Significance
Most humans are infected for their lifetime with Epstein–Barr virus (EBV), which can cause cancer and other EBV-associated diseases. Infected individuals develop strong immune responses to this virus, in particular cytotoxic CD8+ T cells, but viral infection is never cleared nor is EBV eliminated from the body. This suggests that certain viral molecules might prevent effective elimination of EBV-infected cells by CD8+ T cells. EBV is rich in genes coding for microRNAs, many with unknown function. We show that viral microRNAs interfere with recognition and killing of EBV-infected cells by CD8+ T cells. Multiple mechanisms and molecules are targeted by microRNAs to achieve this immune evasion. Therefore, targeting of viral microRNAs may improve antiviral immunity and therapy.
Keywords: adaptive immunity, immune evasion, herpesvirus, CD8 T cells, microRNA
Abstract
Infection with Epstein–Barr virus (EBV) affects most humans worldwide and persists life-long in the presence of robust virus-specific T-cell responses. In both immunocompromised and some immunocompetent people, EBV causes several cancers and lymphoproliferative diseases. EBV transforms B cells in vitro and encodes at least 44 microRNAs (miRNAs), most of which are expressed in EBV-transformed B cells, but their functions are largely unknown. Recently, we showed that EBV miRNAs inhibit CD4+ T-cell responses to infected B cells by targeting IL-12, MHC class II, and lysosomal proteases. Here we investigated whether EBV miRNAs also counteract surveillance by CD8+ T cells. We have found that EBV miRNAs strongly inhibit recognition and killing of infected B cells by EBV-specific CD8+ T cells through multiple mechanisms. EBV miRNAs directly target the peptide transporter subunit TAP2 and reduce levels of the TAP1 subunit, MHC class I molecules, and EBNA1, a protein expressed in most forms of EBV latency and a target of EBV-specific CD8+ T cells. Moreover, miRNA-mediated down-regulation of the cytokine IL-12 decreases the recognition of infected cells by EBV-specific CD8+ T cells. Thus, EBV miRNAs use multiple, distinct pathways, allowing the virus to evade surveillance not only by CD4+ but also by antiviral CD8+ T cells.
Epstein–Barr virus (EBV) is a ubiquitous herpesvirus that infects the majority of the human population worldwide. Although EBV infection persists for life, most carriers remain asymptomatic due to a stringent control by virus-specific immunity. An important component of this immunity is EBV-specific CD8+ T cells, which often expand to high numbers in healthy carriers or after primary infection. Conversely, the absence of EBV-specific CD8+ T cells predicts the emergence of EBV-associated disease in patients after stem cell transplantation or when afflicted with AIDS (1–3). Dangerous EBV-mediated complications can be reversed or prevented by transfer of EBV-specific T cells (4, 5), which further confirms the important role of continuous T-cell control of EBV infection. Among EBV-specific T cells, CD8+ T cells predominate; about 0.05–1% of all CD8+ T cells in healthy donors are typically specific for EBV latent antigens and about twice as many for lytic antigens (6, 7).
EBV predominantly infects B cells and establishes a latent infection before production of progeny virus becomes possible (8). Four distinct programs of EBV latent infection have been defined according to their expression profiles of latent viral genes (9–11). One of these programs, known as latency III or the “growth program,” is characterized by the expression of a restricted set of approximately eight viral proteins, which activate B cells and drive their proliferation, thus increasing the viral reservoir. Latency III is found in EBV-associated malignancies in immunosuppressed patients (9) and likely reemerges continuously in healthy carriers (9, 12), indicating that its control is critical for the health of an EBV carrier. Only at a later stage of infection (13) can the virus enter its lytic phase in infected cells to produce progeny virions, a phase requiring expression of the majority of viral proteins. Some of these lytic-cycle viral proteins are immunoevasins that interfere with CD8+ T-cell recognition: the TAP inhibitor BNLF2a (14, 15); the G protein-coupled receptor BILF1 that associates with MHC class I/peptide complexes, diverts them from the exocytic pathway and the cell surface, and induces their lysomal degradation (16, 17); and the protein BGLF5 that reduces MHC I expression and CD8+ T-cell recognition as a consequence of its generalized host-shutoff function (18, 19). Recently, BDLF3 was identified as an additional lytic-cycle protein that targets MHC molecules for degradation (20). BNLF2a is also expressed early after infection in the prelatent phase (13 for a recent review) and reduces CD8+ T-cell recognition in the first days of infection but does not impair T-cell recognition in established latency (21, 22). It is unclear, though, how latently EBV-infected B cells escape elimination by T cells, in particular during the latency III program that is characterized by a considerable antigenic load and an activated state expected to increase the immunogenicity of B cells (23). In latency III, MHC I, MHC II, and T-cell–coactivating molecules are highly expressed (24, 25), but nonetheless many epitopes are suboptimally recognized by CD8+ T cells (26, 27). Therefore, it is likely that unknown immunoevasive mechanisms operate in these latently infected B cells.
