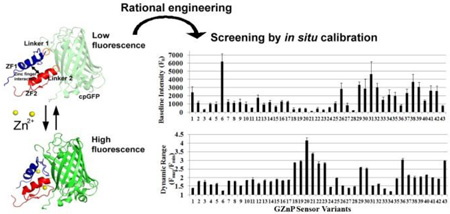
Abstract
Various fluorescent probes have been developed to reveal the biological functions of intracellular labile Zn2+. Here we present Green Zinc Probe (GZnP), a novel genetically encoded Zn2+ sensor design based on a single fluorescent protein (single-FP). The GZnP sensor is generated by attaching two zinc fingers (ZF) of the transcription factor Zap1 (ZF1 and ZF2) to the two ends of a circularly permuted green fluorescent protein (cpGFP). Formation of ZF folds induces interaction between the two ZFs, which induces a change in the cpGFP conformation, leading to an increase in fluorescence. A small sensor library is created to include mutations in the ZFs, cpGFP and linkers between ZF and cpGFP to improve signal stability, sensor brightness and dynamic range based on rational protein engineering and computational design by Rosetta. Using a cell-based library screen, we identify sensor GZnP1 which demonstrates a stable maximum signal, decent brightness (QY = 0.42 at apo state), as well as specific and sensitive response to Zn2+ in HeLa cells (Fmax/Fmin = 2.6, Kd = 58 pM, pH 7.4). The subcellular localizing sensors mito-GZnP1 (in mitochondria matrix) and Lck-GZnP1 (on plasma membrane) display sensitivity to Zn2+ (Fmax/Fmin = 2.2). This sensor design provides freedom to be used in combination with other optical indicators and optogenetic tools for simultaneous imaging and advancing our understanding of cellular Zn2+ function.
Keywords: Zinc, genetically encoded sensor, single fluorescent protein, Rosetta
Graphical Abstract
Zinc is an essential biological ion and suggestions that it may function as a cellular signal have inspired a surge in the development of fluorescent sensors to monitor zinc ions (Zn2+) in cells. Over the last several decades a wide variety of probes have been developed, including small molecule sensors, genetically encoded FRET-based sensors, and hybrid sensors in which small molecule sensors are genetically targeted by crosslinking to a peptide or protein1. Different classes of probes have different strengths and weaknesses. Thus far, most small-molecule fluorescent Zn2+ sensors used in biological studies are intensity-based2. The most significant advantage of these sensors is that they display higher dynamic change in response to Zn2+ compared with ratiometric sensors3. However, applications of small molecular probes are hindered by the inability or challenge of controlling subcellular localization and accumulation within the cell4. On the other hand, genetically encoded Zn2+ biosensors can be targeted to specific subcellular regions such as cytosol5–6, secretory vesicles5, ER6 and Golgi6 by molecular engineering techniques. The current repertoire of genetically encoded Zn2+ probes (ZapCY, eCALWY, eZinCh and CA series) utilizes the shift in FRET efficiencies between two fluorophores in response to Zn2+ binding7. The benefit of this design is that the FRET ratio is less sensitive to sensor concentration, sample thickness and photobleaching, thus this type of sensor is preferred for quantification of labile Zn2+ concentrations. However, many FRET based sensors display a low dynamic range especially when compared to small molecular sensors, and this low dynamic range is often exacerbated when the sensor is targeted to intracellular compartments6, 8–10. The comparatively low dynamic range of many organelle-targeted sensors may be one reason for discrepancies in the quantification of Zn2+ in the ER and mitochondria using different FRET sensor platforms6, 8–10. Furthermore, because most FRET sensors employ two fluorophores their large spectral bandwidth limits their application for multi-analyte imaging.
To complement the existing toolbox and create probes that leverage the high dynamic range and narrow bandwidth of intensity-based sensors with the genetic encodeability and localization specificity of protein-based probes, we developed a new genetically encoded Zn2+ sensor GZnP1 based on a single fluorescent protein (FP). In the design of this probe, we explore the application of Rosetta-based computational design to carry out in silico saturation mutagenesis on select regions of the protein sensor11. Biosensor optimization frequently involves introduction of knowledge-based or random mutations, followed by selection of sensors with desired properties by empirical experimental measurements. This process is slow and laborious and often yields little insight into why particular mutations impact sensor properties for better or worse. Structure-guided mutagenesis was instrumental in identifying regions for targeted improvement in the GCaMP family of Ca2+-sensors3, however it is critical to have structural information and few biosensors have the benefit of a crystal structure. Our sensor design involves fusion of protein domains where the structure of the sensor domain but not the full structure is known. Here we sought test the ability of Rosetta-based computational design to explore the effect of mutations based on a structural model, even in the absence of a structure for the full sensor protein.
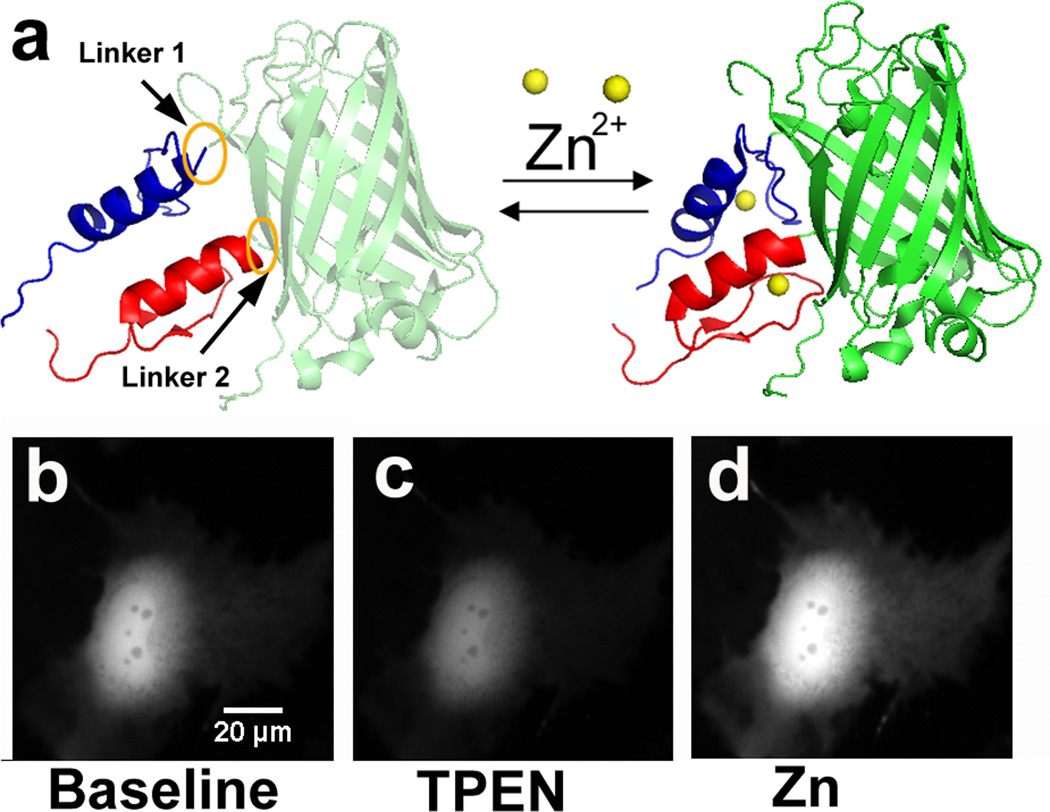
GZnP1 was constructed by attaching two zinc fingers of the yeast transcription factor Zap1 (ZF1 and ZF2) to the two ends of a circularly permuted green fluorescent protein (cpGFP). When Zn2+ is bound, the formation of two zinc finger folds should induce an interaction between the two ZFs, resulting in a subsequent conformational change of cpGFP leading to an increase in fluorescence intensity (Fig. 1a). Through rational and computational design, a stable, sensitive and Zn2+-specific sensor was generated, with a high dynamic range in both the cytosol and when targeted to mitochondria and the plasma membrane. This work has expanded the fluorescent Zn2+ probe toolkit by introducing a new type of genetically encoded sensors for exploring dynamics of labile Zn2+.
