Abstract
Bacterial cold-water disease caused by Flavobacterium psychrophilum is one of the major causes of mortality of salmonids. Three genetic lines of rainbow trout designated as ARS-Fp-R (resistant), ARS-Fp-C (control) and ARS-Fp-S (susceptible) have significant differences in survival rate following F. psychrophilum infection. Previous study identified transcriptome differences of immune-relevant protein-coding genes at basal and post infection levels among these genetic lines. Using RNA-Seq approach, we quantified differentially expressed (DE) long non-coding RNAs (lncRNAs) in response to F. psychrophilum challenge in these genetic lines. Pairwise comparison between genetic lines and different infection statuses identified 556 DE lncRNAs. A positive correlation existed between the number of the differentially regulated lncRNAs and that of the protein-coding genes. Several lncRNAs showed strong positive and negative expression correlation with their overlapped, neighboring and distant immune related protein-coding genes including complement components, cytokines, chemokines and several signaling molecules involved in immunity. The correlated expressions and genome-wide co-localization suggested that some lncRNAs may be involved in regulating immune-relevant protein-coding genes. This study provides the first evidence of lncRNA-mediated regulation of the anti-bacterial immune response in a commercially important aquaculture species and will likely help developing new genetic markers for rainbow trout disease resistance.
World aquaculture industries suffer considerable economic losses annually because of infectious diseases1. Flavobacterium psychrophilum (Fp), a causative agent of Bacterial Cold Water Disease (BCWD), saddleback disease, fry mortality syndrome, or rainbow trout fry syndrome causes significant loss of trout and salmon each year and is a threat to many other salmonids (see review ref. 2). Infection of rainbow trout with Fp results in mortality of up to 30% and several complications in the survivors3. Originally, the pathogen was considered to be endemic to North America but in recent years it has been reported from almost every continent4. Multiple routes of transmission5, wide geographical distribution, the ability of pathogen to cope with harsh survival condition5, limited chemotherapeutic agents, and lack of a commercial vaccine make control measures inefficient. Live-attenuated Fp vaccines can provide protection against BCWD but environmental safety is a concern (see review ref. 6).
Harnessing the host’s immune system by selective breeding is a strategy being pursued to improve farmed fish health7. In order to improve resistance of rainbow trout against Fp, the National Center for Cool and Cold Water Aquaculture (NCCCWA) started a family-based selective breeding program in 2005. A closed genetic line, designated ARS-Fp-R, has undergone multiple generations of selection for increased survival following standardized challenge. This line has improved disease resistance against Fp infection in both laboratory and field settings compared to a susceptible (ARS-Fp-S) and randomly bred control (ARS-Fp-C) lines8. Previously, we performed global expression analysis of protein-coding genes in these genetic lines upon Fp challenge9. The study identified a large number of DE protein-coding genes among genetic lines, a significant proportion of which were genes with described roles in the immune response, especially the innate immune system. We demonstrated transcriptome differences between lines in the absence of infection. However, altered transcriptome abundance of lncRNAs among genetic lines after mock and Fp infection was not addressed.
LncRNAs have appeared as critical regulators of transcription and post-transcriptional events of protein-coding genes10. LncRNAs regulate diverse cellular processes, including disease, immunity, development and cell proliferation11. In mammals, lncRNAs regulate various immune responses including the interferon response, inflammatory processes, and other aspects of innate and adaptive immune responses12,13,14,15,16,17. TLR signaling and inflammatory responses increase the expression of lncRNA-Cox2 that regulates both activation and repression of innate response genes13. LncRNA NeST controls susceptibility to Theiler’s virus and Salmonella infection through epigenetic regulation of the interferon-γ locus17,18. A distinct differential expression profile of lncRNAs in response to microbial infection has been reported in mammals and salmonids, suggesting involvement of a set of lncRNAs in host defense against microbes11,19. To date, most of the studies in the field of lncRNA influence on immune processes are limited to mammalian species, especially human and mouse. To the best of our knowledge, there are no studies exploring the expression of lncRNAs during host defense against bacterial infection in aquaculture finfish. Such studies are difficult as low evolutionary conservation of lncRNAs across species prevents utilization of the information from mammalian species into aquaculture animals.
The overall objective of this study was to identify lncRNAs that are associated with genetic resistance against Fp and to identify immune-relevant protein-coding genes that might be regulated by lncRNAs. To study the expression of lncRNA, we utilized a reference dataset that we recently identified (31,195 lncRNA) in rainbow trout20. Using the abovementioned three genetic lines of rainbow trout, we were able to characterize the transcriptome profile of lncRNAs associated with the early response to Fp infection. We have identified DE lncRNAs between genetic lines of naive animals and in response to infection, identified their genomic co-localization relative to immune-relevant protein-coding genes, and explored their co-expression relationships to suggest possible regulation of immune-relevant protein-coding genes by lncRNAs.
Results and Discussion
Global expression of lncRNA across dataset
Previously, we analyzed mRNA expression in three genetic lines of fish sampled at 1 and 5 days post-Fp challenge9. In our prior analyses, slightly more than half (51.77%) of the RNA-Seq reads aligned to the 46,585 predicted coding mRNAs and thus considerable sequence information remained unaligned and thus enigmatic. In present study, on average, 8.2% of the total RNA-Seq reads aligned to the 31,195 lncRNAs reference (Supplementary Dataset 1A). 94.5% of the reads were uniquely mapped to the reference. On average, each dataset expressed 87.2% of the putative reference lncRNA’s at RPKM cut off ≥0.5. Out of 31,195 reference lncRNAs, only 933 were not expressed in any dataset (RPKM ≥ 0.5). One possible explanation of the low percentages of aligned read to lncRNA reference compared to protein coding mRNAs might be due to the lower lncRNAs expression compared to mRNAs. Recently, we reported that the average RPKM of the most abundant 40,000 transcripts was 3.49 and 15.69 in LncRNAs and protein-coding genes, respectively20. In this study, RNA was sequenced from a whole-body extract, which may be another reason for the low percentage of mappable reads because reference lncRNA dataset was sequenced from about 13 specific tissues. Out of the 933lnc RNAs, only 109 were tissue specific indicating that most of the 933 are very lowly expressed on all tissues.
We utilized pairwise comparisons between different genetic lines and days of infection to identify a sum of 937 DE lncRNA from all comparisons (FDR-P-value < 0.05) (Table 1). Of these, 556 were unique lncRNA showing differential expression in at least one comparison (Supplementary Dataset 2 tab “ALL DEF, non-redundant”). In our previous study using the same genetic lines, ~2,600 DE immune-related and other protein-coding genes were identified in response to Fp infection9. We quantified the number of DE lncRNAs between different genetic lines and infection statuses (total 24 comparisons) and compared the number with that of DE protein-coding genes. Numbers of DE protein-coding genes and lncRNAs showed moderate positive correlation (R2 = 0.40, p = 0.0011) (Table 1). In general, within each pair-wise comparison, fewer differentially regulated lncRNA were identified as compared to DE protein coding transcripts (Table 1). This may, in part, be, due to the overall lower expression level of lncRNA as compared to protein-coding genes21. Numbers of the DE protein-coding genes as well as lncRNAs positively correlated with bacterial load in the body. The susceptible line showed more DE lncRNAs as well as protein-coding genes compared to the resistant and control genetic lines (Table 1). Similarly, more transcripts were expressed on day 5 of infection than on day 1. Correlation between body bacterial load and the number of DE lncRNAs on the 5th day of infection in control, susceptible and resistant genetic lines was strongly positively correlated (R2 > 0.99); however, correlation of body bacterial load with the number of DE protein coding-genes was moderately positive (R2 = 0.34). This finding suggests that, like protein-coding genes, lncRNAs may play a role in the host defense against Fp. Expression trends of seven randomly chosen regulated lncRNAs was verified by real time PCR. A consistent trend (R2 = 0.84) between RNA-Seq and qPCR was observed, albeit with a somewhat lower relative expression measured by qPCR for 6 of the 7 measured lncRNA’s (Supplementary Dataset 1B). Information about primers and the real time PCR cycling program is provided in Supplementary Dataset 1C.