A hallmark of EBV is its array of 44 microRNAs (miRNAs) (28–31), which is the largest number of miRNAs identified in a human pathogen to date. Many EBV miRNAs have no known function. A function of viral miRNAs in innate immunity was suggested earlier by findings showing that some regulate the inflammasome component NLRP3 (32), the natural killer group 2D (NKG2D) ligand MICB (33), and the chemokine CXCL11 (34).
Recently, we found that multiple viral miRNAs limit the control of infected B cells by CD4+ T cells early in EBV infection (35). Several viral miRNAs reduce secretion of IL-12, expression of HLA class II molecules, and expression of lysosomal enzymes important for antigen presentation to CD4+ T cells. EBV miRNAs also regulate many molecules of potential importance in HLA class I presentation and CD8+ T-cell recognition (35).
These findings have led us to determine if viral miRNAs inhibit surveillance of EBV by CD8+ T cells. We have found that viral miRNAs do prevent virus-specific activation of CD8+ T cells and killing of infected B cells; we have also delineated the mechanisms underlying this viral evasion of the immune response.
Results
EBV MiRNAs Support Survival of Infected B Cells in the Presence of CD8+ T Cells from EBV-Positive Donors.
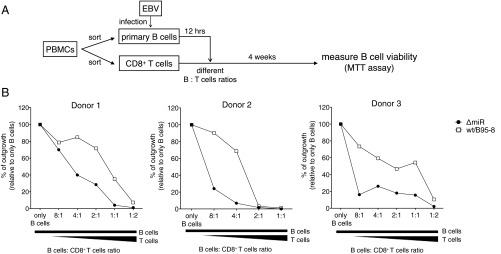
We used in vitro infection of primary human B cells as a simple but representative model of EBV infection (Fig. 1A) to evaluate if EBV miRNAs counteract immune surveillance by EBV-specific CD8+ T cells. B cells (CD19+) isolated from peripheral blood mononuclear cells (PBMCs) from EBV-positive donors were infected with two EBV strains: the laboratory strain B95-8 (WT/B95-8), expressing 13 viral miRNAs, and its derivative ΔmiR, expressing no viral miRNAs (36). Twelve hours later, autologous CD8+ T cells were added, and the cells were cocultured for 4 wk (Fig. 1B), when cell viability was tested in MTT assays. Surviving cells were further analyzed by flow cytometry for their identification (Fig. S1).
Fig. 1.
EBV miRNAs support infected B cells to abrogate CD8+ T-cell responses. (A) Schematic overview of the experimental system. (B) CD19+ B cells were isolated from PBMCs of three different EBV-positive donors and infected with WT/B95-8 EBV or ΔmiR EBV stocks. Twelve hours later, infected B cells were extensively washed to remove free virions. In 96-well microtiter plates 32,000 EBV-infected B cells were seeded per well and CD8+ T cells isolated from the autologous donors were added at different ratios as indicated. After 4 wk, total cell viability was assessed by MTT assay. Outgrowth of B-cell–only conditions (without T cells) was set to 100%. The results shown are based on the mean of six technical replicates per data point.
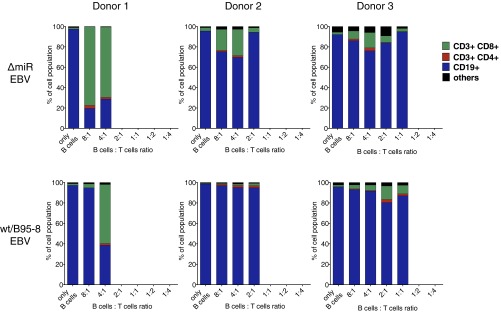
Fig. S1.
