2013; Sherman et al., 2014). Notably, the loss of p62 in HSC promoted constitutive recruitment of SMAD3 to the promoters of Col1a1 and Acta2 in parallel with the defective interaction of VDR with the same promoters (Figures 5G and 5H). Taken together these data establish that the inability of the VDR to bind its promoters in the absence of p62 results in the recruitment of SMAD3 and the hyperactivation of HSC.
p62 interacts with VDR and controls its heterodimerization with RXRα
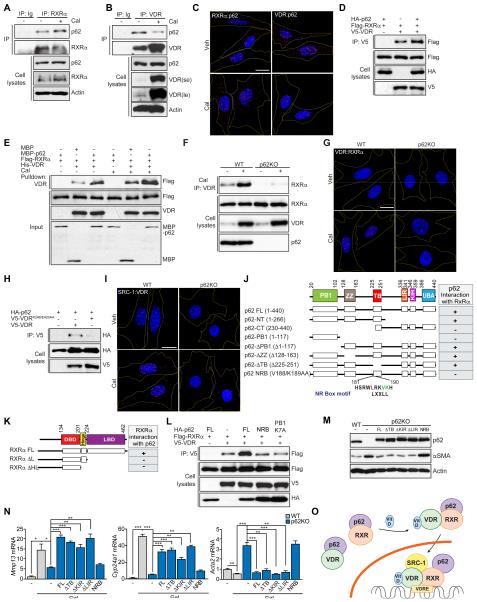
We next examined whether p62 and VDR interact. Since VDR functions as an obligate heterodimer in complex with RXRα (Umesono et al., 1991), we investigated the potential interaction of p62 with both NRs. We transfected HEK293T cells with HA-p62 and V5-tagged VDR, FLAG-tagged RXRα, or the corresponding controls. We found a reproducible interaction between p62 and both VDR and RXRα (Figure S5A). Furthermore, when these experiments were performed with purified recombinant proteins, similar results were obtained, demonstrating that the interaction of p62 with both NRs is direct (Figures S5B and S5C). However, p62 did not interact with LXR, another partner of RXR (Figure S5D), suggesting that p62’s interaction is selective for the VDR:RXR module and are consistent with the lack of effect on the p62 phenotype of the LXR ligand (Figure S4D). Importantly, p62 binding to RXRα and VDR was also detected when endogenous proteins were analyzed in extracts from HSC (Figures 6A and 6B). Furthermore, in situ complex formation of endogenous p62 with RXRα and VDR was also demonstrated by proximity ligation assay (PLA) (Figure 6C and S5E), which gives a positive signal when antibodies recognizing two proteins of interest are within 40 nm of one another. Treatment with Cal did not affect significantly the interaction of p62 with RXRα but weakened the binding of VDR to p62, both in the endogenous and recombinant protein assays (Figures 6A-6C, S5B-S5C and S5E). As observed by PLA, p62 interaction with both nuclear receptors occurred in both cytosol and nucleus (Figure 6C and S5E). The p62-RXRα and p62-VDR nuclear interactions were also confirmed in immunoprecipitates of HSC nuclear extracts (Figures S5F and S5G). As Cal promotes the formation of VDR:RXRα heterodimer that is critical for DNA binding and gene expression, it is possible that p62 binding might impact heterodimer formation. Interestingly, p62 increased the formation of the heterodimer between VDR and RXRα in co-transfection experiments (Figure 6D). Consistent with this, we found that formation of the VDR:RXRα complex was also promoted by p62 under basal conditions and in the presence of Cal when analyzed in vitro with purified recombinant proteins (Figure 6E). We next tested whether p62 was required for endogenous dimer formation in HSC. As previously reported, Cal promoted the formation of the heterodimer, which, importantly, was abrogated by the loss of p62 (Figure 6F). This requirement of p62 for VDR:RXR heterodimerization was also detected in situ by PLA (Figure 6G and S5H). Similar results were obtained by cotransfecting Flag-RXRα and V5-VDR in WT or p62-deficient, generated using the CRISPR/Cas9 system (Linares et al., 2015), HEK293T cells (Figure S5I). These results demonstrate that p62 regulates the binding between VDR and RXRα and provide a potential mechanism for the defective activation of Cal-driven pathways in p62-deficient cells.
Figure 6. p62 regulates the formation of the VDR:RXRα dimer in response to calcipotriol.
