Abstract
In our previous study, NKB/NK3R system has been shown to act at the pituitary level to up-regulate SLα synthesis and secretion in grass carp. However, whether NK3R expression can serve as a regulatory target at the pituitary level and contribute to NKB interactions with other SLα regulators is still unclear. In current study, using grass carp pituitary cells as a model, we have a novel finding that co-treatment of SLα/SLβ with carp TAC3 gene products, could induce a noticeable enhancement in SLα mRNA expression and these potentiating effects occurred with a parallel rise in NK3R transcript level after SLα/SLβ treatment. Interestingly, the stimulatory effects of SLα/SLβ on NK3R gene expression could be further potentiated by co-treatment with IGF-I/-II and simultaneous exposure of carp pituitary cells to SLα/SLβ and IGF-I/-II in the presence of TAC3 gene products was found to markedly elevated SLα mRNA expression (20 fold increase) and this synergistic stimulation was mediated by cAMP/PKA-, PLC/PKC- and Ca2+ -dependent cascades functionally coupled with NK3R activation. These findings suggest that local release of SLα via functional interactions with IGF-I/-II and TAC3/NK3R system may constitute a potent stimulatory signal for SLα gene expression in the carp pituitary via up-regulation of NK3R expression.
Somatolactin (SL), the latest member of growth hormone (GH)/prolactin (PRL) family, is a fish-specific hormone released from the neurointermediate lobe (NIL) of the posterior pituitary1. Two isoforms of SL, SLα and SLβ, have been identified in fish pituitary, e.g., in zebrafish2, salmon3, and grass carp4, and suspected to have overlapping and yet distinct functions5. To date, SL has been shown to be involved in diverse functions in fish models, including chromatophore proliferation and differentiation6, pigment aggregation7, inflation of swim bladder during embryo development5, reproduction3, stress responses8, lipid metabolism9, and osmoregulation10. At the pituitary level, somatolactin secretion and gene expression are known to be under the control by various hypothalamic factors, e.g., dopamine11, GnRH12,13, PACAP4, neurokinin B (NKB)14, MCH15 and octadecdaneuropeptide16. Recently, insulin-like growth factors (IGFs) have also been identified as potent stimulators for SLα and SLβ secretion and gene expression in grass carp pituitary cells17. At the pituitary level, “somatolactin autoregulation” via autocrine/ paracrine effects of SLα and SLβ released locally within the pituitary has also been reported in grass carp7. However, the functional interactions of local release of SL with other SL regulators have not been examined and still awaited for further investigation.
NKB is the gene product of tachykinin 3 (TAC3), which is a member of the tachykinin family with pleiotropic functions, including the control of smooth muscle contraction in the gastrointestinal tract18, fluid secretion in the gut epithelium19, vasodilating effect for blood pressure modulation20 and stimulating effect on sperm movement21. The biological actions of NKB are mediated mainly by the Type 3 neurokinin receptor (NKR), namely NK3R, which is a member of the rhodopsin-type class I group G protein coupled receptor22 and functionally coupled with cAMP/PKA-23, PLC/IP3/PKC-24, and Ca2+ -dependent25 signaling pathways. In recent years, the functional role of NKB in puberty onset26 and human fertility has aroused a lot of interest in the field of reproductive biology, mainly due to the findings that loss-of-function mutations in NKB or its receptor NK3R can lead to hypogonadotropic hypogonadismor and even infertility in human subjects27,28,29. Based on the studies in mammals (e.g., rodent and sheep), NKB was found to be a key regulator for GnRH pulsatility and downstream LH release via NK3R activation in kisspeptin neurons located in the arcuate nuclei within the hypothalamus30,31. Similar investigations have been recently extended to fish models with the novel findings that the TAC3 gene in fish species, e.g., in zebrafish32,33,34, goldfish35, tilapia36 and grass carp14, not only encodes NKB as the gene product, but also the mature peptide of a new member of tachykinin called NKB-related peptide (NKBRP). Similar to NKB, NKBRP was also effective in stimulating LH release, e.g., in zebrafish32, goldfish35 and tilapia36, suggesting the reproductive function of TAC3 gene products was well conserved throughout vertebrate evolution. Based on our recent in vitro studies in grass carp pituitary cells, interestingly, NKB and NKBRP were found to have no effect on LH secretion and LHβ & GtHα gene expression at the pituitary level but rather serve as novel stimulators for prolactin (PRL) and SLα secretion and gene expression via differential activation of NK2R and NK3R expressed in the carp pituitary14. The investigation on SLα regulation by TAC3 gene products in the carp model have become more exciting with the recent demonstration that IGF-I/-II could act in a synergistic manner with TAC3 gene products, namely NKB and NKBRP, to up-regulate SLα gene expression at the pituitary level and this potentiating effect could be paralleled with the concurrent rise in NK3R expression induced by IGF-I/-II treatment. These new findings demonstrated for the first time that NK3R expression at the pituitary level could serve as a regulatory target for modulation of pituitary hormone gene expression in vertebrate species.
In this study, using primary culture of grass carp pituitary cell as a model, the functional interactions between somatolactin autoregulation with IGF-I/-II and TAC3 gene products on SLα gene expression were examined in the carp pituitary with focus on the role of NK3R expression as a regulatory target at the pituitary level. As a first step, co-treatment of SLα/SLβ with either IGF-I/-II alone or TAC3 gene products alone including NKBa and NKBRPa (the gene products of carp TAC3a gene) or with a combination of both were performed to examine their effects on SLα mRNA expression in carp pituitary cells. The potentiating effects observed with SL/IGFs and SL/TAC3 gene product co-treatment on SLα gene expression were correlated with parallel changes of NK3R mRNA expression induced by SLα/SLβ treatment alone or in combination with IGF-I/-II co-treatment. Using a pharmacological approach, the signal transduction mechanisms involved in SLα and SLβ induction of NK3R mRNA expression were elucidated and the functional role of NK3R expression and the post-receptor signaling pathways coupled with NK3R activation in the potentiating effects on SLα mRNA expression observed with cotreatment of IGF-I/-II and TAC3 gene products was also confirmed at the pituitary cell level. Our studies for the first time provide evidence that local release of SLα and SLβ could interact with IGF-I/-II and TAC3 gene products to up-regulate SLα gene expression in the carp pituitary via stimulation of NK3R expression at the pituitary level.