Composition of long-term cocultures of WT/B95-8 or ΔmiR EBV-infected B cells with autologous CD8+ T cells from three different EBV-positive donors. Coculture experiments of CD19+ B cells infected with WT/B95-8 EBV or ΔmiR EBV stocks and autologous CD8+ T cells were performed as shown in Fig. 1. After 4 wk, the compositions of the outgrowing cells were analyzed by FACS: B cells (CD19+), CD8+ T cells (CD8+/CD3+), CD4+ T cells (CD4+/CD3+), or cells negative for all of the four markers are indicated.
In the absence of T cells, we observed robust proliferation of B cells infected with ΔmiR or WT/B95-8 EBV (Fig. 1B). In the presence of T cells, the survival and outgrowth of infected B cells was decreased. A strong reduction of viable cells was achieved by fewer CD8+ T cells for cells infected with ΔmiR EBV than cells infected with WT/B95-8 EBV (Fig. 1B). Flow cytometry analyses showed that B cells represented the viable cells in most of the cultures (Fig. S1). These results indicated that EBV miRNAs protect EBV-infected B lymphocytes from eradication by antiviral CD8+ T cells under these conditions.
EBV MiRNAs Inhibit Recognition, Killing, and Expansion of EBV-Specific CD8+ T Cells.
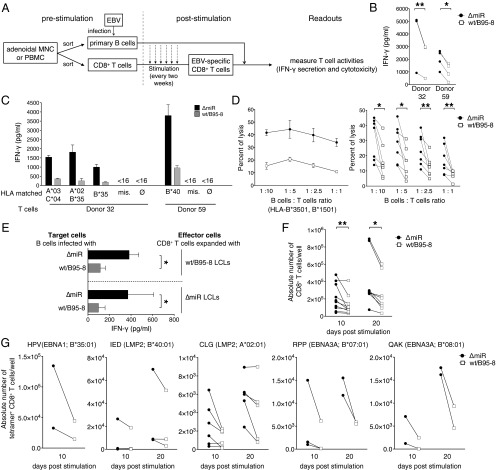
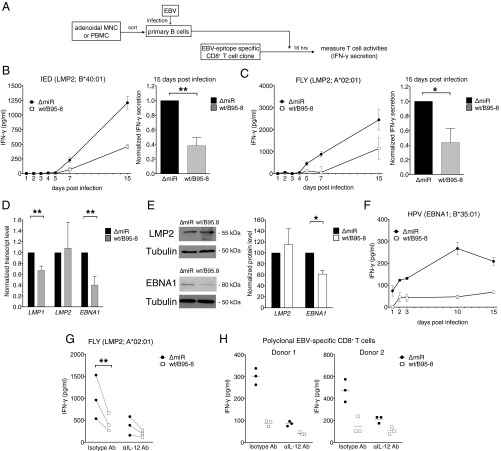
Earlier studies had shown that B-cell survival could be compromised in cells infected with the ΔmiR EBV devoid of miRNAs, because some viral miRNAs contribute to EBV-associated cellular transformation in the early phase of infection (36–38). To evaluate a possible role of viral miRNAs in controlling immune functions of EBV-specific CD8+ T cells, we established polyclonal EBV-specific CD8+ T cells from different donors. Sorted primary CD8+ T cells were stimulated every 2 wk with irradiated lymphoblastoid cell lines (LCLs), which had been established by infecting primary autologous B cells with WT/B95-8 EBV (Fig. 2A). For T-cell effector assays, EBV-specific CD8+ T cells established in this way were cocultured with B cells that had been infected with WT/B95-8 or ΔmiR EBV 15 d earlier. T-cell activation was quantified by measuring IFN-γ concentration in the cell culture supernatants after 16 h or by determining cytolysis of infected cells after 4 h. We observed significantly reduced IFN-γ secretion in response to cells infected with WT/B95-8 relative to cells infected with ΔmiR EBV, both in autologous (Fig. 2B and Table S1) and HLA-matched conditions (Fig. 2C). Importantly, T cells were not activated by HLA-mismatched infected B cells or in B-cell–free cultures, indicating that the observed activation was HLA-restricted and EBV-specific (Fig. 2C). In cytotoxicity assays, we found that the viral miRNAs inhibited killing of infected B cells by EBV-specific CD8+ T cells at all B:T cell ratios and at any HLA-matched conditions tested (Fig. 2D). It did not matter whether EBV-specific CD8+ T-cell cultures had been generated by expansion with ΔmiR or WT/B95-8 EBV-infected B cells; in each case, ΔmiR EBV-infected B cells were better recognized than B cells infected with WT/B95-8 EBV (Fig. 2E).
