Abstract
It is becoming increasingly clear that intracellular signaling can be subject to strict spatial control. As the covalent attachment of a signaling ligand to a nanoparticle (NP) impacts ligand-receptor binding, uptake, and trafficking, nanoconjugation provides new opportunities for manipulating intracellular signaling in a controlled fashion. To establish the effect of nanoconjugation on epidermal growth factor (EGF) mediated signaling, we investigate here the intracellular fate of nanoconjugated EGF (NP-EGF) and its bound receptor (EGFR) by quantitative correlated darkfield/fluorescence microscopy and density-based endosomal fractionation. We demonstrate that nanoconjugation prolongs the dwell time of phosphorylated receptors in the early endosomes and that the retention of activated EGFR in the early endosomes is accompanied by an EGF mediated apoptosis at effective concentrations that do not induce apoptosis in the case of free EGF. Overall, these findings indicate nanoconjugation as a rational strategy for modifying signaling that acts by modulating the temporo-spatial distribution of the activated EGF-EGFR ligand-receptor complex.
Graphical Abstract
Introduction
The selective recognition between cell surface receptors and their ligands is frequently exploited for targeting purposes in nanoparticle (NP) mediated diagnostic and therapeutic applications. Although the ligand is often considered as a mere targeting functionality, the covalent attachment of a ligand to a NP also provides leverage for modifying the cellular response triggered by specific ligand-receptor interactions and for manipulating the ligand function itself.1–7 The current understanding of the impact of nanoconjugation on ligand-receptor interactions and subsequent cellular processes (uptake, trafficking, de-activation) remains, however, rudimentary. We investigate here the effect of nanoconjugation on epidermal growth factor (EGF) mediated signaling. The EGF receptor (EGFR) belongs to the receptor tyrosine kinase (RTK) family of single transmembrane domain receptors and has been investigated in great detail as model system for RTK signaling and transmembrane signaling in general.8–11 According to the textbook model12, 13 of EGFR signaling activation, the binding of EGF to the extracellular domain induces conformational changes that result in the dimerization of the receptor. The dimerization facilitates a cross-phosphorylation of the tyrosine kinase domains and initiates downstream signaling events.
In cells with physiological receptor expression levels, EGF binding to EGFR also triggers endocytosis through a clathrin-dependent pathway, whereas in EGFR overexpressing cells clathrin-independent pathways have been observed especially in the presence of high EGF concentrations.14 Importantly, the activated EGFR remains dimerized with the phosphorylated tyrosine kinase domain exposed in the cytoplasm after internalization into early endosomes.15–17 In this fashion, the receptor continues to signal during intracellular trafficking until it is dephosphorylated by a protein tyrosine phosphatase15, 18, the ligand is removed from the endosome,19 or the phosphorylated EGFR (pEGFR) containing endosome is encapsulated in a multivesicular body.20 The accompanying removal of the phosphorylated receptor tail from the cytoplasm renders the phosphorylated EGFR inaccessible and results in signal termination. In the conventional model, EGFR signaling occurs primarily at the plasma membrane and endocytosis is a key step in signal attenuation14, 16, 21. It is, however, becoming increasingly clear that endocytosis and the subsequent trafficking can also play a more active role in the control of EGFR signaling.15 Although long controversial,22, 23 there is now mounting evidence that endosomes represent signaling platforms for transmembrane receptors and that the signal from receptors in endosomes can differ from those localized at the plasma membrane17, 24–26.
The picture that has emerged recently is that the outcome of the endosomal EGFR signaling sensitively depends on the temporo-spatial distribution of the pEGFR containing endosomes as well as the number of pEGFR (and potentially other EGF binding receptors) embedded in the endosomal membrane. While the former influences and regulates the interplay of the receptor with intracellular effector molecules27, the latter modulates signaling through cooperative interactions between co-localized receptors28, 29. Due to these spatial aspects of the signaling regulation, the disruption of the trafficking of EGF-pEGFR can lead to dramatic changes in the outcome.30 For instance, saturation of the cellular trafficking machinery in EGFR overexpressing epidermoid carcinoma (A431) or basal-like breast cancer (MDA-MB-468) cells after exposure to high concentrations of EGF was found to trigger apoptosis due to an accumulation of EGF-pEGFR in early endosomes.24, 31
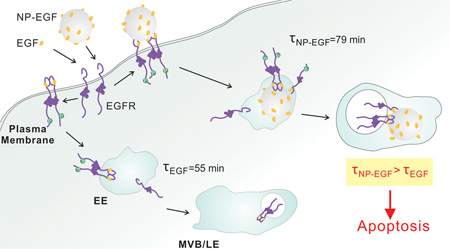
We observed recently that conjugation of EGF to 40 nm Au NPs results in EGF mediated apoptosis at effective ligand concentrations that are much lower than what is required for the free EGF to trigger apoptosis.1 However, the mechanism underlying this intriguing observation remains unclear. It is intuitive to assume that the covalent attachment of a ligand to a NP affects its endocytosis and subsequent trafficking. We, consequently, hypothesized that the temporo-spatial regulation of endosomal EGFR signaling makes the signaling outcome susceptible to intervention via nanoconjugation. In this manuscript we test this hypothesis by characterizing the impact of EGF nanoconjugation on endosomal EGF-pEGFR trafficking. We demonstrate that EGF conjugation to 80 nm diameter Au NPs results in a prolonged dwell time of pEGFR in the early endosomes and that the accumulation of NP tethered phosphorylated receptors in early endosomes goes hand-in-hand with an EGF mediated apoptosis at much lower concentrations than the unconjugated, free peptide.
Materials and methods
Materials
HS–CH2CH2–(CH2CH2O)77–N3 (PEG1) (NANOCS); HS–(CH2)11– (CH2CH2O)6–OCH2–COOH (PEG2) (ProChimia); propargyl dPEG– NHS ester (Quanta Biodesign); recombinant human epidermal growth factor (Life Technologies); 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Life Technologies); L-ascorbic acid (Aldrich); copper(II) sulfate pentahydrate (Aldrich); amantadine (Sigma); monodansyl cadaverine (Sigma); Nystatin (Sigma); genistein (Calbiochem); cochicine (Sigma); nitric acid (EMD) and hydrochloric acid (Fisher) for ICP-MS were used as received. D-Tube dialyzers (3.5 K MWCO) from EMD and Float-A-Lyzer G2 Dialysis Device from spectrum Lab were used for protein and NP-EGF purification. The EGF human ELISA kit and EGFR [pY1068] ELISA kits from Life Technologies were used for EGF surface density and EGFR phosphorylation determination. The EnzChek® caspase-3 Assay from Life Technologies was used in apoptosis assays. EEA1 and Rab7a human ELISA kits from Biomatik were used in endosomal sorting experiments. The BCA kit from Sigma was used to measure the total protein concentration as a reference of cell number. Cell Light® Early Endosome-GFP and Late Endosome-GFP were used to transfect cells with fluorescence.
NP-EGF Preparation and Characterization
Citrate stabilized 80nm Au NPs were synthesized through the Turkevich method. PEG1 (10 mM, 5 µL) and PEG2 (10 mM, 5 µL) were added to 4 mL of 80 nm Au NP colloid to yield a concentration of 25 µM for each PEG and then incubated for around 17 h at room temperature before they were washed three times by centrifugation and resuspension in DI water (2400 rpm, 10 min). The final NP-PEG pellets were resuspended in 770 µL of 0.5X PBS. 2 µL Propargyl-PEG-NHS ester solution in DMSO (100 mg/mL) were added to 100 µL of a 1 mg/mL solution of EGF in 1X PBS, pH 7.4, and incubated at 4 °C for 6 h. This mixture was then dialyzed against 0.5X PBS for 48 h. The obtained propargyl-PEG-EGF (20 µL) was then added to a NP-PEG (770 µL) suspension containing ascorbic acid (500 µM) and CuSO4 (100 µM) as catalyst and incubated at 4 °C overnight. The resulting NP-EGF particles were dialyzed against 0.1X PBS for 48 h and then washed further through centrifugation and resuspension (twice, 2200 rpm, 10 min). DLS, UV-Vis data were measured in 1X PBS at room temperature using a 100-fold dilution of the original NP-EGF pellets. For NP-EGF stability measurements, NP-EGF was resuspended in DMEM with 2 mM L-glutamine to a concentration of 8 pM and incubated at 37 °C for defined time durations. To quantify the EGF surface density, NP-bound EGF-PEG1 was first detached by incubating with PEG2 in great excess for 48h at 37 °C. NPs were subsequently sonicated for 10min and spun down through centrifugation (2400 rpm, 10 min) and the EGF concentration in the supernatant was quantified with an EGF ELISA kit according to the manufacturer’s protocol. Au NP concentrations were determined by UV-Vis.
Cell Binding, Uptake, and ICP-MS Quantification
MDA-MB-468 cells were cultured in 6 well dishes. Cells with a confluency of approx. 70% were then incubated with 1 mL NP-EGF (8 pM), NP-PEG (8 pM), or membrane wrapped NPs (MW-NP; 32 nM) in DMEM, 2 mM L-glutamine buffer at 37 °C for 25 min or 4 h. After that, cells were washed 3 times with ice cold HBSS buffer to remove excess NPs before they were harvested with 0.25% trypsin. Cells were pelleted at 300 g for 5 min and further washed with ice cold 1X PBS for 3 times. Then 200 µL 1X PBS was added to each pellet and cell numbers were determined with a hemacytometer. 1 mL aqua regia was added to the pellets and dried overnight at 50 °C. The samples were then re-dissolved in 2 mL 2% HCl solution. After an additional 250–1000 fold dilution with 2% HCl, the Au content in the samples was quantified with a VG Plasma Quad ExCell ICP-MS.
Phosphorylation Assay
MDA-MB-468 cells were cultured in 6 well dishes. When the confluency reached approx. 70%, cells were starved in serum free DMEM for 16–20 h and then incubated with 1 mL NP-EGF (8 pM) or EGF (1 nM) for 20 min before they were collected and lysed in lysis buffer (10 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 20mM Na4P2O7, 2 mM Na3VO4, 1% Triton 100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate, 1 mM PMFS, 1:20 protease inhibitor cocktail). Cells lysates were then used in the pEGFR ELISA kit as described in the manufacturer’s protocol. Phosphorylation data were plotted as ratio relative to the EGF (1 nM) control.
