Summary
Circadian clocks generate 24-hr rhythms in physiology and behavior. Despite numerous studies, it is still uncertain how circadian rhythms emerge from their molecular and neural constituents. Here, we demonstrate a tight connection between the molecular and neuronal circadian networks. Using fluorescent transcriptional reporters in a Drosophila ex vivo brain culture system, we identified a reciprocal negative regulation between the master circadian regulator CLK and expression of pdf, the main circadian neuropeptide. We show that PDF feedback is required for maintaining normal oscillation pattern in CLK-driven transcription. Interestingly, we found that CLK and neuronal firing suppresses pdf transcription, likely through a common pathway involving the transcription factors DHR38 and SR, establishing a direct link between electric activity and the circadian system. In sum, our work provides evidence for the existence of an uncharacterized CLK-PDF feedback loop that tightly wraps together the molecular oscillator with the circadian neuronal network in Drosophila.
Keywords: CLK, PDF, circadian, Drosophila, transcriptional fluorescent reporters, brain culture
Graphical Abstract
Highlights
-
•
Monitoring circadian transcription ex vivo using fluorescent reporters
-
•
CLK activation in the LNvs provokes downregulation in CLK activity in LNds and DNs
-
•
Reciprocal negative regulation of CLK activity and pdf transcription and signaling
-
•
PDF signaling is required for the normal oscillation pattern in CLK activity
Using fluorescence transcriptional reporters in an ex vivo brain culture, Mezan et al. describe the existence of a reciprocal negative regulation between CLK activity and pdf transcription and signaling. Hence, this work uncovers an uncharacterized regulatory loop in Drosophila pacemaker neurons that integrates essential intra and extracellular circadian factors.
Introduction
Behavior and physiology of most animals follow 24-hr (circadian) rhythms. These rhythms have a molecular basis and depend on self-sustaining transcriptional/post-translational feedback loops (TTFLs) (Darlington et al., 1998, Lee et al., 1998, Rosbash et al., 2007, Zheng and Sehgal, 2008). In Drosophila, CLK and CYC drive circadian oscillations by promoting rhythmic transcription of several key genes, including PER, TIM, and CWO, which repress CLK-CYC-mediated transcription (Allada and Chung, 2010). In addition to transcriptional control, post-transcriptional and post-translational regulatory processes play essential roles in circadian timekeeping (Kadener et al., 2009, Kim et al., 2002, Lerner et al., 2015, Lim and Allada, 2013, Sathyanarayanan et al., 2004, So and Rosbash, 1997, Yu et al., 2009).
The complexity of the circadian system extends beyond the single-cell level. In Drosophila, ∼150 brain neurons express clock gene products. These neurons are organized into a neuronal network. They are clustered in six major subgroups: small and large ventral-lateral neurons (s-LNvs, l-LNvs, and the fifth s-LNv), dorsal-lateral neurons (LNds), and three subgroups of dorsal neurons (DNs1–3). The neuropeptide PIGMENT DISPERSING FACTOR (PDF), the main neuromodulator of the circadian neuronal network, is expressed in the LNvs. PDF is essential for normal circadian activity patterns in light:dark cycles (LD) and for persistent circadian rhythms in constant darkness (DD) (Hyun et al., 2005, Lear et al., 2005, Mertens et al., 2005, Renn et al., 1999). It exerts a widespread effect on the network (Hyun et al., 2005, Im and Taghert, 2010, Shafer et al., 2008).
Drosophila molecular studies postulate that the circadian intracellular TTFL is the main timekeeper. This assumption implies that circadian cells keep time on a cell autonomous basis. This fits well with studies performed in mammals (Nagoshi et al., 2004, Welsh et al., 2004), as well as in Neurospora and cyanobacteria (Brunner and Káldi, 2008, Kitayama et al., 2008). In this context, the main function of the circadian neuronal network is readjusting individual circadian oscillators, hence facilitating resonance or coherence in the network (Abraham et al., 2010, Busza et al., 2007, Depetris-Chauvin et al., 2011, Peng et al., 2003, Tang et al., 2010, Weiss et al., 2014). However, several studies provided evidence for a role of neuronal connectivity in the timekeeping process per se, in flies (Peng et al., 2003, Weiss et al., 2014) (Nitabach et al., 2002, Nitabach et al., 2005 but also see Depetris-Chauvin et al., 2011) and mammals (Bernard et al., 2007, Takahashi et al., 2010).
Nevertheless, in Drosophila, the extent to which the molecular and neuronal circadian networks are intertwined is still not well understood. PDF has a central role in the timekeeping process, as it coordinates phase and amplitude of molecular oscillations of downstream neurons (Collins et al., 2014, Liang et al., 2016, Lin et al., 2004, Nitabach et al., 2006, Peng et al., 2003, Seluzicki et al., 2014, Wu et al., 2008). Moreover, PDF signaling impacts the TTFL, by promoting the stabilization of the proteins TIM and PER (Li et al., 2014, Seluzicki et al., 2014). However, the effect of this regulation on CLK-driven transcription is unclear, and PDF might be merely an output of the dominant pacemaker cells (Depetris-Chauvin et al., 2011, Fernández et al., 2007, Nitabach et al., 2005, Shafer and Yao, 2014). On the other hand, CLK has a key role in development of the pdf-expressing neurons (Allada et al., 2003, Lerner et al., 2015, Park et al., 2000), but nothing is known about the mechanism employed for CLK regulation over pdf expression. Thus, in this context, the interaction between the neuronal network and the molecular oscillator of individual neurons is far from being established.
In mammalian systems, those issues have been addressed using fluorescent reporters (Kuhlman et al., 2003, Nagoshi et al., 2004, Quintero et al., 2003). However, in Drosophila luciferase reporters are more commonly used (Roberts et al., 2015, Sehadova et al., 2009, Stanewsky et al., 2002). In this study, we developed and utilized fluorescent transcriptional reporters for tim and pdf in an ex vivo brain culture setup, which allows us to perturb and monitor circadian transcription with spatiotemporal precision. Using this approach, we found a reciprocal relationship between CLK activity and pdf transcription and signaling. Interestingly, we found that neuronal activity also modulates pdf transcription, likely utilizing a similar pathway as CLK, involving the transcription factors Drosophila hormone receptor-like 38 (DHR38) and stripe (SR). In sum, our results suggest the existence of a tight inter-cellular feedback loop, involving the transcription factor CLK and the neuropeptide PDF, that tightly wraps together the neuronal network and circadian molecular oscillators.