Fig. 2.
EBV miRNAs inhibit recognition and killing of infected B cells as well as expansion of EBV-specific CD8+ T cells. (A) Schematic overview of the experiments shown in the remaining panels of this figure. Polyclonal EBV-specific CD8+ T cells were obtained by repeated stimulation (every two weeks) with autologous irradiated WT/B95-8 EBV-infected LCLs. The T-cell activities of the EBV-stimulated CD8+ T cell were subsequently analyzed with target B cells infected with WT/B95-8 EBV or ΔmiR EBV stocks. (B) Equal numbers of polyclonal EBV-specific CD8+ T cells and autologous B cells infected for 15 d with the indicated EBV strains were cocultured. After 16 h, IFN-γ released from T cells was measured by ELISA. Results of three to four biological replicates are shown for each donor. (C) Polyclonal EBV-specific CD8+ T cells were cocultured with HLA-matched or mismatched EBV-infected B cells and tested as in B. Matched HLA class I alleles are indicated; mis., mismatched; ∅, only T cells; <16, below the threshold of detection (16 pg/mL). HLA allotypes of the donors are listed in Table S1. Data are shown as mean values. Error bars indicate SD of three replicates. (D) Cytotoxic activities of EBV-specific CD8+ T cells directed against HLA-matched infected B cells were analyzed at various B:T cells ratios in calcein release assays after 4 h of coculture as shown in A. A representative experiment with mean values and SD of four replicates (Left) and the overview of seven donors (Right) are shown. (E) CD8+ T cells of donor 115 were repetitively stimulated for 2 mo with irradiated autologous B cells infected with WT/B95-8 or ΔmiR EBV as indicated. The expanded effector T cells were assayed with autologous infected B cells as targets in coculture experiments and tested as in B measuring the IFN-γ released by ELISA. Error bars indicate SD of three biological replicates. (F and G) CD8+ T cells isolated from PBMCs were stimulated on day 0 and 10 with irradiated B cells infected with the indicated EBV strains. Absolute numbers of T cells were determined by flow cytometry 10 d later (on day 10 and 20). (F) Expansion of total CD3+ CD8+ T cells. (G) Expansion of EBV epitope-specific CD8+ T cells, stained with HLA/peptide pentamers as indicated. Results from 3 to 10 different donors are shown. The significance of difference in the T-cell expansion experiments was calculated by Wilcoxon matched-pairs signed rank test. *P < 0.05, **P < 0.01.
Table S1.
HLA class I types of the donors
| Donor | HLA-A | HLA-B | HLA-C |
| 32 | *0201, *0301 | *3501, *1501 | *0102, *0401 |
| 59 | *11, *24 | *35, *40 | n.a. |
| 115 | *02, *23 | *49, *40 | n.a. |
Because clonal expansion is essential for effective antiviral T-cell responses, we also investigated the selective expansion of EBV-specific CD8+ T cells in response to autologous B cells infected with either WT/B95-8 or ΔmiR EBV (Fig. 2F). From PBMCs of 10 donors, we sorted CD8+ T cells and stimulated them twice on day 0 and day 10 with irradiated autologous B cells, which had been infected for 15 d with WT/B95-8 or ΔmiR EBV. Total numbers of CD8+ T cells obtained at days 10 and 20 (after one or two stimulations) were significantly higher after expansion with ΔmiR EBV-infected cells compared with WT/B95-8 EBV-infected cells (Fig. 2F). In the same setting, we also analyzed the expansion of EBV-specific CD8+ T cells that were specific for five different epitopes from EBV proteins latent membrane protein (LMP)2A, EBV nuclear antigen (EBNA)1, and EBNA3A (Fig. 2G). We consistently found increased expansion in response to ΔmiR EBV-infected cells for each of these specificities (Fig. 2G). Together, our data suggest that viral miRNAs in EBV-infected B cells reduce clonal expansion of a wide range of antiviral CD8+ effector T cells.
EBV MiRNAs Inhibit MHC Class I Antigen Processing and Presentation Pathways.