Cell MTT Assay
MDA-MB-468 cells were plated in a 96 well dish at a density of 10,000 cells/well and cultured for 48h. Old growth medium was replaced with 100 µL NP-EGF (8 pM), NP-PEG (8 pM), EGF (1 nM), or EGF (33 nM) in each well and further incubated for 4 h in DMEM with 2 mM L-glutamine at 37°C. Then the buffer was removed and the cells were washed 3 times with prewarmed HBSS buffer and further incubated in complete growth medium for another 20 h. Old growth medium was then removed and 100 µL of phenol red free fresh medium and 10 µL of a MTT solution (12 mM) was added and incubated at 37 °C for 4 h. Then all but 25 µL of the medium was removed and 50 µL of DMSO was added. The samples were incubated at 37 °C for 10 min. The absorbance at 540 nm was measured using a microplate reader. Data were plotted as ratio relative to the control group, in which cells were incubated in complete growth medium.
Cell Apoptosis Measurements
MDA-MB-468 cells were cultured in 6 well dishes at a density of 1×105 cells/mL with 1.6 mL growth medium per well for 48 h. The growth medium was then replaced with 1 mL of fresh medium with or without AG1478 (250 nM) and further incubated for 15 min before the cells were treated with 1 mL of NP-EGF (8 pM), NP-PEG (8 pM), EGF (33 nM), or supernatant from last wash of NP-EGF and further incubated for 4 h in DMEM, 2 mM L-glutamine buffer at 37 °C with or without presence of AG1478 (250 nM). Then, the buffer was removed and the cells were washed 3 times with prewarmed DMEM and further incubated in complete growth medium for another 20 h. Cells were harvested with 0.25% trypsin and collected by centrifugation at 300 g for 5 min. The pellets were washed twice with 1X PBS. 60 µL lysis buffer were added to each pellet and the EnzChek® caspase-3 assay was applied to determine apoptosis activity according to the manufacturer’s protocol in a 96-well plate. Sample sizes were quantified by the total protein concentration through BCA protein assays.
Pharmacological Inhibition of NP Uptake
MDA-MB-468 cells were preincubated with different pharmacological inhibitors at 37 °C in complete DMEM: monodansyl cadeverine (200 µM) for 20 min; amantadine (5 mM) for 30 min; genistein (200 µM) for 60 min; nystatin (50 µg/mL) for 15 min. The cells were then incubated with NP-EGF (8 pM) in the presence of the respective inhibitors for 20 min and washed with HBSS buffer 3 times. 1 mL of the I2/KI aqueous solution (0.34 mM I2, 2.04 mM KI) was added to the culture flask and incubated at room temperature for 2 min and then washed 3 times with HBSS buffer.
Endosomal Sorting Experiment
MDA-MB-468 cells were cultured in 25 cm2 culture flasks in DMEM, 10% FBS and 2 mM L-glutamine at 37 °C in 5% CO2. Cells with a confluency of 80% were incubated with 1.8 mL NP-EGF (8 pM) in DMEM, 2 mM L-glutamine buffer at 4 °C for 2 h. Then, cells were washed 3 times with ice cold HBSS buffer and returned to 37 °C in complete DMEM for specified incubation times. The cells were subsequently washed twice with ice cold HBSS buffer and homogenization buffer (0.25 M sucrose, 10 mM triethanolamine, 10 mM acetic acid, 1 mM EDTA, 2 mM Na3VO4, 1 mM PMFS, 1:20 protease inhibitor cocktail, pH 7.8) before the cells were harvested in 1 mL homogenization buffer with a scraper. 40 strokes through a 23 gauge syringe were applied to the cell suspension. After homogenization, the Au NP containing fractions were pelleted at 800 g for 10 min and then washed with ice cold 1X PBS. For ELISA measurements of EEA1 and Rab7, 250 µL ice cold 1X PBS (pH 7.2) was added to each pellet and the cell suspension was further sonicated for 10 s on ice. The solution was centrifuged at 6000 rpm and the supernatant was collected for the ELISA performed according to the manufacture’s protocol. The concentrations of these two markers were subsequently normalized with the concentration of phosphatidylcholine as a reference of endosome quantity. For ELISA measurement of pEGFR, an I2/KI etching step was added before the homogenization of the cells as described above. After collecting the Au NP containing pellets, 100 µL lysis buffer was added to each pellet and further incubated on ice for 30 min. The solution was centrifuged at 6000 rpm. The supernatant was collected for ELISA measurements and the pellets were collected for ICP-MS measurements to quantify the Au concentration.
Optical Colocalization Experiments
MDA-MB-468 cells were cultured in a glass bottom culture dish in DMEM, 10% FBS and 2 mM L-glutamine at 37 °C in a water vapor saturated, 5% CO2 containing atmosphere for 2 days before they were transfected with Rab5a-GFP or Rab7a-GFP constructs at a confluency of 50% for 20 h. The cells were then incubated with 1 mL NP-EGF (8 pM) containing DMEM, 2 mM L-glutamine buffer at 37 °C for 20 min. Subsequently, the buffer was removed and the cells were transferred into fresh complete growth medium and incubated for specified time durations at 37 °C before they were fixed with 4% PFA for 15 min at room temperature. All optical studies were performed with an Olympus IX71 inverted microscope with a high numerical aperture (NA) water/oil condenser (NA = 1.2–1.4) and 10× air (NA = 0.25) or 60× oil (NA = 0.65) objective. The samples were illuminated with a 100 W tungsten lamp (for dark field) and 100 W mercury lamp (for fluorescence) in conjunction with appropriate filter sets for various dyes. In the case of EGF-Alexa647, EGF-Biotin (20 µg/mL, 12 µL) was mixed with Streptavidin-Alexa647 (1 mg/mL, 2 µL) and incubated at 4 °C overnight to obtain EGF fluorescence conjugates (EGF-Alexa647). Rab5a or Rab7a transfected MDA-MB-468 cells were prepared as described above. 1 mL of 1 nM EGF-Alexa647 was then added to cells in DMEM with 2 mM L-glutamine at 37 °C for 20 min. Subsequently, the cells were washed 3 times with prewarmed 1% BSA containing HBSS buffer, transferred into fresh complete growth medium and maintained in the incubator at 37 °C for different time durations. Then, the cells were washed 3 times with 1% BSA containing HBSS buffer and fixed with 4% PFA for 15 min at room temperature. An Olympus FV1000 scanning confocal microscope with simscan accessory and 488, 633nm excitation lines was used to record fluorescence images. For the colocalization of NP-EGF scattering signals with fluorescence markers, NP-EGF and marker signals were identified based on scattering intensity and size. Subsequently, a new image was generated containing only the identified NPs and fluorescence emitters. Darkfield and fluorescence intensities were rescaled to generate a noticeable color change upon colocalization. In the case of EGF-Alexa647 fluorescence images were background corrected and then combined with the fluorescence marker image. Colocalization percentages were evaluated as fraction of NP-EGF (or EGF-Alexa647) emitters that colocalize with the early/late endosome markers.
Dynamic Colocalization and Data Analysis
Rab5a transfected MDA-MB-468 cells were prepared as described above. 1 mL of NP-EGF (8 pM) was then added and the cells were incubated in DMEM, 2 mM L-glutamine buffer at 37 °C for 20 min. After that, the cells were washed 3 times with pre-warmed DMEM and further incubated in fresh complete DMEM for 4 h at 37 °C in a water vapor saturated atmosphere containing 5% CO2. An Olympus IX71 inverted microscope with a water/oil condenser (NA = 1.2–1.4) and 60× oil (NA = 0.65) objective with a 1.6× magnification changer was used. The experiments were conducted in a cage incubator at 37 °C. The samples were illuminated with a 100 W tungsten lamp (for dark field) and 100 W mercury lamp (for fluorescence) in conjunction with an appropriate filter set for GFP. Only cells with at least moderate expression of Rab5a-GFP and intracellular uptake of NP-EGF were included in the analysis. In a first step, NPs that did not show optical colocalization with GFP tagged endosomes were identified by inspection of the movie and excluded from the subsequent analysis. For those NPs that exhibited colocalization in any single frame, trajectories of NP-EGF and corresponding Rab5a marked endosomes were analyzed by calculating the Pearson product-moment correlation coefficient of the two trajectories. Only trajectories with a Pearson coefficient > 0.5 during the entire movie were considered colocalized.32
RESULTS AND DISCUSSION
One general challenge for the analysis of intracellular signaling is the need for distinguishing signals that originate at the plasma membrane from those that are associated with intracellular endosomes. To accomplish this goal, we apply here a two-pronged approach that utilizes 80 nm Au NPs both as a “load” to facilitate a density-based separation of NP-EGF containing endosomes as well as a bright optical probe to map the temporo-spatial distribution of the nanoconjugated EGF and to characterize its interaction with the cellular trafficking machinery. Au NPs are uniquely suited for this study as they can be synthesized with a narrow size distribution over a broad size range.33–35 The surface properties of Au NPs can be well controlled through established Au-thiol chemistry,36 and the particles have been found to be compatible with diverse biomedical applications.37–45 Furthermore, Au NPs provide large optical scattering cross-sections40, 45–47 that make them useful probes in darkfield microscopy. In this manuscript we combine darkfield microscopy of metal NPs with conventional fluorescence microscopy to simultaneously track and characterize NP containing endosomes in living cells as function of space and time.48 In an independent set of experiments we take advantage of the increase in endosomal density after Au NP uptake to quantify the relative contributions from nanoconjugated EGF in early and late endosomes as a function of time in an entire cell population.
