Abstract
Background
High mobility group-box 3 (HMGB3) has been shown to affect tumor initiation and progression. This research aimed to investigate the role of HMGB3 in gastric cancer (GC) cell proliferation, migration, invasion, chemoresistance, and its potential molecular mechanisms.
Material/Methods
GC MGC803 and BGC823 cells were transfected with siRNA targeting the HMGB3 gene. The expressions of HMGB3 protein in MGC803 and BGC823 cells after transfection were detected by Western blot assays. We detected cell proliferation and cell cycle by MTT and flow cytometry assay. Cell migration and invasion were determined by wound scratch and transwell assay. MGC803 and BGC823 cells were treated with various concentrations of oxaliplatin, cisplatin, and paclitaxel. After 24 hours of drug exposure, we performed MTT assays to investigate chemoresistance in both groups. Western blot assays were used to detect related proteins expression.
Results
Silencing of HMGB3 inhibited cell proliferation and induced G0/G1 phase arrest of GC cells partly via modulating p53 and p21 pathways, and downregulating Bcl-2/Bax ratio. RNA interference of HMGB3 inhibited cell invasion and migration by downregulating MMP2 and MMP9. Silencing of HMGB3 enhanced sensitive to cisplatin and paclitaxel, and reduced sensitive to oxaliplatin.
Conclusions
These findings suggest the importance of HMGB3 in the regulation of growth, migration, and apoptosis of GC, improve our understanding of the mechanisms of GC pathogenesis, and may promote the development of novel targeted therapies.
MeSH Keywords: Cell Migration Inhibition; Cell Proliferation; Drug Resistance; HMGB3 Protein; RNA, Small Interfering; Stomach Neoplasms
Background
Gastric cancer (GC) is the sixth most common cause of tumors worldwide with an age standardized incidence and mortality of 12.1/100,000 and 8.9/100,000 population, respectively [1]. GC is an aggressive disease, and its prognosis remains poor, usually due to the absence of specific symptoms that renders early diagnosis of this cancer difficult [2,3]. The prognosis of stomach cancer is generally poor as this cancer is not very sensitive to commonly used chemotherapies, such as oxaliplatin, cisplatin, and paclitaxel [2]. The onset and progression of GC associated with multifarious molecular and genetic incidents [4]. To explore the significance of the expression of molecular incidents in GC may help us to identify potential therapeutic targets and select suitable treatment options for individual GC patients.
High mobility group-box 3 (HMGB3), also known as HMG2a, is a member of the HMG superfamily and is classified with HMGB1 and HMGB2 into the HMGB subfamily [5]. The HMGB subfamily plays important roles in DNA recombination, repair, replication, and transcription. [6]. HMGB3 is restricted to embryogenesis and is normally diminished or silent in adult tissues [7]. Recently studies have shown that HMGB3 and NPU98 fusion proteins form a new oncogenic gene in leukemia [8]. HMGB3 also participates in recurrence of acute lymphoid leukemia [9]. HMGB3 high-expression has been reported in the progression phase of breast cancer, with HMGB3 acting as an oncogene modulating proliferation, metastasis, and drug resistance [10,11]. In addition, limited studies have been done to evaluate the relationship between HMGB3 expression and solid tumor progression, such as esophageal [12], urinary bladder [13], gastric [4,7], and non-small cell lung cancer [14]. These studies all indicated high expression of HMGB3 protein in solid tumors. In addition, studies have found HMGB3 expression in cancer tissue had positive correlation with clinicopathological features, including the poor prognosis of cancer patients [4,7,12–14]. However, the impact of HMGB3 on proliferation, migration, invasion, and chemoresistance has not completely understood and the mechanism of HMGB3 in GC cells still unclear.
In this work, we evaluated the effects of HMGB3 gene silencing on human GC MGC803 and BGC823 cells in vitro. MGC803 and BGC823 cells with HMGB3 knocked down showed a decreased proliferation and metastasis rate, and the ratio of G0/G1 phase cells in the cell cycle significantly increased. Silencing of HMGB3 could effectively enhance sensitive to cisplatin and paclitaxel, and reduced sensitive to oxaliplatin. Our findings indicate that HMGB3 might be an important protein for stomach cancer progression and proliferation.
Material and Methods
Cell lines and siRNA transfection
All of the cell lines were cultured in RPMI1640 medium supplemented with penicillin (100 U/mL) and streptomycin (100 μg/mL; Hyclone, USA), 10% fetal bovine serum (FBS; Hyclone, USA), and grown in a humidified incubator with 5% CO2 at 37°C. Small interfering RNA (siRNA) against HMGB3 (siR-HMGB3) and negative control siRNA (siR-NC) were synthesized by Sigma (St. Louis, MO, USA). Transfections were performed by Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. The following siRNA sequences were used: siR-HMGB3, 5′-CUGU AUCAAAGUUGUACAUdTdT-3′; siR-NC, 5′-U UCUCCGACGUGUC ACGUTT-3′.