Results
Development of a Fluorescent Circadian Transcriptional Reporter
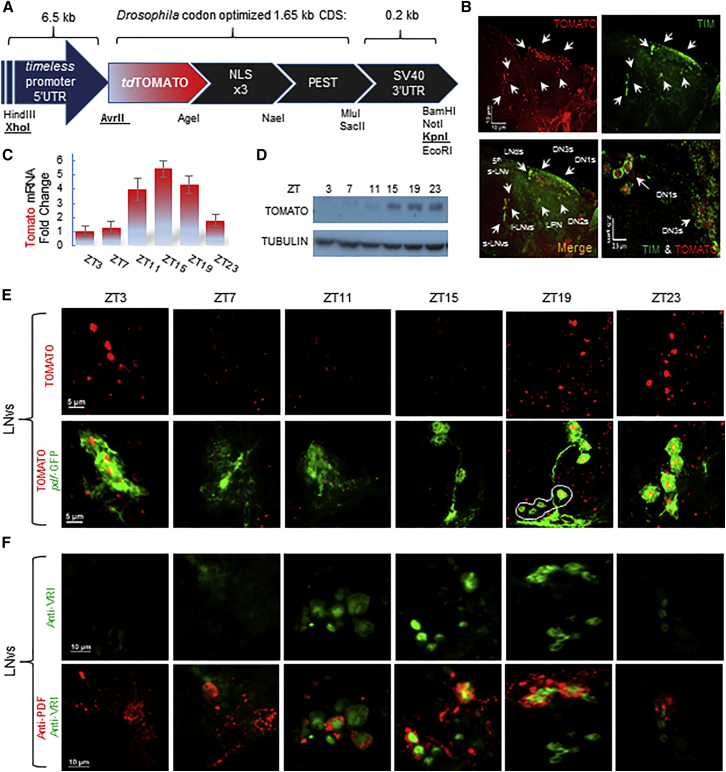
To follow CLK-CYC driven transcription in vivo, we generated a circadian fluorescent transcriptional reporter. It contains Drosophila codon-optimized td-Tomato fluorophore downstream to 6.4 kb of the timeless control region. We fused the td-Tomato to a PEST motif and a nuclear localization signal (NLS) to produce a short-lived, nuclear-localized signal (Figures 1A and S1A). As expected, the reporter is strongly induced by addition of CLK in a system lacking this transcription factor (Drosophila S2 cells; Figure S1B).
Figure 1.
A Fluorescent Circadian Reporter to Assess CLK-Driven Transcription
(A) Scheme of the timTomato transcriptional reporter. Restriction sites are indicated below the scheme. NLS, nuclear localization signal; PEST, mouse ornithine decarboxylase; SV40-3′-UTR, Simian virus 3′ UTR. See also Figure S1A.
(B) The timTOMATO reporter recapitulates TIM (green) spatial expression. Representative pictures from whole-mount immunohistochemistry of reporter brains stained with anti-TIM at ZT17. The nuclear TOMATO signal is surrounded by cytoplasmic TIM signal (bottom right).
(C) Daily oscillations in Tomato mRNA levels measured by qPCR. Error bars represent SD of three biological repeats.
(D) Western blot analysis showing the levels of TOMATO and TUBULIN in timTomato fly head across the day, performed in three biological repeats.
(E) TOMATO neuronal oscillations in the LNvs. Signal of endogenous TOMATO (red) and GFP (green) in brains of UAS-mcd8GFP; timTomato/pdfGAL4 flies that were entrained under 12:12 Light:Dark (LD) conditions, collected, and dissected at the indicated time point. See also Figure S2E.
(F) Oscillations of VRI in cultured brains. Dissected brains of wild-type (Canton S) flies were incubated in culture under 12:12-hr LD conditions for 5 days and then collected and stained for VRI (green) and PDF (red).
See also Figure S2I.
We then generated transgenic flies by random insertion of the timTomato reporter. Five of the lines displayed strong oscillations in TOMATO levels across the day (Figure S1C). We choose one line (#7) with moderate fluorescent intensity, low background, and high specificity for circadian neurons for further experiments (Figure S2A). Immunostaining with anti-TIM and anti-PDF antibodies shows that the reporter is specifically co-expressed with TIM in all circadian neuronal subgroups, including the LNvs (Figures 1B and S2B). The line displays mRNA and protein oscillations in fly head extracts with similar phase and amplitude to endogenous tim gene products (Figures 1C, 1D, S2C, and S2D), likely due to the long maturation time of the tdTomato fluorophore (1 hr at 37°C and probably longer at 25°C). The reporter also recapitulates timeless expression temporally, as we detected synchronized TOMATO oscillations that peak at ZT19 across the circadian neuronal network both in light:dark (LD) as well as in free running conditions (Figures 1E, S2E, and S2F).
To determine whether the timTomato transgene can be used to report acute changes in CLK activity in vivo, we utilized the CLKGR and ClkSV40 transgenes. The UAS-CLKGR transgene directs the expression of a fusion between CLK and the glucocorticoid receptor ligand-binding domain. This fusion protein acts as a dominant negative of CLK (Weiss et al., 2014), but addition of the artificial glucocorticoid analog dexamethasone (Dex) activates the fusion provoking a quick and large increases in CLK-dependent transcription (Kadener et al., 2007, McDonald and Rosbash, 2001, Weiss et al., 2014). We drove expression of CLKGR with the timGAL4 driver (timCLKGR-timTomato flies) and observed a strong upregulation in signal from the reporter, within the circadian cells, in brain of flies incubated with Dex (Figure S2G). Our timTomato reporter also displayed a strong increase in TOMATO in flies carrying a ClkSV40 transgene (Figure S2H). Overall, the timTomato reporter recapitulates timeless spatiotemporal expression.
An Ex Vivo System for Assaying Real-Time Dynamics of CLK Transcriptional Activity
Circadian rhythms are monitored from cultured adult Drosophila brains using luciferase reporters (Roberts et al., 2015, Sehadova et al., 2009, Sellix et al., 2010, Stanewsky et al., 2002). To further establish the use of the brain culture system for long-term studies of the circadian network, we dissected fly brains and cultured them in LD for 5 days. We visualized VRI and PDF levels by post-culture immunostaining and found that VRI oscillations persisted in all circadian neuronal groups with similar phase, peaking at ZT15 (Figures 1F and S2I).
We utilized this brain culture in combination with our reporter to follow CLK-driven transcription immediately after induction of CLK activity in all circadian cells (using timCLKGR-timTomato flies). As expected, addition of Dex strongly upregulates the reporter in all neuronal groups (Movies S1, S2, and S3). The response starts within less than 24 hr, reaching a plateau after approximately 48–60 hr (Figure S3). Our results demonstrate that dynamic changes in CLK-mediated transcription can be manipulated and simultaneously monitored ex vivo using our reporter across the circadian neuronal network.
CLK Activity in the LNvs Determines the Levels of CLK-Driven Transcription across the Circadian Neuronal Network
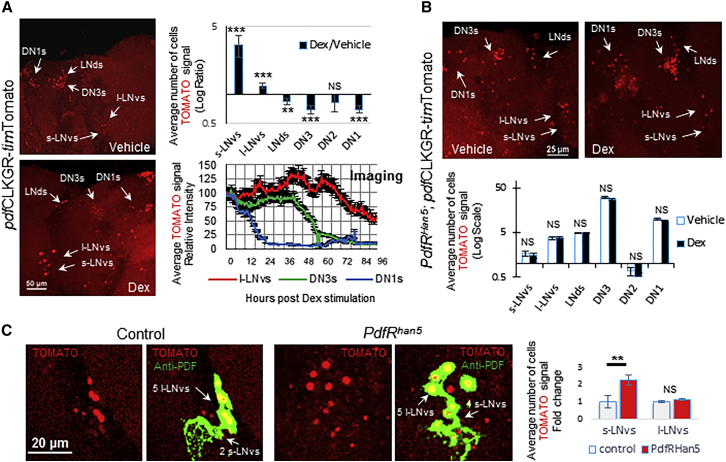
We followed by using our reporter in the brain culture system with the CLKGR transgene to determine the relationship between CLK-driven transcription in the LNvs and the rest of the circadian neuronal network. We generated flies expressing the UAS-CLKGR transgene under the control of the pdfGAL4 driver together with the reporter (pdfCLKR-timTomato flies). The number of timTomato positive cells was used as a proxy for signal intensity. As expected, expression of the dominant-negative protein CLKGR in the LNvs resulted in a low number of cells expressing detectable levels of the reporter at ZT19 (Figure 2A; Vehicle). We observed that a large proportion of the DNs and LNds still displayed strong TOMATO signal. Addition of Dex dramatically increased the number of cells expressing the timTomato reporter in the LNvs. Interestingly, the induction of CLK transcriptional activity in the LNvs suppressed reporter signal in the DNs and LNds (Figure 2A; Dex). The frequency plot of the number of cells expressing TOMATO illustrates this compensatory response (Figure S4A). The activation of CLKGR in the LNvs appeared to consistently inhibit tim transcription in the DNs and in some LNds.