EGF Nanoconjugation and Characterization of NP-EGF
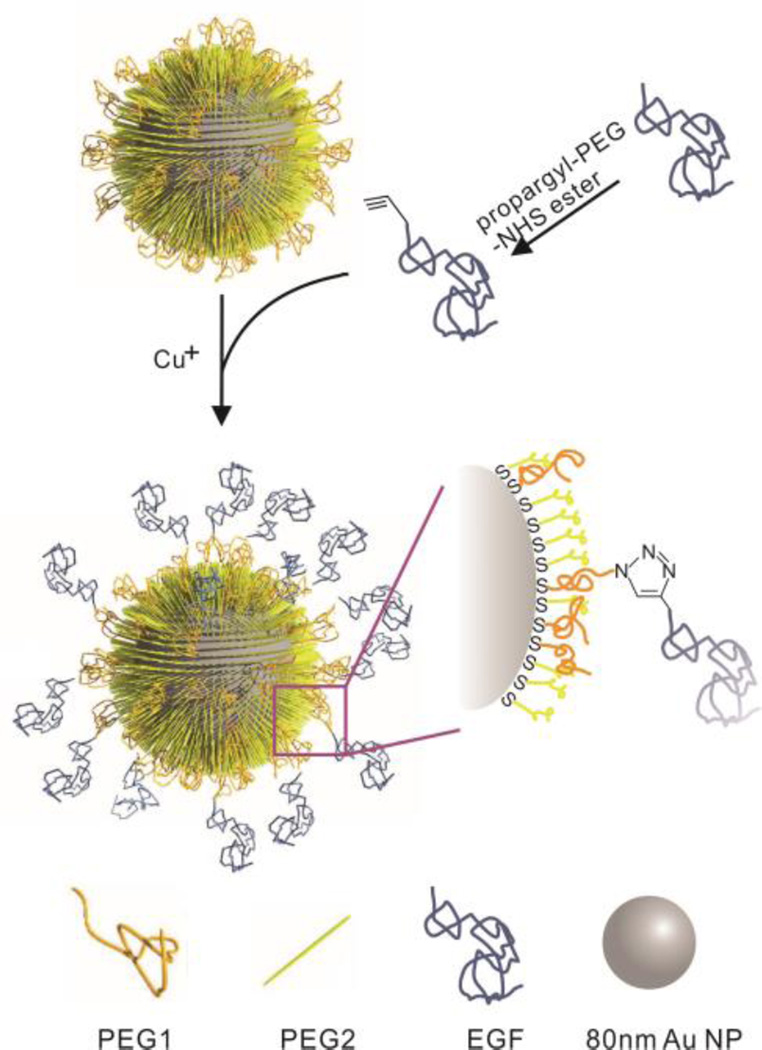
A scheme of the NP design used in this work is given in Fig. 1. To achieve a presentation of EGF with a high degree of conformational flexibility and surface accessibility, we functionalized 80 nm diameter Au NPs with HS-EG77-N3 (PEG1) and HS-(CH2)11-EG6-COOH (PEG2) in a nominal ratio of 1:1. The azido group in PEG1 was used to tether alkyne-modified EGF to the NPs through the CuI catalyzed 1,3-dipolar cycloaddition.49 The shorter PEG2 was introduced to stabilize the NPs and to increase the accessibility of the azido group on the surface. Dynamic light scattering (DLS) confirmed a successful functionalization of the NPs: the hydrodynamic diameter increased from 103.4 ± 0.6 nm for the pegylated NPs (NP-PEG) to 112.3 ± 1.6 nm after EGF conjugation (NP-EGF). The ligand density on the NPs was determined as 124 ± 8 EGF/NP. The zeta potential of NP-EGF was −47.4 ± 5.6 mV. We tested the stability of NP-EGF under the relevant experimental conditions in DMEM at 37 °C by monitoring the average hydrodynamic diameter using DLS. For NP-EGF concentrations of 8 pM and below we did not observe any significant agglomeration within at least 4 h (Fig. S1). As higher NP-EGF concentrations were less stable against agglomeration, we used 8 pM NP-EGF with incubation times ⩽ 4 h in all subsequent cell experiments.
Figure 1.
Schematics of NP-EGF preparation. 80 nm Au NPs are functionalized with PEG1 (=HS-EG77-N3) and PEG2 (=HS-(CH2)11-EG6-COOH) to introduce binding sites and stabilize the NPs, respectively. Functionalized human EGF is then covalently linked to PEG1 through the CuI catalyzed 1,3-dipolar cycloaddition reaction.
Specificity of NP-EGF Uptake and EGFR Activation
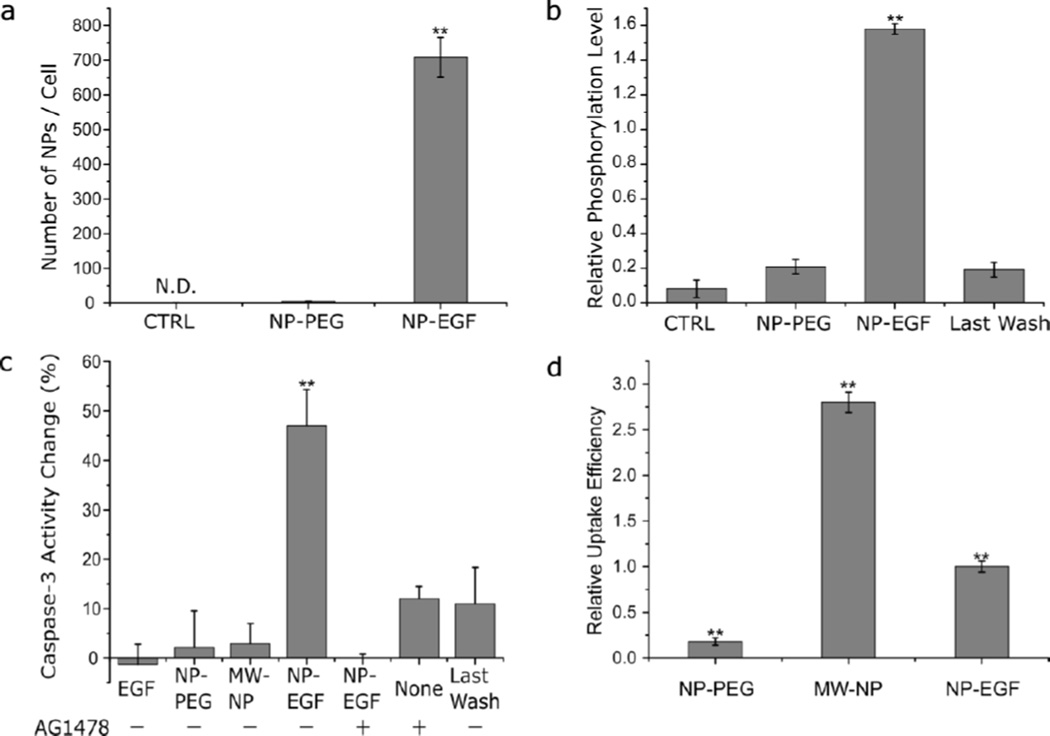
We validated the bioavailability of nanoconjugated EGF under the chosen experimental conditions by quantifying the EGF-specific cellular uptake of NP-EGF and measuring the resulting global EGFR activation. We chose the adenocarcinoma cell line MDA-MB-468 for our studies as it is an already established model for studying free EGF induced apoptosis,31 facilitating an uncomplicated comparison of our data with the free EGF benchmark. We measured the Au concentration in MDA-MB-468 cells after incubation with NP-EGF (8pM) or NP-PEG (8pM) in DMEM for 25 min at 37 °C with inductively coupled plasma mass spectroscopy (ICP-MS). The strategy described in ref.50 was then applied to convert the measured number of Au atoms into a number of 80 nm Au NPs per cell (Fig. 2a). While the Au NP content for NP-PEG was negligible with 5 ± 1 NP/cell, for NP-EGF the average Au NP concentration was 709 ± 56 NP/cell. The control group without NP incubation showed no detectable signal in ICP-MS. The highly selective uptake of NP-EGF confirms that EGF remains bioavailable after nanoconjugation and that the NP-cell interactions are mediated by EGF.
Figure 2.
Characterization of NP-EGF induced uptake, phosphorylation and apoptosis. a. Cellular uptake of NP-PEG and NP-EGF as determined by ICP-MS. MDA-MB-468 cells were incubated with NPs (8 pM) for 25 min. CTRL = no treatment control. Data were collected from 3 independent experiments. b. Phosphorylation level induced by NP-EGF. Cells were treated with (left to right) DMEM (CTRL), NP-PEG (8 pM), NP-EGF (8 pM), and the supernatant from the last wash of NP-EGF for 25 min in the incubator before they were lysed and assayed for phorsphorylated EGFR by ELISA. All data are normalized by the phosphorylation measured for free ligand EGF (1 nM) and were collected from at least 3 independent experiments. c. Change in caspase-3 activity (relative to CTRL) in %. MDA-MB-468 cells were incubated (left to right) with EGF (33 nM) peptide, NP-PEG (8 pM), membrane wrapped Au NP (32 nM), NP-EGF (8pM), supernatant from the last wash of NP-EGF through centrifugation in the presence (+) or absence (−) of AG1478 (250 nM) as indicated. The cells were incubated for 4 h and chased in fresh growth medium for 20 h. Data were collected from 6 independent experiments. d. Relative cellular uptake of NPs with 4 h incubation time. Cells were incubated with NP-PEG (8 pM), MW-NP (32 nM), and NP-EGF (8 pM) for 4h. Data were collected from 3 independent experiments. Error bars represent the standard error of the mean (SEM). Differences between multiple groups were determined by one-way ANOVA with Tukey post-hoc test (** p< 0.01).
Consistent with EGFR activation by nanoconjugated EGF, we detected robust EGFR phosphorylation levels after 25 min of incubation with NP-EGF (8 pM, effective EGF concentration = 8 pM × 124 EGF/NP ≈ 1 nM) (Fig. 2b). In fact, the phosphorylation for NP-EGF exceeded that of the same concentration of free EGF by approximately 60%. NP-PEG or the supernatant of the last wash of NP-EGF cleaning by centrifugation and resuspension showed only background phosphorylation levels, confirming that the increase in phosphorylation is EGF dependent and a result of nanoconjugation.
