Abstract
Muropeptides are a group of bacterial natural products generated from the cell wall in the course of its turnover. These compounds are cell-wall recycling intermediates and also are involved in signalling functions within the bacterium. However, identity of these signalling molecules remains elusive. The identification and characterization of 20 muropeptides from Pseudomonas aeruginosa is described. The least abundant of these metabolites is present at 100 and the most abundant at 55,000 molecules per bacterium. Analysis of these muropeptides under conditions of induction of resistance to a β-lactam antibiotic identified two signaling muropeptides (N-acetylglucosamine-1,6-anhydro-N-acetylmuramyl pentapeptide and 1,6-anhydro-N-acetylmuramyl pentapeptide). Authentic synthetic samples of these metabolites were shown to activate expression of β-lactamase in the absence of any β-lactam antibiotic, hence they serve as chemical signals in this complex biochemical pathway.
Keywords: bacteria, peptidoglycan, antibiotic resistance, β-lactamase
Graphical abstract
A total of 20 muropeptide natural products from Pseuromonas aeruginosa were isolated from the living bacterium and their structures elucidated. The least abundant of these metabolites is present at 100 and the most abundant at 55,000 molecules per bacterium. Two of the natural products were shown to account for the majority of the signalling effect in induction of the antibiotic-resistance response. This signalling is the basis for resistance to β-lactam antibiotics in P. aeruginosa.

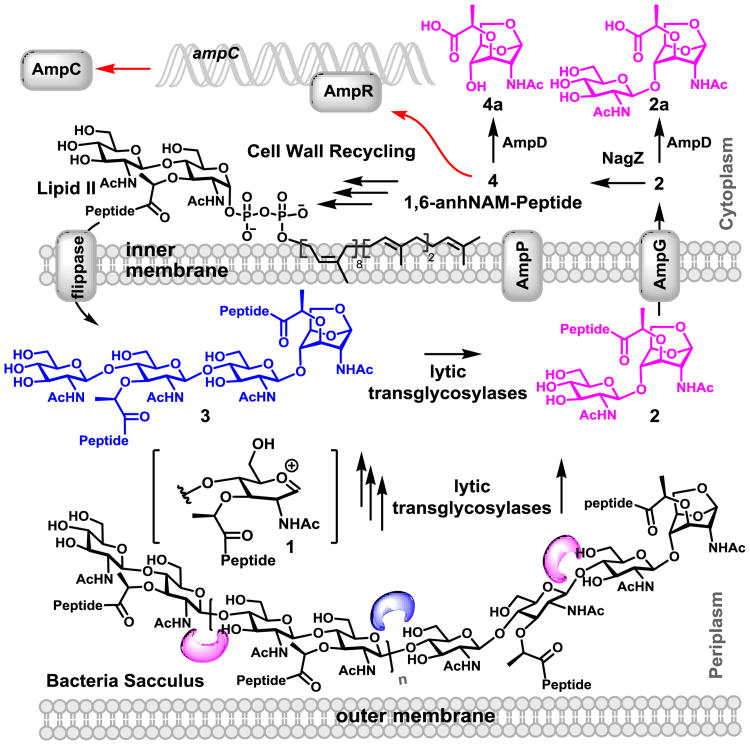
The cell wall (also known as the sacculus) is a complex macromolecular polymer that encases the bacterium. Its major constituent is comprised of repeating N-acetylglucosamine (NAG)-N-acetylmuramic acid (NAM), with a pentapeptide stem attached to the NAM unit.[1] Cell wall is critical for survival of the bacterium, hence, the cell wall and its biosynthetic machinery are targets of antibiotics.[2] Cell wall is synthesized by polymerization of Lipid II,[3, 4] resulting in the NAG-NAM backbone, which is subsequently crosslinked to the neighboring strand through the peptide stem (Fig. 1). In parallel, degradative processes that turn over segments of the assembled cell wall also take place.[4, 5] An important event in this turnover is mediated by a family of enzymes called lytic transglycosylases (LTs), whose reactions generate a series of natural products referred to collectively as muropeptides.
Figure 1.
The reactions of LTs produce the muropeptides containing the 1,6-anhydromuramyl moiety, which are translocated by the permease AmpG (and AmpP in P. aeruginosa). In normal growth of bacteria, 4 would initiate recycling events, regenerating Lipid II. However, as an off shoot of the recycling events, accumulation of 4 would trigger the β-lactamase (AmpC) expression (indicated by red arrows).