Figure 1. GZnP design and in situ response to manipulation of Zn2+.
(a) Model of GZnP consisting of cpGFP and two zinc fingers ZF1 (blue) and ZF2 (red). (b–d) In HeLa cells, the fluorescence intensity of GZnP decreased upon addition of the zinc chelator TPEN (100 µM) (c), and increased upon addition of a high amount of zinc (5 µM pyrithione and 10 µM ZnCl2) (d). All images were scaled to the same intensity range.
Results and Discussion
Design of GZnP sensors
The design of GZnP (green Zn2+ probes) sensors exploited the strategy used to develop the single-FP Ca2+ sensors GCaMPs and GGECOs12–13. A circularly permutated version of GFP (cpGFP) was used as the fluorescent element, because it is more sensitive than the original GFP to analyte binding14. The two tandem zinc fingers of the transcription factor Zap1 (ZF1 and ZF2) were chosen as the Zn2+ sensing domain because they responded robustly to Zn2+ binding in previously developed FRET sensors6. ZF2 and ZF1 were fused to the N- and C-terminus of cpGFP respectively to generate GZnP (Fig. 1a). When Zn2+ binds to ZF1 and ZF2, the formation of two zinc finger folds should induce a finger-finger interaction15, which should induce a subsequent conformational change in the fluorescent protein, resulting in an increase in the fluorescence intensity (Fig. 1a). A first generation of this sensor was constructed and expressed in HeLa cells. As expected, treatment of cells with a specific zinc chelator (N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine, TPEN) induced a decrease in fluorescence, yielding the minimal signal (Fmin), while the fluorescence intensity increased upon perfusion of cells with 5 µM pyrithione (an ionophore) and 10 µM ZnCl2, causing saturation of Zn2+ binding to the sensor and giving rise to the maximal fluorescence signal (Fmax) (Fig. 1b–d).
Engineering of GZnP with optimized properties
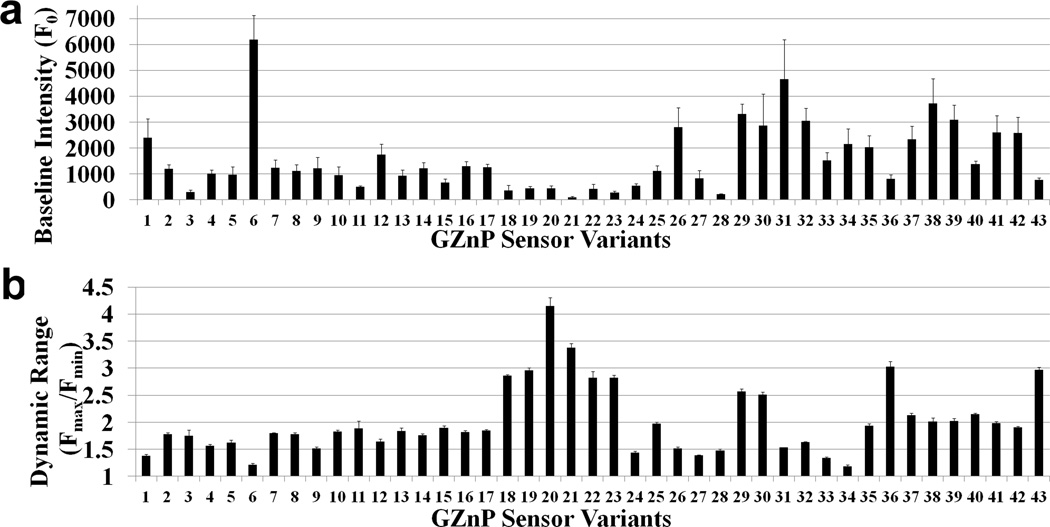
The GZnP sensor is a dynamic, multi-modular protein comprised of three distinct regions: the circularly permuted green fluorescent protein (cpGFP), the Zn2+-binding domain (ZF1 and ZF2), and the connecting linker regions. The relationship between alterations in any of the three regions of the GZnP and the fluorescent output is not yet understood. Therefore, we evaluated mutations in each of the domains to identify which regions yield the most improvement in baseline fluorescence and dynamic range (Fmax/Fmin). We additionally evaluated combined mutations from different regions, such as mutations in the Zn2+ sensing domain with different linker sequences. Using rational mutagenesis and computational protein design we created a small library comprised of 43 sensor variants (Table S1 presents full list of variants) and measured the effect on fluorescence properties. Specifically, Fig. 2 summarizes the baseline fluorescence intensity and dynamic range of the sensors measured by fluorescence microscopy of cells expressing individual sensor variants.
Figure 2. Development and optimization of single-FP zinc sensor GZnP by screening 43 sensor variants.