Apoptotic Efficacy of NP-EGF
To verify that NP-EGF with 80 nm Au NP core induces apoptosis under the chosen experimental conditions, we incubated MDA-MB-468 cells (70% confluency) with NP-EGF (8pM) for 4 h at 37 °C in DMEM. After that, the NP-EGF solution was removed and fresh complete growth medium was added. The cells were further incubated for 20 h before MTT and caspase-351–54 assays were performed to quantify cell viability and apoptosis, respectively. Only NP-EGF treatment resulted in a measurable reduction of the cell viability (Fig. S2); free EGF or NP-PEG controls induced no significant change. The measured relative caspase-3 activities for the different experimental conditions are summarized in Fig. 2c. Importantly, while even high doses of free EGF (33 nM) do not induce apoptosis in MDA-MB-468 cells under our experimental conditions, NP-EGF (8pM) corresponding to an effective EGF concentration of 1 nM is sufficient to induce a significant increase (50%) relative to the no treatment control. As additional controls we included NP-PEG (8 pM) and the supernatant of the last wash of NP-EGF to determine the contribution of bare NPs (no EGF) and of residual free EGF (and copper catalyst), respectively, to the NP-EGF induced apoptosis. Both conditions do not yield any increase in caspase-3 activity. The NP uptake for the NP-PEG is, however, much lower than for NP-EGF (Fig. 2d). We, therefore, also measured the caspase-3 levels obtained with high concentrations of lipid membrane wrapped NPs55, 56 (MW-NP; initial membrane composition: 2% phosphatidylserine (PS), 58% dipalmitoylphosphatidylcholine (DPPC), 40% cholesterol). The membrane wrapped NPs remain stable even when incubated with cells in DMEM in the tens of nM concentration range. At these high concentrations, non-specific adsorptive pinocytosis57 results in elevated intracellular NP concentrations (Fig. 2d, Fig. S3). Importantly, even for Au MW-NP treated cells, whose intracellular Au concentration exceeds that of NP-EGF treated cells by 150%, no significant increase in apoptosis is detected (Fig. 2c).
To obtain further unambiguous experimental proof that the apoptosis detected upon NP-EGF exposure is related to EGFR signaling, we measured the caspase-3 activity of cells treated with the EGFR-specific tyrosine kinase inhibitor tyrphostin AG1478. The inhibition of receptor phosphorylation in cells treated with NP-EGF (8 pM) under otherwise identical conditions decreases the caspase-3 activity to background levels, confirming that EGFR activation is a requirement for NP-EGF induced apoptosis. Together, the apoptosis measurements performed for NP-EGF and the appropriate controls corroborate our hypothesis that nanoconjugation enhances EGF mediated apoptosis and that the underlying mechanism involves EGFR activation.
Mechanism of NP-EGF Uptake
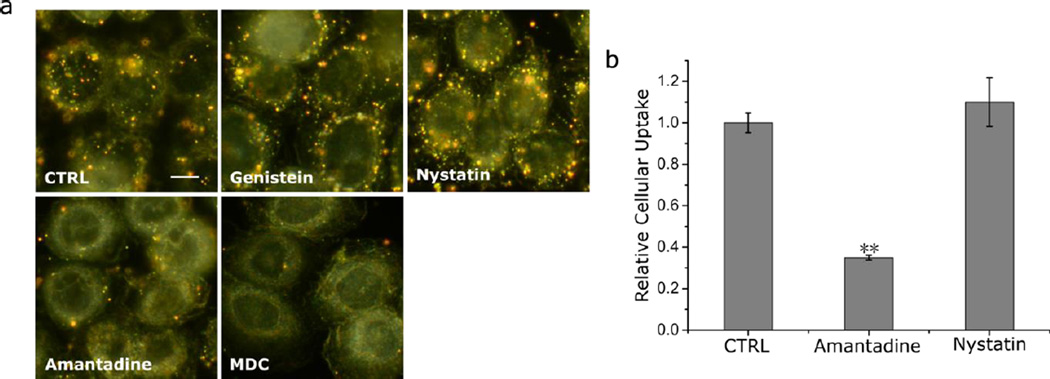
Endocytosis is an integral part of signaling regulation. Cells with physiological levels of receptor expression in environments with physiological EGF concentrations (< 1–2 ng/mL) internalize EGFR via a fast clathrin-mediated endocytosis process.21, 58–61 Alternative, clathrin-independent uptake mechanisms have been observed at higher EGF concentrations and these alternative uptake routes can lead to different temporo-spatial NP-EGF distributions.62–65 In the case of caveolae mediated endocytosis, the uptake kinetics is known to be much slower than for the clathrin-mediated process.21, 31, 66–68 Caveloae mediated endocytosis can also play an important role for the internalization of different NPs and virus particles.69–71 Given the putative relevance of the temporo-spatial distribution of the activated receptor for modulating endosomal signaling, a correct interpretation of the observed apoptosis requires the identification of the internalization mechanism. To that end, we determined the relative contributions from clathrin- and caveolae-mediated NP-EGF-pEGFR endocytosis with well-known pharmacological inhibitors that block specific endocytosis pathways (Fig. 3a). Amantadine and monodansyl cadaverine were used to block clathrin-mediated endocytosis, and nystatin and genistein were applied to inhibit caveolae (lipid raft)- mediated endocytosis. We ensured that only uptaken Au NPs were considered in our comparison of the different inhibitors by removing all surface bound NPs with an aqueous solution of I2/KI (0.34 mM I2, 2.04 mM KI) at room temperature for 2 min prior to any analysis72. The darkfield images in Fig. 3a show that only amantadine and monodansyl cadaverine achieve an obvious reduction of NP uptake but not nystatin and genistein (for positive controls see Fig. S4&S5). For two selected inhibitors (amantadine and nystatin) we validated the results for the optical darkfield imaging studies by quantifying the uptaken Au concentration by ICP-MS (Fig. 3b). Amantadine but not nystatin blocks the NP uptake, confirming that under the chosen experimental conditions a fast clathrin-mediated endocytosis of NP-EGF dominates. We do not rule out the possibility that additional non-clathrin-mediated uptake mechanisms exist, but they play a negligible role under our experimental conditions and are not further investigated.
Figure 3.
Pharmacological inhibition study of clathrin and caveolae dependent endocytosis pathways. a. Darkfield images of MDA-MB-468 cells pre-treated with different inhibitors of clathrin (amantadine, mondansyl cadaverine) or caveolae (genistein, nystatin) dependent endocytosis after incubation with 8 pM NP-EGF for 20 min (ctrl = no treatment). Surface bound NPs were removed by I2/KI etching prior to imaging. Scale bar: 10 µm. b. Relative cellular uptake of NP-EGF for two representative inhibitors: amantadine and nystatin after incubation with 8 pM NP-EGF for 20min and I2/KI etching. Data were collected from 3 independent experiments. Error bars represent the SEM. Differences between multiple groups were determined by one-way ANOVA with Tukey post-hoc test (** p< 0.01).
Isolation and Characterization of NP Containing Endosomes
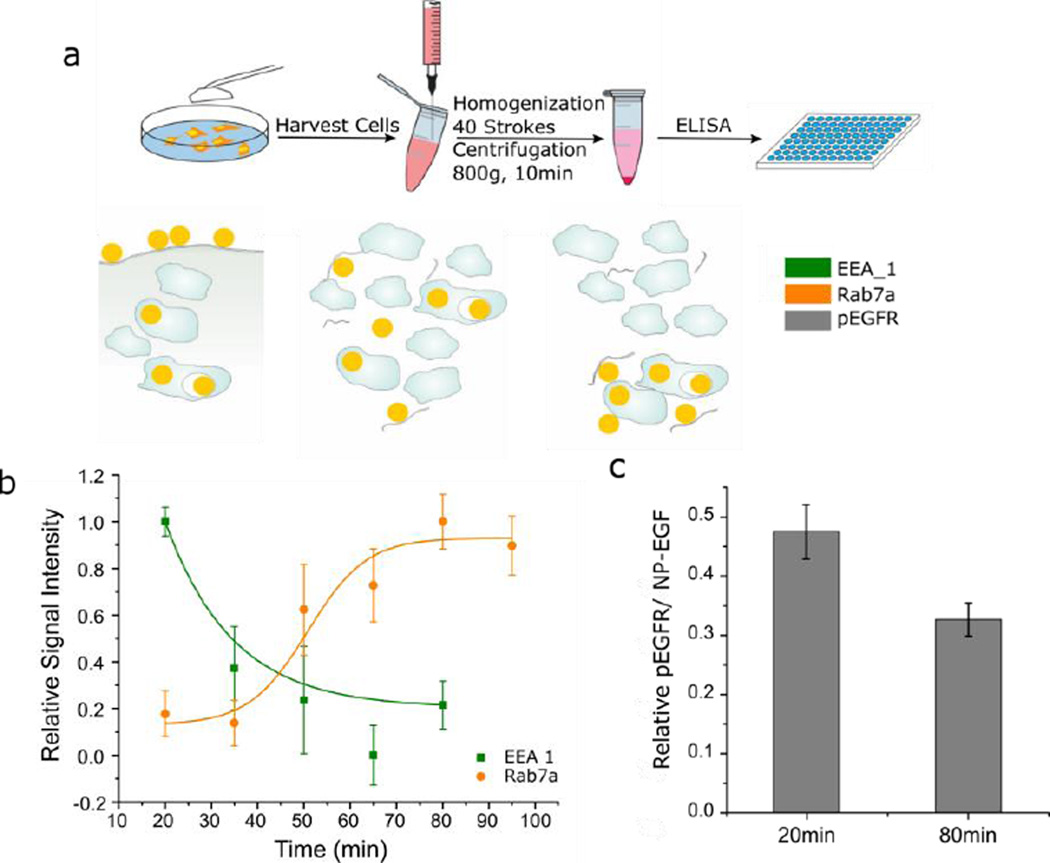
We elucidated the impact of EGF nanoconjugation on the spatial EGFR signaling and its role in apoptosis by characterizing the distribution of NP-EGF on early and late endosomes as a function of time using the endosome isolation technique outlined in Fig. 4a. The outlined approach utilizes the, due to the high density of Au (19.3 g/cm3), higher weight of NP-EGF containing endosomes to separate them from NP-free endosomes by centrifugation at low g-forces.73 A successful isolation of NP containing endosomes makes them amenable to characterization through standard immunoassays. NP-EGF (8pM) was incubated with MDA-MB-468 cells at 4°C for 2h in DMEM before the colloidal solution was removed and endocytosis was induced by increasing the temperature to 37°C in fresh growth medium. The cells were then further incubated for another 5–95 min. Subsequently, the cells were homogenized and the NP containing endosomes were isolated from the cell lysate by centrifugation at 800 g for 10 min and resuspension in 1X PBS buffer. The early endosome antigen 1 (EEA1) and the Ras-related protein Rab7a are frequently used biomarkers for the early and late endosome.74–76 We measured the EEA1 and Rab7a levels of the isolated endosomes as a function of time by ELISA. All EEA1 and Rab7a intensities were normalized by the measured phosphatidylcholine (PC) concentration. PC is a major membrane component77 and served as an internal standard for the relative amount of isolated endosomes. After an initial delay related to thermal equilibration of the cells following their transfer from 4°C to 37°C (Fig. S6), the EEA1 level associated with NP containing endosomes increases due to NP-EGF endocytosis at 37°C and subsequent collection in early endosomes at physiological temperatures. Fig. 4b plots the relative ELISA signal intensities calculated as (I(t) − Imin) /(Imax−Imin) as function of time after the number of NPs in EEA1 positive endosomes has peaked (t = 20 min). I(t) is the normalized ELISA intensity at time t, and Imax and Imin are the minimum and maximum intensities, respectively. Although the measured EEA1 and Rab7a levels from at least six independent experiments show a broad spread, the averages follow clear trends. Consistent with a maturation of NP containing early endosomes into late endosomes, the decrease in EEA1 concentration is accompanied by an increase in Rab7a signal. Importantly, the initial drop in EEA1 by approximately 60% between t = 20 min and t = 35 min, is followed by a much more gradual decrease for t > 35 min. Interestingly, even at t = 80 min, the measured absolute EEA1 levels remain significantly higher than the background obtained from Au NPs mixed with cell lysate (Fig. S7).