Table 1. Comparison of differentially expressed lncRNA and protein coding genes in response to Fp infection.
| Comparison | Day, genetic line and infection status | No. differentially expressed protein-coding genesa | No. differentially expressed lncRNAs |
|---|---|---|---|
| Infected vs PBS | Day 1 R-line (Fp) vs. R-line (PBS) | 515 | 57 |
| Day 5 R-line (Fp) vs. R-line (PBS) | 428 | 36 | |
| Day 1 C-line (Fp) vs. C-line (PBS) | 20 | 0 | |
| Day 5 C-line (Fp) vs. C-line (PBS) | 2201 | 54 | |
| Day 1 S-line (Fp) vs. S-line (PBS) | 1663 | 125 | |
| Day 5 S-line (Fp) vs. S-line (PBS) | 2225 | 196 | |
| Genetic lines (PBS) | Day 1 R-line (PBS) vs. S-line (PBS) | 76 | 24 |
| Day 1 R-line (PBS) vs. C-line (PBS) | 3 | 2 | |
| Day 1 S-line (PBS) vs. C-line (PBS) | 28 | 6 | |
| Day 5 R-line (PBS) vs. S-line (PBS) | 45 | 22 | |
| Day 5 R-line (PBS) vs. C-line (PBS) | 246 | 28 | |
| Day 5 S-line (PBS) vs. C-line (PBS) | 61 | 25 | |
| Genetic lines (Fp) | Day 1 R-line (Fp) vs. S-line (Fp) | 150 | 15 |
| Day 5 R-line (Fp) vs. S-line (Fp) | 1016 | 83 | |
| Day 1 R-line (Fp) vs. C-line (Fp) | 28 | 12 | |
| Day 5 R-line (Fp) vs. C-line (Fp) | 159 | 21 | |
| Day 1 S-line (Fp) vs. C-line (Fp) | 37 | 13 | |
| Day 5 S-line (Fp) vs. C-line (Fp) | 1758 | 5 | |
| Time points | Day 5 vs. Day 1 R-line (PBS) | 1286 | 26 |
| Day 5 vs. Day 1 C-line (PBS) | 294 | 36 | |
| Day 5 vs. Day 1 S-line (PBS) | 376 | 14 | |
| Day 5 vs. Day 1 R-line (Fp) | 334 | 22 | |
| Day 5 vs. Day 1 C-line (Fp) | 2469 | 70 | |
| Day 5 vs. Day 1 S-line (Fp) | 2434 | 45 |
Four different comparisons were made to quantify the differentially expressed transcripts: infected vs. non-infected, one genetic line vs. another without infection, one genetic line vs. another post infection, and day 1 vs. day 5 of infection. Differential expression was considered at fold change ±2 and FDR-corrected p < 0.05. Number of differentially expressed protein coding genes and lncRNAs showed positive correlation (R2 = 0.40, P = 0.0011).
aData are from9.
Recently, we reported tissue specificity of lncRNAs in rainbow trout20. A total of 35 DE lncRNAs were selectively expressed in specific tissues, 10 of them were gill-specific. Out of 13 vital tissues, liver, spleen and head kidney did not have any DE lncRNA. Spleen and head kidney lymphoid organ are mainly involved in generation of antibody response and other humoral components of immune system, but in early phase of BCWD, the first line of defense includes skin, alimentary tract lining, and gill22.
Differential expression of lncRNAs between Fp infected and PBS injected fish
LncRNAs are involved in the host immune response by regulating various immune-related genes12,13,14,15,16,17. In this study, we initially investigated DE lncRNAs associated with Fp injection at days 1 and 5 post-challenge. Pairwise comparison between challenged and time- and line-matched PBS-injected animals identified 327 unique lncRNAs with altered expression (fold change ±2 and FDR-corrected p value < 0.05) (Supplementary Dataset 2 tabs 1–7, and tab “All Fp vs PBS, non-redundant”).
In order to identify lncRNAs that are broadly involved in the response to infection with Fp, we quantified the DE lncRNAs (and their correlated protein-coding genes) that were differentially regulated in all three genetic lines upon infection. On the 5th day of infection, 12 lncRNAs were significantly upregulated (>2-fold) in all three genetic lines (FDR-corrected p- value < 0.05) (Table 2, top panel). These lncRNAs were most highly upregulated in the susceptible line followed by the control and resistant lines. These finding may indicate that these lncRNAs were either upregulated in response to bacterial load or extent of tissue damage caused by bacterial infection. Surprisingly, none of the lncRNAs was downregulated in all three genetic lines.
Table 2.
| LncRNAs upregulated in all three genetic lines (>2 fold) upon infection and their expression correlation with protein coding genes | |||||||
|---|---|---|---|---|---|---|---|
| LncRNA feature ID | Resistant line |
Control line |
Susceptible line |
Correlation with (R2) | |||
| EDGE test: Fold change | FDR p-value correction | EDGE test: Fold change | FDR p-value correction | EDGE test: Fold change | FDR p-value correction | ||
| Omy200117486 | 24.36 | 0 | 41.6 | 0 | 91.18 | 0 | Interferon-induced guanylate-binding protein 1 (0.82) |
| Omy100128008 | 14.95 | 0 | 22.23 | 0 | 46.4 | 0 | Complement protein component C7-1 (c7-1) (0.82) |
| Omy200138656 | 24.3 | 0.000001 | 11.63 | 0.011489 | 28.75 | 0 | Complement C5 (0.66) |
| Omy100149048 | 7.93 | 0.000004 | 5.95 | 0.048727 | 14.15 | 0 | Unknown |
| Omy200107378 | 6.38 | 0.000062 | 11.22 | 0.004248 | 11.22 | 0 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (0.92) |
| Omy200165911 | 3.68 | 0.049287 | 4.5 | 0.040404 | 9.98 | 0 | Unknown |
| Omy100052789 | 5.51 | 0.000564 | 5.13 | 0.047151 | 8.65 | 0 | Unknown |
| Omy200107535 | 3.71 | 0.000147 | 6.16 | 0.012707 | 8.12 | 0 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (0.92) |
| Omy300025398 | 4.37 | 0.006514 | 5.15 | 0.002475 | 4.86 | 0 | Unknown |
| Omy300085997 | 3.68 | 0.000345 | 3.16 | 0.043867 | 4.02 | 0.001759 | Unknown |
| Omy200206941 | 3.16 | 0.000344 | 2.33 | 0.015415 | 3.44 | 0.000002 | Lysozyme C II precursor (0.83) |
| Omy300043066 | 3 | 0.000121 | 3.74 | 0.006748 | 3.03 | 0.000014 | Properdin (0.82) and complement factor b-like (0.89) |
| LncRNAs showing drastic (>100) fold change upon infection in one particular genetic line and associated gene in genome | |||||||
| Feature ID | EDGE test: Fold change | FDR p-value correction | Comparison | Classification of LncRNA | Associated Gene(s) (R2) | ||
| Upregulated upon infection | |||||||
| Omy200018785 | 136.06 | 0.001233 | D1_S_FP vs D1_S_PBS | Intergenic | |||
| Omy200132807 | 121.83 | 0.000167 | D5_S_FP vs D5_S_PBS | Intergenic | |||
| Omy100037031 | 105.28 | 0.012820 | D5_C_FP vs D5_C_PBS | Intergenic | |||
| Downregulated upon infection | |||||||
| Omy200194608 | −168.77 | 0.000001 | D1_S_FP vs D1_S_PBS | Genic, antisense | GSONMG00062425001 si:ch73- protein (0.27) | ||
| Omy200226560 | −121.90 | 0.001972 | D1_R_Fp vs D1_R_PBS | Genic, antisense | GSONMG00065518001 (fatty-acyl reductase-1) (0.36) | ||
| Omy100064313 | −108.56 | 0.001716 | D1_R_Fp vs D1_R_PBS | Intergenic | |||
LncRNAs upregulated in all three genetic lines (>2 fold) on 5th day post Fp challenge and their expression correlation with protein coding genes (top). LncRNAs showing highest fold change (>100-fold) upon Fp infection in at least one genetic line relative to the two other genetic lines and their associated protein coding gene in genome (bottom). Fold change was considered significant if FDR-corrected p value was less than 0.05.
Among DE lncRNAs, 6 lncRNAs showed fold changes >100 fold following Fp challenge (Table 2, bottom panel). Five out of six lncRNAs, all three upregulated (Omy200018785, Omy200132807 and Omy100037031) and two downregulated (Omy200226560 and Omy100064313), exhibited fold change only in one particular ‘genetic line-by-day of infection’ comparison.
Relationship between differentially expressed lncRNAs and immune-related protein-coding genes
LncRNAs can be classified as genic or intergenic based on their physical location in genome relative to protein coding gene20. Classification of all 556 DE lncRNA is given in Supplementary Dataset 1D. Lack of lncRNAs sequence conservation across species21 makes their annotation difficult. In addition, currently there are no enough literature or database resources for rainbow trout and other salmonids to study lncRNAs’ involvement with the host immune system. Therefore, in an effort to implicate association between DE lncRNAs, identified in this study, and the fish defense system, we followed the following criteria based on our prior knowledge of lncRNAs classification and the genetic lines that we used in this study.
Differentially expressed lncRNAs that overlap in position and correlate with expression of immune-related protein-coding genes
Several lncRNAs have been identified that regulate expression of neighboring genes acting in cis configuration23,24. Therefore, we searched for DE lncRNAs that were partially or completely overlapping with protein-coding genes in the trout genome. Out of 556 DE lncRNAs, 92 overlapped with protein-coding loci in sense or antisense orientation (Supplementary Dataset 3). Out of the 92 overlapped genes, 36 genes had hits to KEGG pathways, of them 8 different genes were involved in immunity pathways (such as TNF and mTOR signaling pathways) and 4 genes were associated with microbial diseases (such as Staphylococcus aureus infection and viral carcinogenesis). There were 3 genes common in both sets of these pathways which are Complement component 5, Signal transducer and activator of transcription 3 and Cyclic AMP-responsive element-binding protein 5.