Figure 2.
Activation of CLK-Driven Transcription in the LNvs Downregulates the timTOMATO Reporter in Dorsal Neurons
(A) CLKGR transcriptional activity is induced in the LNvs of PdfGAL4/timTomato;UAS-CLKGR (pdfCLKGR-timTomato) cultured brains by Dex. CLKGR activation decreases TOMATO signal in the DN1s, DN3s, and LNds at ZT19. Left: representative pictures of vehicle (top) and Dex (bottom)-treated brains. Right: quantifications of the response in different neuronal subgroups (top) and kinetic of the response monitored by time-lapse imaging (one frame/30 min) (bottom). NDex = 71, Nvehicle = 80 hemispheres. See also Figure S4A.
(B) Applying Dex to culture brains of PdfRhan5;pdfGAL4/timTomato;UAS-CLKGR flies (PdfRhan5-pdfCLKGR-timTomato) does not stimulate any response in the network at ZT19. Top: representative pictures of vehicle (left) and Dex (right)-treated brains. Bottom: quantification of the response. NDex = 51, Nvehicle = 44 hemispheres.
(C) TOMATO signal is elevated in cultured brains of PdfRhan5;timTomato;UAS-CLKGR (PdfRhan5) relative to timTomato;UAS-CLKGR brains (control). Left: representative pictures of reporter brains immunostained with anti-PDF antibody (green). A fifth l-LNvs nucleus that is PDF negative is also visible. Right: quantification of the response. n = 31, 20 hemispheres, respectively.
Statistical significance determined using two-tailed Student’s t test. NS, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Error bars represent SEM.
To determine the kinetic of this response, we monitored, by live imaging, TOMATO signal from brains of pdfCLKGR-timTomato flies immediately after addition of Dex to the culture (Movie S4; Figures 2A, bottom right, and S4B). We observed a quick repression in TOMATO signal in the DN1s, shortly after addition of Dex. A quick response in this cellular group is consistent with the physical interaction between s-LNvs and DN1s (Seluzicki et al., 2014). The DN3s were also strongly but slowly affected suggesting that the effect is indirect. TOMATO signal in the l-LNvs remained high throughout this time course.
To determine whether this response depends on PDF signaling, we performed the same culture assay utilizing pdfCLKGR-timTomato flies that also carry a mutation in the PDF receptor (PdfRhan5; Hyun et al., 2005). Indeed, the presence of the PdfRhan5 mutation eliminated the difference in TOMATO signal between vehicle-treated and Dex-treated brains in all neuronal subgroups (Figure 2B), demonstrating that this response depends on PDF signaling.
PDF Signaling Regulates Transcription Oscillations in the s-LNvs
Surprisingly, in PdfRhan5;pdfCLKGR-timTomato flies, we did not detect any difference in TOMATO signal between vehicle-treated and Dex-treated brains even in the s-LNvs and l-LNvs (Figure 2B, bottom). This strongly suggests that PDF signaling is required for the cell-autonomous increase in CLKGR-driven transcription induced by addition of Dex. In addition, we observed that introduction of the Pdf receptor mutation lead to a 4-fold increase in TOMATO signal in the s-LNvs of cultured brains (Figure S4C). Importantly, the increase does not depend on the expression of CLKGR (Figure 2C), showing that PDF signaling regulates CLK-mediated transcription, at least in cultured brains. These results indicate that PDF signaling suppresses CLK-transcriptional activity in the main pacemaker neurons.
CLK Negatively Regulates pdf Transcription Post-development
We next decided to investigate whether CLK activity regulates pdf expression. Indeed, at the same culturing conditions described above, treatment of pdfCLKGR brains with Dex resulted in a significant elevation in PDF levels as indicated by quantification of PDF signal intensity in the LNv cell bodies and the number of PDF positive cells (Figure S5A, top right and bottom right).
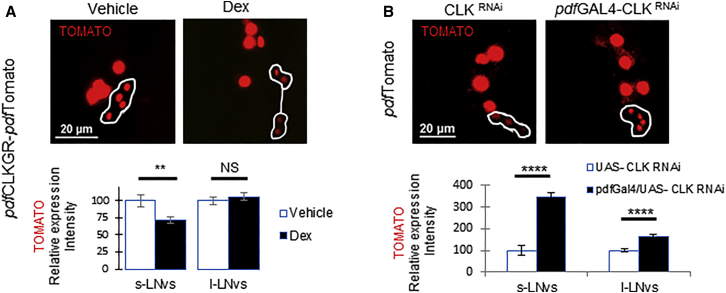
To assess whether CLK regulates pdf at the transcriptional level, we generated transgenic flies carrying a pdfTomato transcriptional reporter, which we built using the same fluorescent protein under the control of a 2.4-kb genomic region from the pdf gene (Figure S5C; Park et al., 2000). The reporter is highly specific, thus making identification of pdf-expressing cells straightforward (Figure S5C, right). We generated pdfCLKGR flies that carry this reporter (pdfCLKGR-pdfTomato). Despite the above-mentioned positive effect on PDF levels, activation of CLKGR by addition of Dex decreased pdf transcription specifically in the s-LNvs, as indicated by the intensity of TOMATO signal (Figure 3A). This result suggests that CLK activity in the LNvs inhibits pdf transcription. To confirm this post-developmental effect of CLK on pdf transcription in vivo, we knocked down Clk in the LNvs of pdfTomato brains using the pdfGAL4 driver (pdfGAL4-pdfTomato-CLKRNAi). In agreement with the result described above, downregulation of Clk in developed pdf-expressing cells resulted in more than 3-fold upregulation in TOMATO intensity in the s-LNvs and with a mild increase in PDF levels (Figures 3B and S5B). Overall, we conclude that CLK suppresses pdf transcription post-development.
Figure 3.
CLK Represses pdf Transcription
(A) CLKGR activation by Dex in cultured brains downregulates expression from the pdfTomato (red) at ZT5. PdfGAL4; pdfTomato/UAS-CLKGR (pdfCLKGR-pdfTomato). Top: representative pictures of vehicle (left) and Dex (right)-treated brains. Bottom: quantification of the response. NDex = 38, Nvehicle = 41 hemispheres. See also Figure S5C.
(B) Expression of Clk RNAi increases pdf transcription (red) at ZT5. Top: representative pictures of pdfTomato/UAS-CLKRNAi brains indicated as CLKRNAi (left) and pdfGAL4;pdfTomato/UAS-CLKRNAi flies indicated as pdfGAL4-CLKRNAi (right). Bottom: quantification of TOMATO intensity in the LNvs. n = 24, 25 hemispheres respectively.
Statistical significance was determined using two-tailed Student’s t test, NS, not significant ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Error bars represent SEM.