Figure 4.
Endosomal sorting experiment. a. Schematics of sample preparation (top row) and NP distribution (bottom row). MDA-MB-468 cells were incubated with NP-EGF (8 pM) for 2 h at 4 °C and then chased in fresh growth medium at 37 °C for the specified time before the cells were harvested. To measure phosphorylation of endosomal EGFR, surface NP-EGF were etched away with I2/KI prior to cell harvesting. 40 strokes through a 23 gauge syringe were applied to the collected cell suspension to break the cellular membrane but retain the endosomal membrane intact. The Au NP containing endosomes were then collected by centrifugation and assayed with ELISA of different markers. b. Relative signal intensity (see text) for early endosome (EEA1, olive) and late endosome (Rab7a, orange) marker as function of time. At least 6 independent experiments were included in the plot. EEA1 values for t = 20 min differ significantly from those at t > 20 min (Paired student’s t test, p < 0.01) and Rab7a values for t ≥ 50 min differ significantly from those at t < 50 min (Paired student’s t test, p < 0.01). c. Endosomal EGFR phosphorylation levels induced per NP-EGF at t = 20 min and t = 80 min. Phosphorylation levels are relative to the t = 5 min chase time phosphorylation and were collected from 3 independent experiments. Error bars represent the SEM.
Although the exact time for the detection of free EGF in the early endosomes depends on ligand concentration and receptor expression levels, a typical timescale lies between t = 5–20 min after exposure.19 The timescale for localization of free EGF in the late endosome is between t = 20–60 min.19 Our data in Fig. 4b indicate that the time scale of early to late endosome maturation in the case of NP-EGF lies at the upper end of this time-range. Only 30 min after the number of NPs in EEA1 positive early endosomes has peaked at t = 20 min, a significant increase in the Rab7a levels is detected. After that, the Rab7a level continues to increase but does not converge until approximately t = 95 min. This together with the detection of NP-EGF in the early endosome even after t = 80 min (Fig. S7) indicates that the dwell time in the early endosomes is significantly longer for NP-EGF than for free EGF. One caveat in our analysis so far is that, although it provides information about the distribution of the Au NPs between early and late endosomes as a function of time, it is unclear if the NP-EGF remains bound to EGFR over the observed timescale and whether the latter remains phosphorylated. Since it is the pEGFR and not the Au NPs per se that is responsible for the hypothesized endosomal apoptotic signaling, we evaluated the pEGFR levels associated with NP-EGF at t = 5, 20 and 80 min. We again used the density-based endosome fractionation approach outlined in Fig. 4a to isolate NP containing endosomes. To eliminate any potential background from plasma-membrane localized pEGFR, we removed in this case all Au NPs on the plasma membrane by etching with an aqueous solution of I2/KI (0.34 mM I2, 2.04 mM KI) at room temperature72 prior to homogenization of the cells. In this way, we ensured that only intracellular NP-EGF is collected by centrifugation, facilitating a selective quantification of endosomal pEGFR. To that end, the collected NP containing endosomes were sonicated for 10 s to disrupt the endosomes. The NPs released from the endosomes were collected by centrifugation and the concentration of pEGFR in the supernatant was quantified by ELISA while the Au concentration in the pellet was determined by ICP-MS. Fig. 4c summarizes the pEGFR levels (normalized by the uptaken NP concentration). The phosphorylation levels for t = 20 min and t = 80 min are given relative to the t = 5 min phosphorylation level. Initially (t = 5 min) the pEGFR concentration is highest in the early endosomes (= 100%) with the phosphorylated C-termini available for signaling in the cytosol. With increasing time the measured phosphorylation gradually decreases due to the maturation of NP-EGF-pEGFR containing endosomes into mature late endosomes (Fig. 4b) or other deactivation processes. After 20 min of incubation the pEGFR level decreases to 47%, and after t = 80 min the pEGFR level in the NP containing endosome fraction is still 33%.
Overall, our analysis of the isolated NP containing endosomes reveals appreciable levels of NP-EGF and pEGFR in the early endosomes for at least 80 min, consistent with the prolonged existence of NP-EGF-pEGFR complexes capable of signaling in the cytoplasm.
Comparison of NP-EGF Trafficking with Free EGF
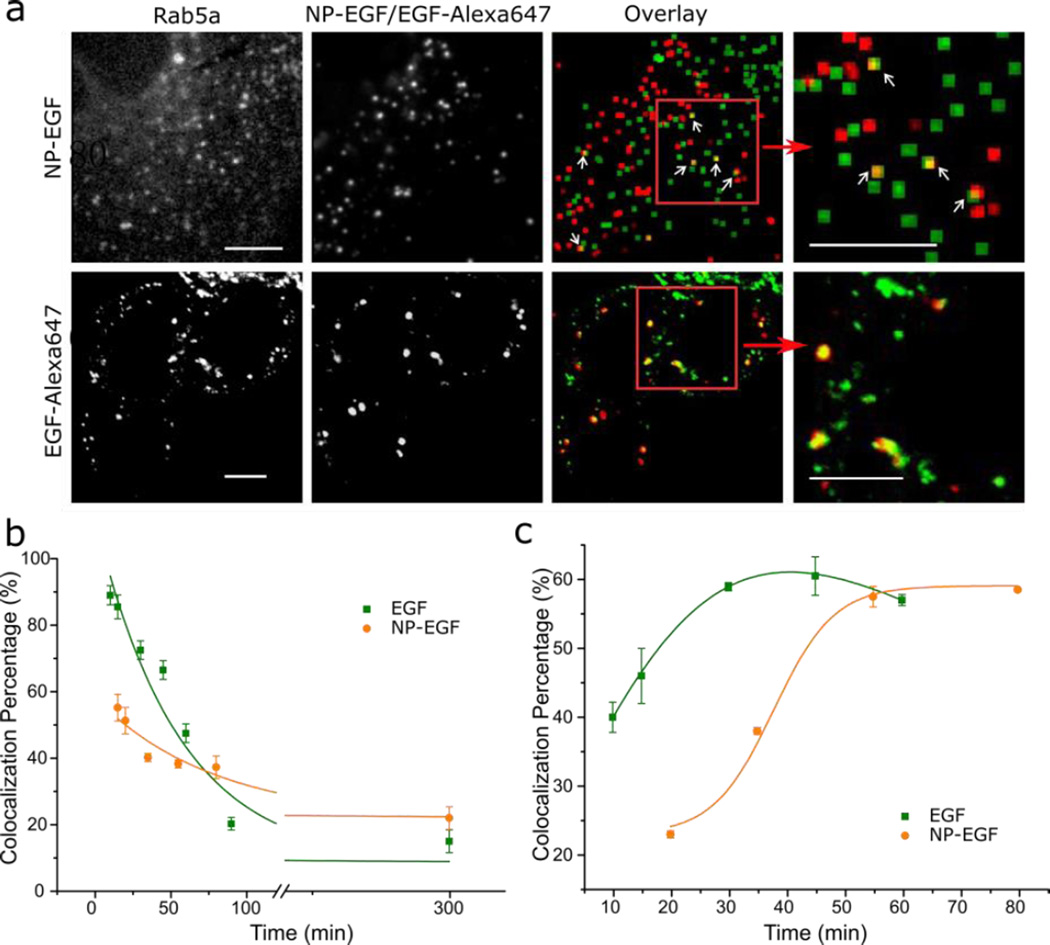
The unambiguous validation of the hypothesis that EGF nanoconjugation prolongs the pEGFR dwell time in early endosomes requires a direct comparison of the NP-EGF trafficking dynamics with that of free EGF at comparable effective concentrations. As the density-based isolation technique is not applicable to the isolation of EGF-pEGFR containing endosomes, we chose a conventional optical colocalization strategy to compare the trafficking dynamics of EGF-pEGFR and NP-EGF-pEGFR. We transfected MDA-MB-468 cells with Rab5a GFP or Rab7a GFP-constructs to label early and late endosomes respectively.75, 76, 78, 79 80 nm Au NPs have large scattering signals and can be easily localized in conventional darkfield microscopy.55 The NP signal in the darkfield microscope can then be correlated with the fluorescence signals of the Rab5a or Rab7a labels to determine the NP-EGF colocalization with early or late endosomes. For free EGF we applied an analogous colocalization strategy based on two-color confocal fluorescence microscopy.
We labeled biotinylated EGF with Alexa647-functionalized streptavidin in a 1:1 ratio (for a validation of the activity of the Alexa647 labeled EGF see Fig. S8). MDA-MB-468 cells transfected with Rab5a and Rab7a GFP-constructs were incubated in DMEM solutions containing NP-EGF (8pM, 1 nM effective EGF concentration) or fluorescent EGF (1nM) for 20 min at 37°C and then chased in fresh DMEM for 0–280 min prior to optical analysis. Representative colocalization images at t = 300 min (20 min incubation + 280 min chase) for NP-EGF and at t= 30 min (20 min incubation + 10 min chase) for free EGF are provided in Fig. 5a. The measured Rab5a and Rab7a colocalization probabilities for NP-EGF and free EGF throughout the entire observation time (incubation + chase) are plotted in Fig. 5b & c.
Figure 5.