Results
Synergistic effects of somatolactin and TAC3a gene products on SLα mRNA expression
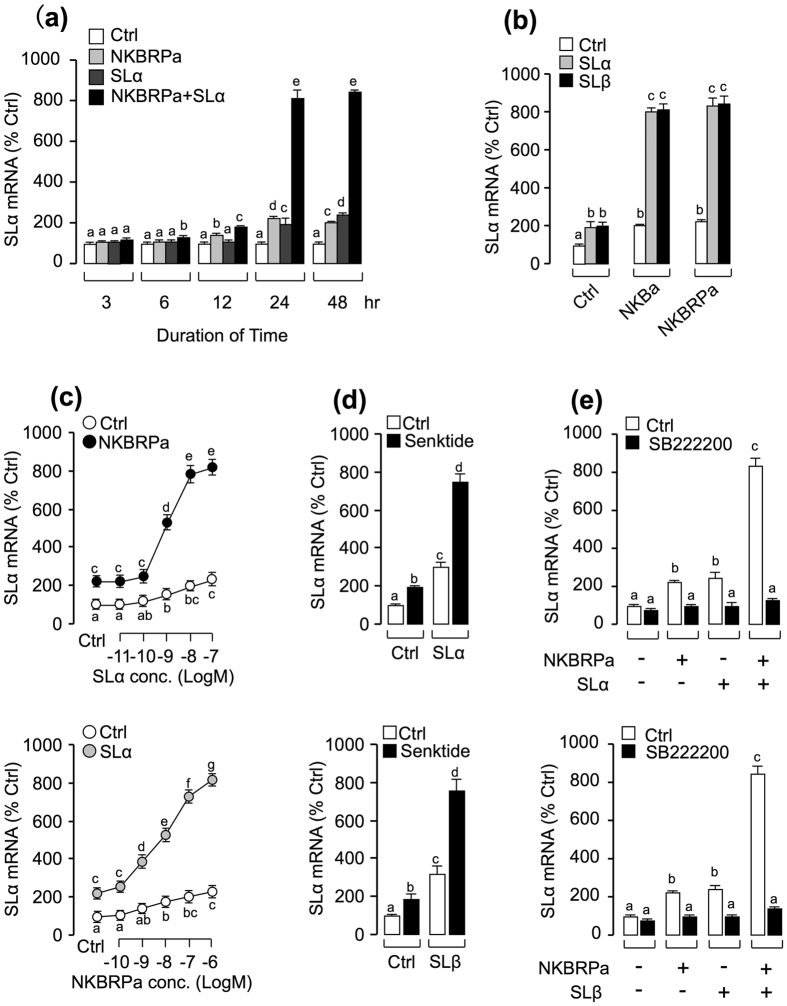
Given that (i) two somatolactin isoforms, SLα and SLβ, have been previously shown to trigger SLα secretion and gene expression at the pituitary level7, and (ii) TAC3a gene products could also stimulate SLα secretion and gene expression via activation of NK3R in carp pituitary cells14, we examined the functional interaction between TAC3a gene products and SLs in their stimulatory activity on SLα gene expression. As shown in Fig. 1a, static incubation with SLα (30 nM) and NKBRPa (1 μM) alone were both effective in elevating SLα mRNA expression in carp pituitary cells in a time-dependent fashion. Interestingly enough, the stimulatory effect on SLα mRNA expression was markedly enhanced (up to 8 fold basal) especially after 24–48 hr of drug treatment with simultaneous exposure to both SLα (30 nM) and NKBRPa (1 μM). In the single-dose experiment with drug treatment fixed at 24 hr, the potentiating effect (up to 8 fold basal) could still be observed with co-treatment of either SLα or SLβ (30 nM) with the carp TAC3a gene products including NKBa (1 μM) and NKBRPa (1 μM), respectively (Fig. 1b). In the parallel experiments, the synergistic action between NKBRPa and SLα was also confirmed by a concentration-response study. As shown in Fig. 1c, NKBRPa (1 μM)-induced SLα mRNA expression was found to be aggravated in a dose-dependent manner with cotreatment of increasing level of SLα (0.01–100 nM). Similar dose-dependence of the potentiating effect was also noted in the reciprocal experiment with cotreatment of SLα (30 nM) with increasing levels of NKBRPa (0.1–1000 nM). In the case of SLβ regulation, IGF-I, SLα and SLβ alone could all trigger SLβ mRNA expression in grass carp pituitary cells, but static incubation with NKBa or NKBRPa were both not effective in elevating SLβ mRNA expression and secretion significantly. In addition, cotreatment with NKBa/NKBRPa and SLα could not potentially increase SLβ transcript levels (Fig.S1). To establish the functional link between SLα potentiation and NK3R at the pituitary level, the NK3R agonist senktide (1 μM) was substituted for NKBa and NKBRPb in the potentiating study with SLα co-treatment. As shown in Fig. 1d, NK3R activation with senktide was found to mimic the synergistic effects of TAC3a gene products on SLα mRNA expression when give together with either SLα (30 nM) or SLβ (30 nM). Besides, co-treatment with NK3R antagonist SB222200 (10 μM) not only could reduce the stimulatory actions on SLα mRNA expression induced by SLα (30 nM)/SLβ (30 nM) and NKBRPa (1 μM) alone, but also significantly suppressed the potentiating effect induced by SLα/SLβ and NKBRPa co-treatment (Fig. 1e). These results indicated that the synergistic effect of SL and TAC3 gene products on SLα mRNA expression is dependent on NK3R activation.
Figure 1. Synergism between NKBRPa and SLα/β in stimulation of SLα gene expression.
(a) Time course of NKBRPa (1 μM), SLα (30 nM), and NKBRPa (1 μM)+ SLα (30 nM) on SLα mRNA expression in carp pituitary cells. (b) Synergistic effect of SLα/β with NKBa and NKBRPa in stimulation of SLα mRNA expression. In this experiment, carp pituitary cells were incubated for 24-hr with SLα/β (30 nM) co-treatment with NKBa, NKBRPa, and senktide (1 μM). (c) Effect of SLα concentration (0.01–100 nM) on basal and NKBRPa (1 μM)-induced SLα mRNA expression in carp pituitary cells. (d) Effects of NKBRPa concentration (0.1–1000 nM) on basal and SLα(30 nM)-induced SLα transcript levels in carp pituitary cells. (e) Synergistic effect of senktide (1 μM) with SLα and SLβ in stimulation of SLα mRNA expression. Receptor specificity for SLα regulation by SLα/β with NKBRPa. In this experiment, carp pituitary cells were challenged for 24-hr with SLα (30 nM)+ NKBRPa (1 μM) or SLβ (30 nM)+ NKBRPa (1 μM) in the presence or absence of NK3R antagonist SB222200 (10 μM). After drug treatment, total RNA was isolated for real-time PCR of SLα mRNA. In the data present (mean ± SEM), the groups denoted by different letters represent a significant difference at p < 0.05 (ANOVA followed by Dunnett’s test).
Up-regulation of NK3R gene expression by SLα/β in grass carp pituitary cells
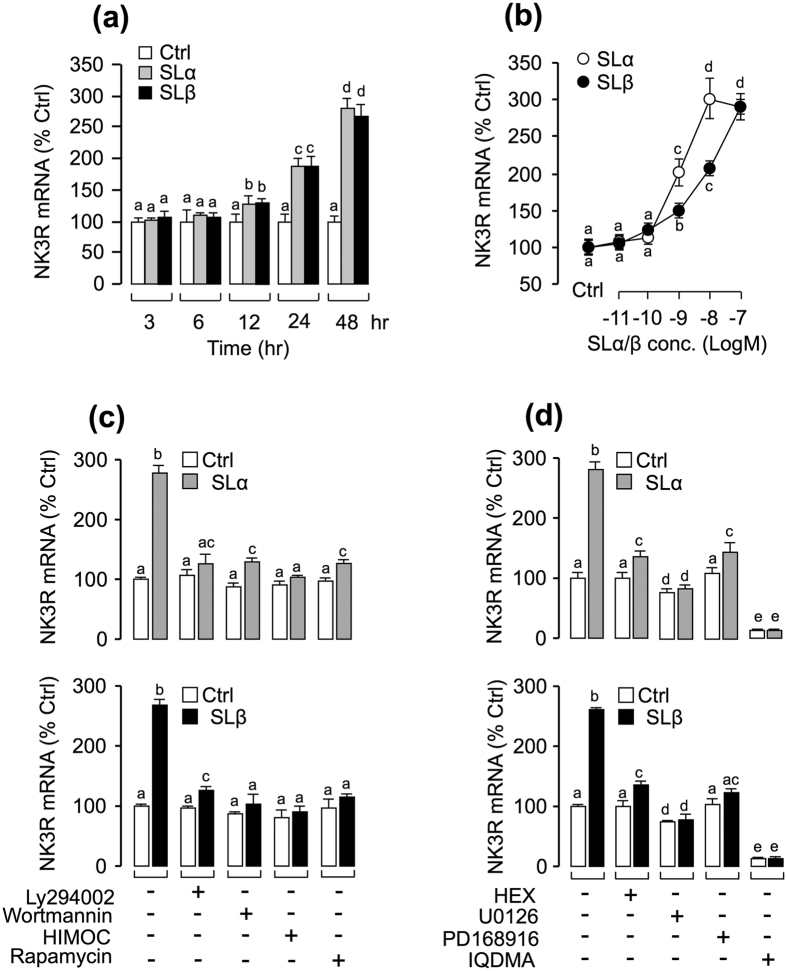
According to our previous study, IGFs could strongly enhance NKB-induced SLα mRNA expression through the up-regulation of NK3R expression in carp pituitary cells. To test the hypothesis that NK3R modulation may have occurred during SL potentiation of NKB-induced SLα expression, primary culture of grass carp pituitary cells were challenged with recombinant carp SLα and SLβ, respectively. Our time-course experiment revealed that SLα/SLβ (30 nM) could both significantly trigger NK3R mRNA expression from 12 hr to 48 hr in a time-dependent fashion (Fig. 2a). In parallel dose-dependent studies, a 24-hr incubation with increasing levels of SLα or SLβ (0.1–100 nM) also elevated NK3R mRNA expression in a dose-dependent manner (Fig. 2b). To further elucidate the signal transduction mechanisms for NK3R regulation by SLs, various pharmacological blockers targeting different pathways were recruited. As shown in Fig. 2c, SLα- or SLβ-induced NK3R transcript expression could be abolished by simultaneous incubation with the PI3K inhibitor wortmannin (1 μM), Akt inhibitor HIMOC (10 μM), or mTOR inhibitor rapamycin (20 nM). Similar results were also observed by preventing PI3K activation using another PI3K inhibitor Ly294002 (10 μM; Fig. 2c). Given that SL activation of MAPK cascades has been reported in grass carp pituitary cells7, the functional role of MAPK cascades on SL-induced NK3R gene expression were also tested in carp pituitaries. As shown in Fig. 2d, SLα- or SLβ -induced NK3R mRNA expression were found to be sensitive to the blockade by the MEK1/2 inhibitor U-0126 (10 μM) or p38MAPK inhibitor PD-169816 (10 μM). In the parallel experiments, SLα- or SLβ -induced NK3R mRNA expression could be attenuated or totally abolished by simultaneous treatment with the JAK2 inhibitor HEX (50 μM; Fig. 2d), and STAT5 inhibitor IQDMA (50 μM; Fig. 2d), respectively.