In order to identify possible relationships between DE lncRNAs and protein-coding genes that physically overlap with them, we compared their expression patterns across 24 different samples that included different genetic lines and infection statuses. Normalized expressions values of the transcripts used to generate clusters are provided in Supplementary Dataset 4. The DE lncRNAs and their overlapping protein-coding genes with a strong expression correlation are listed in Table 3. Overall, we identified 13 protein-coding genes that had strong expression correlation (R2 ≥ 0.70) with their overlapping lncRNAs and 6 of those protein-coding genes had already described role in immune system. Consistent with this observation, previous studies suggested overlapped genomic localization of immunity associated lncRNAs with protein coding genes of immune system25.
Table 3. Correlation between expression patterns of lncRNAs and their overlapping protein-coding genes (R2 > 0.70).
| LncRNA | Size | Neighboring protein-coding genes (ID) | Overlap | Direction relative to LncRNA | Expression correlation type (R2) | Annotation of coding gene | Reference to immunity or pathogenesis function |
|---|---|---|---|---|---|---|---|
| Protein-coding genes with known immune function or association with microbial infection | |||||||
| Omy100063056 | 1,263 | GSONMT00040216001 | Intronic | Antisense | Positive (0.73) | Interferon-induced guanylate-binding protein 1-like | 51,52 |
| Omy200083892 | 1,294 | GSONMT00050654001 | Intronic | Antisense | Positive (0.84) | Tumor necrosis factor receptor superfamily member 9-like (Tnfrsf9) | 9 |
| Omy200080884 | 1,512 | GSONMT00034829001 | Exonic | Antisense | Positive (0.93) | Response gene to complement 32 protein (rgc32) | 53 |
| Omy200206941 | 537 | GSONMT00021084001 | Intronic | Unknown | Positive (0.83) | Lysozyme C II precursor | 29 |
| Omy200107012 | 885 | GSONMT00019341001 | Intronic | Unknown | Positive (0.89) | Stromal interaction molecule 2-like | 54 |
| Omy100228715 | 297 | GSONMT00079494001 | Exonic | Unknown | Positive (0.83) | Unnamed protein product/transcobalamin-1 like | 55 |
| Protein-coding genes with no previously described immunity function | |||||||
| Omy400008156 | 668 | GSONMT00041383001 | Intronic | Unknown | Positive (0.87) | Reticulon-2 like | |
| Omy300038945 | 596 | GSONMT00049537001 | Intronic | Antisense | Positive (0.81) | Cytochrome P450 7B1 | |
| Omy400006181 | 248 | GSONMT00049631001 | Intronic | Unknown | Positive (0.77) | Collagen alpha-1(IX) chain-like | |
| Omy400003716 | 725 | GSONMT00061535001 | Intronic | Sense | Positive (0.87) | Protocadherin 8 (pcdh8) | |
| Omy100224015 | 683 | GSONMT00065518001 | Intronic | Antisense | Positive (0.71) | Fatty acyl-CoA reductase 1 (facr1) | |
| Omy200181316 | 604 | GSONMT00071779001 | Exonic | Unknown | Positive (0.82) | Muscular LMNA-interacting protein | |
| Omy100171980 | 1,292 | GSONMT00073108001 | Exonic | Unknown | Positive (0.82) | Immunoglobulin-like and fibronectin type III domain-containing protein 1 | |
References are provided for protein-coding genes with previously described functions in immunity or association with microbial infection/pathogenesis.
Some lncRNAs showed interesting correlated expression pattern with immune-related protein coding genes post Fp challenge and were selected for the following further discussion.
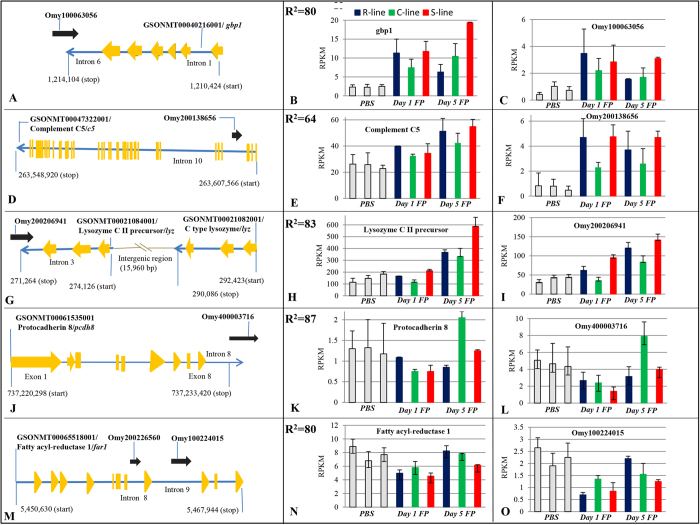
LncRNA Omy100063056 partially overlapped with intron 6 of interferon induced guanylate binding protein-1 like (gbp1) (GSONMT00040216001) in antisense orientation and their expression pattern was positively correlated (R2 = 0.80) (Fig. 1A–C). RPKM (reads per kilobase per million) count showed that both Omy100063056 and gbp1 gene transcript were upregulated on day 1 and 5 post-challenge. Upregulation on day 5 was greater in the susceptible line relative to control and resistant lines. GPB1 gene transcript also shows correlated expression with lncRNA in human26. Previous reports suggested that gbp1 is one of the differentially regulated immune response genes against microbial pathogens in salmon and trout9,27.
Figure 1. Genomic location of selected differentially expressed lncRNAs relative to protein-coding genes with immune-related functions and their expression patterns among PBS injected and day 1 and day 5 post-Fp challenged fish of different genetic lines.
Omy100063056 is partially overlapped with intron 6 of gbp1 in antisense orientation (A) and their expression is positively correlated (R2 = 0.80) (B,C). Omy2001386656 is within intron of complement C5 in antisense orientation (D) and they show correlated expression pattern between PBS and Fp injected fish (R2 = 0.64) (E,F). Omy200206941 partially overlaps with intron of lysozyme CII precursor in antisense orientation (G) and shows correlated expression pattern with the lysozyme CII precursor (R2 = 0.83) (H,I). Omy400003716 partially overlaps with intron of protocadherin 8 in sense orientation (J) and shows strong positive expression correlation with the protocadherin 8 (R2 = 0.87) (K,L). Fatty acyl-reductase 1 has one sense lncRNA in each intron 8 (Omy200226560) and 9 (Omy100224015) (M) and shows positive expression correlation with both the lncRNAs. Expression pattern of fatty acyl reductase 1 and Omy100224015 is given in figure (N,O).
LncRNA Omy200128656 was located in intron 11 of complement C5 (c5) (GSONMT00047322001) gene in antisense orientation and their expression was positively correlated (R2 = 0.64) (Fig. 1D–F). Expression of c5 gene transcript was increased by day 5 post-infection and expression of Omy200128656 was upregulated on days 1 and 5 post-challenge. In human, lncRNA C5T1lncRNA, located in 3′UTR of the C5, showed upregulated expression upon immune stimulation and its knockdown showed corresponding decrease in transcript level of C5 mRNA28. However, unlike in human, lncRNA Omy200128656 in trout is located in intron 11 of the c5 gene.
LncRNA Omy200206941 was partially overlapped with intron 4 of lysozyme CII precursor (lyz) (GSONMT00021084001) gene in antisense orientation and the expression was positively correlated (R2 = 0.83) (Fig. 1G–I). Its expression was also positively correlated with another C type lysozyme (lyz) (GSONMT00021082001) gene transcript located about 18 kb away in the same chromosome (R2 = 0.88). All these three transcripts showed upregulation on day 5 post-challenge. Consistent with this upregulated expression post challenge, it has been established that C type lysozyme is an important component of innate immune system in salmonid fish29. In addition, a neighboring antisense non-coding RNA, LINoCR, is involved in induction of lysozyme locus upon lipopolysaccharide stimulation in chicken30.
LncRNA Omy400003716 partially overlapped with intron 8 of protocadherin 8 (pcdh8) (GSONMT00061535001) in sense orientation and the expression was highly positively correlated (R2 = 0.87) (Fig. 1J–L). RPKM count between PBS and Fp challenged fish showed that both Omy400003716 and pcdh8 gene transcript were downregulated in day 1 post infection relative to naïve and day 5 post-challenged fish.
Two lncRNAs Omy200226560 and Omy100224015 were in intron 8 and 9 of fatty acyl-reductase 1 (far1) (GSONMT00065518001) respectively and they positively correlated with the far1 gene transcript with correlation coefficient (R2) of 0.36 and 0.80 respectively (Fig. 1M–O). These three transcripts showed downregulation on day 1, post challenge relative to PBS injected, and day 5, post-Fp challenged fish.