We screened cellular transcripts targeted by EBV miRNAs and likely critical in fending off antiviral CD8+ T cells. To identify potential targets, we performed high-throughput screening with primary B lymphocytes infected with the different EBV strains and a combination of RNA and RNA induced silencing complexes-immunoprecipitation (RISC-IP) sequencing (35). With this approach, we identified IL12B and three genes (IFI30, the IFN-γ-regulated thiol reductase GILT; LGMN, the asparagine endopeptidase AEP alias legumain; and CTSB, the peptidase cathepsin B) encoding lysosomal enzymes and important for CD4+ T-cell differentiation and antigen processing as direct targets of viral miRNAs (35). Here, we focused on genes consistently inhibited by EBV miRNAs and known to play a role in antigen processing and presentation and cytokine–cytokine receptor interactions or are considered cell adhesion molecules according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway categories (Table 1). A subset of corresponding mRNAs was also enriched in our RISC-IP sequencing analysis (Table 1), indicating that these mRNAs were most likely direct targets of EBV miRNAs. Interestingly, TAP1 and TAP2 were significantly down-regulated in RNA-sequencing (RNA-Seq) experiments, and TAP2 was also found enriched in RISC-IP sequencing (Table 1). The TAP1/TAP2 heterodimer mediates transport of antigenic peptides into the ER lumen, where they are loaded onto MHC class I molecules. The presentation of many EBV epitopes depends on TAP (39), and thus we delineated the mechanisms by which EBV miRNAs regulate it.
Table 1.
Selected genes (cytokine–cytokine receptor interaction, antigen processing and presentation, and cell adhesion molecules) and their regulation by EBV miRNAs in RNA-Seq and RISC-IP experiments
| Gene symbol | Mean of log-twofold change | z score | RISC-IP |
| IL12B | −2.3108 | −2.5002 | Yes |
| CD274 | −0.6644 | −2.3705 | No |
| CD80 | −0.9250 | −2.3533 | No |
| ICAM1 | −0.6553 | −2.3351 | No |
| TAP2 | −0.4012 | −2.1261 | Yes |
| TNF | −0.6389 | −2.0936 | No |
| CD40 | −0.4912 | −2.0739 | Yes |
| CD58 | −0.5088 | −2.0568 | No |
| RFX5 | −0.3576 | −1.9605 | No |
| PSME1 | −0.3338 | −1.9373 | No |
| CTSB | −0.4520 | −1.9370 | Yes |
| PSME2 | −0.4680 | −1.9335 | No |
| CD86 | −0.4486 | −1.8001 | No |
| TAP1 | −0.3426 | −1.7800 | No |
| ALCAM | −0.4300 | −1.6637 | No |
| ICAM3 | −0.5567 | −1.6273 | No |
| ERAP2 | −0.2923 | −1.6048 | No |
| IPO7 | −0.2826 | −1.6870 | Yes |
Genes were identified by mRNA sequencing of WT/B95-8 vs. ΔmiR-infected B cells, ranked by z score, and where indicated, confirmed by RISC-IPs as described in Tagawa and coworkers (35). IPO7 served as a positive control.
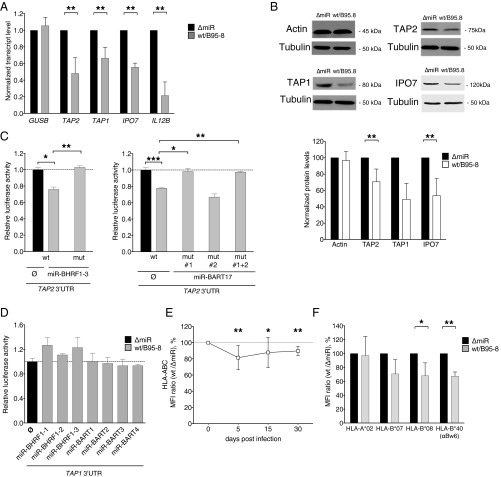
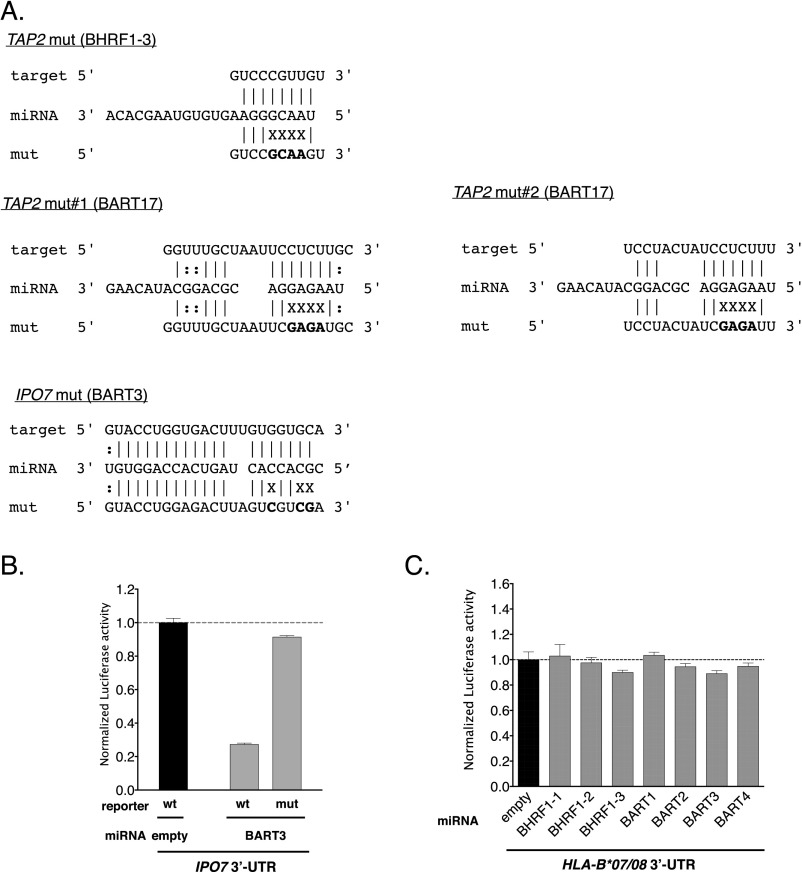