Figure 2. Up-regulation of NK3R gene expression by SLα/β in grass carp pituitary cells.
(a) Time course of carp SLα (30 nM) and SLβ (30 nM) treatment on NK3R mRNA expression in carp pituitary cells. (b) 24-hr incubation with increasing levels of SLα or SLβ (0.01–100 nM) treatment on NK3R mRNA expression in carp pituitary cells. (c) Effects of 24-hr co-treatment with the PI3K inhibitor Ly294002(10 μM) and wortmannin (1 μM), Akt inhibitor HIMOC (10 μM) and mTOR inhibitor rapamycin (20 nM) on SLα (30 nM)- or SLβ (30 nM)-induced NK3R transcript expression in carp pituitary cells. (d) Effects of 24-hr co-treatment with the JAK2 inhibitor HEX (50 μM), STAT5 inhibitor IQDMA (50 μM), MEK1/2 inhibitor U0126 (10 μM) and p38 MAPK inhibitor PD169816 (10 μM) on SLα (30 nM)- or SLβ (30 nM)-induced NK3R mRNA expression. After drug treatment, total RNA was isolated for real-time PCR of NK3R mRNA. In the data present (Mean ± SEM), the groups denoted by different letters represent a significant difference at p < 0.05 (ANOVA followed by Dunnett’s test).
Synergistic effects of IGF and SL on NK3R and SLα gene expression
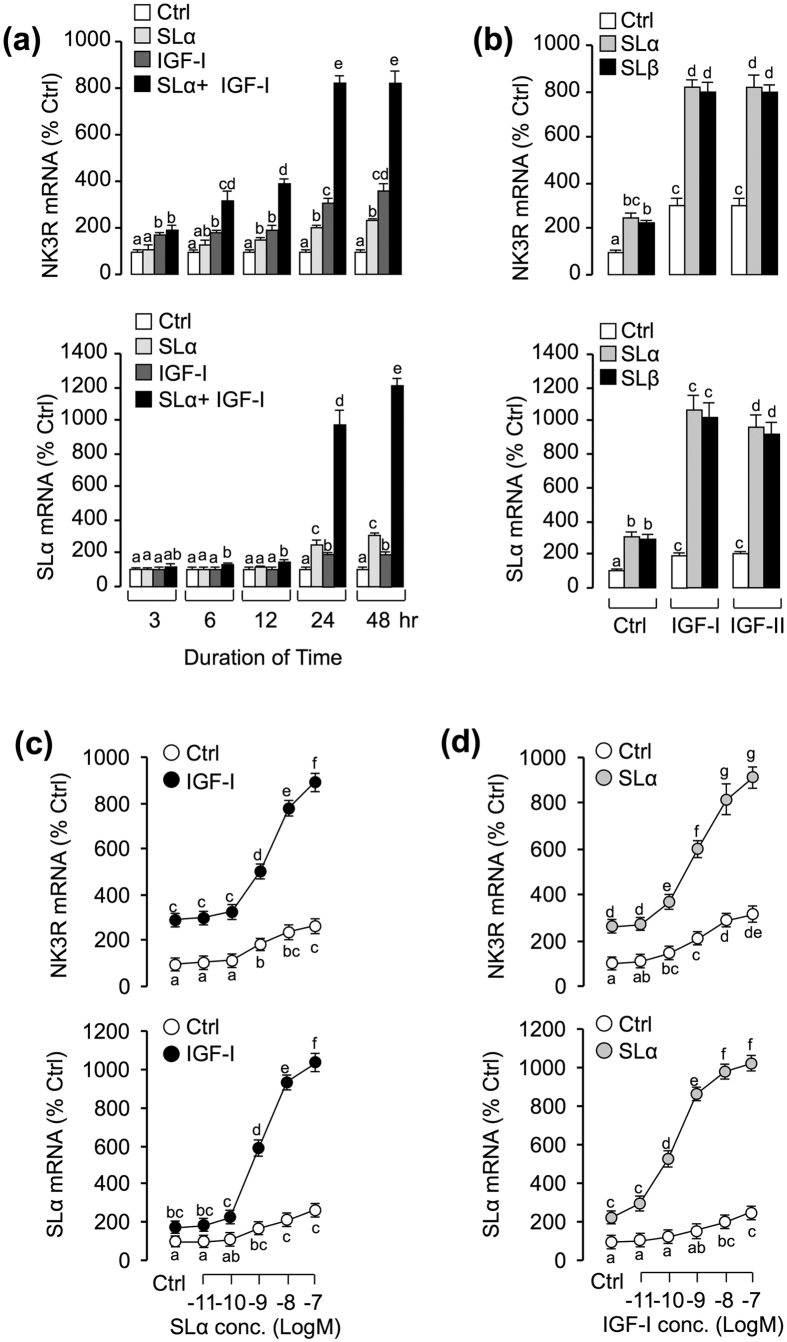
Given our recent studies that IGFs and SLs were both found to be effective in stimulating SLα and NK3R gene expression in carp pituitary cells7,17, the functional interaction between SLs and IGFs in the regulation of SLα and NK3R gene expression were examined in carp pituitary cells. As shown in Fig. 3a, SLα (30 nM) and IGF-I (50 nM) treatment alone could both significantly elevate NK3R and SLα mRNA expression in carp pituitary cells in a time-dependent manner. Interestingly, the stimulatory effects on NK3R and SLα mRNA expression were significantly enhanced (up to 8 fold for NK3R and 10 fold for SLα) especially after 24–48 hr of drug treatment with co-treatment of SLα (30 nM) with IGF-I (50 nM). Following the time-course experiment, a single-dose experiment was performed at 24 hr. As shown in Fig. 3b, the potentiating effect could still be observed with co-treatment of either SLα (30 nM) or SLβ (30 nM) with IGF-I (50 nM) or IGF-II (50 nM), respectively. Besides, the functional interaction between SLα and IGF-I was further confirmed by a dose-response reciprocal reverse experiment. In this case, IGF-I-induced NK3R and SLα mRNA expression were found to be enhanced in a dose-dependent manner with simultaneous treatment with increasing concentrations of SLα (0.01–100 nM; Fig. 3c). The maximal responses occurred at 100 nM SLα (9 fold basal for NK3R and 11 fold basal for SLα). Similar concentration-dependence of the potentiating effect was also observed in the reciprocal experiment with co-treatment of SLα (30 nM) with increasing levels of IGF-I (0.01–100 nM; Fig. 3d).