Strand orientation of Omy200138656, Omy200206941 and Omy300084989 lncRNAs transcripts were confirmed by strand specific PCR relative to their counterpart protein coding loci (Supplementary Dataset 1E).
Differentially expressed lncRNAs that neighbor and correlate with expression of immune-related protein-coding genes
Out of 556 DE lncRNAs, 464 were intergenic without overlap with protein-coding loci in the trout genome. In order to identify the immune-relevant protein-coding genes that were clustered around DE lncRNAs in the genome, we chose protein-coding genes within a 50 kb distance on both sides of DE lncRNAs and performed KEGG pathway analysis of the neighboring protein-coding genes31. Out of 464 DE intergenic lncRNAs, 371 had protein-coding genes within 50 kb distance in the genome. A total of 290 genes neighboring to DE lncRNAs had hits to KEGG pathways, of them 51 different genes were related to immunity pathways, 49 genes were involved in microbial infection processes and 28 genes were common in both sets of these pathways (Supplementary Dataset 5).
In the immune system category, most of the KEGG hits were involved in chemokine signaling, platelet activation, complement system, TNF signaling, T cell receptor signaling, Fc gamma R-mediated phagocytosis, Toll-like receptor signaling, phagosome, cytokine-cytokine receptor interaction, NOD-like receptor signaling, leukocyte trans-endothelial migration and others (Supplementary Dataset 5). Similarly, in the microbial pathogenesis category, hits were involved in the pathogenesis of various viral, bacterial and protozoal infections like tuberculosis, influenza A, herpes simplex infection, amoebiasis, bacterial invasion of epithelial cell, and other microbial infections. Interestingly, almost half of the hits to immune system were involved in signal transduction pathways. Among the neighboring protein-coding genes, expression patterns of 9 were highly positively correlated with that of lncRNA (R2 ≥ 0.70) (Table 4). About half of the protein-coding genes with high correlation in expression patterns with their neighboring lncRNAs were from components of immune system like suppressor of cytokine signaling 3 (SOCS3), complement factor D, ninjurin-1 and ceramide-1 phosphate transfer protein. Previous studies also indicated that many immune relevant lncRNAs are in 5′ or 3′ close proximity of neighboring protein-coding genes15,25.
Table 4. Correlation between expression patterns of lncRNAs and their intergenic neighboring protein-coding genes (within <50 kb and R2 > 0.70).
| LncRNA | Size | Neighboring protein-coding genes (ID) | Distance from LncRNA (KB) | Direction relative to LncRNA | Expression correlation type (R2) | Annotation of coding gene | Reference to immune or pathogenesis function | |
|---|---|---|---|---|---|---|---|---|
| Protein-coding genes with known immune function or association with microbial infection | ||||||||
| Omy200174653 | 519 | GSONMT00031633001 | 5.0 | Unknown/Intergenic | Positive (0.92) | Complement factor D-like | 56 | |
| Omy300084989 | 596 | GSONMT00013116001 | 2.6 | Antisense/Intergenic | Positive (0.71) | Suppressor of cytokine signaling | 57 | |
| Omy300074800 | 493 | GSONMT00003195001 | 1.1 | Unknown/Intergenic | Positive (0.79) | Ninjurin-1 | 58 | |
| Omy200206941 | 537 | GSONMT00021082001 | 18.6 | Unknown/Intergenic | Positive (0.88) | C type lysozyme | 29 | |
| Omy200073559 | 2,093 | GSONMT00017721001 | 3.5 | Unknown/Intergenic | Positive (0.77) | Ceramide-1-phosphate transfer protein-like | 59 | |
| Protein-coding genes with no previously described immunity function | ||||||||
| Omy200061208 | 1057 | GSONMT00041695001 | 0.3 | Unknown/Intergenic | Positive (0.90) | Coiled-coil transcriptional coactivator b | ||
| Omy200112536 | 1,059 | GSONMT00001821001 | 18.2 | Unknown/Intergenic | Positive (0.88) | Serum albumin 1 | ||
| Omy300087476 | 619 | GSONMT00010387001 | 0.9 | Unknown/Intergenic | Positive (0.83) | Neutral amino acid transporter B(0) | ||
| Omy200075445 | 745 | GSONMT00008107001 | 1.3 | Unknown/Intergenic | Positive (0.70) | Hepatocyte nuclear factor 4-beta-like | ||
References are provided for some of the protein-coding genes with previously described functions in immunity or association with microbial infection/pathogenesis.
Differentially expressed lncRNAs that correlate with expression of immune-related protein-coding genes
LncRNAs have ability to work in cis as well as in trans configuration32,33,34 and can regulate protein-coding genes that are distant in position on the same or different chromosome. In order to identify possible expression correlation of lncRNAs with such protein coding genes, we performed clustering of DE lncRNAs and protein-coding genes based on their expression pattern across 24 samples. This clustering identified several protein-coding genes with correlated expression with DE lncRNAs that were distantly located in the genome (Table 5). Most of the proteins in these clusters were related to the innate immune system, mainly the complement system, cytokines and chemokines, and receptors and transcription factors of the innate immune system signal transduction pathways. The list included chemokine CK1, NF-kappa B inhibitor alpha, c-c motif chemokine 19, and several proteins of the complements system such as factor B, properdin, component C7 and C4b-binding protein alpha (Table 5).
Table 5. Correlation between expression patterns of lncRNAs and some distantly located (>50 kb or different chromosome) immune-relevant protein-coding genes.
| LncRNA | Size | Protein-coding genes (ID) | Expression correlation type (R2) | Annotation of coding gene | Reference to immunity or pathogenesis function |
|---|---|---|---|---|---|
| Omy100104455 | 587 | GSONMT00024124001 | Positive (0.96) | Chemokine CK1 | 60 |
| Omy200174653 | 357 | GSONMT00051250001 | Positive (0.92) | C-C motif chemokine 19 precursor | 61 |
| Omy300084989 | 596 | GSONMT00062775001 | Positive (0.83) | C4b-binding protein alpha chain precursor | 62 |
| Omy300041057 | 448 | GSONMT00042009001 | Positive (0.80) | Caspase-8 | 63 |
| Omy300043066 | 715 | GSONMT00001792001 | Positive (0.82) | Properdin | 64 |
| Omy300043066 | 715 | GSONMT00027840001 | Positive (0.89) | Complement factor b-like | 65 |
| Omy200100893 | 357 | GSONMT00051250001 | Positive (0.92) | C-C motif chemokine 19 precursor | 61 |
| Omy200107378 | 522 | GSONMT00016681001 | Positive (0.92) | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 | 45 |
| Omy200107535 | 948 | GSONMT00016681001 | Positive (0.92) | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 | 45 |
| Omy200117486 | 529 | GSONMT00005714001 | Positive (0.82) | Interferon-induced guanylate-binding protein 1 | 51,52 |
| Omy100066751 | 326 | GSONMT00080410001 | Positive (0.84) | Tumor necrosis factor, alpha-induced protein 2 (tnfaip2) | 9 |
| Omy100128008 | 1,232 | GSONMT00070499001 | Positive (0.82) | Complement protein component C7-1 (c7-1) | 66 |
| Omy100063056 | 1,263 | GSONMT00075049001 | Positive (0.85) | Tumor necrosis factor, alpha-induced protein 3 (tnfaip3) | 9 |
| Omy200053140 | 1,510 | GSONMT00071335001 | Negative (0.84) | NF-kappa-B inhibitor alpha | 9 |
References are provided for some of the protein-coding genes with previously described functions in immunity or association with microbial infection/pathogenesis.
Differentially expressed lncRNAs that correlate with expression of several immune-related protein coding genes
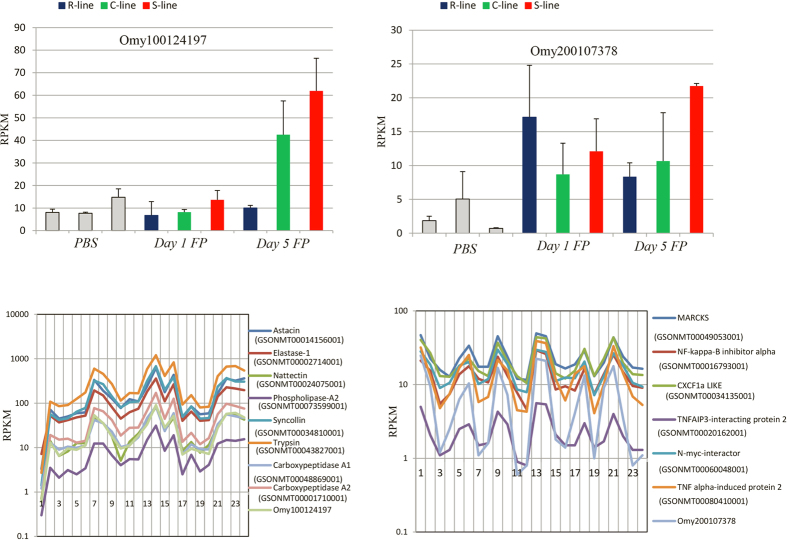
Clustering of DE lncRNAs with protein coding genes based on their expression value identified several protein-coding genes of the immune system correlated with one lncRNA. As an example, lncRNA Omy200107378 was upregulated post Fp challenge and its expression was strongly positively correlated with six different protein coding genes, some of which have already established function in immune system (R2 > 0.98) (Fig. 2). Similarly, expression of Omy100124197 was strongly correlated with 8 different proteins including matrix metallo-proteinase (Astacin) (GSONMT00014156001), elastase-1 (GSONMT00002714001), nattectin (GSONMT00024075001), phospholipase-A2 (GSONMT00073599001), and syncollin (GSONMT00034810001) (R2 > 0.98) (Fig. 2). Role of these correlated proteins in the immune system has already been characterized in different species35,36,37,38,39,40,41. Several studies have also reported correlated expression of several immune related protein-coding genes with a single lncRNA13.