Optical mapping of NP-EGF and free EGF trafficking from early to late endosomes. a. Top: Fluorescence image of Rab5a early endosome marker (left), darkfield image of NP-EGF (second from left) and computed overlay after 20 min exposure to NP-EGF and 5 h further incubation in fresh growth medium (third and fourth from left). Green pixels mark the positions of fluorescence emitters, red of NP-EGF and the overlay is yellow. Bottom: Fluorescence image of Rab5a (green) and EGF-Alexa647 (red) and overlay (yellow) after 20 min exposure to EGF and 30 min further incubation in fresh growth medium (see text). Magnified images for the areas in the read squares are included. Scale bar: 10 µm. b. Optical colocalization (in %) of NP-EGF (orange dot) or EGF-Alexa647 (olive square) with Rab5a positive endosomes as function of time. Data were collected from at least 3 independent experiments with a total of ~120 cells analyzed at each time point. Data were fitted with an exponential decay function: y=A1*exp(−x/t1)+y0. For EGF-Alexa647: τ = 55min with r2= 0.95; for NP-EGF : τ = 79 min with r2= 0.94. c. Optical colocalization of NP-EGF (orange dot) or EGF-Alexa647 (olive square) with Rab7a positive endosomes as function of time. Data were collected from at least 3 independent experiments (~90 cells analyzed). Error bars represent the SEM.
For both NP-EGF and free EGF the colocalization probability between EGF and Rab5a positive early endosomes decrease continuously as a function of time. Simple exponential decay fits yield characteristic time constants of τEGF = 55 min vs. τNP-EGF = 79 min (Fig. 5b). The observed differences in the slope confirm that the dwell time in Rab5a positive endosomes is longer for NP-EGF than for free EGF. The absolute colocalization percentage of NP-EGF with endosomal markers is lower than for EGF-Alexa647 (Fig. 5b), which we attribute to differences in the applied imaging methods. Some cellular scatterers can generate similar scattering patterns as the metal NPs in darkfield, resulting in some localization error for NP-EGF. Furthermore, while confocal fluorescence microscopy (EGF-Alexa647) is compatible with high NA objectives, in widefield darkfield microscopy (NP-EGF) the NA has to be limited to approximately 0.7 to minimize background scattering. As the depth of field increases with decreasing NA,80 the darkfield images represent deeper cell sections than the fluorescence images and can contain surface bound objects, such as extracellular NP-EGFs. Together, these two effects can account for the overall lower absolute colocalization probability for NP-EGF with Rab5a positive endosomes. We applied dynamic colocalization methods to overcome some of the limitations of conventional colocalization microscopy (vide infra).
For both NP-EGF and free EGF, the decrease in Rab5a colocalizing NPs is accompanied by an increase of Rab7a positive endosomes (Fig. 5c), consistent with a maturation from early to late endosome. While free EGF reaches a maximum colocalization (~60%) with Rab7a at t = 35 min, it is not before t = 55 min that NP-EGF plateaus. The delayed transfer from early to late endosome in case of NP-EGF is in line with an overall longer dwell time of the nanoconjugated EGF in the early endosomes.
Independent validation of long NP-EGF dwell times in early endosomes through dynamic colocalization microscopy
The very gradual decrease of optical colocalization between NP-EGF and Rab5a shown in Fig. 5b suggests that even after chase times of several hours a measurable fraction of NP-EGF still remains in the early endosomes. Static colocalization studies (i.e. colocalization in single images) always contain an error from coincidental colocalization. In the case of darkfield microscopy, which is typically performed at lower NAs, and feature-rich samples the error from coincidental colocalization can be significant. We, therefore, independently verified the very long dwell time of at least a sub-set of NP-EGF in early endosomes by performing dynamic colocalization32 studies. The idea underlying this approach is to minimize coincidental colocalization and, thus, to improve the reliability of the detection of interactions between moving objects by correlating entire trajectories of tracked objects. The correlation of entire trajectories greatly improves the ability to reliably detect colocalization using widefield optical microscopies with limited spatial resolution.
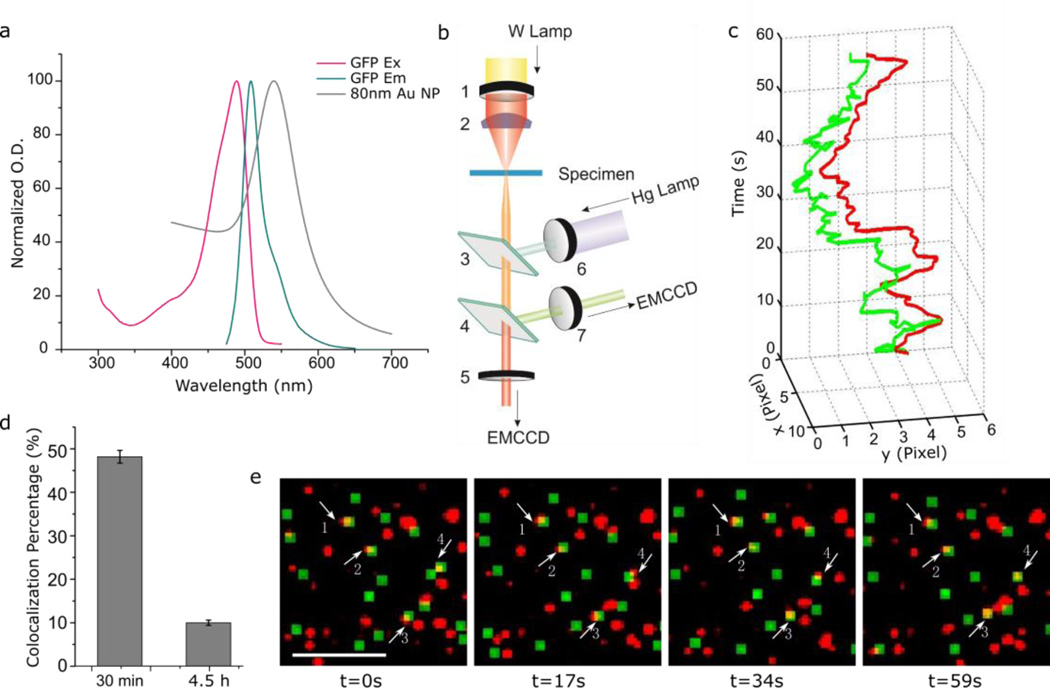
Conventional dynamic colocalization is performed by tracking objects fluorescently labeled with two different dyes on two separate color channels, but the approach can be adjusted for correlating the fluorescence signal of GFP-tagged early endosomes with the elastic scattering signals of NP-EGF (spectral characteristics are provided in Fig. 6a) using the experimental set-up shown in Fig. 6b. Whitelight from a Tungsten lamp passes a bandpass filter (wavelength: 600 nm) and is then injected into the imaging plane at oblique angles using a high numerical aperture (NA = 1.2–1.4) darkfield condenser from one side of the sample. From the opposite side a monochromatic excitation source of 480nm is focused on the imaging plane through a 60× objective (NA = 0.65). The objective collects both the light scattered from NPs in the focal plane as well as the emitted light from fluorescent probes. The collected light is chromatically split using the 580 nm dichroic allowing the collection of fluorescence and NP scattering images on two separate electron multiplying charge coupled devices (EMCCD) simultaneously.
Figure 6.
Quantifying NP-EGF retention in early endosomes through dynamic colocalization. a. Excitation and emission spectra of GFP and UV-Vis spectrum of 80nm NP-EGF. b. Optical setup for correlated fluorescence/darkfield tracking. NP-EGFs are illuminated by a tungsten lamp with a 600 ± 10 nm bandpass filter. The GFP associated with Rab5a is excited at 480 ± 17 nm and the emission is collected at 510 ± 10 nm. The light collected from NPs and GFP is split by a dichroic (transmission/reflection transition at 580 nm) and detected with two separate EMCCDs. 1. 600 ± 10 nm bandpass filter. 2. Oil / Water condenser. 3. 495nm dichroic. 4. 580nm dichroic. 5. 600 ± 10 nm bandpass filter. 6. 480 ± 17 nm bandpass filter. 7. 510 ± 10 nm bandpass filter. c. Representative fluorescence / darkfield trajectories (60s) of colocalized Rab5a–GFP (green) and NP-EGF (red). The trajectories were recorded for particle 4 in (e.). d. Colocalization percentage of NP-EGF with Rab5a determined by dynamic colocalization microscopy at two selected time points (30min and 4.5h). Data were collected from 3 independent experiments of each time point. e. Snapshots of a movie showing colocalization of Rab5a–GFP (green) and NP-EGF (red). Scale bar: 10 µm. Error bars represent the SEM.
For the tracking studies MDA-MB-468 cells were incubated with NP-EGF (4pM) for 20 min and then chased for up to 250 min (total time approximately 4.5 h) at 37 °C. After a defined chase time we recorded movies with a frame rate of 2 s−1 simultaneously on the fluorescent channel (Rab5a marked endosomes) and the darkfield channel (NP-EGF). These movies allowed for a tracking of selected objects throughout the field of view on both the fluorescence and darkfield channel. For all NPs that showed a colocalization with Rab5a at any time throughout the movie we calculated Pearson product-moment correlation coefficients for the coordinates of the associated point-spread-functions on the two monitored channels throughout the entire duration of the movie (~60 s). In this way a quantification of the spatial correlation between NP and endosome trajectories was achieved.32 Fig. 6c contains an example of correlated Rab5a and NP-EGF trajectories recorded at t = 4.5 h. Snapshots (fluorescence and darkfield) at selected time points in a typical dynamic colocalization movie are shown in Fig. 6e. The images demonstrate a continuing colocalization over extended periods of time for selected particles, confirming that these NPs are localized in Rab5a–positive endosomes. The entire movie is provided as Movie S1 in the ESI.
We systematically quantified the colocalization of Rab5a and NP-EGF signal through dynamic colocalization analysis and found that the Rab5a colocalization with NPs dropped from 48% ± 2.5% at t = 30 min to 10% ± 1% at t = 4.5 h (Fig. 6d). This finding corroborates the conclusion from our static colocalization studies that even hours after NP-EGF addition a significant fraction of the particles are still contained in the early endosomes.