Figure 3. Functional interaction between SL and IGF on the regulation of NK3R and SLα gene expression.
(a) Time course of carp SLα, IGF-I and SLα+ IGF-I on NK3R and SLα mRNA expression. In this experiment, carp pituitary cells were incubated from 3 to 48 hr with SLα (30 nM) alone, IGF-I (50 nM) alone and SLα (30 nM)+ IGF-I (50 nM). (b) Synergistic effects of SLα/β and IGF-I/-II on the simulation of NK3R and SLα mRNA expression. In this case, carp pituitary cells were challenged for 24-hr with SLαor SLβ (30 nM) co-treatment with either IGF-I or IGF-II (50 nM). (c) Effect of SLα (0.01–100 nM) treatment on basal and IGF-I (50 nM)-induced NK3R and SLα mRNA expression in carp pituitary cells. (d) Effects of IGF-I (0.01–100 nM) treatment on basal and SLα(30 nM)-induced NK3R and SLα transcript levels in carp pituitary cells. After drug treatment, total RNA was isolated for real-time PCR of NK3R and SLα mRNA. In the data present (mean ± SEM), the groups denoted by different letters represent a significant difference at p < 0.05 (ANOVA followed by Dunnett’s test).
Synergistic effects of SLα, IGF-I and NKBRPa on SLα mRNA expression
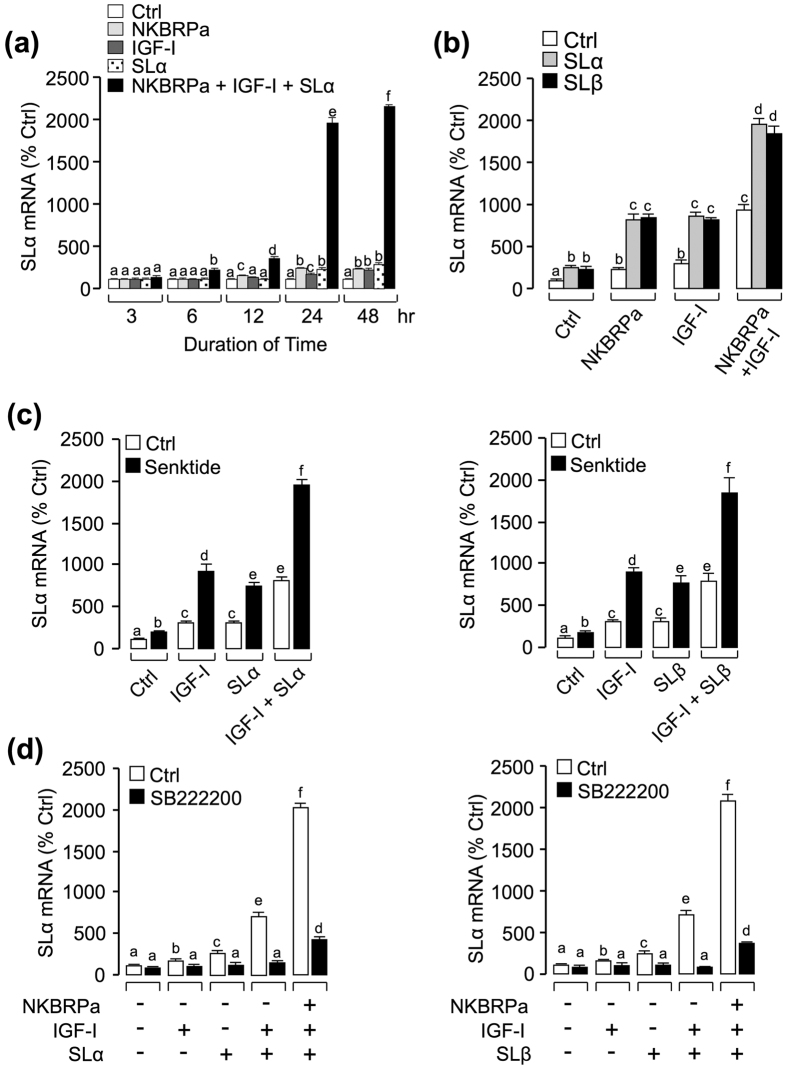
In the current study, the synergism between SL and IGF in stimulation of NK3R mRNA expression was noted in carp pituitary cells. So now we have an interesting idea, what will happen when we use a cocktail containing SL, IGF and NKB to challenge carp pituitary cells ? To answer this question, the cocktail of SLα, IGF-I and NKBRPa was used to test grass carp pituitary cells. As shown in Fig. 4a, IGF-I (50 nM), NKBRPa (1 μM) and SLα (30 nM) treatment alone could stimulate SLα mRNA up to 24 hr, however, co-treatment of the three drugs together could significantly elevate SLα mRNA from 6 hr (up to 2 fold basal) to 48 hr (up to 20 fold basal). In the single-dose experiment with drug treatment fixed at 24 hr, the potentiating effect could still be observed with co-treatment of either SLα (30 nM) or SLβ (30 nM) with NKBRPa (1 μM) and IGF-I (50 nM) (Fig. 4b). To clarify the mechanism responsible for the regulation of SLα mRNA expression by SLα/β co-treated with IGF-I and NKBRPa, a pharmacological approach was performed in carp pituitary cells. As a first step, NK3R agonist senktide (1 μM) was recruited to replace the NKBRPa in the potentiating study with IGF-I (50 nM) and SLα (30 nM). As shown in Fig. 4c, senktide could mimic the synergistic effects of TAC3 gene products on SLα regulation when simultaneous incubation with either SLα/SLβ (30 nM) or SLα (30 nM)+ IGF-I (50 nM)/SLβ (30 nM)+ IGF-I (50 nM). Besides, co-treatment with NK3R antagonist SB222200 (10 μM) not only could reduce the stimulatory actions on SLα mRNA expression induced by IGF-I (50 nM), NKBRPa (1 μM) and SLα/SLβ (30 nM) alone, but also markedly suppressed the potentiating effect induced by SLα/SLβ+ NKBRPa, SLα/SLβ+ IGF-I and SLα/SLβ+ IGF-I+ NKBRPa (Fig. 4d). These findings, taken together, suggested that the synergistic effect of SLα/β, IGF-I and NKBRPa on the stimulation of SLα mRNA expression might be mediated through the activation of NK3R, which is a G protein coupled receptor coupled with activation of AC/cAMP/PKA, PLC/IP3/PKC, and Ca2+ /CaM/CaMK-II cascades.
Figure 4. Functional interactions of SLα/β, IGF-I and NKBRPa/senktide on the regulation of SLα mRNA expression.
(a) Time course of SLα (30 nM), NKBRPa (1 μM), IGF-I (50 nM) and SLα (30 nM)+ NKBRPa (1 μM)+ IGF-I (50 nM) treatment on SLα mRNA expression. (b) Synergistic effects of SLα/β with NKBRPa, IGF-I or NKBRPa+ IGF-I. In this case, carp pituitary cells were incubated for 24-hr with SLα or SLβ (30 nM) co-treatment with NKBRPa (1 μM), IGF-I (50 nM) or NKBRPa (1 μM)+ IGF-I (50 nM), respectively. (c) Synergistic effects of senktide with SLα/β, IGF-I or SLα/β+ IGF-I. In this case, carp pituitary cells were incubated for 24-hr with senktide (1 μM) co-treatment with SLα/β (30 nM), IGF-I (50 nM) or SLα/β(30 nM)+ IGF-I (50 nM), respectively. (d) Receptor specificity for SLα regulation by SLα or SLβ co-treatment with IGF-I or NKBRPa+ IGF-I. In this experiment, carp pituitary cells were incubated for 24-hr with SLα (30 nM)+ IGF-I (50 nM) and SLα (30 nM)+ NKBRPa (1 μM)+ IGF-I (50 nM) or SLβ (30 nM)+ IGF-I (50 nM) and SLβ (30 nM)+ NKBRPa (1 μM)+ IGF-I (50 nM) in the presence or absence of NK3R antagonist SB222200 (10 μM). After drug treatment, total RNA was isolated for real-time PCR of SLα mRNA. In the data present (Mean ± SEM), the groups denoted by different letters represent a significant difference at p < 0.05 (ANOVA followed by Dunnett’s test).