Cell proliferation assay
Cells were seeded into 96-well plates with 100 mL tissue culture medium (3×103 cells/well). At 24, 48, and 72 hour post-transfection with siRNA, 20 μL MTT (5 mg/mL; Solarbio, Beijing, China), was added to the cells. The mixture was further cultured for another 4 hours. Then, the MTT and media were removed and 200 μL of dimethyl sulfoxide (DMSO) (Sigma, St. Louis, USA) was added to each well. After vibration for 10 minutes, the absorbance was measured using a microplate reader at 490 nm.
Cell cycle analysis
To determine the effect of HMGB3 on cell cycle, MGC803 and BGC823 cells were seeded in 6-well cell cultured plates. After siRNA was transfected for 48 hours, all the cells were collected, washed, and centrifuged at room temperature. Cells were fixed with ice cold 70% ethanol and stored at 20°C until further analysis. The cells were washed twice with phosphate-buffered saline and resuspended in 500 μL binding buffer. Subsequently, DNA staining was carried out by 20 mg/mL of RNaseA (Solarbio, Beijing, China), 50 mg/mL of propidium iodide (BD, NJ, USA), and incubated at 37°C for 30 minutes. Cell cycle phases were analyzed by flow cytometry.
Wound healing assay
Cell migration capability was determined by wound healing assay. When transfected cells in 6-well plates were grown to confluence, an artificial wound was created by a 200 μL sterile pipette tip and then cultured for another 24 hours. The cells were observed, and photographed were captured using a microscope at time 0 and 24 hours. Cell relative motility was calculated by measuring the distance travelled by the cells. Analysis of the wound closure was performed by NIH Image program.
Transwell migration and invasion assay
Migration assay in vitro were performed using transwell-24 plates (Corning, NY, USA) containing a polycarbonate membrane filter with an 8 μm pore size without Matrigel coating. At 48 hours post-transfection, approximately 2×105 cells were added into the upper chambers and incubated at 37°C in medium without FBS. RPMI1640 medium containing 10% FBS was added to the lower chamber as a chemoattractant. After 12 hours, the cells that did not migrate through the pores of the upper surface of the membrane were removed by cotton swabs, and the invaded cells were stained with 0.1% crystal violet. For the invasion assay, a membrane well-coated with Matrigel was used for the upper chamber, with culture time for 24 hours. Cells were visually counted in five randomly selected fields under an inverted microscope (200×).
Cell viability assay
Two groups were generated for the subsequent experiments: a siR-NC transfected group (siR-NC), and a siRHMGB3 transfected group (siR-HMGB3). Briefly, 3×103 cells/well were plated in 96-well plates. After siRNA was transfected for 48 hours, cells were exposed to various concentrations of chemotherapeutic drugs (oxaliplatin: 0, 1, 5, 10, and 20 μg/mL; cisplatin: 0, 1.25, 2.5, 5, and 10 mg/L; paclitaxel: 0, 1, 5, 10, and 50 mg/mL). After 24 hours of drug exposure, 20 μL MTT solution (5 mg/mL) was added to the medium and placed for 4 hour at 37°C until the formazan crystals formed, then medium was removed, and 200 μlL DMSO was added to each well. The absorbance was measured using a microplate reader at 490 nm. The absorption value is directly proportional to the number of living cells in the medium: cell inhibition (%) = [1-(Atreated – Azero)/(Acontrol – Azero)] ×100, where “Atreated” was the absorbance of cells treated with drugs, “Acontrol” was the absorbance of cells treated without drugs and “Azero” was the background absorbance.
Western blot assay
Total proteins from cells were extracted using RIPA lysis buffer (Beyotime, Haimen, China). The concentrations of total proteins were determined using bicinchoninic acid (BCA) protein assay (Beyotime, Haimen, China). Proteins were electrophoresed via 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred onto polyvinylidene fluoride (PVDF) (GE Healthcare, UK). The PVDF membranes were blocked with 5% skimmed milk powder for two hours, and then incubated overnight with antibodies against HMGB3 (abcam, UK), BCL-2 (Boster, Wuhan, China), Bax (Boster, Wuhan, China), p53 (Bioss, Beijing, China), p21 (Bioss, Beijing, China), MMP2 (Proteintech, USA), MMP9 (Proteintech, USA), and β-actin (Proteintech, USA). The membranes were washed three times with TBST at room temperature. Then the PVDF membrane was further incubated with horseradish peroxidase (HRP)-conjugated corresponding secondary antibody at room temperature for two hours. All the immunoblots were visualized by enhanced chemiluminescence (ECL; Beyotime, Haimen, China). Antibody against β-actin served as an endogenous reference.
Statistical analysis
Statistical analyses in this study were performed using SPSS 17.0 software. All data were presented as mean ± standard deviation. The significance between different samples was analyzed by one-way analysis of variance (ANOVA). For all analyses, the level of statistically significance was set at p<0.05.