Figure 2. Top two bar graphs show expression patterns of lncRNAs Omy100124197 and Omy200107378 among PBS injected, and day 1 and day 5 post-Fp challenged fish in three genetic lines.
Respective bottom expression line graphs show expression level of these lncRNAs with different protein-coding genes across 24 samples consisting of different genetic lines and infection statuses. Expression clusters were generated by the Multi-experiment Viewer (MeV) program using a cut off R2 minimum of 0.98.
LncRNAs expression of naïve fish in different genetic lines
Three genetic lines of rainbow trout used in this study had significant differences in infection susceptibility to Fp as a result of selective breeding9. To investigate differences in transcription between lines, we quantified the DE lncRNAs among genetic lines on day 1 following PBS injection. Pairwise comparison identified 32 DE lncRNAs among different genetic lines. Two lncRNAs were DE between the resistant and control lines, 6 lncRNAs between control and susceptible lines, and 24 lncRNAs were DE between resistant and susceptible lines (Supplementary Dataset 2). In our previous study, we identified differences in transcriptome abundance of protein-coding genes among naïve genetic lines9. The numbers of DE lncRNAs were smaller but consistent with the numbers of DE protein-coding genes among different naïve genetic lines (Table 1). Expression analysis identified an interesting pattern of transcriptome differences among genetic lines, which correlated with infection susceptibility. LncRNAs Omy200019549, Omy200132559, Omy200160814, Omy200075485 and Omy300048239 were most highly expressed in the resistant line, followed by control and susceptible lines. In contrast, Omy300052204, Omy200142923, Omy200118054 and Omy200165975 were upregulated in the susceptible line relative to the resistant and control lines (Fig. 3). These DE lncRNAs between genetic lines may contribute to differences in infection susceptibility among genetic lines. In consistent with our findings, genetic variation in lncRNAs was shown to be associated with human disease resistance/susceptibility42,43.
Figure 3. Comparison of transcriptome abundance of selected lncRNAs among naïve fish in all genetic lines.
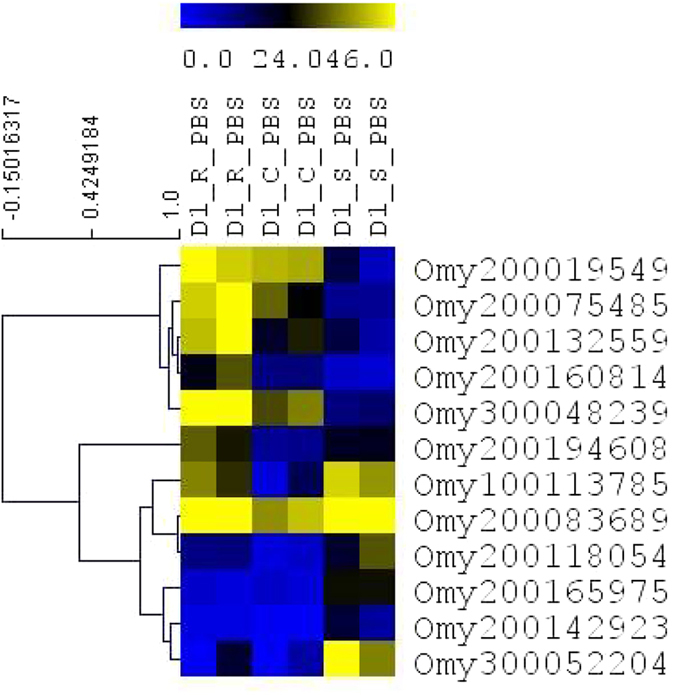
Genes are hierarchically clustered based on their expression pattern. D1 indicates day 1 post challenge and PBS indicates PBS injection. C, R and S represent control, resistant and susceptible genetic lines of the fish.
Difference in transcriptome abundance of lncRNAs among genetic lines after infection
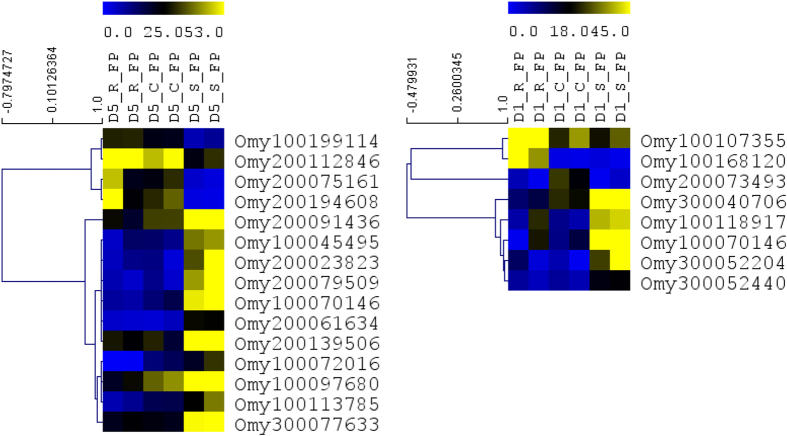
Induction and activation of adaptive and some of the innate immune components requires pathogen entry into the host suggesting that basal naïve transcriptome level may not be sufficient enough to explain the differences in the ability of the control, susceptible, and resistant fish lines to clear the pathogen. Therefore, we reasoned that, in addition to differences in naïve lncRNA abundance, the genetic lines had altered ability to express immune-relevant transcripts following pathogen challenge. To investigate this point, we quantified DE lncRNAs among genetic lines on days 1 and 5 following Fp infection. Pairwise comparison identified 149 DE lncRNAs between genetic lines combined from the 1st and 5th days of infection (Table 1 and Supplementary Dataset 2). On 5th day of infection, there were 83 lncRNAs DE between resistant and susceptible lines; 21 lncRNAs between resistant and control lines, and 5 lncRNAs between control and susceptible lines. On 1st day of infection, these numbers were 15, 12 and 13 respectively (Supplementary Dataset 2). Similarly, on the 1st day of infection majority of the lncRNAs were upregulated on susceptible line relative to two other genetic lines. The expression number of DE’s correlated with the gradient of bacterial load between the three genetic lines: S > C > R. Previous report also indicated correlation of lncRNAs expression with microbial load44. Figure 4 shows abundance of selected hierarchically clustered lncRNAs among genetic lines after infection with Fp. On the 5th day of infection, most of the lncRNAs were upregulated in the susceptible line compared to control and resistant lines, with only Omy200112846, Omy200075161, Omy200194608 and Omy100199114 exhibiting opposite trend in expression level (Fig. 4).
Figure 4. Comparison of transcriptome abundance of selected lncRNAs among genetic lines after infection with Fp.
Genes are hierarchically clustered based on their expression pattern. D1 and D5 indicate day 1 and day 5 of sampling after injection. Fp indicates Fp injection. C, R and S represent control, resistant and susceptible genetic lines of the fish.
LncRNA transcriptome change as the disease progress from day 1 to day 5
During the course of infection, the host can utilize different immune components at different stages of disease, which requires change in expression of immune-relevant genes. We reasoned that if lncRNAs regulate the immune system, their transcriptome changes, like that of protein-coding genes, would change as the disease progresses. Pairwise comparison between day 1 and day 5 post-Fp challenge identified 137 lncRNAs whose expression was significantly changed during two time points (Supplementary Dataset 2). This finding is consistent with previous report demonstrating change in the number of differentially regulated lncRNAs at different ISAV infection time points in Atlantic salmon19. Figure 5 shows abundance of selected hierarchically clustered lncRNAs between day 1 and day 5 of Fp injection in each genetic line. As expected, some of the lncRNAs that showed altered expression between day 1 and day 5 post-challenges had strong expression correlation with immune relevant protein coding genes. LncRNAs Omy200174653 had altered expression on day 5 relative to day 1 post challenge in susceptible lines and a strong positive correlation with complement factor D (Table 4). Similarly, Omy100066751 and Omy200107535 exhibited a strong positive expression correlation with tumor necrosis factor alpha-induced protein 2 (tnfaip2) and nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (NFKB2) (R2 = 0.92), respectively (Table 5). NFKB2 is a transcription factor required to maintain normal level of antigen specific antibody production in response to antigen challenge45. It is noteworthy that Omy200107535 was one of the 12 lncRNAs that were upregulated on day 5 post challenge relative to naïve fish in all three genetic lines (Table 2). This change in expression pattern of lncRNAs during the course of infection suggests that these lncRNAs may play a role in adjustment of immunity depending on severity and stage of the disease. In addition, these DE lncRNAs might play a role in host pathogen interaction or pathogen life cycle during the course of infection as suggested in previous studies46.