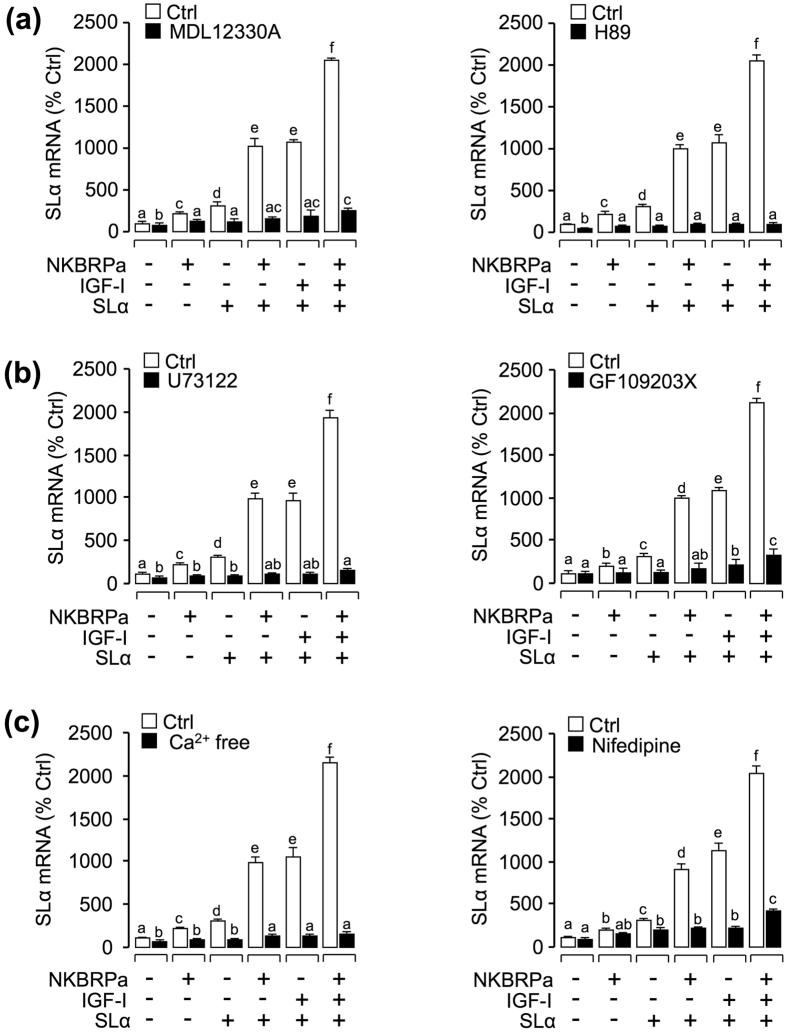
To clarify the signal transduction for the synergistic regulation of SLα mRNA expression, various pharmacological inhibitors/blockers targeting different pathways of NK3R were used. As a first step, the possible involvement of cAMP-dependent pathway was examined at the pituitary cell level. As shown in Fig. 5a, the AC-inhibitor MDL12330A (10 μM) or PKA inhibitor H89 (10 μM) could block the synergistic effects of SLα, IGF-I and NKBRPa on the induction of SLα mRNA expression. To shed light on the role of PLC-dependent cascade in the synergistic actions, SLα regulation by SLα, IGF-I and NKBRPa were tested with inhibitors for individual components of this pathway. In this case, the synergistic effects of SLα, IGF-I and NKBRPa on SLα gene expression were observed to be suppressed/abolished by simultaneous incubation with the PLC inhibitor U73122 (10 μM) or PKC inhibitor GF109203X (10 μM), respectively (Fig. 5b). To examine the possible role of Ca2+ -dependent cascade in SLα regulation by the cocktail containing SLα, IGF-I and NKBRPa, the synergistic effects were also tested with various inhibitors for Ca2+ pathway. In this case, this cocktail-induced SLα mRNA expression were found to be attenuated/abolished by incubation with Ca2+ free medium or co-treatment with the voltage sensitive calcium channel (VSCC) inhibitor nifedipine (10 μM), respectively (Fig. 5c).
Figure 5. Functional role of AC/PKA, PLC/PKC and Ca2+ -dependent signaling pathways in the regulation of SLα mRNA expression.
In this experiment, carp pituitary cells were incubated for 24 hours in the absence (control) or presence of (a) AC inhibitor MDL12330A (10 μM) and PKA inhibitor H89 (10 μM), (b) PLC inhibitor U73122 (10 μM) and PKC inhibitor (10 μM), or (c) Ca2+ free medium and VSCC blocker nifedipine (10 μM) in combination of NKBRPa (1 μM), SLα (30 nM), SLα (30 nM)+ NKBRPa (1 μM), SLα (30 nM)+ IGF-I (50 nM), or SLα (30 nM)+ NKBRPa (1 μM)+ IGF-I (50 nM). After drug treatment, total RNA was isolated for real-time PCR of SLα mRNA. In the data present (mean ± SEM), the groups denoted by different letters represent a significant difference at p < 0.05 (ANOVA followed by Dunnett’s test).
Discussion
At present, except for a single study in carp pituitary cells showing that SLα and SLβ play a stimulatory role in autocrine/paracrine regulation of SLα secretion and synthesis in grass carp7, little is known about the functional role of SLs at the pituitary level. Since (i) carp SLα and SLβ were found to be effective in triggering SLα secretion, protein production and gene expression in carp SLα cells7, (ii) carp NK3R was specifically expressed in SLα cells within the NIL lobe of the carp pituitary14, we speculate that SLs may play a role on NK3R regulation at the pituitary level, which may have a functional impact on SLα expression in carp pituitary cells. In the present study, using grass carp pituitary cells as a model, we demonstrated for the first time that SLα and SLβ can up-regulate NK3R gene expression in time- and dose-dependent manner via direct actions at the pituitary level. In fish models, SL receptor has been identified as a member of the Type I GHR family37, and its activation can lead to JAK2 recruitment and subsequent activation of STATs, MAPK, and PI3K pathway in grass carp pituitary cells7. To test the possible involvement of these signaling cascades in SL-induced NK3R, a pharmacological approach using the inhibitors for the respective pathways was used. In carp pituitary cells, the stimulatory effects on NK3R mRNA expression induced by SLα or SLβ treatment were either totally abolished or partially suppressed by the JAK2 inhibitor HEX, STAT5 blocker IQDMA, PI3K inhibitors wortmannin and Ly294002, Akt inhibitor HIMOC, MEK1/2 blocker U-0126, and P38 MAPK inhibitor PD169816. These results, together with our previous findings that SLα and SLβ treatment could trigger rapid phosphorylation of STAT5, Akt, MEK1/2, ERK1/2, MKK3/6, and P38 MAPK in carp pituitary cells7, suggest that SL induction of NK3R gene expression at the pituitary cell level is mediated through activation of JAK2/STAT5, PI3K/Akt, MEK/ERK1/2, and MKK3/6/P38 MAPK cascades.