DHR38 and SR Links CLK Activity to pdf Transcription
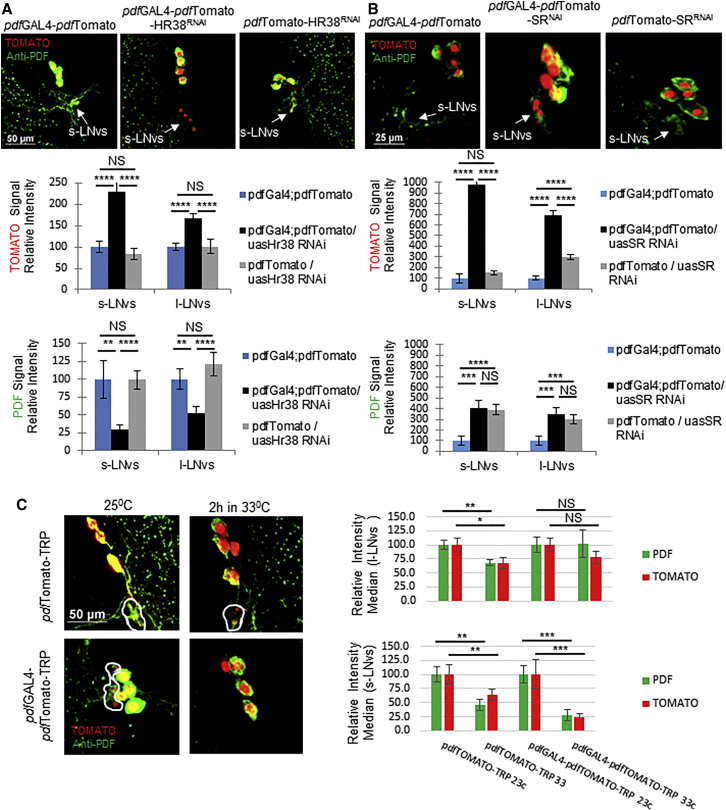
To determine the mechanism by which CLK regulates pdf expression, we identified putative regulators of pdf transcription using a yeast one-hybrid (Y1H) screen. For doing so, we generated five partially overlapping fragments from the 2.5-kb pdf promoter (Figure S6) and utilized a previously described library of 650 Drosophila transcription factors (Hens et al., 2011). This assay identified 27 putative regulators of pdf (Table 1). We then focused on candidates that are strongly enriched in the s-LNvs and known to be activated by the Mef2 and CLK transcription factors (Abruzzi et al., 2011, Kula-Eversole et al., 2010, Nagoshi et al., 2010, Sivachenko et al., 2013). Mef2 has been shown to mediate between CLK and the plasticity of the terminals in the pdf-expressing cells. We found two such candidates DHR38 and SR (Figure S6). We assessed the effect of these factors on pdf transcription by expressing RNAi transgenes in the pdf-expressing cells of flies which also carry the pdfTomato reporter (pdfGAL4-pdfTomato-RNAi). Interestingly, downregulation of dhr38 and sr led to a strong upregulation of pdf transcription (Figures 4A and 4B), suggesting that both transcription factors inhibit pdf transcription. Consistently with this result, downregulation of sr lead to an increase in PDF levels (Figures 4B and S7C). However, downregulation of dhr38 either in the pdf or tim-expressing cells reduced PDF levels, suggesting that this transcription factor regulates PDF expression at different levels (Figures 4A, S7A, and S7B).
Table 1.
Putative Regulators of pdf Identified by Yeast One-Hybrid Assay
| Bait Position Relative to TSS | Interacting Gene Identified | |
|---|---|---|
| 1 | −2447 to −1756 | CG17612 |
| 2 | CG4575 | |
| 3 | −1967 to −1368 | Abd-B |
| 4 | ecd | |
| 5 | CG31835 | |
| 6 | CG9571 | |
| 7 | CG14117 | |
| 8 | CrebA | |
| 9 | −1526 to −923 | ttk |
| 10 | drm | |
| 11 | crol | |
| 12 | sna | |
| 13 | CG4282 | |
| 14 | −1130 to 535 | drm |
| 15 | rib | |
| 16 | sd | |
| 17 | Snoo | |
| 18 | −690 to −1 | ush |
| 19 | opaa | |
| 20 | sra | |
| 21 | D1 | |
| 22 | woc | |
| 23 | Hr38a | |
| 24 | Jra ¦ kay | |
| 25 | Rfx | |
| 26 | CG18446 | |
| 27 | sqza |
27 genes identified as putative regulators of pdf. Bait position relative to the pdf promoter transcription start site (TSS) is indicated for each gene.
Gene that was previously identified as direct CLK targets and enriched in the LNvs. Interestingly, all four bind pdf most proximal bait fragment (see also Figure S6).
Figure 4.
pdf Transcription Is Regulated by Electrical Activity, Likely through Dhr38 and sr
Flies carrying the pdfGAL4;pdfTomato transgenes were crossed with flies carrying the UAS-hr38RNAi (A) UAS-srRNAi (B) and UAS-TrpA1 (C). Brains were dissected at ZT5 and immunolabeled with anti-PDF antibody before visualization of TOMATO (red) and PDF (green).
(A and B) Top: representative pictures of the indicated genotypes from left to right: pdfGAL4;pdfTomato, pdfGAL4;pdfTomato/UAS-RNAi, pdfTomato/UAS-RNAi. Bottom: quantification of TOMATO (upper) and PDF (lower) signal intensities in the cell body. For (A), n = 32, 33, 36 hemispheres, respectively. For (B), n = 30, 41, 37 hemispheres respectively.
(C) Left: representative pictures of the indicated genotypes: control pdfTomato/UAS-TRPA1 brains (upper) and pdfGAL4;pdfTomato/UAS-TRPA1 brains (lower), incubated for 2 hr at 25°C (left) or 33°C (right). Right: quantification of TOMATO (red) and PDF (green) signal intensities in the cell body of l-LNvs (upper) and s-LNs (lower). n = 35, 44, 53, 42 hemispheres, respectively.
See Figure S7D. Statistical significance was determined using two-tailed Student’s t test, NS, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Error bars represent SEM.
Neuronal Activity Modulates pdf Expression
Interestingly, dhr38 and sr were also identified as activity regulated genes in Drosophila (Fujita et al., 2013; X. Chen and M. Rosbash, personal communication) suggesting neuronal firing could regulate pdf levels. Thus, we determined whether pdf transcription is regulated by neuronal firing by using the heat-activated cation channel dTrpA1 (Rosenzweig et al., 2005) in the pdf-expressing cells of flies that also carry the pdfTomato reporter (pdfGAL4-pdfTomato-TRP). DHR38 protein levels have been shown to reach a maximal expression within 2 hr of stimulation of the dTrpA1 channel (Fujita et al., 2013). Thus, we determined the changes in TOMATO expression and PDF immunoreactivity in the LNvs of flies that were entrained at 25°C and stimulated at 33°C for 2 hr (Figure 4C). We observed that both experimental and control groups respond to the heat stimulation by downregulating TOMATO and PDF levels (Figure 4C), plausibly due to the partial overlap in expression of the endogenous dTrpA1 channel—RNA and protein—with the PDF-expressing cells and the s-LNvs specifically (Das et al., 2016, Kula-Eversole et al., 2010). Nevertheless, the decrease in TOMATO and PDF signal intensity, relative to the basal levels at 25°C, was significantly stronger in flies overexpressing the channel in the s-LNvs (Figure 4C), and the expression of the TRP channel strongly contributed to downregulate pdf expression in the s-LNvs at 33°C (Figure S7D). We conclude that neuronal activity of the LNvs suppresses pdf expression in the s-LNv. Our previous results strongly suggest that this effect is mediated by DHR38 and SR.