Figure 5. Comparison of transcriptome abundance of selected lncRNAs between day 1 and day 5 of Fp injection in each genetic line.
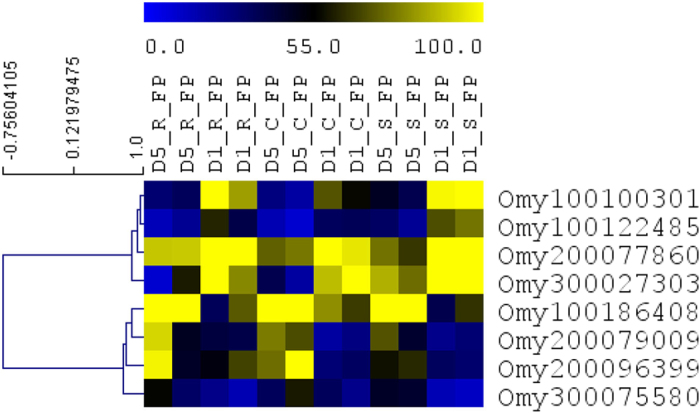
Genes were hierarchically clustered based on their expression pattern. D1 and D5 indicate day 1 and day 5 of sampling after injection and Fp indicates Fp injection. C, R and S represent control, resistant and susceptible genetic lines of the fish.
Sequence homology with lncRNAs in Atlantic salmon
Recently differentially regulated lncRNAs in response to infectious salmon anemia virus (ISAV) has been characterized in Atlantic salmon19. Out of 556 DE lncRNA in trout genetic lines in various comparisons, 23 showed significant sequence homology with Atlantic salmon lncRNAs that were associated with ISAV infection (query cover > 50%, sequence identity > 90% and E value < 1e-10) (Supplementary dataset 6). Interestingly, out of 23 conserved lncRNA, 17 showed regulated expression in Fp injected fish relative to PBS injected naïve animals; and remaining 6 were differently regulated between genetic lines and time points of infection comparison (Supplementary dataset 2). It is worth mentioning that one of the conserved lncRNA, Omy300043066 had strong positive expression correlation with properdin and complement factor b like protein in trout (Table 5) and was one of the 12 lncRNAs that were upregulated during infection in all three genetic lines relative to their PBS injected fish (Table 2). All of the 23-conserved lncRNA were regulated in salmon in response to ISAV, indicating potential role in general immunity rather than being bacterial or virus specific.
Novel lncRNAs in resistant and susceptible genetic lines
Novel lncRNAs were detected in each genetic line separately by running sequence reads through our previously described lncRNA discovery pipeline20. 589 susceptible-specific and 631 resistant-specific novel lncRNAs were predicted. FASTA files are available at http://www.animalgenome.org/repository/pub/MTSU2015.1014/. Correlation analyses of gene expression showed only 9 lncRNAs in moderate correlation (R2 ≥ 0.70) with protein coding genes. However, none of these proteins was overlapped with lncRNA or had previously described role in immune system (Supplementary dataset 7). While identification of these lncRNAs were limited to each genetic line, their multiple group ANOVA analysis of gene expression (genetic line X infection status X time point) showed a complex expression pattern (Supplementary dataset7-PCA). Interestingly, two lncRNA (dis_R_00048342 and dis_R_00050098) showed resistant-line specific gene expression regardless of the infection status or the time points. Similarly, three lncRNA (dis_S_00030301, dis_S_00043616 and dis_S_00083595) were susceptible-line specific (Supplementary dataset 7). On the other hand, 20 lncRNAs showed explicit expression after Fp infection, regardless of the time of infection or the genetic line. In addition, three lncRNA showed explicit expression between day 1 and day 5 of infection (Supplementary dataset 7). These finding may suggests that genomic selection for BCWD over three generations may have introduced novel genomic variations or genomic reorganization of some lncRNA loci and altered expressions of lncRNAs.
Conclusion
Thus far, studies on host response to microbial infection in salmonids have given significant attention to changes in protein-coding gene expression. However, lncRNAs have emerged as key regulators of host defense against a wide variety of pathological processes including microbial infection11,12,13,14,15,16,17,19. Manipulation of individual lncRNAs is sufficient to change the expression of hundreds of immune response genes13, and variation in expression of other lncRNA’s alter host susceptibility to different microbial pathogens17. In the present study, we quantified DE lncRNAs in response to Fp infection, which is an important cause of morbidity and mortality in salmon and trout2. This study is novel as we characterized the expression signature of lncRNAs on a genome-wide scale in response to one of the major bacterial infection of a salmonid fish. To our knowledge, regulation of lncRNA during bacterial pathogen challenge has not previously been studied in any aquaculture/fish species.
Using transcriptome-wide datasets of protein-coding genes and lncRNAs across 24 samples, we were able to identify potential immune-relevant and other protein-coding genes correlating with DE lncRNAs. This study identified correlation between the genomic physical proximity and coordinated expression of a large number of immune related and other protein coding genes with that of lncRNAs during BCWD in rainbow trout. In this study, most of the DE lncRNAs (sense and antisense) had significant positive expression correlation (R2 > 0.70) with their overlapped and/or neighboring protein coding genes. These results are consistent with human ENCODE project results that showed particularly striking positive correlation of lncRNAs with the expression of antisense coding genes21. In trans-acting lncRNAs, the ENCODE project observed that lncRNAs are more positively than negatively correlated with protein-coding genes, a finding consistent with our observation of more frequent positive than negative correlation with distantly located protein coding genes. The positive correlation between lncRNA and protein coding genes suggest potential for co-expression10.
This study has characterized DE lncRNAs in response to an initial phase of BCWD (day 1 and 5 post-challenge) and has explored expression correlation of lncRNAs with immune relevant protein coding gene that may play crucial role in pathogenesis or immunity during the early phase of the disease in rainbow trout. Further mechanistic study of the underlying biological relationship between correlated DE lncRNAs and proteins of innate immune system will help understand regulation of pathogenesis/immunity at this crucial phase of infection in juvenile rainbow trout.
Materials and Methods
Ethics statement
Fish were maintained at the NCCCWA and all experimental protocols and animal procedures were approved and carried out in accordance with the guidelines of NCCCWA Institutional Animal Care and Use Committee Protocols #053 and #076.
Experimental animals and RNA-Seq experimental design
Three rainbow trout genetic lines ARS-Fp-R, ARS-Fp-C, and ARS-Fp-S used in this study were developed at National Center for Cool and Cold Water Aquaculture (NCCCWA) rainbow trout breeding program. These genetic lines differ significantly to their susceptibility to Fp infection as a result of genetic selection8 and we have previously reported the challenge experiment utilized in this study9. Briefly, fifty randomly selected fish from each genetic line were assigned to four challenge tanks (total 12 tanks for three genetic lines). At the time of challenge, average body weight was 1.1 g and fish age was 49 days post-hatch. For each genetic line, fish in two tanks were injected with Fp (experimental group) and fish in the other two tanks were injected with PBS (control group). Fish were injection challenged with either 4.2 × 106 CFU Fp suspended in 10 μl of chilled PBS or PBS alone, and survival was monitored daily for 21 days9. For RNA extraction, five individuals were sampled from each tank on days 1 and 5 post infections. Survival at 21 days post-challenge injection was monitored during the experiment. Post-challenge bacterial load in the body was measured in a subset of fish by qPCR and was expressed in terms of Fp genome equivalents (GE).
RNA extraction, library preparation, and sequencing
Tissue sampling, RNA extraction, library preparation and sequencing were done as described previously9. Briefly, total RNA was extracted and equal amounts of RNA from five fish were pooled from each of the 12 tanks at each of the two time points (total of 24 pools, n = 120 fish total). cDNA libraries were prepared using Illumina’s TruSeq Stranded mRNA Sample Prep kit following the manufacturer’s instructions. The 24 indexed and barcoded libraries were randomly divided into three groups (eight libraries per group) and sequenced in three lanes of an Illumina HiSeq 2000 (single-end, 100 bp read length). RNA-Seq reads are available at the NCBI Short Read Archive (BioProject ID PRJNA259860, accession number SRP047070).