(A and B) Endogenous interaction of RXRα (A) or VDR (B) with p62 in response to calcipotriol (Cal). Cell lysates and immunoprecipitates were analyzed for the levels of specified proteins; se: short exposure; le: long exposure. (C) Proximity ligation assay (PLA) of RXRα–p62 and VDR– p62 interactions (red) in WT HSC treated with vehicle (DMSO) or 100 nM Cal for 24 hr. (D) Cell lysates and V5-tagged immunoprecipitates of HEK293T cells transfected with the indicated cDNAs in expression vectors were analyzed by immunoblotting. (E) Recombinant His-VDR was incubated with FLAG-RXRα in the presence of recombinant MBP-p62 in response to Cal and interactions were analyzed by immunoblot in His-beads pulldown. (F) Cell lysates and endogenous VDR-immunoprecipitates of WT and p62KO HSC treated with or without Cal were analyzed by immunoblotting. (G) PLA of VDR:RXRα heterodimer (red) in WT and p62KO HSC treated with vehicle (DMSO) or 100 nM Cal for 24 hr. (H) Cell lysates and V5-tagged immunoprecipitates of HEK293T cells transfected with the indicated cDNAs in expression vectors were analyzed by immunoblotting. (I) PLA of SRC-1:VDR (red) in WT and p62KO HSC treated with vehicle (DMSO) or 100 nM Cal for 24 hr. (J) Schematic representation of domain structure of p62 and a summary of the interactions of RXRα with the different domains of p62. (K) Schematic representation of domain structure of RXRα and a summary of the interactions of p62 with the different domains of RXRα. (L) Cell lysates and V5-tagged immunoprecipitates of HEK293T cells transfected with the indicated cDNAs in expression vectors were analyzed by immunoblotting. (M) Immunoblot analysis of αSMA, p62 and Actin in WT, p62KO and p62KO HSC reconstituted with the indicated constructs and treated with vehicle (DMSO) or Cal for 24 hr. (N) qPCR analysis of mRNA of Mmp13, Cyp24a1, and Acta2 in the reconstitution experiment as in (M). (O) Model for the role of p62 in the regulation of VDR:RXR heterodimerization. Scale bars, 20 μm. Results are presented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001. See also Figure S5.
p62 interacts with VDR through the same region that is required for binding of the co-activator SRC-1. Thus, the mutation of K246 and E420 to alanine in VDR, which had been shown to abolish VDR binding to SRC-1 (Zhang et al., 2011), also disrupted the interaction of p62 with VDR (Figure 6H). These results suggest that p62 is released from VDR upon Cal treatment to allow SRC-1 to be recruited to the VDR:RXR heterodimer. Consistent with this model, since the VDR:RXR heterodimer is not formed in p62KO HSC, SRC-1 cannot be recruited (Figure 6I and S5J). We next sought to identify the region(s) of p62 responsible for the interaction with these NRs and established that p62 interacts with RXRα through the p62’s N-terminal region encompassing amino acids 1-266 (Figures 6J and S5K). Further mapping of that region identified the sequence between the ZZ and TB domains (167-230) as responsible for the interaction with RXRα. Careful analysis of the amino acid sequence of this region detected a putative NRbox motif reminiscence of the LXXLL consensus that mediates the interaction of NRs with co-regulatory proteins (Heery et al., 1997) (amino acids 181-190; Figure 6J). Importantly, site directed mutagenesis of V188 and K189 to alanine completely abolished p62-RXRα interaction (Figures 6J and S5L). In addition, we mapped the domain of RXRα mediating interaction with p62. NRs share a common structural organization with a variable N-terminal domain, a conserved DNA binding domain (DBD), and a C-terminal ligand-binding domain (LBD) linked by a flexible hinge peptide (Rastinejad et al., 1995). Results shown in Figures 6K and S5M demonstrated that RXR’s LBD was required for p62 interaction. We next mapped the VDR-p62 interaction and identified the PB1 domain as the one sufficient for this binding (Figure S5N). In agreement with the notion that p62 binding promotes the formation of the VDR:RXR dimer, mutation in the NRbox sequence that abolished p62 binding to RXRα also inhibited the stimulatory effect on VDR:RXR heterodimerization (Figure 6L). Likewise, a PB1 mutant also inhibited the formation of the heterodimer, consistent with p62 binding to VDR through this domain (Figure S5N). To further test the functional relevance of the p62-VDR:RXRα module, we reconstituted p62KO HSC with p62 FL or with the NRB mutant. Whereas p62 FL was able to rescue the VDR defect as well as the profibrotic phenotype of the p62KO HSC, as determined