The reactions of all eight Escherichia coli LTs have been studied in vitro.[6-8] These enzymes generate a transient oxocarbenium species (1) at the muramyl moiety of the peptidoglycan, which results in the cleavage of the β-1,4-glycosidic bond between a NAM and a NAG (Fig. 1), giving rise to the 1,6-anhydromuramyl moiety (2 and 3). Some LTs perform this reaction at the ends of the peptidoglycan, the so called exolytic reaction, giving rise to the NAG-anhNAM disaccharides (2). Others perform the reaction in the middle of the peptidoglycan, the endolytic reaction, which gives rise to a longer backbone for the sugar (3). These released muropeptides are translocated to the cytoplasm by the permease AmpG (and possibly by AmpP in Pseudomonas aeruginosa).[9] Once in the cytoplasm, the muropeptides enter the cell-wall recycling process, regenerating Lipid II.[4, 8, 10] Alternatively, other muropeptides are involved in signaling functions, leading to disparate responses such as antibiotic resistance, virulence and inflammation.[11, 12] The functions in resistance to β-lactam antibiotics involve binding to the gene regulator AmpR, which allows for transcription of the gene ampC for the Gram-negative AmpC β-lactamase (Fig. 1).[13] β-Lactamase hydrolyzes β-lactams antibiotics, which inhibit the action of pencillin-binding proteins by mimicking the structure of the terminal d-Ala-d-Ala in the stem peptides of the peptidoglycan.
The processes that muropeptides mediate are not fully understood due to impediments such as the minute quantities, rapid metabolic flux and complexity of the structures. Once the structures are elucidated, they need to be prepared in the laboratory for validation of the assigned structure and for the conduct of biochemical studies. Here, we report the identification, characterization, and quantification of several muropeptides from periplasm of P. aeruginosa, an opportunistic human pathogen.
Sample preparation is important, as dilution of the minute quantities and contamination could confound analysis. Initially osmotic shock was used for liberation of the periplasmic content that contains the muropeptides. This would have separated the periplasmic and cytoplasmic metabolites before attempt at isolation of muropeptides. Unfortunately, 10-30% cytoplasmic contamination was noted in these samples. The results were also not reproducible, and muropeptides could merely be identified near the detection limit of 0.4 pmol by our instrumentation.
The muropeptides that enter the cytoplasm via the permease AmpG (or AmpP) are expected to undergo rapid metabolic flux (Fig. 1). This assertion was documented by preparing spheroplasts of P. aeruginosa PAO1. The cytoplasmic content from the lysed spheroplasts were analyzed by LC/MS for muropeptides. None could be detected, suggesting that the cytoplasmic muropeptides were rapidly metabolized to Lipid II, with concentrations below our detection limit. Hence, the whole bacterium was grown and lysed, an approach that proved to be reliable and reproducible. The muropeptides that were generated under these conditions could only have come from the intact periplasm in the whole bacterium. After sample preparation (SI), the LC/MS analyses were performed for detection and identification of muropeptides.
The muropeptide content of the whole cells of P. aeruginosa PAO1 was compared in the absence and presence of half of the minimal-inhibitory concentration (MIC) of antibiotic cefoxitin, a β-lactam that interferes with cell-wall synthesis.[14] Cefoxitin at sub-MIC levels activates the expression of β-lactamase efficiently, leading to antibiotic resistance in P.aeruginosa.[14] This is believed to be mediated by a messenger function of a muropetide.[12, 15, 16] Hence, one or more of the muropeptides listed in Fig. 2A is expected to serve as the signaling molecule for antibiotic resistance.
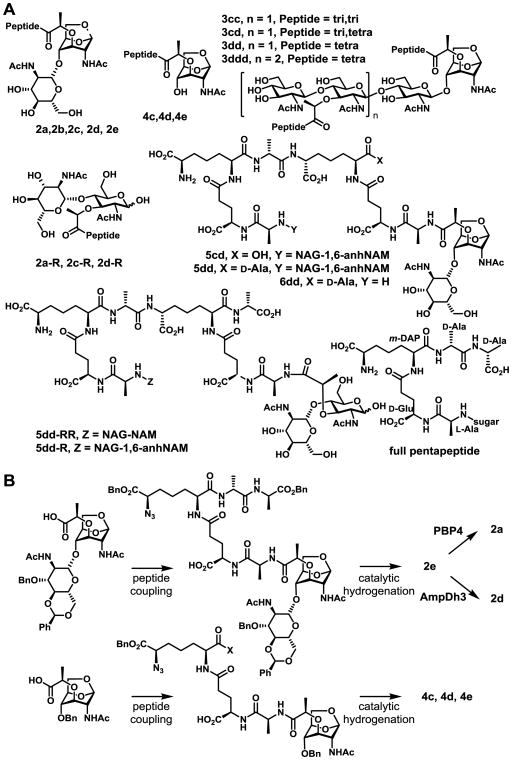
Figure 2.