First, we verified the regulation of TAP1/2 expression by viral miRNAs. Fifteen days post infection expression of TAP1 and TAP2 was reduced in B cells infected with WT/B95-8 EBV compared with ΔmiR EBV both at the level of transcript (Fig. 3A) and protein (Fig. 3B). As a control, we verified that IPO7 (Importin-7), a known target of EBV miR-BART3 (40), was also down-regulated (Fig. 3 A and B). Because RISC-IP (Table 1) in combination with the in silico target algorithm TargetScan (41) predicted that the 3′UTR of TAP2 was directly targeted by EBV miRNAs, we performed dual luciferase reporter assays to test this assumption. We cotransfected HEK293T cells with a luciferase reporter plasmid containing the 3′UTR of TAP2 and single expression plasmids, each of which encoded one viral primary miRNA.
Fig. 3.
EBV miRNAs reduce TAP and MHC class I levels in infected B cells. (A) Transcript levels of TAP2, TAP1, IPO7, and IL12B were assessed by quantitative RT-PCR in EBV-infected B cells 15 d post infection (dpi). IPO7 is a known target of viral miRNAs and is used here as positive control. GUSB was used as negative control. Transcript levels were quantified relative to the mean of the housekeeping genes HPRT1 and HMBS (35) and were normalized to the transcript level of ΔmiR EBV-infected cells. Data are shown as mean values and SD of seven donors. (B) Protein levels of TAP1 and TAP2 were assessed by Western blot analyses in EBV-infected B cells 15 dpi. β-Actin served as negative and IPO7 as positive controls. Representative examples (Top) and protein levels relative to Tubulin (Bottom) are shown. The results were normalized to the protein levels of ΔmiR EBV-infected cells, set to 100%. Data are shown as mean values and SD of three to seven donors. (C and D) EBV miRNAs directly regulate TAP2 but not TAP1. HEK293T cells were cotransfected with miRNA expression vectors and dual luciferase reporter plasmids carrying a wild-type or mutated 3′UTR of TAP2 (C). For the analysis of TAP1 (D), all viral miRNAs present in WT/B95-8 EBV were tested with the exception of miR-BART15, which is barely expressed in our infection model (35). Sequence details of the 3′UTRs are contained in Fig. S2. The luciferase activities were normalized to lysates from cells cotransfected with the wild-type 3′UTR reporter and an empty plasmid in place of the miRNA expression plasmid. Data are shown as mean values and SD of three to four replicates. mut, mutated 3′UTR; WT, wild-type 3′UTR; ∅, empty plasmid. (E and F) Cell surface expression levels of total HLA class I (Left) and specific HLA class I allotypes (Right) of B cells infected with the indicated EBV strains for 15 d were measured by flow cytometry. Ratios (%) of WT/B95-8 divided by ΔmiR EBV-infected B cells are shown. Data are shown as mean values and SD of experiments with 5 to 10 different donors. *P < 0.05, **P < 0.01, ***P < 0.001.