Results
Silencing of HMGB3 expression inhibits cell proliferation
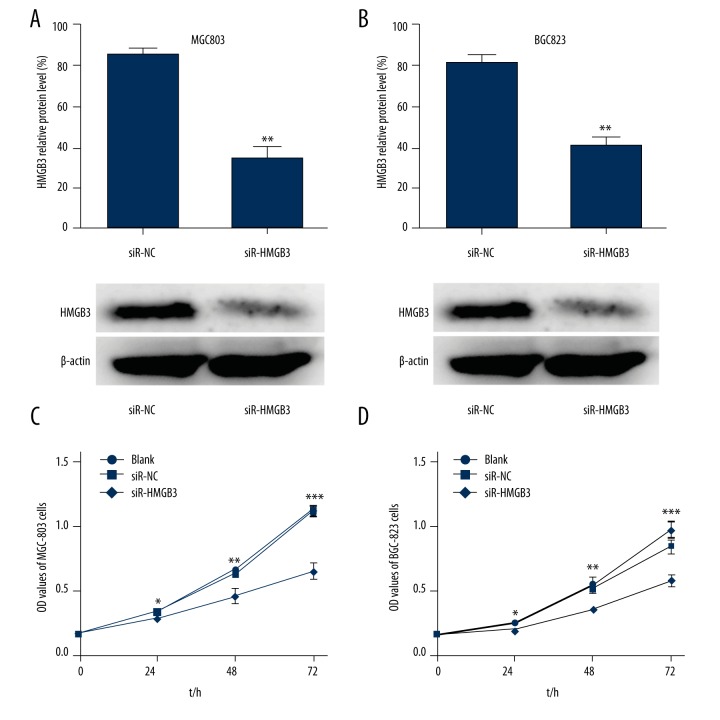
HMGB3 knockdown was performed by transfecting specific HMGB3-siRNA into GC cells. The protein expression level of HMGB3 evaluated by Western blot was significantly reduced in MGC803 cells (p<0.01, Figure 1A) and BGC823 cells (p<0.01, Figure 1B) transfected with siR-HMGB3 compared with siR-NC. To elucidate the effects of the HMGB3 on MGC803 and BGC823 GC cells proliferation, MTT assay was used to count the number of viable cells. Compared to cells transfected with the siR-NC, the proliferation properties of MGC803 cells (p<0.05), and BGC823 cells (p<0.05) were significantly inhibited in cells transfected with siR-HMGB3 at 24, 48, and 72 hours (Figure 1C, 1D). Results indicated that after HMGB3 was silenced, cell proliferation rate was significantly reduced. HMGB3 played an important role in cell proliferation.
Figure 1.
Silencing of HMGB3 expression inhibited cell proliferation. RNA interference was used to knockdown HMGB3 in the MGC803 cells (A) and BGC823 cells (B). The expression of HMGB3 in the blank, siR-NC, and siR-HMGB3 group cells were detected by Western blot assay. (C, D) Cellular proliferation ability was assayed by MTT assay. * p<0.05, ** p<0.01, and *** p<0.001 vs. siR-NC group. Blank – non-transfected; siR-NC – control siRNA; siR-HMGB3 – HMGB3 siRNA; HMGB3 – high mobility group-box 3.
Silencing of HMGB3 induces cell cycle arrest
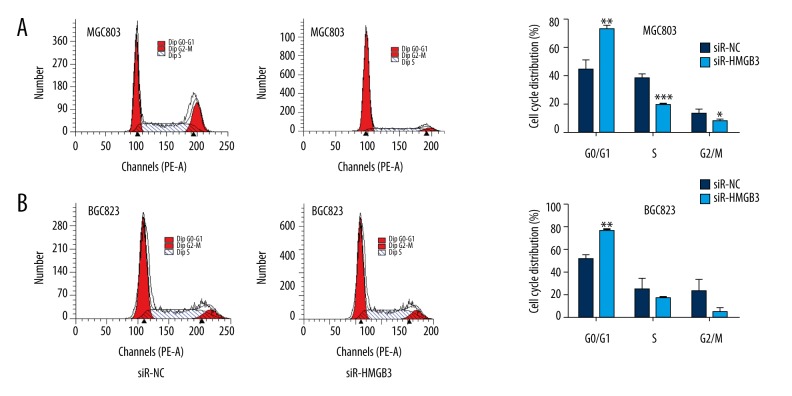
To investigate a potential mechanism for HMGB3 regulation of cell proliferation, we further examined the effect of knockdown HMGB3 on the cell cycle using flow cytometry. Results showed that the proportion of MGC803 cells in the G0/G1 phase increased significantly in the siR-HMGB3 group cells compared with the NC group cells (p<0.01, Figure 2A). Similarly, down expressing of HMGB3 also significantly increased the proportion of BGC823 cells in the G0/G1 phase (p<0.01, Figure 2B). These data showed that HMGB3 knockdown induced G0/G1 phase arrest in GC cells.
Figure 2.
Silencing of HMGB3 expression induced GO/G1 cell cycle arrest in GC MGC803 cells (A) and BGC823 cells (B). * p<0.05, ** p<0.01, and *** p<0.001 vs. siR-NC group. Blank – non-transfected; siR-NC – control siRNA; siR-HMGB3 – HMGB3 siRNA; HMGB3 – high mobility group-box 3.