(a) Baseline fluorescence intensity for the 43 sensor variants. (b) Sensor dynamic range (Fmax/Fmin) was compared among 43 sensor variants. (n ≥ 5 cells, the error bar represents standard error of mean)
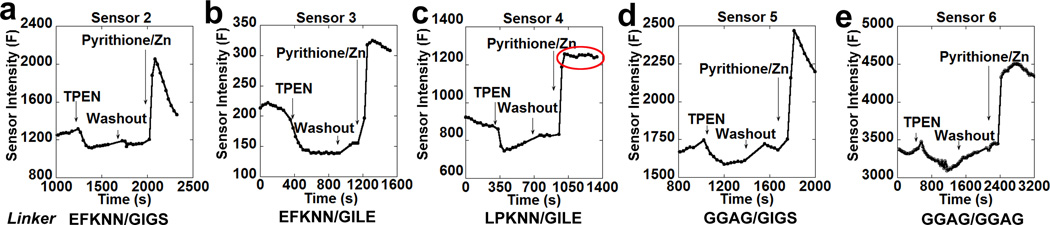
The first parameter we explored was the nature of the cpGFP that comprises the fluorophore portion of the sensor. Sensor variant 1 and 2 utilized the cpGFP from GCaMP313 and GGECO116, respectively. Although these two versions of cpGFP only differ in two residues (K58I, L112Q), in situ titrations showed that the dynamic range of sensor 2 was significantly higher than sensor 1 (Fig. S1), thus cpGFP from GGECO1 was utilized for further development of the GZnP sensor. However, the in situ titration of sensor 2 revealed that the sensor florescence intensity reached a maximal peak followed by an abrupt drop under conditions of Zn2+ saturation (Fmax) (Fig. 3a). We speculated that this could be caused by instability in the Zn2+-induced interaction between ZF1 and ZF2. Because the linker regions between the Zn2+ binding domains and cpGFP can determine how the formation of the zinc finger complex affects the conformational change of cpGFP, we tested the effect of different linkers on the stability of the maximal fluorescence signal. We tested linkers of different lengths and numbers of glycine residues to confer flexibility, as well as more restrictive linker residues (LE, LP) such as those used in GCaMP5G17. The resulting sensors were tested in HeLa cells. For linkers predicted to be more flexible, the maximum fluorescence signal rapidly dropped after reaching the peak (Fig. 3a, Fig. 3d, Fig. 3e and Table S1), while more restrictive linkers maintained the maximal signal at a stable level (Fig. 3b and c). These data suggest that the linker regions can affect the stability of the signal, and more conformationally restricted linkers may be preferred for this particular sensor platform. The restricted linker used here is the same one used in the single-FP Ca2+ sensor GCaMP5G17, suggesting that previous work and structural information from Ca2+ sensor development can serve as a guide in the creation of Zn2+ sensors.
Figure 3. Effects of linker regions on in situ response of GZnP sensors.
Representative traces of sensor responses to cellular zinc perturbation with TPEN (100 µM) and pyrithione (5 µM)/ZnCl2 (10 µM) for sensor mutants 2 (a), 3 (b), 4 (c), 5 (d) and 6 (e) with different linker regions. Sensor 2 and 5 showed unstable maximal signal with treatment of zinc, while 4 demonstrated the most stable maximal intensities (indicated by the red circle).
To further optimize the sensor dynamic range, Rosetta protein modeling software was used to identify mutations that were predicted to 1) lower the Rosetta-derived energy to stabilize the Zn2+-bound structure, 2) optimize interactions in the ZF1 and ZF2 interface by increasing the stability of either the ZF1 or ZF2 domain or increase the solubility of either domain. GZnP has not been structurally characterized therefore we focused on mutations within the zinc binding domain, for which there is an NMR structure (1zw8)18. In silico saturation mutagenesis was carried out at each position along the Zn2+ binding domain excluding the Zn2+-ligating residues, using two protocols to explore the sequence and conformational space at each position. The first method involved the standard fixed backbone approach, where all backbone atoms were fixed to NMR coordinates while side chains were varied (Table S2, “fixbb”). Mutations were selected if Rosetta predicted a lower energy side-chain compared to the wild type protein and the resulting side chain yielded favorable design features, such as an increase in buried surface area. The second method involved backbone relaxation followed by design (fast relax followed by fixed backbone mode, Table S2, “relax+fixbb”). This second approach yielded two additional mutations that were not identified using the fixed backbone protocol alone (Table S2). In addition, the interface between ZF1 and ZF2 was optimized by redesigning all interface residues using the fixed backbone protocol and the relaxation combined with fixed backbone protocols. Sequence positions neighboring the Zn2+ binding site were challenging to design; Rosetta identified glycine as the lowest energy residue for both position 18 and 33 (positions 49 and 64 respectively in PDB 1zw8) using fixed backbone sequence design. The native residues at positions 18 and 33 are phenylalanine and glutamine respectively. Replacing either side-chain with glycine would result in removal of a significant number of potentially stabilizing atomic contacts with Zn2+-binding histidine residues. Analysis of other Zn2+-binding domains revealed glycine is not observed at either position 18 or 33. We therefore evaluated the contributions from individual Rosetta energy terms and used the Lennard-Jones repulsive term (LJrep) as a filter, removing any mutations that would likely clash with neighboring positions, as indicated by a LJrep score greater than 10 (see Methods for more detail). The Rosetta electrostatic energy term was used to identify residue substitutions at each position (Tables S3 and S4) leading to suggested mutations of F18K or M and Q33T (Table S2, “electrostatic energy term”, Fig. 4a).
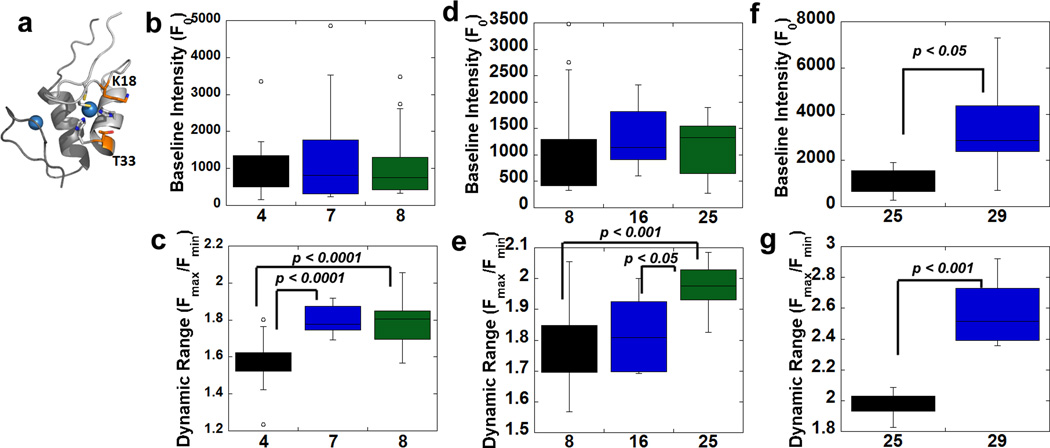
Figure 4. Introduction of mutations to improve GZnP sensor properties.