PDF Signaling Positively Modulates pdf Expression
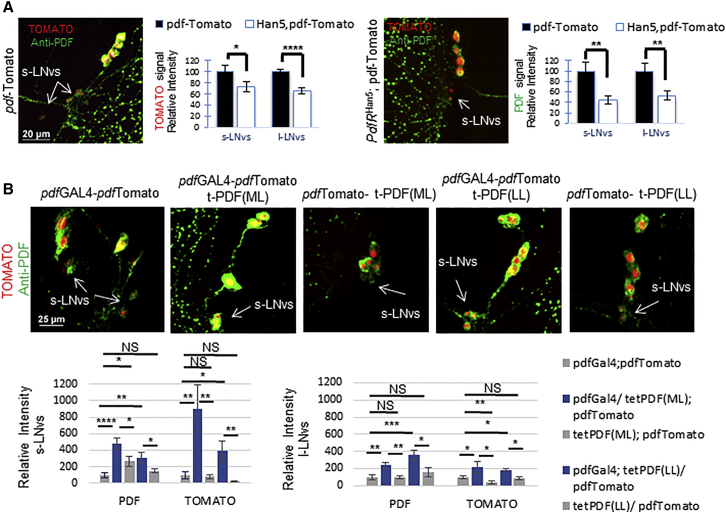
Our results indicate a strong and mutual regulation between Clk and pdf. This predicts that PDF should regulate pdf transcription, at least indirectly. To test this, we utilize the pdfTomato reporter to evaluated pdf expression in PdfRhan5 mutant flies. We found that both pdf transcription and PDF immunostaining were strongly downregulated in PdfRhan5 flies (Figure 5A, left and right, accordingly), suggesting that PDF signaling is necessary for continuous pdf expression. To determine whether continuous activation of PDF signaling will increase pdf transcription, we utilized the tethered-PDF (t-PDF) technology (Choi et al., 2009, Choi et al., 2012). Indeed, expression of t-PDF transgene in the pdf-expressing cells leads to a significant increase in pdf transcription, compared to controls, as assessed by the pdfTomato reporter (Figure 5B). Together, the results presented in this section demonstrate that PDF signaling positively modulate pdf transcription.
Figure 5.
PDF Signaling Positively Regulates pdf Expression
(A) PdfRHan5 flies exhibit reduced levels of pdf transcription and PDF neuropeptide. Representative pictures of a fly brain carrying the pdfTomato transgene (left) or with mutation in PDFR (PdfRhan5; pdfTomato) (right). Reporter brains were dissected at ZT5, immunolabeled for PDF, and visualized for TOMATO (red) and PDF (green). Quantification of TOMATO (left) and PDF (right) intensities in the LNvs cell body of the whole sampled population are shown (n = 30, 38 hemispheres respectively).
(B) Flies expressing the tethered-PDF transgenes under the control the pdfGAL4 driver exhibit elevated levels of pdf transcription. PdfGAL4;pdf-Tomato flies were crossed with UAS-tethered-PDF flies carrying short (ML) or long poly-linker (LL). Brains were dissected at ZT5, immunolabeled with anti-PDF antibody, and visualized for TOMATO (red) and PDF (green). Shown (top) are representative pictures of fly brain carrying the following genotypes (left to right): pdfGAL4;pdfTomato, pdfGAL4/UAS-tet-PDF(ML);pdfTomato, UAS-tet-PDF(ML);pdfTomato, pdfGAL4;pdfTomato/UAS-tet-PDF(LL), pdfTomato/UAS-tet-PDF(LL). Bottom: quantification of TOMATO and PDF in the s-LNvs (left) and l-LNvs (right). n = 23, 28, 24, 25, 20 hemispheres, respectively.
Statistical significance was determined using one-tailed Student’s t test. NS, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Error bars represent SEM.
Discussion
Here, we utilized an ex vivo brain culture system combined with recently developed fluorescent transcriptional reporters to analyze interactions between the neuronal network and the molecular oscillator in the Drosophila circadian system. We found that activation of CLK in the LNvs inhibits CLK activity in the other circadian groups. We showed that CLK and PDF regulate each other and that neural activity regulates pdf transcription, probably through a common pathway involving the direct CLK targets Hr38 and sr. We also found that PDF signaling is required for pdf transcription, adding additional complexity to this cycle. Together, this study identified a tight connection between the core molecular circadian pacemaker, neuronal activity, and PDF.
We present an important technical innovation: the use of fluorescent reporters in dissected brains in culture to study circadian regulation. Our reporter system can be used to evaluate pdf and CLK-dependent transcription at single-cell resolution. This type of system has not been used in Drosophila. Using our tim reporter, we were able to visualize and follow dynamics in CLK activity across the circadian network. Our VRI staining demonstrated that long-term ex vivo brain culture supports coherent molecular oscillations in all neuronal groups. As reported (Ayaz et al., 2008), we also observed some degree of abnormality in PDF staining of these cultures.
Our data revealed that PDF signaling negatively regulates CLK activity in the s-LNvs (Figures 2C and S4C). The timTomato reporter is the best tool available to specifically evaluate CLK-driven transcription in the s-LNvs. As, TIM and PER levels are under strong post-transcriptional control, which can also be regulated by PDF signaling (Li et al., 2014, Seluzicki et al., 2014). The latter is analogous to the one described for Vasoactive Intestinal Polypeptide (VIP) and PER1 in the mammalian suprachiasmatic nucleus (SCN) (Maywood et al., 2006). The effect of PDF signaling on CLK-driven transcription in the s-LNvs can likely be extended to the other circadian cells (Figures 2A and 2B). However, further studies are needed to determine the mechanism of this compensatory effect. This is mainly due to the opposite effects that CLK-driven transcription has on pdf transcription and overall PDF levels.
The lack of ability to activate the CLKGR transgene (Figure 2B) could be due to several reasons, including: (1) the tim promoter is already active up to its maximal capacity; (2) CLKGR cannot be activated by the addition of Dex in the PdfRHan5 mutants; and (3) a combination of both options. It is also possible that PDF signaling somehow interferes with Dex induction of CLKGR, although we did not find experimental support for this possibility. Since we show that in the absence of PDF signaling there is a 4-fold increase in signal originated from the timTomato reporter in s-LNvs (Figure S4C) and since similar increase was observed independently of CLKGR expression (Figure 2C), we can conclude that in the absence of PDF signaling endogenous CLK activity is higher. We speculate that the observed increase in endogenous CLK activity overrides the ability to activate CLKGR in the PdfRHan5 flies.