(A) Chemical structures of detected muropeptides. (B) The chemo/enzymatic syntheses of six muropeptides.
As shown in Fig. 2A, compounds are numbered according to the nature of sugar, 2 for NAG-1,6-anhNAM and 4 for 1,6-anhNAM. For the peptide component, a has no peptide, b, c, d, and e carry di-, tri-, tetra- and pentapeptide, respectively (full pentapeptide is l-Ala-γ-d-Glu-m-DAP-d-Ala-d-Ala; bottom right of Fig. 2A). Compounds 3 are (NAG-NAM-peptide)n-NAG-1,6-anhNAM-peptide and compounds 5 are for cross-linked species. For example, compound 5dd indicates cross-linked peptide between two tetrapeptides. Compounds with reducing-end sugars, which lack the 1,6-anhNAM, were also detected as minor components and are designated with the letter R.
Fig. 3A is the LC/MS total-ion chromatogram (TIC) of the pseudomonal sacculus turnover products in the presence of the purified E. coli LT MltA. In contrast to this typical in vitro analysis with the isolated sacculus, samples from the whole-cell did not reveal any discernable muropeptides (Fig. 3B). This made necessary the preparation of authentic standards for comparison to the LC/MS extracted-ion chromatograms (EICs) of individual metabolite (Figs. 3C-3F).
Figure 3.

Analysis of P. aeruginosa muropeptides. The LC/MS TICs of (A) sacculus digested by MltA, of (B) whole-cell analysis. EICs of (C) 2a, (D) 4c, (E) 4d, and (F) 2d of whole-cell sample, (G) the TIC of authentic synthetic standards mixed together.
Four muropeptides, 2e,[17] 4c, 4d, and 4e[18, 19] were synthesized. A few of these authentic samples were also converted to new species by known enzymatic reactions (Figs. 2B and S1). For example, 2e was converted to 2d by the use of penicillin-binding protein 4 (PBP4),[15] and 2e to 2a using AmpDh3,[19] both purified recombinant enzymes from P. aeruginosa. Figs. 3C-3F show EICs of the detected metabolites 2a, 2d, 2e, 4c, and 4d, and their comparison to the authentic standards (Fig. 3G). Analysis was further done with comparison of MS and MS/MS with authentic samples, as exemplified in Fig. S1. For structure assignment of metabolites whose authentic standards were not available, the method that was developed previously by our laboratory to analyze turnover products of sacculus by LTs and PBP4 was used.[6, 15]
Quantification was done by integrating peak areas from EICs of the corresponding m/z values of the individual muropeptide. This was converted to concentration using standard curves generated with the authentic 2e. The concentration was converted to numbers of molecules (of each compound) per bacterium (Table 1; SI). Standard curves for 2e, 4c, 4d, 4e and 7 (β-methoxy-NAG-NAM (pentapeptide)-NAG-NAM (pentapeptide))[20] were very similar within 7% variation of each other (Chart S1 and Fig. S2). The collection of our synthetic standards covers distinctive chemical structures of >95% of the detected muropeptides. So, 2e was chosen as a representative synthetic standard for quantification.
Table 1.