by the expression of αSMA and VDR target genes, the NRB mutant failed to do so (Figures 6M and 6N). We next tested the potential contribution of other signaling pathways known to be regulated by p62, such as NF-κB, NRF2 and autophagy, to this phenotype using well-established p62 mutants that dissect the different p62 functions (Moscat and Diaz-Meco, 2009). We analyzed the following mutants: ΔKIR (it disrupts p62 interaction with Keap1 that controls NRF2); ΔTB (it disrupts p62 interaction with TRAF6 that activates NF-κB) and ΔLIR (it abolishes binding to LC3 that is essential for autophagy). Importantly, those three mutants behaved functionally identically to p62 FL when reconstituted into p62KO HSC at comparable expression levels (Figures 6M and 6N). These results indicate that the ability of p62 to regulate these pathways is not relevant for the fibrotic phenotype, which is fully dependent on the ability of p62 to impact VDR function. Collectively, these results support a model according to which p62 interacts in the cytosol with VDR and RXR and upon ligand, p62 is released from the VDR and favors the formation of the VDR:RXR heterodimer in the cytosol, which is rapidly translocated to the nucleus where the coactivator SRC-1 is recruited and the complex binds to DNA for the regulation of gene expression (Figure 6O).
p62 deficiency in HSC impairs the anti-fibrotic response to calcipotriol in vivo
From the above results we concluded that p62 deficiency enhanced the activation phenotype of HSC by making them resistant to VDR actions in vitro. To determine whether this is also critical in vivo, we treated WT and GFAP-p62KO mice with CCl4, in combination or not with Cal, as described (Sherman et al., 2014, Figure 7A). Treatment with CCl4 led to a more potent fibrotic and inflammatory response in GFAP-p62KO mice than in identically treated WT mice as described above (Figures 7B-7E). More importantly, whereas the response to CCl4 was strongly attenuated by Cal treatment in WT mice, GFAP-p62KO mice did not show a significant response to Cal (Figures 7B-7E). To determine whether the loss of p62 in HSC affects the response to vitamin D, we examined expression of bona fide direct VDR targets. Notably, the ability of Cal to stimulate expression of Cyp24a1, Mmp13 and Mmp10 was completely blunted in CCl4-treated GFAP-p62KO as compared to identically treated WT mice (Figure 7F).
Figure 7. p62 deficiency in HSC impairs Vitamin D-mediated repression of fibrosis.
(A) Schematic diagram of CCl4-induced fibrosis model and reversion with calcipotriol (Cal). Eight week-old mice were i.p. injected with CCl4 (0.5 ml/kg) three times per week during 4 weeks. Cal (20 μg/kg) was administered via oral gavage five times during the last week of CCl4 treatment. Mice are sacrificed 3 days after the last injection. (B) Sirius red staining of livers of WT (n=6) and GFAP-p62KO (n=7) mice treated as in (A). Scale bars, 100 μm. (C) Sirius red positive area and Ishak score of WT and GFAP-p62KO livers. (D) Immunoblot analysis of αSMA and Actin in WT and GFAP-p62KO livers. (E and F) qPCR analysis of mRNA of HSC activation markers (E) and VDR targets (F) in livers described in (A). Results are presented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001.
DISCUSSION
The complexity of liver cancer makes the design of more targeted therapeutic strategies for its treatment extremely difficult. Earlier studies on p62 and liver pathophysiology showed that whole body p62 ablation attenuated the formation of benign adenomas in autophagy deficient livers (Komatsu et al., 2007). In contrast, our results show that whole body p62 deficiency enhanced the formation of malignant liver tumors. These findings were also different from those we obtained recently by examining the effect of hepatocyte-selective p62 ablation on HCC development (Umemura et al., 2016), which demonstrated very clearly that elevated expression of p62 in hepatocytes enhances HCC induction. Our analysis of these apparent contradictions revealed that p62 acts as a suppressor of HCC development in HSC. Since p62 ablation in hepatocytes reduced HCC induction by DEN plus HFD, the same protocol whose ability to induce HCC was enhanced by whole body p62 ablation, we conclude that the tumor suppressive effect of p62 in HSC is dominant to its tumor promoting effect in hepatocytes. Importantly, our results provide strong evidence that HSC play an important role in liver carcinogenesis and that their activation provide a strong tumor promoting microenvironment.