Silencing of HMGB3 expression reduces cell migration
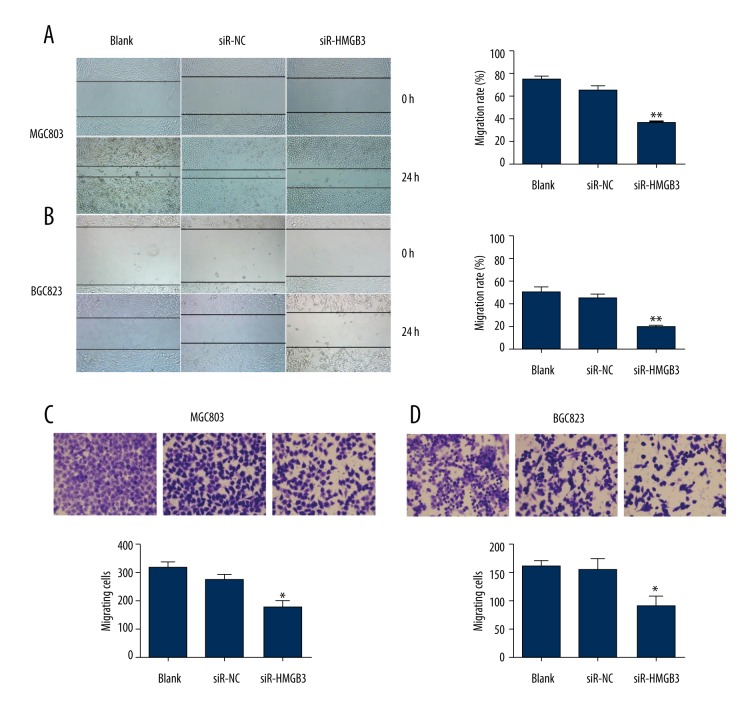
As cell motility is a measurement of the metastatic potential of cancer [15], the wound healing assay and the transwell migration assay were performed to test the effect of HMGB3 in GC cell migration (Figure 3). In the wound assay, the results showed that the area change for wound-healing in the siR-HMGB3 group of MGC803 cells (p<0.01, Figure 3A) and BGC823 cells (p<0.01, Figure 3B) were reduced compared to the siR-NC group cells. Transwell assay results revealed that the mobility of MGC803 cells (p<0.05, Figure 3C) and BGC823 cells (p<0.05, Figure 3C) was evidently suppressed after treatment with siR-HMGB3, compared with the control group. Taken together, these results indicated that HMGB3 may promote the cell migration ability of GC cell lines.
Figure 3.
Suppression of cell migration induced by HMGB3 small interfering RNA (siRNA). The migration of GC cells was detected by wound healing assay. MGC803 cells (A) and BGC823 cells (B) were monitored and photographed under electron microscope at 0 and 24 hours. Migration rate was quantified by NIH Image program. MGC803 cells (C) and BGC823 cells (D) migratory ability detected by transwell assay. * p<0.05 and ** p<0.01 vs. siR-NC group. Blank – non-transfected; siR-NC – control siRNA; siR-HMGB3 – HMGB3 siRNA; HMGB3 – high mobility group-box 3.
Silencing of HMGB3 expression reduces cell invasion
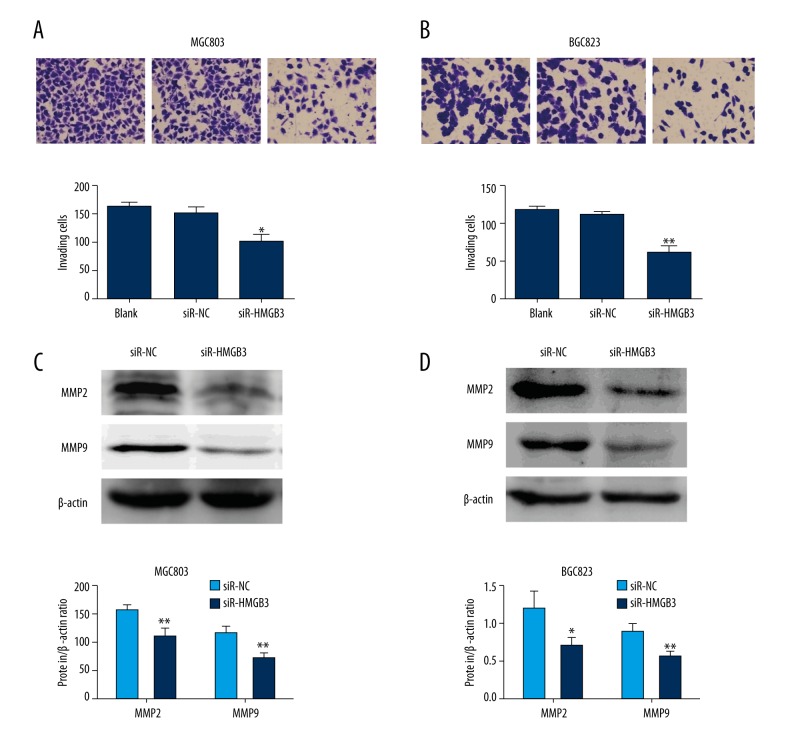
We performed transwell invasion assay to examine the role of HMGB3 in GC cell invasion. In contrast with blank and negative control cells, the number of siR-HMGB3 group MGC803 cells (p<0.05, Figure 4A) and BGC823 cells (p<0.01, Figure 4B) that migrated through the Matrigel was significantly lower. These results demonstrated that lower-expression of HMGB3 inhibited the invasion ability of GC cell. To analyze the potential mechanism underlying HMGB3-specific silencing-mediated migration and invasion block, MMP2 and MMP9 proteins was examined in the GC cell lines. As shown in Figure 4C and 4D, MMP2 and MMP9 protein levels were reduced in both GC cell lines with downregulation of HMGB3.