Detected muropeptides from whole-cell analysis (in molecules per bacterium ×104).a
| Muropeptide | Wild-type | Inducedb | p-valuec |
|---|---|---|---|
| NAG-1,6-anhNAM | |||
| 2a | 1.5 ± 0.07 | 0.3 ± 0.01 | 0.01* |
| 2b | 0.2 ± 0.01 | 0.08 ± 0.01 | 0.004* |
| 2c | 0.8 ± 0.05 | 0.3 ± 0.02 | 0.02* |
| 2d | 5.5 ± 0.3 | 1.0 ± 0.3 | 0.002* |
| 2e | 0.01 ± 0.005 | 0.1 ± 0.02 | 0.04* |
| 1,6-anhNAM | |||
| 4c | 0.3 ± 0.06 | 0.1 ± 0.003 | 0.0003* |
| 4d | 0.4 ± 0.01 | 0.07 ± 0.003 | 0.01* |
| 4e | N.D.d | 0.08 ± 0.01 | 0.04* |
| (NAG-NAM)n-NAG-1,6-anhNAM | |||
| 3cc | 0.1 ± 0.01 | 0.02 ± 0.006 | 0.003* |
| 3cd | 0.2 ± 0.02 | 0.03 ± 0.007 | 0.03* |
| 3dd | 0.9 ± 0.06 | 0.09 ± 0.03 | 0.002* |
| 3ddd | 0.1 ± 0.02 | 0.01 ± 0.003 | 0.049* |
| NAG-1,6-NAM-crosslinked-1,6-NAM-NAG | |||
| 5cd | 0.03 ± 0.004 | 0.02 ± 0.001 | 0.2 |
| 5dd | 0.3 ± 0.01 | 0.07 ± 0.007 | 0.001* |
| 6dd | 0.04 ± 0.001 | 0.02 ± 0.004 | 0.07 |
| Reduced end | |||
| 2a-R | 0.07 ± 0.01 | N.D.d | 0.02* |
| 2c-R | 0.07 ± 0.03 | 0.02 ± 0.002 | 0.11 |
| 2d-R | 0.5 ± 0.1 | 0.1 ± 0.03 | 0.06 |
| 5dd-RR | 0.1 ± 0.002 | 0.01 ± 0.002 | 0.0002* |
| 5dd-R | 0.02 ± 0.002 | 0.009± 0.002 | 0.047* |
| Totale | 11.1 ± 0.8 | 2.4 ± 0.5 | |
Average of two runs.
PAO1 was exposed to cefoxitin at 512 μg/mL.
p-values from Student t-test.
Numbers with *are significantly different (p < 0.05) between the wild-type and induced sample.
not detected.
total-detected muropeptides.
The most abundant muropeptide in wild-type PAO1 strain is NAG-1,6-anhNAM-tetrapeptide (2d). The di-, tri-, and pentapeptide variants (2b, 2c, and 2e) are also found, along with 2a (with no peptide). These are reaction products of LTs, mostly from the exolytic activity. The discovery of compounds with the core 1,6-anhNAM-peptides (4c and 4d) suggests the existence of the N-acetylglucosaminidase activity in P. aeruginosa. The presence of such an enzyme (FlgJ) in the periplasm was recently documented in Salmonella enterica.[21] This activity in P. aeruginosa might be mediated by PA1085, which has an identity of 31% and a similarity of 46% at the amino-acid level to FlgJ from S. enterica (Fig. S3).[21] To our knowledge this is the first documentation of a periplasmic N-acetylglucosaminidase reaction product in P. aeruginosa.[21] Oligomeric sugars (up to hexamers) with tetrapeptide (3dd and 3ddd) or a mix of tetra and tripeptide (3cd and 3cc) were also found. These are products of the endolytic reactions of LTs.[6]
Cross-linked muropeptides such as 5cd, 5dd, and 6dd were also found. As minor components, muropeptides containing a sugar with a reducing end (2a-R, 2c-R, 2d-R, 5dd-RR, and 5dd-R) were also detected. This indicates that the reactive oxocarbenium species partitions between either entrapment of the internal C6-hydroxyl or of a water molecule, or there exists a yet-to-be identified hydrolytic glycosidase in this organism. The ratio of the two types of products (non-reducing to reducing) is ∼14:1.
Same sample preparation and analyses were carried out with P. aeruginosa PAO1 exposed to a cell-wall-active antibiotic cefoxitin at half of the MIC (i.e., 512 μg/mL), hence a non-lethal concentration.[14] The exposure to the antibiotic alters the pool of muropeptides, where one or more is understood to enter the cytoplasm via AmpG (or AmpP) permease and upregulate production of β-lactamase, the antibiotic-resistance determinant.[9, 12] The induction of resistance was confirmed by the β-lactamase assay using nitrocefin.