Of note, myofibroblasts and cancer-associated fibroblasts (CAFs) have been proposed to be key mediators of the crosstalk between malignant cancer cells and their microenvironment (Erez et al., 2010). However, the exact mechanisms by which fibroblasts are activated to impact tumorigenesis of the epithelial cell compartment remain largely unknown. Here we show that HCC samples from human patients show that, in addition to the well established increased parenchymal expression of p62 as previously reported (Denk et al., 2006), there is also a dramatic reduction in p62 expression in HSC. The importance of this observation is supported by the fact that genetic inactivation of p62 in mouse HSC leads to increased tumorigenesis accompanied by fibrosis whose characteristics resemble NASH. Therefore, liver cancer progression is impacted by p62 in opposite manners depending on where it is genetically inactivated. The tumor suppressive activity of p62 in HSC is most likely due to its ability to prevent HSC activation, resulting in reduced production of tumor promoting chemokines and cytokines, including IL-6, that are known to be produced by this cell population. However, it remains to be formally established whether collagen production by activated HSC and the deposition of collagen fibers, which can result in eventual damage to hepatocytes, also plays a role is promotion of liver tumorigenesis. Despite these uncertainties, our results show that enhanced HSC activation brought about by p62 ablation is an important promoter of liver tumorigenesis. Our results are consistent with the previous suggestion that activation of pancreatic stellate cells promotes the malignancy of pancreatic cancer, and interferes with its therapy (Sherman et al., 2014). They differ, however, from the results of two other studies in which acute ablation of pancreatic stellate cells promotes the growth of pancreatic cancer (Ozdemir et al., 2014; Rhim et al., 2014). However, the methods used for stellate cell ablation in these studies may have been too drastic and have therefore led to stellate cell death rather than reversion. Indeed, targeting of stellate cells with Vitamin D analogues, which induces their reversion to a quiescent state, not only prevents liver fibrosis but also enhances the anti-tumor effects of classical chemotherapy in pancreatic cancer (Sherman et al., 2014). Curiously, we found that the loss of p62 in HSC severely abolished the anti-fibrogenic function of the VDR. This is due to a direct interaction between p62 and VDR, demonstrating that in addition to its ability to interact with cytoplasmic signaling intermediates, p62 can also interact with and modulate the activity of transcriptional factors. Previous results from our and other laboratories have shown how p62 regulates NF-κB and NRF2, but the mechanism in these cases was by impacting the signaling machineries that regulate their activation (Duran et al., 2008; Inami et al., 2011; Ling et al., 2012). In the case of the VDR we report a direct interaction with p62 that is critical for the VDR-RXR dimer to bind promoters in its target genes. This is consistent with the notion that p62 not only functions in the cytosol but also is located in the nucleus (Pankiv et al., 2010), where it directly impacts transcription. These results are reminiscent from the well-established role of p62 in promoting the dimerization of the Rag proteins for the activation of mTORC1 or of TRAF6 for the activation of NF-κB (Duran et al., 2011; Sanz et al., 2000). Here we report that p62 promotes the anti-fibrotic and anti-inflammatory role of the VDR-RXR dimer, which is impaired during NASH and HCC in our HSC-specific KO mice. An important corollary of our findings is that p62 levels should be monitored when designing clinical approaches involving targeting the VDR in fibrosis and cancer. In this regard, several clinical trials have been designed and are underway using VDR activators. Our data predicts that those patients with low levels of p62 in HSC will be worse responders, which means that p62 levels in this cell population should be utilized as a criterion for patient stratification.
EXPERIMENTAL PROCEDURES
A detailed description of the Experimental Procedures utilized in this work can be found in the Supplemental Experimental Procedures.
Mice
WT and p62KO mice were previously described (Duran et al., 2004). Lrat-Cre mice were previously described (Mederacke et al., 2013) and generously provided by Dr. Schwabe. All animal handling and experimental procedures performed in this study were approved by the Institute Institutional Animal Care and Use Committee (IACUC) at the SBP Medical Discovery Institute.
Cell culture
HEK293T cells are from ATCC. HEK293T p62KO (sgp62) cells were previously described (Linares et al. 2015). WT and p62KO HSCs were isolated from 10-week-old mice by in situ pronase, collagenase perfusion and single-step nycodenz gradient as previously described (Kisseleva et al. 2012). Human stellate cells (hTERT HSCs) were a kind gift from Dr. David Brenner (University of California San Diego).