Differential gene expression analysis of lncRNAs
Complete description of lncRNA reference dataset with their discovery pipeline has been recently described20. From this discovery datasets, a stringently selected set of lncRNAs (31,195) were used as a reference for gene expression analysis. For differential gene expression analysis, sequencing reads from each library were mapped to the lncRNA reference using a CLC genomics workbench. Mapping conditions were, mismatch cost = 2, insertion/deletion cost = 3, minimum length fraction = 0.9 and similarity fraction = 0.9. The expression value of lncRNAs was calculated in terms of RPKM (reads per kilobase per million). EDGE (extraction and analysis of differential gene expression) tests were performed to identify DE genes between various groups, e.g. infected vs. non-infected, day 1 vs. day 5, and one genetic line vs. other with or without Fp injection47. To control false discovery due to multiple testing, p-values were FDR-corrected. LncRNA was considered significant at a fold-change cutoff value of ±2 and a corrected p-value of less than 0.05.
Validation of RNA-Seq data by qPCR
From DE lncRNAs in the RNA-Seq study, 7 were randomly selected from the DE day 5 susceptible line for experimental validation using individual (unpooled) samples. RNA isolation, cDNA synthesis and primer design were completed using the same technique as described previously9. Briefly, RNAs were treated with Optimize™ DNAase I (Fisher Bio Reagents, Hudson, NH) to eliminate genomic DNA. One microgram of the purified RNA was converted to cDNA using the Verso cDNA Synthesis Kit (Thermo Scientific, Hudson, NH) according to the manufacturer protocol. Reverse transcription was performed using My Cycler™ Thermal Cycler (Bio Rad, Hercules, CA) at 42 °C for 30 min (one cycle amplification) followed by 95 °C for 2 min (inactivation). Blend of random hexamer and oligo (dT) primer (3:1 V/V), at a final concentration of 25 ng/μL, was used to prime the reverse transcription reaction.
The Bio-Rad CFX96™ Real Time System (Bio-Rad, Hercules, CA) in conjunction with SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA) was used to quantify the amount of the expressed gene of interest in PBS and Fp injected whole-body fish homogenates. Each primer was used at a concentration of 0.1 nM/μL and cDNA template was used at a concentration of 0.006 μg/μL. Cycling temperatures were set up according to the manufacturer’s protocol and different annealing temperatures were used depending on primers. Fold change in gene expression was calculated as described previously9. Briefly, β-actin (Accession: AJ438158) was used as endogenous reference to normalize each target lncRNA. qPCR data were quantified using delta delta Ct (ΔΔCt) method48. Ct-values of β-actin were subtracted from Ct-values of the target gene to calculate the normalized value (ΔCt) of the target lncRNA in both the calibrator samples (PBS-injected) and test samples (Fp-injected). The ΔCt value of the calibrator sample was subtracted from the ΔCt value of the test sample to get the ΔΔCt value. Fold change in gene expression in the test sample relative to the calibrator sample was calculated by the formula 2−ΔΔCt and the normalized target Ct values in each infected and non-infected group was averaged. Correlation between gene expression fold-change measured by qPCR and RNA-Seq was performed by Pearson correlation. All statistics were performed with a significance of P < 0.05.
Gene clustering and gene expression correlation
Sequencing reads from all 24 libraries (samples) were mapped to a combined reference sequence consisting of all lncRNAs, that we previously identified20, and mRNAs that were identified in the rainbow trout genome49. Expression of lncRNAs and protein-coding genes was measured in terms of RPKM. The expression value of each transcript in each sample was normalized using the scaling method50. Mean was chosen as normalization value and median mean was chosen as reference. Five percent of the data on both sides of the tail were trimmed. Normalized expression values of transcripts in each sample were used to cluster protein-coding genes and lncRNAs using algorithms in Multi-experiment Viewer (MeV). Clusters were generated with a minimum correlation coefficient of 0.92. During clustering, 30% of the sequences with flat expression values over samples were excluded from cluster generation to prevent uninteresting cluster generation. Correlation in expression of lncRNAs and neighboring/overlapped protein-coding genes was performed in Excel using regression analysis using normalized expressions values of the transcripts.
Discovery of novel lncRNAs in resistant and susceptible genetic lines
Novel lncRNA were identified according to Al-Tobasei et al.20. Briefly, sequencing reads from each genetic lines (resistance, control and susceptible) were aligned to a rainbow trout reference genome using TopHat49. Cufflinks, Cufflinks compare and Cufflinks Merge were used to predict transcripts in each genetic line. Transcripts shorter than 200 nt were filtered out using in house perl script. Transcripts which had open reading frame (ORF) longer than 100 amino acids were removed. In addition, if ORF of the transcript is longer than 35% of the transcript length, the transcript was filtered out even if the ORF is shorter than 100 amino acids. Subsequently, transcripts were searched against NR protein database (updated on May 2016) using BLASTx, and any transcripts with sequence homology to existing proteins were removed. To remove any remaining protein coding transcripts, coding potential calculator (CPC) was applied to the transcripts (Index value < −1.0). Other classes of non-coding RNAs (e.g. rRNA, tRNA, snoRNA, miRNA, siRNA and others) in the dataset were removed by blasting (BLASTn) the transcripts against multiple RNA databases including genomic tRNA database. Finally, any single exon transcripts within 500 nts of protein coding gene was removed. After these filtration steps, remaining transcripts were considered as putative lncRNAs. To identify lncRNAs specific to a particular genetic line, lncRNAs from one genetic line were compared with lncRNAs from other two genetic lines. Resistant and susceptible specific lncRNA were reported.
Additional Information
How to cite this article: Paneru, B. et al. Differential expression of long non-coding RNAs in three genetic lines of rainbow trout in response to infection with Flavobacterium psychrophilum. Sci. Rep. 6, 36032; doi: 10.1038/srep36032 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We acknowledge technical contributions from Travis Moreland and Joel Caren. This work was supported by Agricultural Research Service CRIS Project 1930-32000-005 “Integrated Approaches for Improving Aquatic Animal Health in Cool and Cold Water Aquaculture,” CRIS Project 1930-32000-009 “Utilization of Genomics for Improving Production Traits in Cool and Cold Water Aquaculture” and Agriculture and Food Research Initiative competitive grants no. 2012-67015-30217 (to Gregory D. Wiens) and No. 2014-67015-21602 (to Mohamed Salem) from the USDA National Institute of Food and Agriculture. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US, Department of Agriculture. USDA is an equal opportunity employer.
Footnotes
Author Contributions Y.P., G.D.W. and M.S. conceived and designed the experiments. Y.P. and G.D.W. performed the experiment. B.P., R.A.T., Y.P., G.D.W. and M.S. analyzed the data. R.A.T. contributed reagents/materials/analysis tools; B.P. and M.S. wrote the paper. All authors reviewed and approved the manuscript.