The same number of bacteria and the same conditions were used in both cases; hence, the values of the two columns of Table 1 can be compared to each other. The analysis showed that the total muropeptide (molecules/bacterium) was significantly reduced (p-value < 0.05 by Student's t-test) in the induced vs the uninduced case: 24,000 vs. 111,000 (Table 1). The most abundant muropeptide in the uninduced sample was 2d (NAG-1,6-anhNAM-tetrapeptide). Muropeptide 2e (NAG-1,6-anhNAM-pentapeptide) was enriched at 46-fold upon antibiotic induction (1000 in 24,000 molecules vs. 100 in 111,000 molecules). That is to say that the β-lactam antibiotic inhibits the targeted PBP, whose lack of activity leaves its peptidoglycan substrate in the sacculus intact. This observation in living bacteria agrees with the finding of the in vitro sacculus analysis of the induced P. aeruginosa.[15, 16] Compound 4e was detected only in the induced sample. This observation suggests that as the concentration of 2e increased upon induction, the compound was likely turned over by the aforementioned N-acetylglucosaminidase to produce 4e. Other than 2e and 4e, the rest of muropeptides detected in the induced sample were similar to those in the uninduced. It is not immediately obvious as to why the total quantity of muropeptides is lower in the induced sample (one fifth of the uninduced). This likely reflects the altered cell-wall modifications of the bacterium in the presence of the sub-lethal concentration of the antibiotic.
The obvious question now becomes whether exogenously added authentic muropeptides 2e or 4e could cause induction of antibiotic resistance in the absence of antibiotic. We investigated this first with the wild-type P. aeruginosa PAO1 strain. Addition of muropeptide 2e or 4e at upwards of 500 μg/mL failed to induce β-lactamase expression (Table 2). Going with the premise that the Gram-negative outer membrane is a formidable barrier to penetration of most small molecules into the periplasm, we procured a mutant strain defective in its outer membrane. The strain P. aeruginosa Z61 has the full complement of the genes necessary for induction of β-lactamases, but it expresses a mutant version of the β-lactamase with diminished activity.[22] Nonetheless, using the nitrocefin assay we observed a 4.7 fold increase of induction of β-lactamase at quarter-MIC level of cefoxitin. The same experiment performed with the bacteria exposed to 100 μg/mL muropeptides 2e or 4e resulted in 1.4 or 1.7 induction, respectively (Table S1). This is a large excess of these compounds, but we used them so, as we had expected that the exogenously added compounds would undergo turnover by periplasmic enzymes. Hence, the two metabolites—both produced in response to the exposure of bacteria to the antibiotic inducer cefoxitin—collectively account for most of the induction observed by cefoxitin. We note that compound 2a and 4a (metabolites without a peptide stem; products of the reaction of AmpD) as negative controls. As expected, under the same conditions, induction was not observed. Activity of AmpD (Fig. 1) is at the crossroads of induction of resistance vs. cell-wall recycling (reversal of induction).
Table 2.
Induction of β-lactamase activity by three inducers.a
| inducer | Wild-type (PAO1) | Mutant (Z61)b | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Conc. (μg/mL) | β-lactamase inductionc | Conc. (μg/mL) | β-lactamase inductionc | |||
|
|
|
|||||
| Cefoxitind | 512 | 136e | 1555/11.4 | 0.0156 | 4.7e | 4.6/0.98 |
| 2e | 500 | 1.0 | 19.1/19.1 | 100 | 1.4e | 27.5/20.0 |
| 4e | 500 | 1.0 | 19.1/19.1 | 100 | 1.7e | 5.5/3.3 |
| 2ad | -f | 10g | 1.0 | 4.1/4.0 | ||
| 4ad | -f | 100 | 1.0 | 3.6/3.5 | ||
Average of two runs.
The strain is defective in its outer membrane.
The number (the left column) is calculated by lactamase activity in the presence of an inducer divided by that without an inducer (the right column) under the same condition.The enzyme activity was expressed as the nanomole of nitrocefin hydrolyzed per min per mg of protein for wild-type and picomole/min/mg for mutant.
Cefoxitin and compounds 2a/4a were used as positive and negative controls for induction, respectively.
β-Lactamse activities were significantly different (p < 0.05) between induced and uninduced samples.
Not measured.
due to limited supply of compound, we could only assess the effect at the lower concentration.