Figure 4.
HMGB3 downregulation inhibited invasion of GC cells. A transwell assay was performed to examine the invasion capacity of MGC803 cells (A) and BGC823 cells (B) after transfection with siR-HMGB3 or siR-NC. The changes of MMP2 and MMP9 protein levels were assessed by Western blot assays (C, D). * p<0.05 and ** p<0.01 vs. siR-NC group. Blank – non-transfected; siR-NC – control siRNA; siR-HMGB3 – HMGB3 siRNA; HMGB3 – high mobility group-box 3.
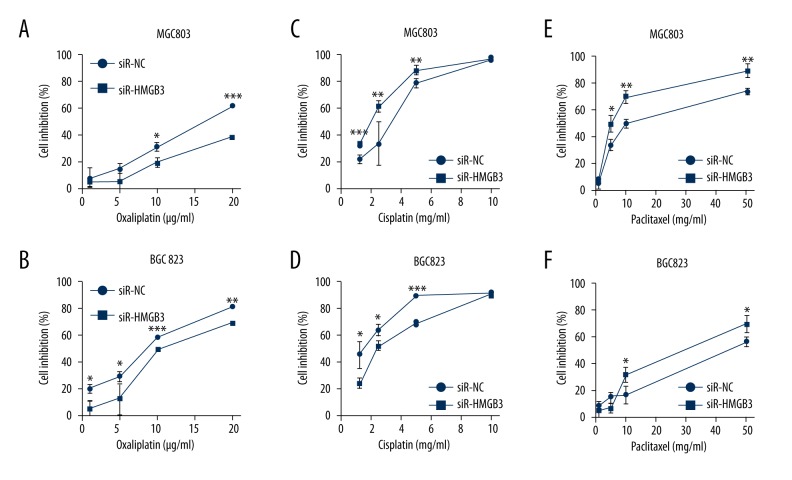
Silencing of HMGB3 expression influences the chemosensitivity to cisplatin, paclitaxel, and oxaliplatin
The fact that high-expression of HMGB3 foreboded a poor prognosis, especially for advanced GC patients who are more likely to receive chemotherapy or chemoradiotherapy, suggested that HMGB3 might conduce drug tolerance [4,7,12–14]. The cell viability assays were used to investigate this question. We found by MTT assays that MGC803 and BGC823 cells treated with combined oxaliplatin and HMGB3-targeted siRNA (siR-HMGB3) showed lower growth inhibition than cells treated with combined oxaliplatin and the negative control siRNA (Figure 5A, 5B). As shown in Figure 5C and 5D, cells that were treated with cisplatin, and cells in the siR-HMGB3 group showed obvious inhibition compared with cells in the NC group. Incubation with paclitaxel and siR-HMGB3 transfected cells exhibited enhanced paclitaxel sensitivity compared with siR-NC transfected MGC803 and BGC823 cells (Figure 5E, 5F). Our results demonstrated that inhibition of HMGB3 in GC cells significantly increased their tolerance toward oxaliplatin, and reduced their tolerance toward paclitaxel and oxaliplatin.
Figure 5.
(A, B) Knockdown of HMGB3 resulted in increased growth inhibition by oxaliplatin. HMGB3-transfected MGC803 cells (A) and BGC823 cells (B) were significantly more resistant to oxaliplatin than siR-NC-transfected cells as revealed by MTT cell viability assays. (C, D) Knockdown of HMGB3 resulted in increased chemosensitivity by cisplatin. MGC803 cells (E) and BGC823 cells (F) were treated with paclitaxel, cells in the siR-HMGB3 group showed a significantly greater growth inhibition than cells in the siR-NC group. * p<0.05, ** p<0.01, and *** p<0.001 vs. siR-NC group. siR-NC – control siRNA; siR-HMGB3 – HMGB3 siRNA; HMGB3 – high mobility group-box 3.