In our recent in vitro studies, we have demonstrated that (i) SLα and SLβ could both elevate SLα mRNA expression in carp pituitary cells7, and (ii) carp TAC3a gene products, namely NKBa and NKBRPa, could up-regulate SLα gene expression via activation of NK3R expressed in the carp pituitary14. In our initial attempt of investigate the functional interactions between SLs and TAC3 gene products on SLα expression, we have the novel findings that co-treatment of SLα/SLβ with either NKBa or NKBRPa, respectively, could trigger a synergistic effect on SLα mRNA expression in a time- and dose-dependent manner. These potentiating effect could be mimicked by replacing TAC3 gene products with the NK3R agonist senktide and blocked by simultaneous incubation with the NK3R antagonist SB222200, suggesting the possible dependence of the synergistic effect on NK3R expression at the pituitary level. Together with our current finding of SLα/SLβ up-regulation of NK3R gene expression in carp pituitary cells, it raises the possibility that SL treatment may enhance the stimulatory effect of TAC3 gene products on SLα gene expression by increasing NK3R expression in the carp pituitary. In previous studies, the interactions of glucagon with GH have been reported in different species, e.g., in rat38 and grass carp39. In rat hepatocytes, cotreatment of glucagon and GH is known to have a potentiating effect on the stimulation of IGF-I mRNA expression38. In grass carp, our previous studies have also shown that glucagon could potentiate GH-induced IGF-I gene expression via up-regulation of GHR expression in carp hepatocytes39. Apparently, potentiation of the bioactivity of various members of the GH family lineage in fish species can occur by functional interactions with GPCR activation by increasing receptor expression mediated the respective stimulating influence at the cellular level.
In cancer cell models, e.g., pancreatic cancer cells, functional crosstalk of the post-receptor signaling between IGF-I/Insulin receptor with GPCR (e.g., neurotensin receptor and type I angiotensin receptor) has been reported40. In wound repairing of the rabbit cornea, co-treatment of IGF-I with SP is known to have a potentiating effect on the migration of corneal epithelial cells41,42. In our current study with carp pituitary cells, SLα and SLβ not only could potentiate SLα mRNA expression induced by TAC3 gene products, but also notably enhance the stimulatory effect caused by IGF-I/-II cotreatment on SLα mRNA expression. Interestingly enough, simultaneous treatment of carp pituitary cells of SLα/SLβ with IGF-I and NKBRPa was found to markedly increase basal levels of SLα mRNA up to 20 fold basal, which was much higher than the corresponding SLα responses induced by SLα/SLβ cotreatment with either IGF-I/-II (up to 8 fold basal) or TAC3 gene products NKBa and NKBRPa (up to 8 fold basal), respectively. This notable increase in the potentiating effect caused by simultaneous treatment of SLα, IGF-I and NKBRPa together also occurred with a novel finding in carp pituitary cells, in which cotreatment with SLα/SLβ could potentiate the stimulatory effect of IGFs on NK3R gene expression at the pituitary level. Similar to our results with SL cotreatment with TAC3 gene products, the highly potent synergistic effect on SLα gene expression induced by simultaneous treatment of the three stimulators together could be mimicked by substituting the NK3R agonist senktide for NKBRPa and blocked by cotreatment with the NK3R antagonist SB222200. These findings strongly suggest that the potentiating effect caused by the three SL stimulators is highly depended on NK3R expression in the carp pituitary. NK3R is a member of the rhodopsin-type class I type G-protein coupled receptors (GPCRs), and in mammals its activation can trigger intracellular signaling via Go and Gq/1125,43 followed by cAMP production44,45, PLC-dependent PI hydrolysis23,24, mobilization of IP3-sensitive intracellular Ca2+ ([Ca2+]i)24 and extracellular Ca2+ ([Ca2+]e) entry via voltage-dependent Ca2+ channels25,46. In carp pituitary cells, our recent studies have also demonstrated that NKBa/NKBRPa could stimulate SLα gene expression by NK3R activation via AC/cAMP/PKA, PLC/IP3/PKC, and Ca2+ /CaM/CaMK-II pathways14. Consistent with these previous findings, blocking the respective post-receptor signaling pathways using the AC inhibitor MDL12330A, PKA inactivator H89, PLC blocker U73122, PKC inhibitor GF109203X, removal of [Ca2+]e with a Ca2+ -free culture medium, and inactivating voltage-sensitive Ca2+ channel using nifedipine were all effective in inhibiting/blocking the highly potent synergistic effect on SLα gene expression caused by simultaneous stimulation with SLα, IGF-I and NKBRPa. These results, as a whole, provide evidence that SLα and SLβ can synergize with IGF-I/-II potentiation on SLα gene expression induced by TAC3 gene products by up-regulation of NK3R expression in the carp pituitary.
In summary, we have demonstrated for the first time that SLα and SLβ could act at the pituitary cell level to potentiate the stimulatory effects of TAC3 gene products and IGF-I/-II on SLα gene expression via up-regulation of pituitary NK3R expression. The stimulation on NK3R gene expression probably was mediated through the JAK2/STAT5, MAPK and PI3K/Akt cascades. In this study, we also have a novel findings that simultaneous treatment with SL, IGF and TAC3 gene product together could serve as a highly potent stimulatory signal for SLα gene expression and this stimulatory effect was dependent on NK3R expression in the carp pituitary and involved the activation of post-receptor signaling cascades, namely the AC/cAMP/PKA, PLC/PKC and Ca2+ -dependent pathways, coupled to NK3R stimulation. Since SL autoregulation via local release of SLα and SLβ has been recently demonstrated in the carp pituitary7. Our findings suggest that SLα and SLβ released at the pituitary level may act in an autocrine/paracrine manner to modulate the pituitary sensitivity to the synergistic stimulation on SLα expression triggered by IGF-I/-II and TAC3 gene products via up-regulation of NK3R expression in the carp pituitary.
Materials and Methods
Animals
One-year-old (1+) grass carps (Ctenopharyngodon idellus) with body weight ranging from 2.0 to 3.0 kg were bought from local markets and maintained in well aerated 250L aquaria at 20 ± 2 °C under a 12L:12D photoperiod. Given the carps at this stage are pre-pubertal and sexual dimorphism is not apparent, fish of mixed sexes were used for preparation of pituitary cell cultures. All animal experiments were conducted in accordance with the guidelines and approval of the respective Animal Research and Ethics committees of the University of Hong Kong and Huazhong Agricultural University.
Reagents
Recombinant proteins of grass carp SLα and SLβ were expressed in E. coli, purified and functionally characterized as described previously7. The two hormones were dissolved in PBS and stored frozen at −80 °C as 0.1 mM stocks in small aliquots. Grass carp NKBa and NKBRPa were synthesized from GenScript using the automated solid-phase method, and the carboxyl-terminus of individual peptides was amidated. These peptides were dissoved in DMSO, and stored frozen at −80 °C as 1 mM stocks in small aliquots. Human IGF-I and IGF-II were purchased from Sigma and dissolved in double-distilled deionized water and stored as 0.1 mM stocks in small aliquots at −80 °C. Pharmacological agents, including MDL12330A, H89, GF109203X, U73122, nifedifine, Ly294002, wortmannin, rapamycin, HIMOC, U0126, PD168916, HEX, and IQDMA were acquired from Calbiochem, while senktide and SB222200 were purchased from Tocris. Similar to the peptides, these pharmacological agents were prepared as a high concentration frozen stock in small aliquots and diluted with pre-warmed culture medium to appropriate concentrations 15 min prior to drug treatment.
Primary culture of grass carp pituitary cells
Grass carp pituitary cells were prepared by trypsin/DNase digestion method as described previously47. Briefly, pituitaries were excised from grass carp and diced into 0.5-mm fragments using a McILwain tissue chopper. After 30-min trypsin digestion with constant shaking at 28 °C, pituitary fragments were suspended in Ca2+ -free MEM supplemented with DNase I (0.01 mg/ml, Sigma). Pituitary cells were dispersed by gently trituration and filtered through a sterile nylon mesh (pore size: 20 μM) to remove undigested fragments/debris. After that, the cells were harvested by centrifugation at 1000 rpm for 10 min and re-suspended in MEM medium. Total cell yield and percentage viability were estimated by cell counting in the presence of trypan blue using a hematocytometer.