This study reports the nature and quantities of 20 muropeptides from P. aeruginosa. The levels of muropeptide 2e became elevated by 46-fold on exposure of bacteria to sub-MIC levels of a good inducer, cefoxitin. We also observed muropeptide 4e only in the induced cells. This study discloses that authentic synthetic samples of muropeptides 2e and 4e could serve as inducers of β-lactam-antibiotic resistance in the absence of antibiotic. These experiments clearly document that at least muropeptides 2e and 4e are chemical elicitors of induction of antibiotic resistance.
Supplementary Material
Acknowledgments
This work was supported by grant GM61629 from the NIH (to SM) and in-part by NSF IIP-1237818 [PFI-AIR: CREST-I/UCRC-Industry Ecosystem to Pipeline Research] (KM), Florida International University (FIU) Bridge Funding (KM), FIU Herbert Wertheim College of Medicine Graduate Assistantship (SD).
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Prof. Kalai Mathee, Email: matheek@fiu.edu.
Prof. Shahriar Mobashery, Email: mobashery@nd.edu.
References
- 1.Fisher JF, Mobashery S. In: Practical Handbook of Microbiology. 3rd. Goldman E, Green LH, editors. CRC Press; 2015. p. 221. [Google Scholar]; Vollmer W, Blanot D, de Pedro MA. FEMS Microbiol Rev. 2008;32:149. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 2.Bush K. In: Ann N Y Acad Sci. Bush K, editor. Vol. 1277. Wiley-Blackwell; 2013. [Google Scholar]
- 3.Qiao Y, Lebar MD, Schirnier K, Schaefer K, Tsukamoto H, Kahne D, Walker S. J Am Chem Soc. 2014;136:14678. doi: 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Heijenoort J. Microbiol Mol Biol Rev. 2007;71:620. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park JT, Uehara T. Microbiol Mol Biol Rev. 2008;72:211. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi H, Boggess B, Mobashery S. J Am Chem Soc. 2013;135:3311. doi: 10.1021/ja309036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheurwater E, Reid CW, Clarke AJ. Int J Biochem Cell Biol. 2008;40:586. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]; Yunck R, Cho H, Bernhardt TG. Mol Microbiol. 2016 doi: 10.1111/mmi.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Heijenoort J. Microbiol Mol Biol Rev. 2011;75:636. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong KF, Aguila A, Schneper L, Mathee K. BMC Microbiol. 2010;10:328. doi: 10.1186/1471-2180-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson JW, Fisher JF, Mobashery S. Ann N Y Acad Sci. 2013;1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasubramanian D, Kumari H, Mathee K. Pathog Dis. 2015;73:1. doi: 10.1111/2049-632X.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boudreau MA, Fisher JF, Mobashery S. Biochemistry. 2012;51:2974. doi: 10.1021/bi300174x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher JF, Mobashery S. Bioorg Chem. 2014;56:41. doi: 10.1016/j.bioorg.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs C, Frère JM, Normark S. Cell. 1997;88:823. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]; Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Hoiby N, Mathee K. Antimicrob Agents Chemother. 2005;49:4567. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore DM. Clin Microbiol Rev. 1995;8:557. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee M, Hesek D, Blázquez B, Lastochkin E, Boggess B, Fisher J, Mobashery S. J Am Chem Soc. 2015;137:190. doi: 10.1021/ja5111706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyá B, Dotsch A, Juan C, Blazquez J, Zamorano L, Haussler S, Oliver A. PLoS Pathog. 2009;5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hesek D, Lee M, Zhang W, Noll BC, Mobashery S. J Am Chem Soc. 2009;131:5187. doi: 10.1021/ja808498m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M, Zhang W, Hesek D, Noll BC, Boggess B, Mobashery S. J Am Chem Soc. 2009;131:8742. doi: 10.1021/ja9025566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Lee M, Hesek D, Lastochkin E, Boggess B, Mobashery S. J Am Chem Soc. 2013;135:4950. doi: 10.1021/ja400970n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee M, Hesek D, Shah IM, Oliver AG, Dworkin J, Mobashery S. ChemBioChem. 2010;11:2525. doi: 10.1002/cbic.201000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herlihey FA, Moynihan PJ, Clarke AJ. J Biol Chem. 2014;289:31029. doi: 10.1074/jbc.M114.603944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angus BL, Carey AM, Caron DA, Kropinski AM, Hancock RE. Antimicrob Agents Chemother. 1982;21:299. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]; Balibar CJ, Grabowicz M. Antimicrob Agents Chemother. 2016;60:845. doi: 10.1128/AAC.01747-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.