Discussion
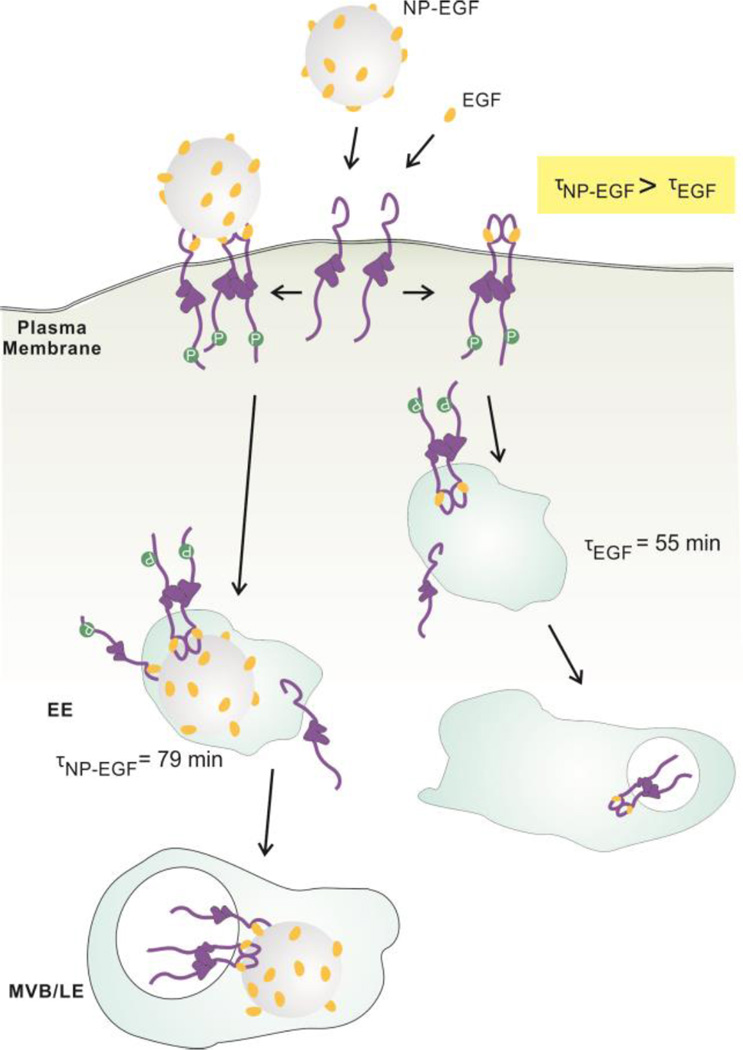
In order to determine the impact of nanoconjugation on the intracellular spatial and temporal distribution of EGF-pEGFR containing endosomes we have investigated NP-EGF uptake and trafficking using two orthogonal experimental approaches. The first took advantage of the increase in density associated with Au NP uptake to isolate NP-EGF-pEGFR containing endosomes. The second approach was based on correlated darkfield/fluorescence imaging to map the spatial distribution of NP-EGF-pEGFR containing endosomes as a function of time. These studies revealed that EGF conjugated to a 80 nm Au NP core enters the investigated MDA-MB-468 cells through a clathrin-dependent pathway and that the NP core achieves a retention of bound pEGFR in the early endosomes. The impact of nanoconjugation on EGF trafficking and EGF-pEGFR signaling is schematically summarized in Fig. 7. Even after a short (20 min) exposure to a relatively low concentration (8 pM) of NP-EGF (effective EGF concentration ≈ 1 nM) a non-negligible fraction of NP-EGF was found to remain in early endosomes for at least 5 hours. The observed slow trafficking of NP-EGF after internalization is consistent with previous findings that particle uptake and trafficking speed depend on the stiffness of the NP core, with stiffer particles traveling slower.81, 82 Vesicle fusion is essential in endosome maturation83–85 and, therefore, also plays an important role in EGFR degradation. As the ability to deform aids vesicle fusion82, the stiff metal core of NP-EGF can perturb and slow-down trafficking of NP-EGF containing vesicles. In a simplified model, one can expect the transmembrane receptors to degrade faster if the internalized vesicles can rapidly fuse. In the case of NP-EGF the long retention in the early endosomes provides a platform for the NP tethered EGF-pEGFR to initiate cytoplasmic signaling over extended periods of time. Since accumulation of pEGFR in early endosomes has been identified as origin of EGF induced apoptosis in the case of the free ligand,24 we attribute the enhanced EGF mediated apoptosis after nanoconjugation to the prolonged dwell time of the NP-EGF-pEGFR in the early endosomes. Consistent with this model, inhibition of EGFR phosphorylation by addition of receptor tyrosine kinase suspended the apoptotic effect of NP-EGF.
Figure 7.
Intracellular trafficking of NP-EGF and free EGF. NP-EGF and EGF bind to EGFR on the plasma membrane, activate EGFR and then induce clathrin-mediated endocytosis that trafficks EGF and the bound receptor from the early endosomes (EE) to the multivesicular bodies (MVB) of the late endosome. EGF nanoconjugation induces a longer average dwell time (79 min) for NP-EGF-pEGFR in the early endosomes, prolonging endosomal EGFR signaling and inducing apoptosis.
Nanoconjugation induced apoptosis is not unique to EGF but has also been observed for other ligands, such as for transferrin in Ramos B-cell lymphomas.2 Interestingly, NP-EGF requires significantly lower threshold concentrations to be effective in inducing apoptosis than nanoconjugated transferrin. Exposure to NP-EGF (8 pM), corresponding to an EGF concentration of 6 ng/mL (1 nM), for 4h is sufficient to induce a significant increase in caspase-3 activity, whereas transferrin conjugated NPs require 200 µg/mL (2.5 µM).2 Transferrin functionalized NPs are internalized through a specialized pathway, which involves only a transient interaction with early endosomes, avoids the lysosome and, instead, results in an accumulation of the particles in acidified compartments that are EEA1 negative. The apoptosis observed for transferrin functionalized NPs was attributed to the multivalent presentation of the ligand.2 The intracellular fate of NP-EGF differs starkly form that of nanoconjugated transferrin as nanoconjugated EGF traffics through EEA1 positive early endosomes to the lysosomes. However, the multivalent ligand presentation will certainly also contribute to nanoconjugation enhanced apoptosis in the case of EGF. For instance, the resulting higher binding affinity will be instrumental in achieving an increased concentration of pEGFR in the early endosomes, where the apoptotic signaling is believed to originate.24 Furthermore, a multivalent engagement of EGFR can impact the signalling outcome. Future studies will test and quantify the relative contributions from nanoconjugation mediated spatial regulation of endosomal and cell-surface4 EGFR signaling, and test the impact of multivalency on enhancing EGF-mediated apoptosis.
Conclusions
We have shown that nanoconjugation prolongs the dwell time of phosphorylated receptors in early endosomes and that this deceleration of pEGFR trafficking is accompanied by an EGF mediated apoptosis at effective concentrations that do not induce apoptosis in the case of free EGF. Our findings are consistent with the hypothesis that spatial regulation of intracellular signaling through trafficking represents an important control mechanism,16, 28, 29, 86–88 and that nanoconjugation represents an alternative strategy for manipulating endosomal signaling. One conceivable application of this approach is the treatment of apoptosis evasion of cancer cells89 through NPs loaded with EGF that efficiently enrich at the tumor site due to the enhanced permeability and retention effect (EPR)90–92. Nanoconjugation of benign ligands, such as EGF, could pave the way to new anti-cancer strategies that reduce or entirely avoid harmful side effects associated with conventional aggressive chemotherapies.
Supplementary Material
Acknowledgments
This work was financially supported by the National Institutes of Health through grant R01CA138509. We thank Ali Khanehzar for providing colloidal nanoparticles.
Footnotes
Electronic Supplementary Information (ESI) available: DLS data of NP-EGF in growth medium; MTT cell viability assay; Validation of MW-NP uptake; Positive controls for pharmacological inhibitors; EEA1 background for NP-EGF incubated with cell lysate; Phosphorylation for EGF-Alexa647; Live cell dynamic colocalization movie of MDA-MB-468 cells expressing Rab5a–GFP (green) 4.5 h after exposure to 8 pM NP-EGF (red). See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Wu L, Yu X, Feizpour A, Reinhard BM. Biomat. Sci. 2014;2:156–166. doi: 10.1039/C3BM60142K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J, Tian SM, Petros RA, Napier ME, DeSimone JM. J. Am. Chem. Soc. 2010;132:11306–11313. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H, Fung SY, Xu SY, Sutherland DP, Kollmann TR, Liu MY, Turvey SE. Acs Nano. 2015;9:6774–6784. doi: 10.1021/nn505634h. [DOI] [PubMed] [Google Scholar]
- 4.Paviolo C, Chon JWM, Clayton AHA. Small. 2015;11:1638–1643. doi: 10.1002/smll.201402701. [DOI] [PubMed] [Google Scholar]
- 5.Kah JCY, Grabinski C, Untener E, Garrett C, Chen J, Zhu D, Hussain SM, Hamad-Schifferli K. Acs Nano. 2014;8:4608–4620. doi: 10.1021/nn5002886. [DOI] [PubMed] [Google Scholar]
- 6.Patel S, Yin PT, Sugiyama H, Lee KB. Acs Nano. 2015;9:6909–6917. doi: 10.1021/acsnano.5b00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 8.Yarden Y, Sliwkowski MX. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 9.Fischer OM, Hart S, Gschwind A, Ullrich A. Biochem. Soc. Trans. 2003;31:1203–1208. doi: 10.1042/bst0311203. [DOI] [PubMed] [Google Scholar]
- 10.Wiley HS, Shvartsman SY, Lauffenburger DA. Trends Cell Biol. 2003;13:43–50. doi: 10.1016/s0962-8924(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 11.Chen WS, Lazar CS, Poenie M, Tsien RY, Gill GN, Rosenfeld MG. Nature. 1987;328:820–823. doi: 10.1038/328820a0. [DOI] [PubMed] [Google Scholar]
- 12.Schlessinger J. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 13.Schlessinger J. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 14.Sorkin A, Goh LK. Exp. Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 15.Tomas A, Futter CE, Eden ER. Trends Cell Biol. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorkin A, von Zastrow M. Nat. Rev. Mol. Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villaseñor R, Nonaka H, Del Conte-Zerial P, Kalaidzidis Y, Zerial M. eLife. 2015;4:06156. doi: 10.7554/eLife.06156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiganis T. Iubmb Life. 2002;53:3–14. doi: 10.1080/15216540210811. [DOI] [PubMed] [Google Scholar]
- 19.Burke P, Schooler K, Wiley HS. Mol. Biol. Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Pennock S, Chen XM, Wang ZX. Mol.Cell. Biol. 2002;22:7279–7290. doi: 10.1128/MCB.22.20.7279-7290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Dev. Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Brankatschk B, Wichert SP, Johnson SD, Schaad O, Rossner MJ, Gruenberg J. Sci. Signal. 2012;5:215. doi: 10.1126/scisignal.2002351. [DOI] [PubMed] [Google Scholar]