Measurement of carp SLα and NK3R mRNA expression
Grass carp pituitary cells were seeded in poly-D-lysine coated 24-well culture plates at a density of ~2.5 × 106 /ml/well. On the following day, drug treatment was initiated by replacing the old medium with MEM containing appropriate levels of test substances. After drug treatment, total RNA was extracted from individual well using Trizol and reversely transcribed by Superscript II (50 Unit, Invitrogen). The RT samples obtained were subjected to qPCR using a LightCycler SYBR Green I Kit (Roche) with primers specific for grass carp SLα (Forward Primer: 5′-ACCCACT GTACTTCAATCTCC-3′; Reverse Primer: 5′-CGTCGTAACGATCAAGAGTAG-3′) and NK3R (Forward Primer: 5′-GCCAAGAGAAAGGTTGTGAAGA-3′; Reverse Primer: 5′-GTGTACATGCTGCTCTGGCG-3′), respectively. PCR cycling parameters for SLα and NK3R mRNA detection were set at 94 °C for 3 min followed by 35 cycles of amplification with denaturation at 94 °C for 30 sec, annealing at 52 °C for SLα mRNA or 56 °C for NK3R mRNA for 30 sec, and extension at 72 °C for 30 sec. Signal detection was routinely set for 20 seconds at 84 °C for SLα and 86 °C for NK3R, respectively. In these studies, serial dilutions of plasmid DNA containing the ORF of SLα (GeneBank no: EF372074) and NK3R (GenBank no: JQ254913) cDNA were used as the standards for data calibration. Parallel qPCR measurement of β-actin was also conducted in individual experiment to serve as the internal control.
Data transformation and statistical analysis
For real-time PCR measurement of NK3R and SLα mRNA, standard curves with a dynamic range of ≥105 and a correlation coefficient ≥0.95 were used for data calibration with RotorGene-Q software 1.7 (Qiagen) under unsupervised mode. Since no significant changes were noted for β-actin mRNA levels between different experiment groups in our studies, SLα and NK3R mRNA data were simply transformed as a percentage of the mean value in the control group without drug treatment (as “%Ctrl”). The data presented (as Mean ± SEM) were pooled results from 6–8 separate experiments and analyzed with ANOVA followed by Dunnett’s test using Prism 6.0 and differences between groups were considered as significant at P < 0.05.
Additional Information
How to cite this article: Hu, G. et al. Novel Functional Role of NK3R Expression in the Potentiating Effects on Somatolactin α Autoregulation in grass carp pituitary cells. Sci. Rep. 6, 36102; doi: 10.1038/srep36102 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Profs. Haoran Lin and Yong Zhang (Sun Yatsen University, China) for sending us the zebrafish NKB and NKBRP for our initial studies. We also specially thank Ms. Wendy Ko (The University of Hong Kong) for her help in the experimental preparation. The project was supported by GRF Grants (17128215, 781113 & 780312) and NSFC/RGC Joint Grant (N_HKU 732/12) from Research Grant Council, Hong Kong and HMRF Grant (13142591) from Food and Health Bureau, Hong Kong (to AOLW). Financial support was also provided from NSFC Grants (31602130 for G.F.H) and the Fundamental Research Funds for the Central Universities to G.F.H. (2015BQ039).
Footnotes
Author Contributions G.F.H. and M.L.H. were involved in experimental work and data analysis. G.F.H. and O.L.W. were involved in project planning, data analysis and manuscript writing.
References
- Ono M. et al. cDNA cloning of somatolactin, a pituitary protein related to growth hormone and prolactin. Proc. Natl. Acad. Sci. USA 87, 4330–4334 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. et al. Cloning of somatolactin alpha and beta cDNAs in zebrafish and phylogenetic analysis of two distinct somatolactin subtypes in fish. J. Endocrinol. 182, 509–518 (2004). [DOI] [PubMed] [Google Scholar]
- Benedet S., Bjornsson B. T., Taranger G. L. & Andersson E. Cloning of somatolactin alpha, beta forms and the somatolactin receptor in Atlantic salmon: seasonal expression profile in pituitary and ovary of maturing female broodstock. Reprod. Biol. Endocrinol. 6, 42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Ko W. K., Lerner E. A., Chan K. M. & Wong A. O. Grass carp somatolactin: I. Evidence for PACAP induction of somatolactin-alpha and -beta gene expression via activation of pituitary PAC-I receptors. Am. J. Physiol. Endocrinol. Metab. 295, E463–E476 (2008). [DOI] [PubMed] [Google Scholar]
- Zhu Y., Song D., Tran N. T. & Nguyen N. The effects of the members of growth hormone family knockdown in zebrafish development. Gen. Comp. Endocrinol. 150, 395–404 (2007). [DOI] [PubMed] [Google Scholar]
- Fukamachi S. et al. Conserved function of medaka pink-eyed dilution in melanin synthesis and its divergent transcriptional regulation in gonads among vertebrates. Genetics 168, 1519–1527 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q. & Wong A. O. Signal transduction mechanisms for autocrine/paracrine regulation of somatolactin α secretion and synthesis in carp pituitary cells by somatolactin α and β. Am. J. Physiol. Endocrinol. Metab. 304, E176–E186 (2013). [DOI] [PubMed] [Google Scholar]
- Khalil N. A., Hashem A. M., Ibrahim A. A. & Mousa M. A. Effect of stress during handling, seawater acclimation, confinement, and induced spawning on plasma ion levels and somatolactin-expressing cells in mature female Liza ramada. J. Exp. Zool. A Ecol. Genet. Physiol. 317, 410–424 (2012). [DOI] [PubMed] [Google Scholar]
- Sasano Y., Yoshimura A. & Fukamachi S. Reassessment of the function of somatolactin alpha in lipid metabolism using medaka mutant and transgenic strains. BMC Genetics 13, 64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo R., Suetake H., Suzuki Y., Aoyama J. & Tsukamoto K. Profiles of mRNA expression for prolactin, growth hormone, and somatolactin in Japanese eels, Anguilla japonica: The effect of salinity, silvering and seasonal change. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 164, 10–16 (2013). [DOI] [PubMed] [Google Scholar]
- Kakizawa S., Kaneko T. & Hirano T. Effects of hypothalamic factors on somatolactin secretion from the organ-cultured pituitary of rainbow trout. Gen. Comp. Endocrion. 105, 71–78 (1997). [DOI] [PubMed] [Google Scholar]
- Bhandari R. K. et al. Seasonal changes of responses to gonadotropin-releasing hormone analog in expression of growth hormone/prolactin/somatolactin genes in the pituitary of masu salmon. Gen. Comp. Endocrinol. 130(1), 55–63 (2003). [DOI] [PubMed] [Google Scholar]
- Onuma T., Ando H., Koide N., Okada H. & Urano A. Effects of salmon GnRH and sex steroid hormones on expression of genes encoding growth hormone/rolactin/somatolactin family hormones and a pituitary-specific transcription factor in masu salmon pituitary cells in vitro. Gen. Comp. Endocinol. 143(2), 129–141 (2005). [DOI] [PubMed] [Google Scholar]
- Hu G. F., He M. L., Ko W. K., Lin C. Y. & Wong A. O. Novel pituitary actions of TAC3 gene products in fish model: -Receptor specificity and signal transduction for prolactin and somatolactin alpha regulation by neurokinin B (NKB) and NKB-related peptide in carp pituitary cells. Endocrinology 155(9), 3582–3596 (2014). [DOI] [PubMed] [Google Scholar]