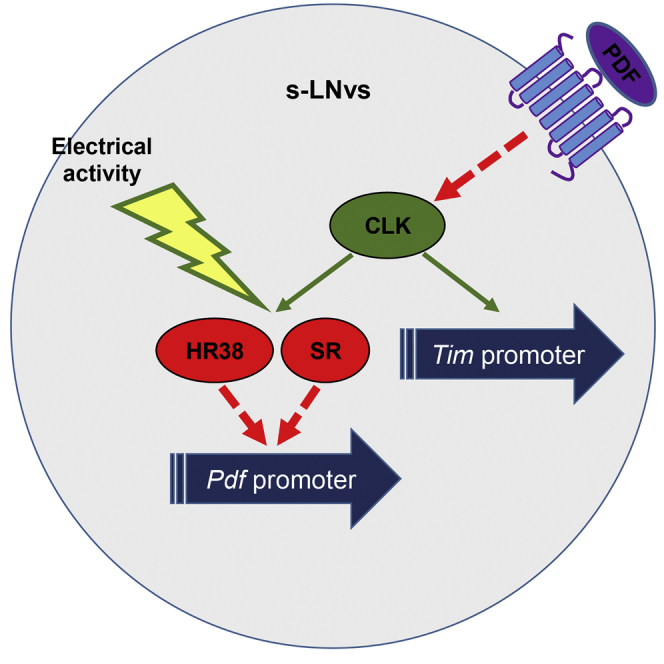
Previous studies support the notion that CLK positively regulate the development of the pdf cells (Allada et al., 2003, Lerner et al., 2015, Park et al., 2000, Zhao et al., 2003). However, those studies rely on the broad expression of CLK protein. Here, we show that specific induction of CLK activity in the adult main pacemaker cells, as well as silencing CLK post-development of the pdf cells using the pdfGAL4 driver, result in repression of pdf transcription (Figure 6, model). This suggests opposite effects of CLK on pdf transcription either pre and post-development of the LNvs or within and outside the LNvs. As PDF signaling positively regulates pdf expression, the continuous expression of pdf by the LNvs is a consequence of a balance between CLK and PDF activities. Neuronal electrical activity might have a crucial role in this balance, as it can suppresses pdf transcription in the main pacemaker cells through Hr38 and sr. The effect of CLK on pdf seems to also involve those factors that are direct CLK and MEF2 transcriptional targets (Abruzzi et al., 2011, Sivachenko et al., 2013) and are in the LNvs (Kula-Eversole et al., 2010) and represses pdf transcription (Figure 6, model: red inhibitory arrow).
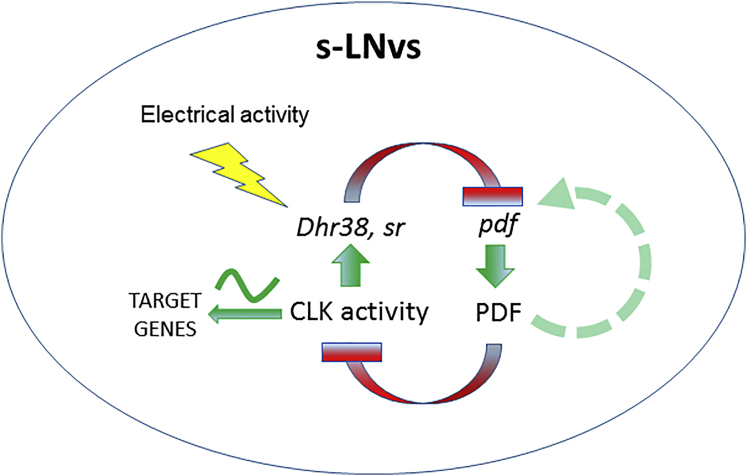
Figure 6.
A Model for the Integration of PDF in the Molecular Clockwork
CLK transcriptional activity and neuronal activity indirectly regulates pdf levels (red inhibitory arrow) likely through a common pathway involving hr38 and sr (green arrow). PDF feedbacks into the s-LNvs to resist its own inhibition by suppressing CLK activity (red inhibitory arrow) and inducing its own transcription (dotted green arrow). The model predicts that post-translational control over PDF expression/processing/secretion regulates pdf transcription.
Although CLK, HR38, SR, and neuronal activity downregulate pdf transcription, their effects on PDF neuropeptide levels are intriguing. Knockdown of hr38 (Figure 4A) and activation of CLKGR (Figures 3A and S5A) yielded opposite effects on the pdfTomato reporter and PDF levels (up and down or down and up, respectively). This suggests that hr38 is involved in the regulation of pdf by CLK but also that there might be a more complex post-translational effects on PDF. However, neural stimulation downregulates both pdf transcription and protein levels, reinforcing the idea of additional regulatory factors and possibly post-translational control over PDF expression/processing/secretion. Hence, our model predicts that both neuronal firing and CLK activity are indirectly regulating pdf transcription, not only through promoting activities of targets genes such as dhr38 and sr, but also through the post-translational control over PDF (Sivachenko et al., 2013) since, as we show here, PDF feeds back to regulate pdf transcription (model, green dotted arrow). This positive auto-regulation introduces complexity to the CLK-PDF feedback loop, and it is in agreement with a previously proposed model for PDF auto-receptor signaling in the LNvs (Choi et al., 2012). This intriguing feature of the loop should be addressed in future studies in order to extend our understanding of how post-translational mechanisms for PDF regulation affect its own transcription. The regulation of PDF signaling on pdf transcription can be also predicted from the strong and mutual regulation between Clk and pdf; however, we favor a more direct effect of PDF signaling on pdf transcription (model, green dotted arrow).
We have identified other putative regulators of pdf (Table 1). Their influence on pdf transcription remains to be determined. Interestingly, the transcription factor TTK is expressed in all TIM positive circadian neurons except the LNvs (Nagoshi et al., 2010), suggesting it might be an important endogenous regulator of pdf spatial expression.
PDF signaling was shown to be required for normal pattern of oscillations in CLK-driven transcription under constant darkness (Peng et al., 2003). Based on the upregulation of timTomato in PDFR mutant, we postulate that in the pacemaker cells PDF feedback is required for normal pattern of oscillations in CLK-driven transcription under light:dark cycles (Figure 6, model: inhibitory red arrow). Since ZT19 is the peak of TOMATO expression in the s-LNvs (Figure 1E, white circle), upregulation at this ZT cannot be a consequence of a phase shift; thus, it must reflect an increase in the amplitude of TOMATO oscillation. This demonstrate that in the absence of PDF signaling, the LNvs lose control of their own circadian molecular clockwork (Collins et al., 2014, Peng et al., 2003), likely because it is overridden by signals from the LNds and/or the DNs (Weiss et al., 2014, Yao and Shafer, 2014).
In sum, our findings challenge the notion that PDF is a merely output of the circadian system. Indeed, we demonstrate the existence of an uncharacterized and essential CLK-PDF-CLK regulatory loop in the LNvs that integrates PDF signaling into the single-cell molecular oscillator. The question of whether PDF feeds back to the same cells that secrete it within the subgroup of s-LNvs or whether its role as a communication agent is also applied within this subgroup remains to be answered in order to determine the final impact of this loop on the concept of cellular autonomy in the fly brain.
Experimental Procedures
Cloning of Reporter Constructs and DNA Baits of the pdf Promoter
To generate the timTomato reporter construct, a Drosophila codon optimized TdTomato-NLSx3-PEST coding sequence was fused with a SV40 3′ UTR in a pCaSpeR4 vector downstream to a 6.4-kb DNA fragment containing the promoter and 5′ UTR stretching into to the second exon ATG of the timeless gene. To generate the pdfTomato reporter, we utilized a similar approach but using a previously a 2.45-kb fragment containing the genomic region upstream to the pdf gene transcription start site inserted in a pattB based vector. To generate DNA baits of the pdf promoter for Drosophila transcription factor screen (Y1H), we used a carrier vector containing the 2.45-kb promoter of the pdf gene as a template to generate five overlapping PCR fragments approximately 600 bp long (see the Supplemental Experimental Procedures).
Fly Strains
The reporter transgenic fly lines were generated by BestGene using the P-element-mediated germline transformation for tim reporter and PhiC31 integrase method into the pattP2 site for pdf reporter.
TimGAL4, pdfGAL4,pdfRHan5, UAS-CLKGR; UAS-Dcr2, Hr38RNAi, sr RNAi, Clk RNAi,UAS-t-PDF, UAS-TrpA1, and ClkSV40 fly lines were previously described (Choi et al., 2009, Choi et al., 2012, Dietzl et al., 2007, Hyun et al., 2005, Kaneko and Hall, 2000, Lerner et al., 2015, Perkins et al., 2015, Renn et al., 1999, Sivachenko et al., 2013, Weiss et al., 2014).