Histological Analysis
Livers were isolated, rinsed in ice-cold PBS, fixed in 10% neutral buffered formalin for 24 h, dehydrated, and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin (H&E), Sirius red and oil red. Fibrosis was scored using the Ishak modified histological activity index (HAI) scoring system in a blinded fashion by a pathologist.
Human liver samples
The human samples used in this study were obtained under an approved Institutional Review Board protocol of the Mayo Clinic with written patient consent. De-identified samples were sent to the SBP Medical Discovery Institute and used for histological analysis. The study was approved by the Ethics Committee of the SBP Medical Discovery Institute.
RNA Analysis
Total RNA from mouse tissues and cultured cells was isolated using the TRIzol reagent (Life Technologies) and the RNeasy Mini Kit (QIAGEN), followed by DNase treatment.
Statistical Analysis
All the statistical tests are justified for every figure. Data are presented as the mean ± SEM. Significant differences between groups were determined using a Student’s t test (two-tailed unpaired) when the data met the normal distribution tested by D’Agostino test. If the data did not meet this test, a Mann-Whitney test was used. Two-way ANOVA test was used for in vitro experiments, as indicated. The significance level for statistical testing was set at p<0.05.
Supplementary Material
HIGHLIGHTS.
p62 levels are reduced in hepatic stellate cell (HSC) in human HCC samples
Loss of p62 in HSC results in increased fibrosis, inflammation and HCC
p62 is critical for VDR:RXR heterodimerization and inhibition of HSC activation
Enhanced HSC activation by p62 loss impairs VDR signaling and promotes HCC
SIGNIFICANCE.
HSC are central players in Nonalcoholic Steatohepatitis (NASH), which is conducive to HCC. There are not effective treatments for NASH or HCC, and it is difficult to identify patients at high risk to develop HCC. The identification of factors accounting for NASH development and HCC is a major gap in the field. Here we identify p62 as an HSC repressor and a non-cell autonomous tumor suppressor. p62 interaction with VDR:RXR is required for their HSC inhibitory functions. Studies in vitro, in HSC-specific p62-deficient mice, and in human HCC samples establish that p62 loss in the stroma is a common feature of HCC, and predict that therapies aimed at activating VDR effects on HSC will be impaired by p62 deficiency.
In hepatocellular carcinomas (HCC), p62 is increased in hepatocytes but decreased in hepatic stellate cells (HSC). Duran et al. show that loss of p62 in HSC promotes HCC development via reducing the vitamin D receptor (VDR)-RXR interaction, leading to impaired repression of fibrosis and inflammation by VDR.
ACKNOWLEDGEMENTS
Research was supported by grants from NIH (R01DK108743, R01CA172025 to J.M.; R01CA192642 M.T.D.-M.; 5P30CA030199 to M.T.D-M. and J.M.; CCSPG Pilot Project Grant under award 5P30CA030199 to J.M.; R01CA163798 and R01CA118165 to M.K.), and the Superfund Basic Research Program P42ES010337 to M.K. We thank Robert F. Schwabe (Department of Medicine, Columbia University) for providing the LRat-Cre mouse line, and Dr. Jun Xu (University of California San Diego) for helping in the preparation of primary hepatic stellate cells. We thank Diantha LaVine for the artwork, and Guillermina Garcia (Histology Core), Brian James (Genomics Core), and the personnel of the Cell Imaging, Animal Facility, and Viral Vectors Shared Resources at SBP for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.T.D.-M. and J.M. devised and coordinated the project. A.D., E.D.H and M.R.-C. performed all the experiments; E.A.C. provided pathologist expertise for histological analysis; S.S. and S.R. participated in the generation and analysis of RNA-Seq data; L.R.R. provided human samples; T.K. and M.K. provided expertise in fibrosis and HCC mouse models; M.T.D.-M., and J.M. designed the experiments and wrote the manuscript with help from A.D., E.D.H., M.R.-C., and M.K.; M.T.D.-M., and J.M. provided funding.
ACCESSSION NUMBER
The Gene Expression Omnibus accession number for all the RNA-Seq data reported in this paper is GSE78760.