(a) Mutations near the Zn2+ binding site of ZF2 alter the baseline fluorescence and dynamic range of the sensor. ZF1 and ZF2 bind Zn2+ and form the Zn2+ sensing domain. The Zn2+ sensing domain is shown in cartoon representation, with ZF1 in dark gray and ZF2 in light gray. The two mutations designed to alter the Zn2+ sensing domain, F18K and Q33T, are shown in stick representations. Zn2+ ions are shown as blue spheres. (b) There are no differences in baseline brightness among sensor variants 4, 7 (F18M) and 8 (F18K). (c) Mutations F18M or F18K in zinc finger ZF2 increased the dynamic range. (d) There are no differences in baseline brightness among sensor variants 8, 16 (three sfGFP mutations) and 25 (sfGFP mutations/Q33T). (e) Introduction of mutation Q33T in zinc finger ZF2 (variant 25) greatly increased dynamic range. (f) Shortening of Linker 2 increased sensor baseline brightness. (g) Shortening of Linker 2 increased sensor dynamic range. Data in (b)–(g) are represented as a box and whisker plot. The box represents 50% of the data with the bottom and top of the box denoting the first and third quartiles, and the median marked by a line. The whiskers mark the minimum and maximum data points that fall within 1.5-times the interquartile distance. Data points that fall above or below 1.5-times the interquartile distance (lower quartile minus 1.5-times interquartile distance or upper quartile plus 1.5-times interquartile distance) are considered outliers and are denoted as circles.
The F18K/M and Q33T mutations predicted by Rosetta to maintain stability around the ZF2 Zn2+ binding site were introduced into the sensor and the optical properties were evaluated in cells (Fig. 4). F18M (variant 7) and F18K (variant 8) exhibited increased dynamic range (1.8 vs. 1.6-fold) without affecting sensor brightness (Fig. 4b–c). The introduction of both mutations (F18K and Q33T) in sensor variant 25 further raised the dynamic range to 2-fold (Fig. 4e) without affecting the sensor baseline brightness (Fig. 4d). We also tested other mutations predicted by Rosetta to increase stability of ZF1 or ZF2 individually or alter the solubility of these domains (Table S1 and S2). However, these mutations did not give rise to improved sensor properties (variants 37, 38, 39, 40, 41 and 42). Overall, in silico mutagenesis revealed that Rosetta can identify mutations that yield an improved dynamic range, particularly when mutations are designed to enhance interactions within the ligand binding domain, as opposed to merely increasing stability or solubility of isolated domains.
In addition to using Rosetta to scan the mutational space of the Zn2+-binding domain, we sought to leverage insights gleaned from the structure of GCaMP that suggested blocking solvent access to the cpGFP chromophore could enhance sensor brightness3. To decrease the distance between the Zn2+ sensing domain and cpGFP, we reduced the length of linker 1 and linker 2. When linker 2 (at the N terminus of cpGFP) was shortened by deleting two residues (variant 29), both sensor brightness and dynamic range were improved (Fig. 4f & g). Shortening of linker 1 (variant 26) also increased sensor brightness, but the sensor dynamic range was greatly reduced at the same time (Fig. S2).
Finally, we also tested superfolder GFP (sfGFP) mutations, which previously had been shown to improve the brightness and sensitivity of the GCaMP-HS Ca2+ sensor19; however, most sfGFP mutations failed to change either of the GZnP sensor properties (Fig. S3). The only exception was K99V (variant 22), which alone or in combination with other sfGFP mutations (variant 18) significantly increased the dynamic range, although this was because the baseline fluorescence decreased significantly (Fig. S3). This position (K99V) is the same position (A206K) in original GFP that has been implicated in fluorescent protein dimerization19–20. Although three sfGFP mutations (S165R, N240T, Y174N) did not change sensor properties, we still include these mutations into the sensor because they might stabilize the protein folding, as seen in sfGFP21.
Among 43 sensor variants, we surprisingly discovered that those mutations that can improve sensor properties are all localized near to the N terminus of cpGFP, suggesting that this region is critical in determining the sensor conformational change in response to Zn2+. The combination of knowledge-guided and computational design resulted in identification of two sensors with increased brightness (variant 29 and 30, 38% higher than the original sensor 1) and higher dynamic range (variant 29 and 30, 87% higher than 1) (Fig. 3a & b). We named one of the optimized sensor variants (29) GZnP1, which is a bright sensor with a 2.6-fold fluorescence increase in response to Zn2+ perturbations.
Sensor variant 22 displayed the highest dynamic range (Fmax/Fmin = 4.2), however, its baseline intensity is very low, which requires high intensity illumination as well as sensitive microscope detectors. Sensor variant 6 is very bright, but its dynamic response is very low (Fig. 2) and the sensor signal is not stable (Fig. 3e). Our main goal is to create a probe that can be easily used on a regular widefield fluorescence microscope without supersensitive detectors, and can be applied in biological samples susceptible to phototoxicity. Thus, we selected the bright sensor GZnP1 (variant 29) with a 2.6-fold dynamic response to Zn2+ for further characterization.
In vitro characterization
To further understand the biophysical features of GZnP1, the sensor protein was purified and characterized in vitro. For complete characterization, we measured the apparent dissociation constant (Kd’) for Zn2+, the pH dependence of the Kd’, the specificity for Zn2+ over other biologically relevant metal ions, and the photophysical properties (quantum yield, absorption and emission properties) in the presence and absence of Zn2+. GZnP1 showed a fluorescence turn-on response in vitro (Fig. 5a & Fig. 5b). The quantum yield (QY) of GZnP1 increased upon Zn2+ binding (0.57 in the Zn2+ bound state versus 0.42 in the apo state) (Fig. 5b, Table 1). The QY of the Zn2+-bound state is comparable to the Ca2+-bound state in the single-FP Ca2+ sensor GCaMP2 (QY = 0.53) and GCaMP3 (QY = 0.65)17. The in vitro dynamic range can be calculated by dividing the intensity of the emission peak of the Zn2+-saturated sensor by that of the apo sensor, which yields ~ 2.6. We can also estimate the theoretical dynamic range based on the product of changes in quantum yield (QYZn/QYapo) and absorbance peak at 488 nm (AbZn/Abapo), so the dynamic range is (0.57*0.27)/(0.42*0.16)= 2.3 (Table 1). These estimates of in vitro dynamic range are consistent with the in situ dynamic range (Fig. 4g).
Figure 5. In vitro characterization of GZnP1 sensor.