Drosophila Adult Brain Culture
Flies were dissected (leaving trachea and imaginal discs) in 1 × PBS on a clean surface at room temperature. Each brain was immediately placed in a separate well of a 96-well plate containing 150 μl Drosophila Shield and Sang M3 insect medium (10% FBS, insulin [10 μg/mL], and a 1:100 dilution of antibiotic solution). For activation of CLKGR in culture, the medium was prepared with a supplement of 10 mg/mL dexamethasone (D4902; Sigma), kept in 100% ethanol, and diluted 1:500 (Dex) or with a 1:500 dilution of 100% EtOH only (vehicle). Plates were incubated at 25°C for 96 hr under LD conditions.
Post-culture Procedure
Brains were taken out from the well, cleaned in ice-cold 1 × PBS, and then fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature. Brains were then washed with ice-cold 1 × PBS before downstream application (visualization or immunolabeling) (see the Supplemental Experimental Procedures).
Live Imaging
Dissected brains were completely cleaned from trachea to avoid floating of the sample. Brains were then placed on a 12-μm cell-culture insert (Millipore) inside a 35-mm culturing dish containing 800 μl of culture medium as previously described (Ayaz et al., 2008) and entrained for 24 hr at 25°C in LD. Medium was supplemented with Dex or vehicle before time-lapse imaging was performed (one frame/30 or 40 min).
Confocal Microscopy
NIKON eclipse Ti confocal microscope was used to perform time-lapse imaging (Olympus ×20 super Plan Flour, 0.5 numerical aperture [NA], long distance 8,200 mm, air lens) and visualization (Olympus ×40 Plan Fluor 1.3 NA, 240 mm, ×60 Plan Apo 0.9 NA, 150 mm, oil lenses) of fluorescence.
Images Analysis and Statistics
For relative intensity quantification, background was manually determined and subtracted. Both the sum and the average signal intensity of an auto or manually detected ROIs were calculated. Signal was summed across all confocal planes surrounding the neuronal cell bodies belong to a certain sub-neuronal group in the brain. The average signal of the whole population of samples was then calculated, and statistical significance was determined. For cell-number-based quantification, microscope settings were set as permissive for weak signal (high excitation energy and low detection threshold). The number of cells in a certain neuronal group was determined based on the existence of a positive (weak or strong) TOMATO or PDF signal. TOMATO signal in the s-LNvs of reporter brains was quantified from brains over-stained for PDF and by evaluation of anatomical location. Significance was than determined using a two-tailed Student’s t test.
Transfection in S2 Cells
S2 cells were maintained using standard procedures and transfected using 1.5 μl Trans-IT insect transfection reagent (Mirus). The timTomato and timYFP (Lerner et al., 2015) were transfected in equal molarity (400 and 200 ng, respectively) with 100 ng pActin-CLKV5 or pMT-ClkSV40. DNA was adjusted to 0.5 μg with pBluescript. 12 hr post-transfection, Cu+2 was added to a final concentration of 1 mM. 48 hr post-induction, cells were washed and imaged.
Yeast One Hybrid
The yeast one-hybrid screens were conducted as described in Hens et al. (2011) and further detailed in Hens et al. (2012).
Author Contributions
S.M. and S.K. conceived the experiments and wrote the manuscript. S.K. supervised the work. S.M. designed and performed the experiments and analyzed the data. J.D.F. and B.D. performed the Y1H screen.
Acknowledgments
We thank Paul Hardin, Justin Blau, Michael Nitabach, and Michael Rosbash for reagents and Manny Benish for supporting confocal microscopy. S.K. is funded by the European Research Council Starting Grant (ERC no. 260911) and by the Israeli Science Foundation Personal Grant (ISF no. 840/2014). B.D. is funded by EPFL and AgingX (SystemsX.ch).
Published: October 11, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.09.048.
Supplemental Information
References
- Abraham U., Granada A.E., Westermark P.O., Heine M., Kramer A., Herzel H. Coupling governs entrainment range of circadian clocks. Mol. Syst. Biol. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abruzzi K.C., Rodriguez J., Menet J.S., Desrochers J., Zadina A., Luo W., Tkachev S., Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R., Chung B.Y. Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R., Kadener S., Nandakumar N., Rosbash M. A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude. EMBO J. 2003;22:3367–3375. doi: 10.1093/emboj/cdg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz D., Leyssen M., Koch M., Yan J., Srahna M., Sheeba V., Fogle K.J., Holmes T.C., Hassan B.A. Axonal injury and regeneration in the adult brain of Drosophila. J. Neurosci. 2008;28:6010–6021. doi: 10.1523/JNEUROSCI.0101-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard S., Gonze D., Čajavec B., Herzel H., Kramer A. Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus. PLoS Comput. Biol. 2007;3:e68. doi: 10.1371/journal.pcbi.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner M., Káldi K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol. Microbiol. 2008;68:255–262. doi: 10.1111/j.1365-2958.2008.06148.x. [DOI] [PubMed] [Google Scholar]
- Busza A., Murad A., Emery P. Interactions between circadian neurons control temperature synchronization of Drosophila behavior. J. Neurosci. 2007;27:10722–10733. doi: 10.1523/JNEUROSCI.2479-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Fortin J.P., McCarthy Ev., Oksman L., Kopin A.S., Nitabach M.N. Cellular dissection of circadian peptide signals with genetically encoded membrane-tethered ligands. Curr. Biol. 2009;19:1167–1175. doi: 10.1016/j.cub.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Cao G., Tanenhaus A.K., McCarthy E.V., Jung M., Schleyer W., Shang Y., Rosbash M., Yin J.C.P., Nitabach M.N. Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in drosophila. Cell Rep. 2012;2:332–344. doi: 10.1016/j.celrep.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B., Kaplan H.S., Cavey M., Lelito K.R., Bahle A.H., Zhu Z., Macara A.M., Roman G., Shafer O.T., Blau J. Differentially timed extracellular signals synchronize pacemaker neuron clocks. PLoS Biol. 2014;12:e1001959. doi: 10.1371/journal.pbio.1001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith K., Ceriani M.F., Staknis D., Gekakis N., Steeves T.D., Weitz C.J., Takahashi J.S., Kay S.A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Das A., Holmes T.C., Sheeba V. dTRPA1 in Non-circadian Neurons Modulates Temperature-dependent Rhythmic Activity in Drosophila melanogaster. J. Biol. Rhythms. 2016;31:272–288. doi: 10.1177/0748730415627037. [DOI] [PubMed] [Google Scholar]
- Depetris-Chauvin A., Berni J., Aranovich E.J., Muraro N.I., Beckwith E.J., Ceriani M.F. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr. Biol. 2011;21:1783–1793. doi: 10.1016/j.cub.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Fernández M.P., Chu J., Villella A., Atkinson N., Kay S.A., Ceriani M.F. Impaired clock output by altered connectivity in the circadian network. Proc. Natl. Acad. Sci. USA. 2007;104:5650–5655. doi: 10.1073/pnas.0608260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Nagata Y., Nishiuchi T., Sato M., Iwami M., Kiya T. Visualization of neural activity in insect brains using a conserved immediate early gene, Hr38. Curr. Biol. 2013;23:2063–2070. doi: 10.1016/j.cub.2013.08.051. [DOI] [PubMed] [Google Scholar]
- Hens K., Feuz J.-D., Isakova A., Iagovitina A., Massouras A., Bryois J., Callaerts P., Celniker S.E., Deplancke B. Automated protein-DNA interaction screening of Drosophila regulatory elements. Nat. Methods. 2011;8:1065–1070. doi: 10.1038/nmeth.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hens K., Feuz J.-D., Deplancke B. A high-throughput gateway-compatible yeast one-hybrid screen to detect protein-DNA interactions. Methods Mol. Biol. 2012;786:335–355. doi: 10.1007/978-1-61779-292-2_20. [DOI] [PubMed] [Google Scholar]