REFERENCES
- Anstee QM, Day CP. The genetics of NAFLD. Nature reviews Gastroenterology & hepatology. 2013;10:645–655. doi: 10.1038/nrgastro.2013.182. [DOI] [PubMed] [Google Scholar]
- Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nature reviews Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- Denk H, Stumptner C, Fuchsbichler A, Muller T, Farr G, Muller W, Terracciano L, Zatloukal K. Are the Mallory bodies and intracellular hyaline bodies in neoplastic and non-neoplastic hepatocytes related? J Pathol. 2006;208:653–661. doi: 10.1002/path.1946. [DOI] [PubMed] [Google Scholar]
- Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153:601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The Signaling Adaptor p62 Is an Important NF-[kappa]B Mediator in Tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- El-Serag HB. Hepatocellular carcinoma. The New England journal of medicine. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nature reviews Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- Galavotti S, Bartesaghi S, Faccenda D, Shaked-Rabi M, Sanzone S, McEvoy A, Dinsdale D, Condorelli F, Brandner S, Campanella M, et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2013;32:699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. The Journal of cell biology. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito I, Waku T, Aoki M, Abe R, Nagai Y, Watanabe T, Nakajima Y, Ohkido I, Yokoyama K, Miyachi H, et al. A nonclassical vitamin D receptor pathway suppresses renal fibrosis. J Clin Invest. 2013;123:4579–4594. doi: 10.1172/JCI67804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva T, Brenner DA. Fibrogenesis of parenchymal organs. Proceedings of the American Thoracic Society. 2008;5:338–342. doi: 10.1513/pats.200711-168DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva T, Cong M, Paik Y, Scholten D, Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto H, et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci U S A. 2012;109:9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shen C, Nakamura E, Ando K, Signoretti S, Beroukhim R, Cowley GS, Lizotte P, Liberzon E, Bair S, et al. SQSTM1 Is a Pathogenic Target of 5q Copy Number Gains in Kidney Cancer. Cancer Cell. 2013;24:738–750. doi: 10.1016/j.ccr.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Duran A, Reina-Campos M, Aza-Blanc P, Campos A, Moscat J, Diaz-Meco MT. Amino Acid Activation of mTORC1 by a PB1-Domain-Driven Kinase Complex Cascade. Cell reports. 2015 doi: 10.1016/j.celrep.2015.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J, Peng B, Fleming JB, Wang H, et al. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V, et al. Non-Cell-Autonomous Tumor Suppression by p53. Cell. 2013;153:449–460. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. Feedback on fat: p62-mTORC1-autophagy connections. Cell. 2011;147:724–727. doi: 10.1016/j.cell.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. p62: a versatile multitasker takes on cancer. Trends Biochem Sci. 2012;37:230–236. doi: 10.1016/j.tibs.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285:5941–5953. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomao M, Remotti H, Vaughan R, Siegel AB, Lefkowitch JH, Moreira RK. The steatohepatitic variant of hepatocellular carcinoma and its association with underlying steatohepatitis. Hum Pathol. 2012;43:737–746. doi: 10.1016/j.humpath.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. Embo J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, Collisson EA, Connor F, Van Dyke T, Kozlov S, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Aburatani H. Exploration of liver cancer genomes. Nature reviews Gastroenterology & hepatology. 2014;11:340–349. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- Stumptner C, Fuchsbichler A, Zatloukal K, Denk H. In vitro production of Mallory bodies and intracellular hyaline bodies: the central role of sequestosome 1/p62. Hepatology. 2007;46:851–860. doi: 10.1002/hep.21744. [DOI] [PubMed] [Google Scholar]
- Toffanin S, Friedman SL, Llovet JM. Obesity, inflammatory signaling, and hepatocellular carcinoma-an enlarging link. Cancer Cell. 2010;17:115–117. doi: 10.1016/j.ccr.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S, Duran A, et al. p62, Upregulated during Preneoplasia, Induces Hepatocellular Carcinogenesis by Maintaining Survival of Stressed HCC-Initiating Cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia T, Kim JY, Abu-Baker S, Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA, Metallo CM, Diaz-Meco MT, et al. Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell. 2014;26:121–135. doi: 10.1016/j.ccr.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Jung Y, Omenetti A, Witek RP, Choi S, Vandongen HM, Huang J, Alpini GD, Diehl AM. Fate-mapping evidence that hepatic stellate cells are epithelial progenitors in adult mouse livers. Stem Cells. 2008;26:2104–2113. doi: 10.1634/stemcells.2008-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, et al. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol. 2011;18:556–563. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.