(a) Absorption and emission spectra of GZnP1 in Zn2+-chelated (solid lines, generated by treating the sensor with the Zn2+ specific chelator (Tris(2-pyridylmethyl)amine, TPA28) and saturated (100 µM Zn2+; dotted lines) solutions. Absorption and emission maximum are at 501 nm and 513 nm, respectively. (b) Measurement of sensor quantum yield (QY) for GZnP1 treated with Zn2+ and chelators. (c) GZnP1 was specific for Zn2+ compared to 4 µM Ca2+, Mg2+, Co2+, Ni2+, Fe2+, but also showed a slight response to 4 µM Cu2+, Mn2+ and Cu+. (d) The apparent dissociation constant decreased with increasing pH.
Table 1.
Summary of biophysical characteristics of GZnP1
| quantum yield (QY) | absorbance at λmax (497 nm) |
absorbance at 488 nm |
|
|---|---|---|---|
| GZnP1 (Zn2+) | 0.57 (QYZn) | 0.31 | 0.27 (AbZn) |
| GZnP1 (TPA) | 0.42 (QYapo) | 0.18 | 0.16 (Abapo) |
| GZnP1 (TPEN) | 0.42 (QYapo) | 0.19 | 0.16 (Abapo) |
Absorbance at λmax for GZnP1 in Zn2+- bound and Zn2+-free state was normalized to the absorbance at 280 nm for each sample.
GZnP1 was specific for Zn2+ over other ions, although there was some response to high levels (4 µM) of Cu+ (Fig. 4c, Fig. S4). Because the intracellular Cu+ concentration has been suggested to be less than femtomolar22, we believe that the fluorescence signal is unlikely to be affected by intracellular Cu+. The Kd’ for Zn2+ was found to be 58 pM at pH 7.4, and was found to decrease with increasing pH (Fig. 4d & Fig. S5). The fluorescence intensity of GZnP1 is also dependent on pH, with a pKa of GZnP1 around 7.4 (7.3 for the Zn2+ saturated state and 7.8 for the apo state) (Fig. S6).
Development of mitochondria targeted Zn2+ sensor mito-GZnP1 and membrane targeted Zn2+ sensor Lck-GZnP1
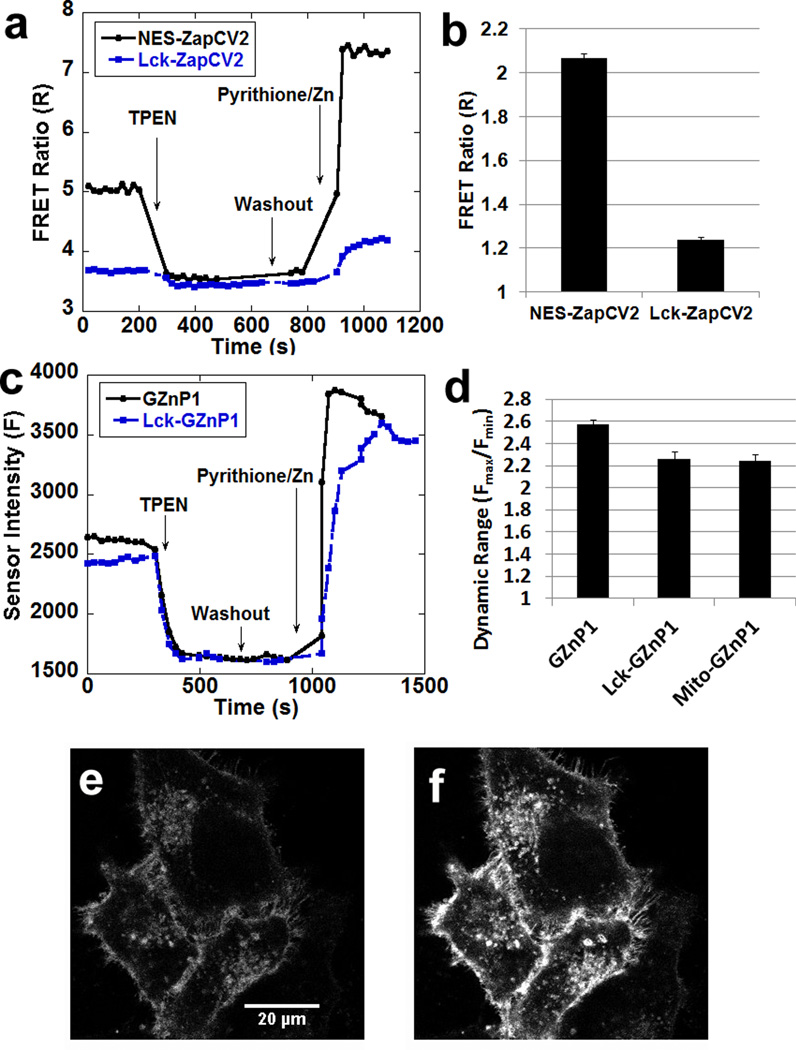
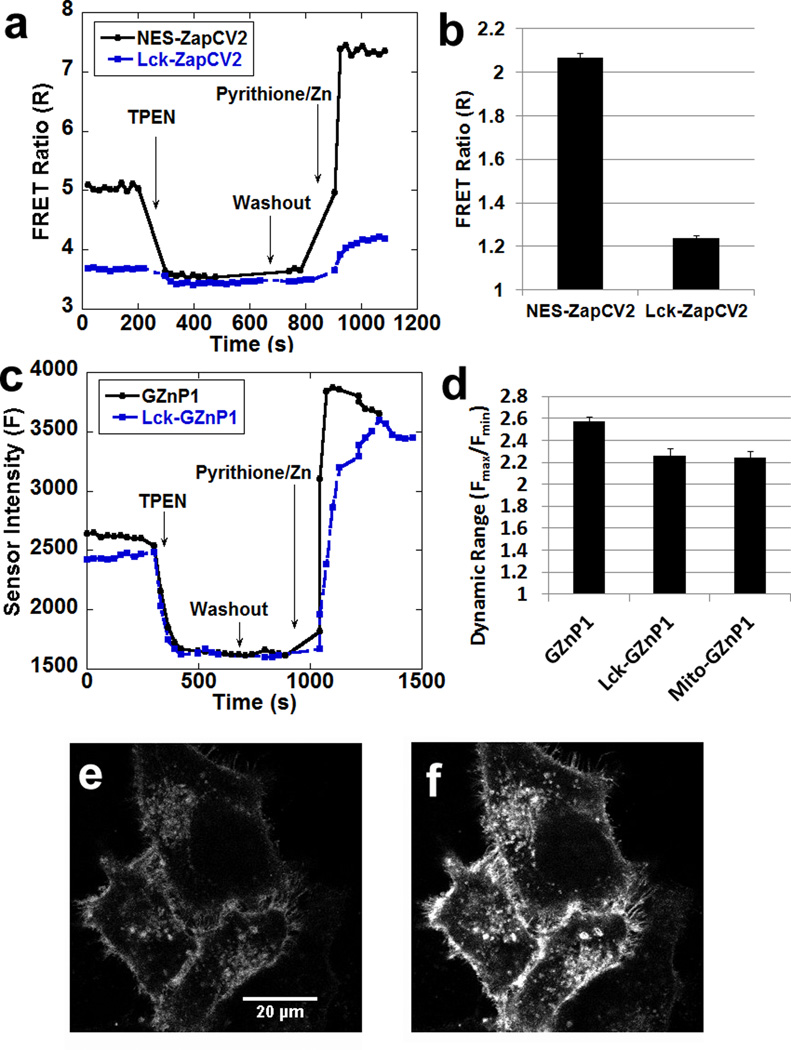
Although genetic targeting is one of the major advantages of genetically encoded sensors, it is well documented that FRET-based sensors often suffer from reduced dynamic range (Rmax/Rmin) when targeted to specific subcellular locations6, 9–10, 23. For example, the dynamic ranges of FRET sensors (ZapCY16, and eZinCh-210) were reduced when these sensors were targeted to mitochondria. Our previously reported FRET sensor ZapCY1 displayed 26% reduction in dynamic range from 4.3 in the cytosol to 3.2 in mitochondria6, while the recently reported eZinch-2 displayed 27–46% reduction in dynamic range from 2.6 in cytosol to 1.4–1.9 in mitochondria (estimated from published cellular calibration data)10. We created mito-GZnP1, which displayed only 15% reduction in dynamic range from 2.6 in cytosol to 2.2 in mitochondria (Fig. S7). In addition, we also compared the dynamic range of sensors attaching to the membrane. Due to restricted flexibility, we hypothesize that localizing FRET-sensors onto membrane may lower sensor dynamic range. Indeed, targeting our best cytosolic zinc sensor ZapCV2 to the plasma membrane by attaching the membrane targeting motif (Lck24) resulted in a 40% decrease in the dynamic range from 2 to 1.2 (Fig. 6a & b). On the other hand, targeting Lck-GZnP1 gave rise to a robust response with dynamic range of 2.2 (Fig. 6c & Fig. 6d). Thus, the Lck-GZnp1 sensor will be useful for detecting Zn2+ dynamics adjacent to plasma membrane.
Figure 6. GZnP1 retains properties more robustly than FRET sensor ZapCV2 when targeted to cellular compartments.
(a) Representative traces of cytosolic FRET Zn2+ sensor ZapCV2 and plasma membrane targeted sensor Lck-ZapCV2 in response to Zn2+ perturbation in cells. (b) The dynamic range of Lck-ZapCV2 is greatly reduced compared to ZapCV2. (c) Representative traces of cytosolic GZnP1 and plasma membrane targeting sensor Lck-GZnP1 in response to Zn2+ in cells. (d) Dynamic range of Lck-GZnP1 and mito-GZnP1 is only slightly reduced compared to GZnP1. n >= 14 cells, the error bar represents standard error of mean. (e–f) The fluorescence intensity was reduced in HeLa cells expressing Lck-GZnP1 treated with 100 µM TPEN (e), then the sensor brightness was increased with treatment of 1.25 µM pyrithione and 20 µM ZnCl2 (f).
Conclusions