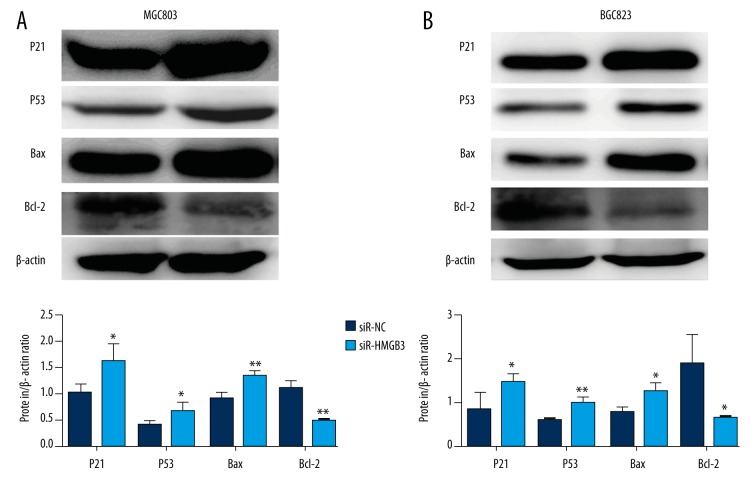
Effect of HMGB3 knockdown on p21 and other proteins expression
In order to study the molecular mechanisms of HMGB3, Western blot assay was performed to examine the expression of several proteins in the present study. As shown in Figure 6A and 6B, p21, p53, and Bax protein levels were upregulated and Bcl-2 protein levels were downregulated in MGC803 and BGC823 cell lines with low-expression of HMGB3.
Figure 6.
(A, B) Protein expression levels of p21, p53, Bax, and Bcl-2 were detected by Western blot assays and compared by quantitative analysis of the gray value. * p<0.05 and ** p<0.01 vs. siR-NC group. siR-NC – control siRNA; siR-HMGB3 – HMGB3 siRNA; HMGB3 – high mobility group-box 3.
Discussion
Overexpression of HMGB3 is found in many human cancers, which correlate with clinicopathological characteristics, as well as poor prognosis [4,7,12–14]. Elgamal et al. reported that regulation of HMGB3 by tumor suppressor gene miR-205 inhibited proliferation and infiltration of breast cancer cells [11]. Li et al. demonstrated that in human urinary bladder cancer cells, downregulation of HMGB3 expression resulted in the inhibition of cell migration and proliferation [13]. Furthermore, recently studies reported that HMGB3 was identified as a major target of miR-513b, and that downregulation of HMGB3 inhibits gastric cell proliferation [4,7]. In this study, we analyzed the role of HMGB3 in stomach cancer cells in vitro.
We found that the knockdown of HMGB3 in MGC803 and BGC823 cells were capable of inhibiting cell proliferation and inducing cell cycle arrest. This indicated that HMGB3 may promote cell proliferation through cell cycle progression. It is well-known that p53 plays an important role in the regulation of cell cycle arrest, senescence, apoptosis, and autophagy [16]. P21 is a transcriptional target of p53 and plays a key role in cell cycle arrest [17]. In this study, the expressions of p53 and p21 proteins were both significantly upregulated in silencing-HMGB3 cells. Thus, our results indicated that HMGB3 knockdown resulted in cell cycle arrest at the G0/G1 phase, possibly through the modulation of the p53 and pP21 signaling pathways. One of the major determinants of cell survival is the balance between the anti-apoptotic and pro-apoptotic members of the Bcl-2 family [18]. In the Bcl-2 protein family, pro-apoptotic factor Bax and anti-apoptotic factor Bcl-2 are the active regulators and effectors, and the ratio between Bcl-2 and Bax (Bcl-2/Bax) affects apoptosis induction [19,20]. In the meantime, we evaluated Bcl-2 and Bax after HMGB3 knockdown and found that Bax increased and Bcl-2 reduced when compared with the control group. These findings suggest that silencing of HMGB3 may inhibit cell proliferation through increased cell apoptosis. In other words, silencing of HMGB3 inhibited the cell proliferation and induced G0/G1 phase arrest of GC cells partly via modulating p53 and p21 pathways, and downregulating Bcl-2/Bax ratio.
Our experiments demonstrated that the downregulation of HMGB3 markedly reduced the capacity for invasion and migration. MMP2 and MMP9 can degrade collagen IV and correlates with metastatic and invasive properties, advanced stage, and poor prognosis [21,22]. Li et al. [13] reported that HMGB3 was decreased in HMGB3-silenced urinary bladder cancer cells. We found that MMP2 and MMP9 were downregulated in HMGB3-silenced GC cells. Our results show that HMGB3 promoted cell migration and invasion by upregulating MMP2 and MMP9.
Improved chemotherapeutics are the key to better prognosis for GC patients, but chemoresistance is a hallmark of many cancers, including GC [23,24]. Previous studies demonstrated that the elevated expression of HMGB1 was correlated positively with drug resistance [25,26].