- 23.Sousa LP, Lax I, Shen HY, Ferguson SM, De Camilli P, Schlessinger J. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4419–4424. doi: 10.1073/pnas.1200164109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rush JS, Quinalty LM, Engelman L, Sherry DM, Ceresa BP. J. Biol. Chem. 2012;287:712–722. doi: 10.1074/jbc.M111.294470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, Mahoney JP, Steyaert J, Rasmussen SGF, Sunahara RK, El-Samad H, Huang B, von Zastrow M. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy JE, Padilla BE, Hasdemir B, Cottrell GS, Bunnett NW. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17615–17622. doi: 10.1073/pnas.0906541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ceresa B. ch. 3. In: Gunduz MGE, editor. Breast Cancer-Carcinogenesis, Cells Growth and Signalling Pathways. InTech; 2011. p. 49. [Google Scholar]
- 28.Gandhi H, Worch R, Kurgonaite K, Hintersteiner M, Schwille P, Bökel C, Weidemann T. Biophys. J. 2014;107:2515–2527. doi: 10.1016/j.bpj.2014.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurgonaite K, Gandhi H, Kurth T, Pautot S, Schwille P, Weidemann T, Bökel C. J Cell Sci. 2015;128:3781–3795. doi: 10.1242/jcs.170969. [DOI] [PubMed] [Google Scholar]
- 30.Mosesson Y, Mills GB, Yarden Y. Nat. Rev. Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 31.Hyatt DC, Ceresa BP. Exp. Cell Res. 2008;314:3415–3425. doi: 10.1016/j.yexcr.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vercauteren D, Deschout H, Remaut K, Engbersen JFJ, Jones AT, Demeester J, De Smedt SC, Braeckmans K. ACS Nano. 2011;5:7874–7884. doi: 10.1021/nn2020858. [DOI] [PubMed] [Google Scholar]
- 33.Ziegler C, Eychmuller A. J. Phys. Chem. C. 2011;115:4502–4506. [Google Scholar]
- 34.Zhao PX, Li N, Astruc D. Coord. Chem. Rev. 2013;257:638–665. [Google Scholar]
- 35.Daniel MC, Astruc D. Chem. Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 36.Sapsford KE, Algar WR, Berti L, Gemmill KB, Casey BJ, Oh E, Stewart MH, Medintz IL. Chem. Rev. 2013;113:1904–2074. doi: 10.1021/cr300143v. [DOI] [PubMed] [Google Scholar]
- 37.Pissuwan D, Niidome T. Nanoscale. 2015;7:59–65. doi: 10.1039/c4nr04350b. [DOI] [PubMed] [Google Scholar]
- 38.Haine AT, Niidome T. J. of Photopolym. Sci. Technol. 2015;28:705–710. [Google Scholar]
- 39.Alkilany AM, Murphy CJ. J. Nanopart. Res. 2010;12:2313–2333. doi: 10.1007/s11051-010-9911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy CJ, Gole AM, Stone JW, Sisco PN, Alkilany AM, Goldsmith EC, Baxter SC. Accounts Chem. Res. 2008;41:1721–1730. doi: 10.1021/ar800035u. [DOI] [PubMed] [Google Scholar]
- 41.Shukla R, Bansal V, Chaudhary M, Basu A, Bhonde RR, Sastry M. Langmuir. 2005;21:10644–10654. doi: 10.1021/la0513712. [DOI] [PubMed] [Google Scholar]
- 42.Dykman L, Khlebtsov N. Chem. Soc. Rev. 2012;41:2256–2282. doi: 10.1039/c1cs15166e. [DOI] [PubMed] [Google Scholar]
- 43.Lee SE, Chen Q, Bhat R, Petkiewicz S, Smith JM, Ferry VE, Correia AL, Alivisatos AP, Bissell MJ. Nano Lett. 2015;15:4564–4570. doi: 10.1021/acs.nanolett.5b01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SE, Lee LP. Curr. Opin. Chem. Biol. 2010;14:623–633. doi: 10.1016/j.cbpa.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stokes N, Cortie MB, Davis TJ, McDonagh AM. Plasmonics. 2012;7:235–243. [Google Scholar]
- 46.Dreaden EC, Alkilany AM, Huang XH, Murphy CJ, El-Sayed MA. Chem. Soc. Rev. 2012;41:2740–2779. doi: 10.1039/c1cs15237h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraire JC, Masseroni ML, Jausoro I, Perassi EM, Diaz Añel AM, Coronado EA. ACS nano. 2014;8:8942–8958. doi: 10.1021/nn501575c. [DOI] [PubMed] [Google Scholar]
- 48.Wang HY, Wu LX, Reinhard BM. ACS Nano. 2012;6:7122–7132. doi: 10.1021/nn302186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolb HC, Finn MG, Sharpless KB. Angew. Chem.-Int. Edit. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 50.Chithrani BD, Ghazani AA, Chan WCW. Nano Letters. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 51.Porter AG, Janicke RU. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 52.Thornberry NA, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 53.Cohen GM. Biochem. J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jun YW, Sheikholeslami S, Hostetter DR, Tajon C, Craik CS, Alivisatos AP. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17735–17740. doi: 10.1073/pnas.0907367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu X, Feizpour A, Ramirez NGP, Wu L, Akiyama H, Xu F, Gummuluru S, Reinhard BM. Nat. Commun. 2014;5:4136. doi: 10.1038/ncomms5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu X, Xu F, Ramirez N-GP, Kijewski SDG, Akiyama H, Gummuluru S, Reinhard BM. ACS Nano. 2015;9:4182–4192. doi: 10.1021/acsnano.5b00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lloyd JB, Williams KE. Biochem. Soc. Trans. 1984;12:527–528. doi: 10.1042/bst0120527. [DOI] [PubMed] [Google Scholar]
- 58.Chinkers M, McKanna JA, Cohen S. J. Cell Biol. 1979;83:260–265. doi: 10.1083/jcb.83.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haigler HT, McKanna JA, Cohen S. J. Cell Biol. 1979;81:382–395. doi: 10.1083/jcb.81.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamazaki T, Zaal K, Hailey D, Presley J, Lippincott-Schwartz J, Samelson LE. J. Cell Sci. 2002;115:1791–1802. doi: 10.1242/jcs.115.9.1791. [DOI] [PubMed] [Google Scholar]
- 61.Orth JD, Krueger EW, Weller SG, McNiven MA. Cancer Res. 2006;66:3603–3610. doi: 10.1158/0008-5472.CAN-05-2916. [DOI] [PubMed] [Google Scholar]
- 62.Hansen CG, Nichols BJ. J. Cell Sci. 2009;122:1713–1721. doi: 10.1242/jcs.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mayor S, Pagano RE. Nat. Rev. Mol. Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henriksen L, Grandal MV, Knudsen SLJ, van Deurs B, Grøvdal LM. PLoS One. 2013;8:e58148. doi: 10.1371/journal.pone.0058148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Engelman JA, Chu C, Lin A, Jo H, Ikezu T, Okamoto T, Kohtz DS, Lisanti MP. Febs Letters. 1998;428:205–211. doi: 10.1016/s0014-5793(98)00470-0. [DOI] [PubMed] [Google Scholar]
- 67.Mineo C, Gill GN, Anderson RG. J. Biol. Chem. 1999;274:30636–30643. doi: 10.1074/jbc.274.43.30636. [DOI] [PubMed] [Google Scholar]
- 68.Rejman J, Bragonzi A, Conese M. Mol. Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 69.Pelkmans L, Helenius A. Curr. Opin. Cell Biol. 2003;15:414–422. doi: 10.1016/s0955-0674(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 70.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Biochem. J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nam HY, Kwon SM, Chung H, Lee S-Y, Kwon S-H, Jeon H, Kim Y, Park JH, Kim J, Her S. J. Control. Release. 2009;135:259–267. doi: 10.1016/j.jconrel.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 72.Cho EC, Xie JW, Wurm PA, Xia YN. Nano Lett. 2009;9:1080–1084. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- 73.Tjelle TE, Brech A, Juvet LK, Griffiths G, Berg T. J. Cell Sci. 1996;109:2905–2914. doi: 10.1242/jcs.109.12.2905. [DOI] [PubMed] [Google Scholar]
- 74.Mu FT, Callaghan JM, Steelemortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EPC, Toh BH. J. Biol. Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 75.Feng Y, Press B, Wandingerness A. J. Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vanlandingham PA, Ceresa BP. J. Biol. Chem. 2009;284:12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leventis PA, Grinstein S. In: Ann. Rev. Biophys. Rees DC, Dill KA, Williamson JR, editors. Vol. 39. 2010. pp. 407–427. [DOI] [PubMed] [Google Scholar]
- 78.Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Nat. Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- 79.Gorvel JP, Chavrier P, Zerial M, Gruenberg J. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 80.Shillaber CP. Photomicrography in Theory and Practice. New York: John Wiley and sons; 1944. [Google Scholar]
- 81.Sun H, Wong EH, Yan Y, Cui J, Dai Q, Guo J, Qiao GG, Caruso F. Chem. Sci. 2015;6:3505–3514. doi: 10.1039/c5sc00416k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hartmann R, Weidenbach M, Neubauer M, Fery A, Parak WJ. Angew. Chem.-Int. Edit. 2015;54:1365–1368. doi: 10.1002/anie.201409693. [DOI] [PubMed] [Google Scholar]
- 83.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 84.Futter CE, Pearse A, Hewlett LJ, Hopkins CR. J. Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huotari J, Helenius A. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Casaletto JB, McClatchey AI. Nat. Rev. Cancer. 2012;12:387–400. doi: 10.1038/nrc3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moore CA, Milano SK, Benovic JL. Annu. Rev. Physiol. 2007;69:451–482. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- 88.Shaul PW. Annual review of physiology. 2002;64:749–774. doi: 10.1146/annurev.physiol.64.081501.155952. [DOI] [PubMed] [Google Scholar]
- 89.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 90.Fang J, Nakamura H, Maeda H. Adv. Drug Deliv. Rev. 2011;63:136–151. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 91.Cho K, Wang X, Nie S, Shin DM. Clin. Cancer Res. 2008;14:1310–1316. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 92.Brannon-Peppas L, Blanchette JO. Adv. Drug Deliv. Rev. 2012;64:206–212. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.