We designed and characterized a new sensor platform distinct from prior FRET-based configurations. By inserting a pair of zinc finger domains into a single fluorescent protein (cpGFP), Zn2+-induced interaction between two zinc fingers can be coupled with the deprotonation of the chromophore of cpGFP. This design enables rapid detection of fluorescence intensity changes upon Zn2+ binding. To optimize sensor responses, we explored two different versions of cpGFP, investigated the effects of different linkers on sensor stability and dynamic range, tested mutations that were previously suggested to optimize the performance of single FP Ca2+ sensors (such as mutations to promote folding and maturation of the FP found in sfGFP), and used Rosetta to model and design mutations in the Zn2+ binding domain. A substantial bottleneck in the development and optimization of genetically encoded sensors is the lack of a general framework for exploring parameters that influence dynamic range and overall optical properties. In this work, we demonstrate that Rosetta-based computational design can be effectively used to screen the mutational space of the ligand binding domain, as Rosetta-identified mutations increased dynamic range from 1.4 to 2.6. Furthermore, we found that strategies used for structure-guided optimization of the GCaMP family sensors also led to improved properties of the GZnP platform. For example, the linkers used in GCaMP5 stabilized the fluorescence signal of Zn2+-saturated GZnP1 (Fig. 3), and shortening of linker regions to provide more protection against solvent access to the chromophore as suggested by the GCaMP crystal structures improved both the sensor dynamic range and brightness (Fig. 4f & Fig. 4g). The resulting GZnP sensor exhibited a robust dynamic range in the cytosol (Fmax/Fmin = 2.6), mitochondria (Fmax/Fmin = 2.2), and on the plasma membrane (Fmax/Fmin = 2.2). The in vitro biophysical characterization showed that the Zn2+ dependent fluorescence increase for GZnP1 was attributable to changes in both quantum yield and absorbance peak, while the single-FP Ca2+ sensor GCaMP sensors have identical quantum yield at both metal saturation and chelation state25, suggesting that the underlying photophysical process of GZnP1 might be different from the GCaMP sensors. The single-FP sensor platform possesses a number of potential advantages over FRET-based sensors. First, single-FP sensors can be used on a simpler imaging system compared to two-FP sensors (by measuring FRET) because the signal can be acquired at only a single wavelength. Second, development of single-FP Ca2+ sensors suggested that single-FP sensors have a larger dynamic range compared with FRET sensors using a similar recognition element3. Third, single-FP sensors can be easily used for simultaneous multi-analyte imaging with the single excitation/emission pair, thus avoiding bleed-through issues. On the other hand, single-FP sensors have a few limitations. As an intensity-based sensor, artifacts can arise from variations in sensor protein expression, light source and cell shape and thickness. Another challenge for using GZnP1 (or other single-FP sensors) is the sensitivity to pH changes, which could arise from 1) modulation of fluorescence intensity by pH (Fig. S6), or 2) modulation of Zn2+ binding to the ZF domains as a function of pH (Fig. 5d & Fig. S5). Thus, interpretation of GZnP1 signals should also consider if cellular pH is perturbed. In spite of these limitations, the single-FP sensor is still a robust platform for monitoring subcellular metal dynamics and expands the toolbox for monitoring labile Zn2+.
METHODS
GZnP Sensor construction
Plasmids were constructed by molecular cloning and verified by sequencing. Briefly, GZnP sensors were generated by inserting cpGFP (from GCaMP3 or GGECO1) between the first and second zinc fingers of Saccaromyces cerevisiae Zap1 (corresponding to residues 575 – 643). Site directed mutagenesis was used to introduce mutations to the designed position. Mitochondria and membrane sensors were cloned using a mitochondrial targeting motif8 and membrane targeting motif24 as previously described.