But we know little about the role of HMGB3 in tumor resistance. A recent study found that silencing of HMGB3 indeed partially phenocopied the effects of miR-27b in decreasing tamoxifen resistance in breast cancer cells [10]. In this study, GC cells were treated with oxaliplatin, cisplatin, and paclitaxel, respectively. Our experiments showed that HMGB3 knockdown contributed to increased cisplatin and paclitaxel sensitivity. This was consistent with the role of HMGB1 in chemoresistance reported in cancer cell lines. However, our results indicated that HMGB3 knockdown contributed to increased chemoresistance to oxaliplatin. Oxaliplatin is a member of the third generation platinum-based drugs, widely used in metastatic gastrointestinal tumor treatment [27–29]. The research on oxaliplatin resistance mechanisms has focused on DNA repair, reduced apoptosis, autophagy, and drug detoxification [26,27,30]. The antitumor effect of oxaliplatin and cisplatin are likely achieved by interfering with DNA transcription and replication through covalently binding to DNA strands [27–29]. In addition, oxaliplatin and cisplatin induce apoptosis through cytoplasmic effects, and they often, but not always, exhibit cross-resistance [30]. In spite of their similarity, however, oxaliplatin and cisplatin have fundamental differences, and HMGB3 plays the opposite role in GC cells. Our results support the hypothesis that HMGB3 is an important but complex mediator of GC chemoresistance. To date, many genes have already been discovered related to anti-neoplastic resistance. In our study, drug susceptibility testing represents preliminary research, and these findings need to be further explored and validated. Because HMGB3 is highly homologous with HMGB1, it may have similar effects with regard to tumor initiation and progression [31]. Overexpression of HMGB1 is found in the majority of gastric adenocarcinoma types, which correlate with poor prognosis [25,32]. Other studies have found that downregulating the protein expression of HMGB1 significantly reduced cellular proliferation, invasion, and metastatic ability in GC cells [33,34]. Furthermore, in many cancer types, HMGB1 has been reported to inhibit anticancer drug-induced apoptosis and promote autophagy, both of which are believed to be important mechanisms for the development of multidrug resistance [33]. These mechanisms may also hold true for HMGB3, though further research is needed.
Conclusions
This research provides evidence that HMGB3 promotes tumor growth, invasion, and regulates chemotherapy sensitivity of MGC803 and BGC823 cells in vitro. HMGB3 may provide oppotunities for individualized treatment and promote the development of novel targeted therapies.
Footnotes
Statement
The authors declare that there are no conflicts of interest.
Source of support: This study was supported by the Project of Clinical Capabilities Construction Program for Liaoning Provincial Hospitals (grant number: No. LNCCC-D28-2015)
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Montenegro R, Clark P, Howarth A, et al. BET inhibition as a new strategy for the treatment of gastric cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.9766. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song B, Yan J, Liu C, et al. Tumor suppressor role of miR-363-3p in gastric cancer. Med Sci Monit. 2015;21:4074–80. doi: 10.12659/MSM.896556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang HR, Luo XQ, Xu G, et al. High mobility group-box 3 overexpression is associated with poor prognosis of resected gastric adenocarcinoma. World J Gastroenterol. 2012;18:7319–26. doi: 10.3748/wjg.v18.i48.7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemeth MJ, Curtis DJ, Kirby MR, et al. Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. 2003;102:1298–306. doi: 10.1182/blood-2002-11-3541. [DOI] [PubMed] [Google Scholar]
- 6.Štros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–13. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Zhao G, Wang F, et al. Upregulation of miR-513b inhibits cell proliferation, migration, and promotes apoptosis by targeting high mobility group-box 3 protein in gastric cancer. Tumour Biol. 2014;35:11081–89. doi: 10.1007/s13277-014-2405-z. [DOI] [PubMed] [Google Scholar]
- 8.Petit A, Ragu C, Della-Valle V, et al. NUP98-HMGB3: A novel oncogenic fusion. Leukemia. 2010;24:654–58. doi: 10.1038/leu.2009.241. [DOI] [PubMed] [Google Scholar]
- 9.Staal F, de Ridder D, Szczepanski T, et al. Genome-wide expression analysis of paired diagnosis-relapse samples in ALL indicates involvement of pathways related to DNA replication, cell cycle and DNA repair, independent of immune phenotype. Leukemia. 2010;24:491–99. doi: 10.1038/leu.2009.286. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Wu Y, Liu A, Tang X. MiR-27b is epigenetically downregulated in tamoxifen resistant breast cancer cells due to promoter methylation and regulates tamoxifen sensitivity by targeting HMGB3. Biochem Biophys Res Commun. 2016;477:768–73. doi: 10.1016/j.bbrc.2016.06.133. [DOI] [PubMed] [Google Scholar]