- Tanaka M. et al. Melanin-concentrating hormone reduces somatolactin release from cultured goldfish pituitary cells. J. Endocrinol. 203, 389–398 (2009). [DOI] [PubMed] [Google Scholar]
- Azuma M. et al. The octadecaneuropeptide stimulates somatolactin release from cultured goldfish pituitary cells. J. Neuroendocrinol. 25(3), 312–321 (2013). [DOI] [PubMed] [Google Scholar]
- Jiang Q., Ko W. K. & Wong A. O. Insulin-like growth factor as a novel stimulator for somatolactin secretion and synthesis in carp pituitary cells via activation of MAPK cascades. Am. J. Physiol. Endocrinol. Metab. 301, E1208–E1219 (2011). [DOI] [PubMed] [Google Scholar]
- Lecci A., Capriati A., Altamura M. & Maggi C. A. Tachykinins and tachykinin receptors in the gut, with special reference to NK2 receptors in human. Auton. Neurosci. 126–127, 232–249 (2006). [DOI] [PubMed] [Google Scholar]
- Shimizu Y., Matsuyama H., Shiina T., Takewaki T. & Furness J. B. Tachykinins and their functions in the gastrointestinal tract. Cell. Mol. Life Sci. 65, 295–311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelrahman A. M. & Pang C. C. Effect of substance P on venous tone in conscious rats. J. Cardiovasc. Pharmacol. 45, 49–52 (2005). [DOI] [PubMed] [Google Scholar]
- Ravina C. G. et al. A role for tachykinins in the regulation of human sperm motility. Hum. Reprod. 22, 1617–1625 (2007). [DOI] [PubMed] [Google Scholar]
- Shigemoto R., Yokota Y., Tsuchida K. & Nakanishi S. Cloning and expression of a rat neurokinin-K receptor cDNA. J. Biol. Chem. 265, 623–628 (1990). [PubMed] [Google Scholar]
- Nakajima Y., Tsuchida K., Negishi M., Ito S. & Nakanishi S. Direct linkage of three tachykinin receptors to stimulation of both phosphatidylinositol hydrolysis and cyclic AMP cascades in transfected Chinese hamster ovary cells. J. Biol. Chem. 267, 2437–2442 (1992). [PubMed] [Google Scholar]
- Mizuta K. et al. Expression and coupling of neurokinin receptor subtypes to inositol phosphate and calcium signaling pathways in human airway smooth muscle cells. Am. J. Physiol. Lung Cell Mol. Physiol. 294, L523–L534 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawaja A. M. & Rogers D. F. Tachykinins: Receptor to effector. Int. J. Biochem. Cell Biol. 28, 721–738 (1996). [DOI] [PubMed] [Google Scholar]
- Topaloglu A. K. Neurokinin B signaling in puberty: human and animal studies. Mol. Cell. Endocrinol. 324, 64–69 (2010). [DOI] [PubMed] [Google Scholar]
- Guran T. et al. Hypogonadotropic hypogonadism due to a novel missense mutation in the first rxtracellular loop of the neurokinin B receptor. J. Clin. Endocr. Metab. 94, 3633–3639 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaloglu A. K. et al. TAC3 and TACR3 mutations in familial hypo- gonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat. Genet. 41, 354–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. et al. TAC3 and TACR3 defects cause hypothalamic congenital hypogonadotropic hypogonadism in humans. J. Clin. Endocrinol. Metab. 95, 2287–2295 (2010). [DOI] [PubMed] [Google Scholar]
- Lehman M. N., Coolen L. M. & Goodman R. L. Minireview: Kisspeptin/Neurokinin B/Dynorphin (KNDy) Cells of the Arcuate Nucleus: A Central Node in the Control of Gonadotropin-Releasing Hormone Secretion. Endocrinology 151, 3479–3489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro V. M. New insights into the control of pulsatile GnRH release: the role of Kiss1/neurokinin B neurons. Front Endocrinol. 3, 48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biran J., Palevitch O., Ben-Dor S. & Levavi-Sivan B. Neurokinin Bs and neurokinin B receptors in zebrafish-potential role in controlling fish reproduction. Proc. Natl. Acad. Sci. USA 109, 10269–10274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S. et al. Cloning and expression of tachykinins and their association with kisspeptins in the brain of zebrafish. J. Comp. Neurol. 520, 2991–3012 (2012). [DOI] [PubMed] [Google Scholar]
- Zhou W. et al. The evolution of tachykinin/tachykinin receptor (TAC/TACR) in vertebrates and molecular identification of the TAC3/TACR3 system in zebrafish (Danio rerio). Mol. Cell. Endocrinol. 361, 202–212 (2012). [DOI] [PubMed] [Google Scholar]
- Qi X. et al. Goldfish neurokinin B: Cloning, tissue distribution, and potential role in regulating reproduction. Gen. Comp. Endocrinol. 221, 267–277 (2015). [DOI] [PubMed] [Google Scholar]
- Biran J. et al. Direct regulation of gonadotropin release by neurokinin B in tilapia (Oreochromis niloticus). Endocrinology 155(12), 4831–4842 (2014). [DOI] [PubMed] [Google Scholar]
- Fukamachi S. & Meyer A. Evolution of receptors for growth hormone and somatolactin in fish and land vertebrates: lessons from the lungfish and sturgeon orthologues. J. Mol. Evol. 65, 359–372 (2007). [DOI] [PubMed] [Google Scholar]
- Kachra Z., Yang C. R. & Posner B. I. The augmentation of insulin-like growth factor-I messenger ribonucleic acid in cultured rat hepatocytes: activation of protein kinase-A and -C is necessary, but not sufficient. Endocrinology 134(2), 702–708 (1994). [DOI] [PubMed] [Google Scholar]
- Brown G. F. Novel aspects of grass carp GHR gene regulation. The University of Hong Kong (2009). [Google Scholar]
- Rozengurt E., Sinnett-Smith J. & Kisfalvi K. Crosstalk between insulin/insulin-like growth factor-1 receptors and G protein-coupled receptor signaling systems: a novel target for the antidiabetic drug metformin in pancreatic cancer. Clin. Cancer. Res. 16, 2505–2511 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Ofuji K., Chikama T. & Nishida T. Combined effects of substance P and insulin-like growth factor-1 on corneal epithelial wound closure of rabbit in vivo. Curr. Eye Res. 16, 275–278 (1997). [DOI] [PubMed] [Google Scholar]
- Nakamura M., Chikama T. & Nishida T. Up-regulation of integrin α5 expression by combination of substance P and insulin-like growth factor-1 in rabbit corneal epithelial cells. Biochem. Bioph. Res. Co. 246, 777–782 (1998). [DOI] [PubMed] [Google Scholar]
- Quartara L. & Maggi C. A. The tachykinin NK1 receptor. Part I: ligands and mechanisms of cellular activation. Neuropeptides 31, 537–563 (1997). [DOI] [PubMed] [Google Scholar]
- Palanche T. et al. The neurokinin A receptor activates calcium and cAMP responses through distinct conformational states. J. Biol. Chem. 276, 34853–34861 (2001). [DOI] [PubMed] [Google Scholar]
- Lecat S., Bucher B., Mely Y. & Galzi J. L. Mutations in the extracellular amino- terminal domain of the NK2 neurokinin receptor abolish cAMP signaling but preserve intracellular calcium responses. J. Biol. Chem. 277, 42034–42048 (2002). [DOI] [PubMed] [Google Scholar]
- Mau S. E., Witt M. R., Saermark T. & Vilhardt H. Substance P increases intracellular Ca2+ in individual rat pituitary lactotrophs, somatotrophs, and gonadotrophs. Mol. Cell. Endocrinol. 126, 193–201 (1997). [DOI] [PubMed] [Google Scholar]
- Wong A. O., Ng S., Lee E. K., Leung R. C. & Ho W. K. Somatostatin inhibits (D-Arg6, Pro9-NEt) salmon gonadotropin-releasing hormone- and dopamine D1- stimulated growth hormone release from perifused pituitary cells of Chinese grass carp, Ctenopharyngodon idellus. Gen. Comp. Endocrinol. 110, 29–45 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.