Rosetta-based computational protein design
The Rosetta protein modeling suite was used to carry out in silico saturation mutagenesis along the Zn2+ sensing domain11 (structure: 1zw8.pdb18) to identify mutations that were predicted to stabilize the ZF interface, stabilize each ZF domain individually, improve the solubility of the Zn2+-binding domain or alter side-chain packing around Zn2+-binding residues. Fixed backbone Monte Carlo simulations were used, where side-chain conformations were sampled from rotamer libraries (command-line: fixbb.macosgccrelease –resfile <input_resfile> –s 1zw8.pdb –ex1 –ex2 –ex3 –extrachi_cutoff 0 –database ~path-to-database –overwrite). In the sensor the two zinc fingers of the Zn2+-binding domain are separated and attached to different termini of the cpGFP, therefore for Rosetta modeling the Zn2+ sensing domain was separated into two Zn2+ finger folds, ZF1 and ZF2. Because the structure for the reconstituted Zn2+-binding domain could differ from the native binding domain characterized in 1zw8, in addition to the fixed backbone design we also tested the Rosetta relax protocol. In the Rosetta relax protocol the protein structure is refined by iterating between repacking side-chains and gradient-based minimization of torsion angles to relieve atomic clashes. The relaxed structure differed from 1zw8.pdb by 1.7 Å (Command Line: relax.macosgccrelease –database ~path-to-database –s 1zw8.pdb –use_input_sc –constrain_relax_to_start_coords –relax:fast). Fixed backbone design was then performed on the relaxed structure. The two different protocols resulted in identification of different mutations (Table S2 and S3). In order to optimize interactions at the interface between ZF1 and ZF2 we simultaneously optimized all residues at the interface. Positions 23 and 306 were found to be coupled in silico, both mutating together to T23L+L306S or T23W+L306F using fixbb and relax+fixbb respectively.
The amino acid positions immediately adjacent to the Zn2+-ligating residues were difficult to design using either protocol; Rosetta consistently predicted glycine as the lowest energy residue, even though mutation to glycine would abolish a number of potentially stabilizing contacts with the neighboring histidine residues. Evaluating the contributions from individual Rosetta energy terms revealed that selection of glycine at positions 18 and 33 resulted from decreases in atomic clashes, indicated by the low Lennard-Jones repulsive energy scores (LJrep), as well as decreases in the Dunbrack score, measuring the backbone-dependent probability of finding a given side-chain conformation. Both terms favor the selection of a glycine residue, and unfortunately overwhelm other energy terms for these two positions. To overcome these limitations, we used the Lennard-Jones repulsive term (LJrep) as a filter, removing any mutations which likely would clash with neighboring positions indicated by an LJrep score greater than 10. No mutations displayed a Dunbrack score exceeding the WT residue by more than 1 energy unit. We therefore did not generate a filter based on the Dunbrack score. Because positions 18 and 33 are immediately adjacent to the Zn2+-binding domain, we additionally did not consider residues known to bind metal ions (histidine, cysteine, aspartic acid, glutamic acid, asparagine). Other residues aligning with positions 18 and 33 in canonical Zn2+-binding domains are often polar or charged, thus substitutions with the lowest electrostatic energies were selected (Table S4 and S5).
PyMOL was used to evaluate the linker regions between the ZF1 and ZF2 domains and the GFP (Schrödinger Inc., Portland, OR). Distances between ZF1 or ZF2 and GFP were evaluated by adding residues to the N-terminus or C-terminus using the Residue building functions in PyMOL.
In Vitro Characterization of Kd’ and metal specificity
For in vitro studies, the sensor was cloned into pBAD and expressed in Top-10 Escherichia coli upon addition of 0.2% arabinose. Sensor protein was purified using Ni2+ ion affinity chromatography and the His tag was removed with TEV protease treated in 20 mM Tris, 100 mM NaCl, pH8 buffer at room temperature overnight (sensor to TEV protease ratio was 10:1). Zinc titrations were performed in HEPES buffer (150 mM Hepes, 100 mM NaCl, 0.5 mM TCEP and 10% glycerol, pH 7.4) with 1 µM sensor protein and different Zn2+ concentrations as described previously6. Purified sensor protein was titrated with Zn2+ to determine the fluorescence intensity as a function of Zn2+ concentration. The sensor responsiveness to Zn2+ is reported as an apparent dissociation constant (Kd’), which reflects the Zn2+ concentration midway between the Fmin and Fmax. Metal selectivity was measured as previously described6. The in vitro fluorescence measurements used for Zn2+ titrations and metal specificity studies were made on a Tecan Safire-II fluorescence plate reader using the following parameters: excitation: 488 nm, emission: 515 nm, and the emission bandwidth was 5 nm.
Biophysical characterization of GZnP1
All biophysical experiments were done in 30 mM MOPS, pH 7.4, 100 mM KCl, 1 mM TCEP. To compare the behavior in high vs. low Zn2+ conditions, the buffer was supplemented with 100 µM ZnCl2 or a Zn2+ chelator (100 µM TPA or 100 µM TPEN). The protein was equilibrated in each buffer for at least 20 minutes to establish a stable signal before recording the absorption spectrum using a Cary 500 UVVIS-NIR spectrophotometer. The quantum yield was determined using fluorescein as a reference26–27. The absorption at 488 nm was recorded for fluorescein in 0.1 M NaOH, for GZnP1 in 100 µM ZnCl2 and for GZnP1 in the presence of two different Zn2+ chelators (100 µM TPA or 100 µM TPEN) and dilution series were generated. The fluorescence spectrum for each sample was then recorded using a PTI-fluorimeter (excitation at 488 nm, emission 500 – 600 nm, 1 nm steps, 1 s integration time, 0.2 nm slits). After subtracting the buffer background signal, the fluorescence signal for each sample was integrated and plotted vs. the absorption at 488 nm. The steepness of the resulting linear plot for each dilution series reports on the quantum yield, and a quantum yield of 0.92 for fluorescein in 0.1 M NaOH was used as the reference.