- Hyun S., Lee Y., Hong S.T., Bang S., Paik D., Kang J., Shin J., Lee J., Jeon K., Hwang S. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Im S.H., Taghert P.H. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J. Comp. Neurol. 2010;518:1925–1945. doi: 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S., Stoleru D., McDonald M., Nawathean P., Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S., Menet J.S., Sugino K., Horwich M.D., Weissbein U., Nawathean P., Vagin V.V., Zamore P.D., Nelson S.B., Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Hall J.C. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kim E.Y., Bae K., Ng F.S., Glossop N.R.J., Hardin P.E., Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- Kitayama Y., Nishiwaki T., Terauchi K., Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 2008;22:1513–1521. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman S.J., Silver R., Le Sauter J., Bult-Ito A., McMahon D.G. Phase resetting light pulses induce Per1 and persistent spike activity in a subpopulation of biological clock neurons. J. Neurosci. 2003;23:1441–1450. doi: 10.1523/JNEUROSCI.23-04-01441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula-Eversole E., Nagoshi E., Shang Y., Rodriguez J., Allada R., Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc. Natl. Acad. Sci. USA. 2010;107:13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear B.C., Merrill C.E., Lin J.M., Schroeder A., Zhang L., Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae K., Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Lerner I., Bartok O., Wolfson V., Menet J.S., Weissbein U., Afik S., Haimovich D., Gafni C., Friedman N., Rosbash M., Kadener S. Clk post-transcriptional control denoises circadian transcription both temporally and spatially. Nat. Commun. 2015;6:7056. doi: 10.1038/ncomms8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Guo F., Shen J., Rosbash M. PDF and cAMP enhance PER stability in Drosophila clock neurons. Proc. Natl. Acad. Sci. USA. 2014;111:E1284–E1290. doi: 10.1073/pnas.1402562111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Holy T.E., Taghert P.H. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science. 2016;351:976–981. doi: 10.1126/science.aad3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C., Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 2013;16:1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- Lin Y., Stormo G.D., Taghert P.H. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J. Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood E.S., Reddy A.B., Wong G.K.Y., O’Neill J.S., O’Brien J.A., McMahon D.G., Harmar A.J., Okamura H., Hastings M.H. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- McDonald M.J., Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Mertens I., Vandingenen A., Johnson E.C., Shafer O.T., Li W., Trigg J.S., De Loof A., Schoofs L., Taghert P.H. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Nagoshi E., Sugino K., Kula E., Okazaki E., Tachibana T., Nelson S., Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat. Neurosci. 2010;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach M.N., Blau J., Holmes T.C. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Nitabach M.N., Sheeba V., Vera D.A., Blau J., Holmes T.C. Membrane electrical excitability is necessary for the free-running larval Drosophila circadian clock. J. Neurobiol. 2005;62:1–13. doi: 10.1002/neu.20053. [DOI] [PubMed] [Google Scholar]
- Nitabach M.N., Wu Y., Sheeba V., Lemon W.C., Strumbos J., Zelensky P.K., White B.H., Holmes T.C. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J. Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Helfrich-Förster C., Lee G., Liu L., Rosbash M., Hall J.C. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci. USA. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Stoleru D., Levine J.D., Hall J.C., Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.S. The transgenic RNAi project at Harvard medical school: Resources and validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero J.E., Kuhlman S.J., McMahon D.G. The biological clock nucleus: a multiphasic oscillator network regulated by light. J. Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn S.C., Park J.H., Rosbash M., Hall J.C., Taghert P.H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Roberts L., Leise T.L., Noguchi T., Galschiodt A.M., Houl J.H., Welsh D.K., Holmes T.C. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr. Biol. 2015;25:858–867. doi: 10.1016/j.cub.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Bradley S., Kadener S., Li Y., Luo W., Menet J.S., Nagoshi E., Palm K., Schoer R., Shang Y., Tang C.H. Transcriptional feedback and definition of the circadian pacemaker in Drosophila and animals. Cold Spring Harb. Symp. Quant. Biol. 2007;72:75–83. doi: 10.1101/sqb.2007.72.062. [DOI] [PubMed] [Google Scholar]
- Rosenzweig M., Brennan K.M., Tayler T.D., Phelps P.O., Patapoutian A., Garrity P.A. The Drosophila ortholog of vertebrate TRPA1 regulates thermotaxis. Genes Dev. 2005;19:419–424. doi: 10.1101/gad.1278205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan S., Zheng X., Xiao R., Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Sehadova H., Glaser F.T., Gentile C., Simoni A., Giesecke A., Albert J.T., Stanewsky R. Temperature entrainment of Drosophila’s circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron. 2009;64:251–266. doi: 10.1016/j.neuron.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Sellix M.T., Currie J., Menaker M., Wijnen H. Fluorescence/luminescence circadian imaging of complex tissues at single-cell resolution. J. Biol. Rhythms. 2010;25:228–232. doi: 10.1177/0748730410368016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A., Flourakis M., Kula-Eversole E., Zhang L., Kilman V., Allada R. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol. 2014;12:e1001810. doi: 10.1371/journal.pbio.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer O.T., Yao Z. Pigment-dispersing factor signaling and circadian rhythms in insect locomotor activity. Curr. Opin. Insect Sci. 2014;1:73–80. doi: 10.1016/j.cois.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer O.T., Kim D.J., Dunbar-Yaffe R., Nikolaev V.O., Lohse M.J., Taghert P.H. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachenko A., Li Y., Abruzzi K.C., Rosbash M. The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron. 2013;79:281–292. doi: 10.1016/j.neuron.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So W.V., Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R., Lynch K.S., Brandes C., Hall J.C. Mapping of elements involved in regulating normal temporal period and timeless RNA expression patterns in Drosophila melanogaster. J. Biol. Rhythms. 2002;17:293–306. doi: 10.1177/074873002129002609. [DOI] [PubMed] [Google Scholar]
- Takahashi J.S., Ko C.H., Yamada Y.R., Welsh D.K., Buhr E.D., Liu A.C., Zhang E.E., Ralph M.R., Kay S.a., Forger D.B. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010;8:e1000513. doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.H., Hinteregger E., Shang Y., Rosbash M. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron. 2010;66:378–385. doi: 10.1016/j.neuron.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R., Bartok O., Mezan S., Malka Y., Kadener S. Synergistic interactions between the molecular and neuronal circadian networks drive robust behavioral circadian rhythms in Drosophila melanogaster. PLoS Genet. 2014;10:e1004252. doi: 10.1371/journal.pgen.1004252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh D.K., Yoo S.-H., Liu A.C., Takahashi J.S., Kay S.A. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Cao G., Nitabach M.N. Electrical silencing of PDF neurons advances the phase of non-PDF clock neurons in Drosophila. J. Biol. Rhythms. 2008;23:117–128. doi: 10.1177/0748730407312984. [DOI] [PubMed] [Google Scholar]
- Yao Z., Shafer O.T. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Zheng H., Price J.L., Hardin P.E. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol. Cell. Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Kilman V.L., Keegan K.P., Peng Y., Emery P., Rosbash M., Allada R. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113:755–766. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]
- Zheng X., Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.