References
- Asche F., Håvard H., Ragnar T. & Sigbjørn T. The salmon disease crisis in Chile. Marine Resource Economics 24, 405–411 (2009). [Google Scholar]
- Nematollahi A., Decostere A., Pasmans F. & Haesebrouck F. Flavobacterium psychrophilum infections in salmonid fish. J Fish Dis 26, 563–574 (2003). [DOI] [PubMed] [Google Scholar]
- Kent L., Groff J., Morrison J., Yasutake W. & Holt R. Spiral swimming behavior due to cranial and vertebral lesions associated with Cytophaga psychrophila infections in salmonid fishes. Diseases of Aquatic Organisms 6, 11–16 (1989). [Google Scholar]
- Carson L. & Schmidtke J. Characteristics of Flexibacter psychrophilus isolated from Atlantic salmon in Australia. Diseases of Aquatic Organisms 21, 157–161 (1995). [Google Scholar]
- Brown L., Cox W. & Levine R. Evidence that the causal agent of bacterial cold-water disease Flavobacterium psychrophilum is transmitted within salmonid eggs. Diseases of Aquatic Organisms 29, 213–218 (1997). [Google Scholar]
- Gómez E., Méndez J., Cascales D. & Guijarro J. A. Flavobacterium psychrophilum vaccine development: a difficult task. Microb Biotechnol 7, 414–423, doi: 10.1111/1751-7915.12099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjedrem T. Selection and breeding programs in aquaculture. Dordrecht: Springer (2005). [Google Scholar]
- Wiens G. D. et al. On-farm performance of rainbow trout (Oncorhynchus mykiss) selectively bred for resistance to bacterial cold water disease: effect of rearing environment on survival phenotype. Aquaculture 388, 128–136 (2013). [Google Scholar]
- Marancik D. et al. Whole-body transcriptome of selectively bred, resistant-, control-, and susceptible-line rainbow trout following experimental challenge with Flavobacterium psychrophilum. Front Genet 5, 453, doi: 10.3389/fgene.2014.00453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927, doi: 10.1101/gad.17446611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X. et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio 1, doi: 10.1128/mBio.00206-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H. et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 42, 10668–10680, doi: 10.1093/nar/gku713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S. et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792, doi: 10.1126/science.1240925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313, doi: 10.1126/science.1251456 (2014). [DOI] [PubMed] [Google Scholar]
- Hu G. et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol 14, 1190–1198, doi: 10.1038/ni.2712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia F. et al. Dynamic transcription of long non-coding RNA genes during CD4+ T cell development and activation. PLoS One 9, e101588, doi: 10.1371/journal.pone.0101588 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J. A. et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152, 743–754, doi: 10.1016/j.cell.2013.01.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S. P., Collins P. L., Williams C. L., Boothby M. R. & Aune T. M. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol 189, 2084–2088, doi: 10.4049/jimmunol.1200774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltana S., Valenzuela-Miranda D., Aguilar A., Mackenzie S. & Gallardo-Escarate C. Long noncoding RNAs (lncRNAs) dynamics evidence immunomodulation during ISAV-Infected Atlantic salmon (Salmo salar). Sci Rep 6, 22698, doi: 10.1038/srep22698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tobasei R., Paneru B. & Salem M. Genome-Wide Discovery of Long Non-Coding RNAs in Rainbow Trout. PLoS One 11, e0148940, doi: 10.1371/journal.pone.0148940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research 22, 1775–1789, doi: 10.1101/gr.132159.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madetoja J., Nyman P. & Wiklund T. Flavobacterium psychrophilum, invasion into and shedding by rainbow trout Oncorhynchus mykiss. Diseases of Aquatic Organisms 43, 27–38, doi: 10.3354/dao043027 (2000). [DOI] [PubMed] [Google Scholar]
- Ørom U. A. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58, doi: 10.1016/j.cell.2010.09.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Sun S. & Lee J. T. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143, 390–403, doi: 10.1016/j.cell.2010.09.049 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- NE, I. I. et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nature communications 5, 3979, doi: 10.1038/ncomms4979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriocanal M., Carnero E., Segura V. & Fortes P. Long Non-Coding RNA BST2/BISPR is Induced by IFN and Regulates the Expression of the Antiviral Factor Tetherin. Front Immunol 5, 655, doi: 10.3389/fimmu.2014.00655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries K. M. et al. Immune response genes and pathogen presence predict migration survival in wild salmon smolts. Mol Ecol 23, 5803–5815, doi: 10.1111/mec.12980 (2014). [DOI] [PubMed] [Google Scholar]
- Messemaker T. C. et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun 17, 85–92, doi: 10.1038/gene.2015.54 (2016). [DOI] [PubMed] [Google Scholar]
- Saurabh S. & Sahoo P. K. Lysozyme: an important defence molecule of fish innate immune system. Aquaculture Research 39, 223–239 (2008). [Google Scholar]
- Lefevre P., Witham J., Lacroix C. E., Cockerill P. N. & Bonifer C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell 32, 129–139, doi: 10.1016/j.molcel.2008.07.023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A. C. & Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35, W182–W185, doi: 10.1093/nar/gkm321 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K. M., Mayer C., Postepska A. & Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24, 2264–2269, doi: 10.1101/gad.590910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Ramadass A., Serra Barros A., Chow N. & Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445, 666–670, doi: 10.1038/nature05519 (2007). [DOI] [PubMed] [Google Scholar]
- Jeon Y. & Lee J. T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133, doi: 10.1016/j.cell.2011.06.026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Ferreira M. et al. Structural and biological characterization of Nattectin, a new C-type lectin from the venomous fish Thalassophryne nattereri. Biochimie 93, 971–980, doi: 10.1016/j.biochi.2011.03.001 (2011). [DOI] [PubMed] [Google Scholar]
- Saraiva T. C. et al. Nattectin a fish C-type lectin drives Th1 responses in vivo: licenses macrophages to differentiate into cells exhibiting typical DC function. Int Immunopharmacol 11, 1546–1556, doi: 10.1016/j.intimp.2011.05.012 (2011). [DOI] [PubMed] [Google Scholar]
- Parks W. C., Wilson C. L. & López-Boado Y. S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol 4, 617–629, doi: 10.1038/nri1418 (2004). [DOI] [PubMed] [Google Scholar]
- Bach J. P. et al. The secretory granule protein syncollin localizes to HL-60 cells and neutrophils. J Histochem Cytochem 54, 877–888, doi: 10.1369/jhc.5A6792.2006 (2006). [DOI] [PubMed] [Google Scholar]
- Belaaouaj A. et al. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med 4, 615–618 (1998). [DOI] [PubMed] [Google Scholar]
- Belaaouaj A., Kim K. S. & Shapiro S. D. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science 289, 1185–1188 (2000). [DOI] [PubMed] [Google Scholar]
- Belaaouaj A. Neutrophil elastase-mediated killing of bacteria: lessons from targeted mutagenesis. Microbes Infect 4, 1259–1264 (2002). [DOI] [PubMed] [Google Scholar]
- Kumar V. et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet 9, e1003201, doi: 10.1371/journal.pgen.1003201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One 7, e35145, doi: 10.1371/journal.pone.0035145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar K., Hanisch C., Palma Vera S. E., Einspanier R. & Sharbati S. Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci Rep 6, 19416, doi: 10.1038/srep19416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caamaño J. H. et al. Nuclear factor (NF)-kappa B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J Exp Med 187, 185–196 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaria V. & Pasha A. Long Non-Coding RNAs in Infection Biology. Front Genet 3, 308, doi: 10.3389/fgene.2012.00308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M. et al. Transcriptome assembly, gene annotation and tissue gene expression atlas of the rainbow trout. PLoS One 10, e0121778, doi: 10.1371/journal.pone.0121778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D. & Livak K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- Berthelot C. et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nature communications 5, 3657, doi: 10.1038/ncomms4657 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M. & Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). [DOI] [PubMed] [Google Scholar]
- Itsui Y. et al. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50, 1727–1737, doi: 10.1002/hep.23195 (2009). [DOI] [PubMed] [Google Scholar]
- Anderson S. L., Carton J. M., Lou J., Xing L. & Rubin B. Y. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256, 8–14, doi: 10.1006/viro.1999.9614 (1999). [DOI] [PubMed] [Google Scholar]
- Tang R., Zhang G. & Chen S. Y. Response gene to complement 32 protein promotes macrophage phagocytosis via activation of protein kinase C pathway. J Biol Chem 289, 22715–22722, doi: 10.1074/jbc.M114.566653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmann M. K. et al. Stromal interaction molecules 1 and 2 are key regulators of autoreactive T cell activation in murine autoimmune central nervous system inflammation. J Immunol 184, 1536–1542, doi: 10.4049/jimmunol.0902161 (2010). [DOI] [PubMed] [Google Scholar]
- Flach C. F. et al. Broad up-regulation of innate defense factors during acute cholera. Infect Immun 75, 2343–2350, doi: 10.1128/IAI.01900-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprong T. et al. Deficient alternative complement pathway activation due to factor D deficiency by 2 novel mutations in the complement factor D gene in a family with meningococcal infections. Blood 107, 4865–4870, doi: 10.1182/blood-2005-07-2820 (2006). [DOI] [PubMed] [Google Scholar]
- Arnold C. E. et al. A critical role for suppressor of cytokine signalling 3 in promoting M1 macrophage activation and function in vitro and in vivo. Immunology 141, 96–110, doi: 10.1111/imm.12173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein C. et al. Ninjurin1 Promotes TLR4 Signaling and Contributes to Systemic Inflammation. Am J Respir Cell Mol Biol, doi: 10.1165/rcmb.2014-0354OC (2015). [DOI] [PubMed] [Google Scholar]
- Rivera I. G. et al. Sphingomyelinase D/ceramide 1-phosphate in cell survival and inflammation. Toxins (Basel) 7, 1457–1466, doi: 10.3390/toxins7051457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutyser E., Richmond A. & Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol 78, 14–26, doi: 10.1189/jlb.1204712 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aritomi K. et al. Altered antibody production and helper T cell function in mice lacking chemokines CCL19 and CCL21-Ser. Microbiol Immunol 54, 691–701, doi: 10.1111/j.1348-0421.2010.00266.x (2010). [DOI] [PubMed] [Google Scholar]
- Blom A. M. A cluster of positively charged amino acids in the alpha-chain of C4b-binding protein (C4BP) is pivotal for the regulation of the complement system and the interaction with bacteria. Scand J Clin Lab Invest Suppl 233, 37–49 (2000). [PubMed] [Google Scholar]
- Beisner D. R., Ch’en I. L., Kolla R. V., Hoffmann A. & Hedrick S. M. Cutting edge: innate immunity conferred by B cells is regulated by caspase-8. J Immunol 175, 3469–3473 (2005). [DOI] [PubMed] [Google Scholar]
- Götze O. & Müller-Eberhard H. J. The role of properdin in the alternate pathway of complement activation. J Exp Med 139, 44–57 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade C. et al. Deficiency in complement factor B. N Engl J Med 369, 1667–1669, doi: 10.1056/NEJMc1306326 (2013). [DOI] [PubMed] [Google Scholar]
- Merle N. S., Church S. E., Fremeaux-Bacchi V. & Roumenina L. T. Complement System Part I - Molecular Mechanisms of Activation and Regulation. Front Immunol 6, 262, doi: 10.3389/fimmu.2015.00262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.