Cellular Imaging
Sensor constructs were transiently transfected into HeLa cells and imaged 48 h post-transfection. Imaging experiments were performed on a Zeiss Axiovert 200M inverted fluorescence microscope configured as previously described6. Images were collected with a 40× 1.3NA oil objective, using a neutral density of 1.3, and 300 msec exposure time and data were collected using Metafluor software (Universal Imaging). Images of Lck-GZnP1 were taken using a Nikon A1R laser scanning confocal microscope with 60× objective. All images were background corrected by generating a region of interest (ROI) on a blank area of the coverslip and subtracting the fluorescence intensity in the GFP channel. Cells were imaged in phosphate-free HHBSS, pH = 7.4 to prevent precipitation of Zn2+. To carry out in situ calibrations, cells were treated with 100 µM cell permeable zinc chelator TPEN to obtain the fluorescence intensity at minimum Zn2+ (Fmin) and 1.25 µM pyrithione and 20 µM ZnCl2 to establish the sensor intensity at maximum Zn2+ (Fmax). For calibration in mitochondria, cells were treated with 100 µM TPEN to give the minimum signal and 5 µM pyrithione and 20 µM ZnCl2 to establish the maximum signal.
Statistical analysis
Statistical analysis was performed using the t-test in the KaleidaGraph program. The error bars used in column graph represent the standard error of the mean.
Supplementary Material
Acknowledgments
Funding Sources
We would like to acknowledge the following sources for general financial support: K99/R00 EB017289 (to YQ), NIH R01 GM084027 (to AEP). The Nikon A1R microscope was acquired through the generous support of NIST-CU Cooperative Agreement award number 70NANB15H226. This work was funded by the US Department of Energy's Bioenergy Technologies Office (DOE-BETO), Contract No. DE-AC36-08GO28308 with the National Renewable Energy Laboratory.
Footnotes
ASSOCIATED CONTENT
SUPPORTING INFORMATION
Supporting information includes Tables S1–S4, Figures S1 – S7. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no conflict of interest.
REFERENCES
- 1.Dean KM, Qin Y, Palmer AE. Visualizing metal ions in cells: An overview of analytical techniques, approaches, and probes. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Que EL, Domaille DW, Chang CJ. Metals in neurobiology: probing their chemistry and biology with molecular imaging. Chem Rev. 2008;108:1517–1549. doi: 10.1021/cr078203u. [DOI] [PubMed] [Google Scholar]
- 3.Akerboom J, Rivera JD, Guilbe MM, Malave EC, Hernandez HH, Tian L, Hires SA, Marvin JS, Looger LL, Schreiter ER. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J Biol Chem. 2009;284:6455–6464. doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin Y, Miranda JG, Stoddard CI, Dean KM, Galati DF, Palmer AE. Direct Comparison of a Genetically Encoded Sensor and Small Molecule Indicator: Implications for Quantification of Cytosolic Zn. ACS Chem Biol. 2013 doi: 10.1021/cb4003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinkenborg JL, Nicolson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat Methods. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin Y, Dittmer PJ, Park JG, Jansen KB, Palmer AE. Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc Natl Acad Sci U S A. 2011;108:7351–7356. doi: 10.1073/pnas.1015686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter KP, Young AM, Palmer AE. Fluorescent sensors for measuring metal ions in living systems. Chem Rev. 2014;114:4564–4601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JG, Qin Y, Galati DF, Palmer AE. New sensors for quantitative measurement of mitochondrial Zn(2+) ACS Chem Biol. 2012;7:1636–1640. doi: 10.1021/cb300171p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chabosseau P, Tuncay E, Meur G, Bellomo EA, Hessels A, Hughes S, Johnson PR, Bugliani M, Marchetti P, Turan B, Lyon AR, Merkx M, Rutter GA. Mitochondrial and ER-targeted eCALWY probes reveal high levels of free Zn2+ ACS Chem Biol. 2014;9:2111–2120. doi: 10.1021/cb5004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hessels AM, Chabosseau P, Bakker MH, Engelen W, Rutter GA, Taylor KM, Merkx M. eZinCh-2: A Versatile, Genetically Encoded FRET Sensor for Cytosolic and Intraorganelle Zn(2+) Imaging. ACS Chem Biol. 2015;10:2126–2134. doi: 10.1021/acschembio.5b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 13.Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Sierra AM, Ai HW, Campbell RE. Identification of sites within a monomeric red fluorescent protein that tolerate peptide insertion and testing of corresponding circular permutations. Photochem Photobiol. 2008;84:111–119. doi: 10.1111/j.1751-1097.2007.00206.x. [DOI] [PubMed] [Google Scholar]
- 15.Qiao W, Mooney M, Bird AJ, Winge DR, Eide DJ. Zinc binding to a regulatory zinc-sensing domain monitored in vivo by using FRET. Proc Natl Acad Sci U S A. 2006;103:8674–8679. doi: 10.1073/pnas.0600928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. An expanded palette of genetically encoded Ca(2)(+) indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, Gordus A, Orger MB, Portugues R, Engert F, Macklin JJ, Filosa A, Aggarwal A, Kerr RA, Takagi R, Kracun S, Shigetomi E, Khakh BS, Baier H, Lagnado L, Wang SS, Bargmann CI, Kimmel BE, Jayaraman V, Svoboda K, Kim DS, Schreiter ER, Looger LL. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Feng LS, Matskevich V, Venkataraman K, Parasuram P, Laity JH. Solution structure of a Zap1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae. J Mol Biol. 2006;357:1167–1183. doi: 10.1016/j.jmb.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Muto A, Ohkura M, Kotani T, Higashijima S, Nakai J, Kawakami K. Genetic visualization with an improved GCaMP calcium indicator reveals spatiotemporal activation of the spinal motor neurons in zebrafish. Proc Natl Acad Sci U S A. 2011;108:5425–5430. doi: 10.1073/pnas.1000887108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 21.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 22.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 23.Allen MD, Zhang J. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Commun. 2006;348:716–721. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- 24.Shigetomi E, Kracun S, Sofroniew MV, Khakh BS. A genetically targeted optical sensor to monitor calcium signals in astrocyte processes. Nat Neurosci. 2010;13:759–766. doi: 10.1038/nn.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akerboom J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, Tolo J, Gordus A, Orger MB, Severi KE, Macklin JJ, Patel R, Pulver SR, Wardill TJ, Fischer E, Schuler C, Chen TW, Sarkisyan KS, Marvin JS, Bargmann CI, Kim DS, Kugler S, Lagnado L, Hegemann P, Gottschalk A, Schreiter ER, Looger LL. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brouwer AM. standards for photoluminescence quantum yield measurements in solution. Pure Appl. Chem. 2011;83:6. [Google Scholar]
- 27.Shen JS. Richard Thermal lens measurement of absolute quantum yields using quenched fluorescent samples as references. Chemical Physics Letters. 1989;155:4. [Google Scholar]
- 28.Huang Z, Zhang XA, Bosch M, Smith SJ, Lippard SJ. Tris(2-pyridylmethyl)amine (TPA) as a membrane-permeable chelator for interception of biological mobile zinc. Metallomics. 2013;5:648–655. doi: 10.1039/c3mt00103b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.