- 11.Elgamal OA, Park JK, Gusev Y, et al. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PloS One. 2013;8:e76402. doi: 10.1371/journal.pone.0076402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Zou Z, Gao J, et al. Increased expression of HMGB3: A novel independent prognostic marker of worse outcome in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:345–52. [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Cai Y, Zhao H, et al. Overexpression of HMGB3 protein promotes cell proliferation, migration and is associated with poor prognosis in urinary bladder cancer patients. Tumour Biol. 2015;36:4785–92. doi: 10.1007/s13277-015-3130-y. [DOI] [PubMed] [Google Scholar]
- 14.Song N, Liu B, Wu JL, et al. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 2013;34:2599–603. doi: 10.1007/s13277-013-0807-y. [DOI] [PubMed] [Google Scholar]
- 15.Zhu F, Zhang Z, Wu G, et al. Rho kinase inhibitor fasudil suppresses migration and invasion though down-regulating the expression of VEGF in lung cancer cell line A549. Med Oncol. 2011;28:565–71. doi: 10.1007/s12032-010-9468-5. [DOI] [PubMed] [Google Scholar]
- 16.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong MJ, Stang MT, Liu Y, et al. Interferon regulatory factor 1 (IRF-1) induces p21 WAF1/CIP1 dependent cell cycle arrest and p21 WAF1/CIP1 independent modulation of survivin in cancer cells. Cancer Lett. 2012;319:56–65. doi: 10.1016/j.canlet.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu C, Wu A, Zhu H, et al. Melatonin is involved in the apoptosis and necrosis of pancreatic cancer cell line SW-1990 via modulating of Bcl-2/Bax balance. Biomed Pharmacother. 2013;67:133–39. doi: 10.1016/j.biopha.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Duo J, Ying GG, Wang GW, Zhang L. Quercetin inhibits human breast cancer cell proliferation and induces apoptosis via Bcl-2 and Bax regulation. Mol Med Rep. 2012;5:1453–56. doi: 10.3892/mmr.2012.845. [DOI] [PubMed] [Google Scholar]
- 20.Bi D, Yang M, Zhao X, Huang S. Effect of cnidium lactone on serum mutant P53 and BCL-2/BAX expression in human prostate cancer Cells PC-3 tumor-bearing BALB/C nude mouse model. Med Sci Monit. 2015;21:2421–27. doi: 10.12659/MSM.893745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, Mi Y, Ma Y, Jin W. LEF1 regulates glioblastoma cell proliferation, migration, invasion, and cancer stem-like cell self-renewal. Tumour Biol. 2014;35:11505–11. doi: 10.1007/s13277-014-2466-z. [DOI] [PubMed] [Google Scholar]
- 22.La Rocca G, Pucci-Minafra I, Marrazzo A, et al. Zymographic detection and clinical correlations of MMP-2 and MMP-9 in breast cancer sera. Br J Cancer. 2004;90:1414–21. doi: 10.1038/sj.bjc.6601725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–92. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 24.Gillet JP, Gottesman MM. Overcoming multidrug resistance in cancer: 35 years after the discovery of ABCB1. Drug Resist Updat. 2012;15:2–4. doi: 10.1016/j.drup.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Yin Y, Li W, Deng M, et al. Extracellular high mobility group box chromosomal protein 1 promotes drug resistance by increasing the expression of P-glycoprotein expression in gastric adenocarcinoma cells. Mol Med Rep. 2014;9:1439–43. doi: 10.3892/mmr.2014.1961. [DOI] [PubMed] [Google Scholar]
- 26.Zhang R, Li Y, Wang Z, et al. Interference with HMGB1 increases the sensitivity to chemotherapy drugs by inhibiting HMGB1-mediated cell autophagy and inducing cell apoptosis. Tumour Biol. 2015;36:8585–92. doi: 10.1007/s13277-015-3617-6. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, Zhang Z, Zhang Y, et al. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol Ther. 2015;16:511–17. doi: 10.1080/15384047.2015.1017691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saif MW, Choma A, Salamone SJ, Chu E. Pharmacokinetically guided dose adjustment of 5-fluorouracil: A rational approach to improving therapeutic outcomes. J Natl Cancer Inst. 2009;101:1543–52. doi: 10.1093/jnci/djp328. [DOI] [PubMed] [Google Scholar]
- 29.Chao Y, Yeh KH, Chang C, et al. Phase II study of weekly oxaliplatin and 24-h infusion of high-dose 5-fluorouracil and folinic acid in the treatment of advanced gastric cancer. Br J Cancer. 2004;91:453–58. doi: 10.1038/sj.bjc.6601985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tesniere A, Schlemmer F, Boige V, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene. 2010;29:482–91. doi: 10.1038/onc.2009.356. [DOI] [PubMed] [Google Scholar]
- 31.Chuangui C, Peng T, Zhentao Y. The expression of high mobility group box 1 is associated with lymph node metastasis and poor prognosis in esophageal squamous cell carcinoma. Pathol Oncol Res. 2012;18:1021–27. doi: 10.1007/s12253-012-9539-3. [DOI] [PubMed] [Google Scholar]
- 32.Akaike H, Kono K, Sugai H, et al. Expression of high mobility group box chromosomal protein-1 (HMGB-1) in gastric cancer. Anticancer Res. 2007;27:449–57. [PubMed] [Google Scholar]
- 33.Zhang J, Kou YB, Zhu JS, et al. Knockdown of HMGB1 inhibits growth and invasion of gastric cancer cells through the NF-κB pathway in vitro and in vivo. Int J Oncol. 2014;44:1268–76. doi: 10.3892/ijo.2014.2285. [DOI] [PubMed] [Google Scholar]
- 34.Song B, Song WG, Li ZJ, et al. Effect of HMGB1 silencing on cell proliferation, invasion and apoptosis of MGC-803 gastric cancer cells. Cell Biochem Funct. 2012;30:11–17. doi: 10.1002/cbf.1811. [DOI] [PubMed] [Google Scholar]