The expression of exogenous miR-BHRF1–3 significantly decreased the luciferase activity of the TAP2 reporter (Fig. 3C, Left). A mutation within the 3′UTR in the seed-matching region abolished this inhibition completely, demonstrating that TAP2 is a direct target of miR-BHRF1–3 (Fig. 3C). Similarly, miR-BART17, which is expressed in EBV field strains but not in the WT/B95-8 strain, targeted the 3′UTR of the TAP2 directly at one of two predicted sites (Fig. 3C; predicted seed sequences are provided in Fig. S2A). In contrast, we did not observe a regulation of the TAP1 3′UTR by any viral miRNA present in WT/B95-8 EBV (Fig. 3D). This result suggested that TAP1 may not be a direct target of EBV miRNAs, consistent with our RISC-IP data (Table 1). A parallel dual luciferase reporter assay performed for IPO7 served as a positive control together with miR-BART3 in these assays (Fig. S2B).
Fig. S2.
Predicted miRNA target sites, their mutations, and luciferase assays of selected targets. (A) Partial sequences of 3′UTRs of selected transcripts analyzed in Fig. 3C and in B below are shown with corresponding miRNAs and mutations within the 3′UTRs in the reporter vectors. Complementarities are based on in silico predictions according to the RNAhybrid algorithm and depicted as Watson–Crick (‘|’) or G:U (‘:’) pairs. Nonmatching nucleotide residues are indicated (X). (B and C) HEK293T cells were cotransfected with miRNA expression plasmids and luciferase reporter plasmids carrying either a wild-type or mutated 3′UTR of IPO7 (B) or the 3′UTR of HLA-B*07/B*08 (C). The luciferase activities were normalized to lysates from cells cotransfected with the wild-type 3′UTR reporter and an empty plasmid in place of a miRNA plasmid. Data are shown as mean values and SD of three replicates. Mut, mutated 3′UTR; WT, wild-type 3′UTR; ∅, empty plasmid.
Next, we quantified the levels of classical HLA class I (HLA-A, -B, and -C) cell-surface expression on WT/B95-8 or ΔmiR EBV-infected B cells during the course of infection. Steady-state surface levels of HLA class I molecules are a function of TAP activities, as HLA class I molecules lacking peptides are unstable. We consistently observed a slight reduction by 10–20% of overall surface MHC class I molecules in cells infected with WT/B95-8 relative to ΔmiR EBV during the entire observation period (Fig. 3E). By assaying individual HLA class I alleles, we found that HLA-B*07, B*08, and B*40 allotypes were reduced by 20–30%, whereas HLA-A*02 levels were not reduced (Fig. 3F). This finding is consistent with the known preference of HLA-A*02 (but not of the other allotypes investigated here) to bind highly hydrophobic peptides, some of which reach the ER independently of TAP (42). Dual luciferase reporter assays were performed for HLA-B*07 and B*08, but direct targeting by miRNAs could not be demonstrated (Fig. S2C).
EBV MiRNAs Control Multiple Facets of Viral Immune Evasion.
These results suggested that EBV miRNAs impose allele-specific controls of HLA molecules, namely affecting HLA-B allotypes. We therefore asked if HLA-B allotype-restricted antigen presentation is directly controlled by viral miRNAs (Fig. 4). We cocultured infected B cells and CD8+ T-cell clones specific for the IED or the FLY epitope, both of which are derived from the viral LMP2 protein (Fig. 4A). Presentation of the B*40:01-restricted IED epitope is dependent on active TAP transportation (39), whereas the HLA-A*02:01–restricted epitope FLY is highly hydrophobic and presented TAP-independently (26). In a time course experiment with B cells infected with either WT/B95-8 or ΔmiR EBV, we observed reactivity of the clonal T cells as early as 5–7 d post infection (Fig. 4 B and C). EBV miRNAs significantly reduced the activation of the IED-specific T-cell clone (Fig. 4B) as expected from the down-regulation of the TAP complex and subsequent reduction of HLA-B molecules. Surprisingly, the activation of the FLY-specific T-cell clone was also strongly reduced (Fig. 4C) even though FLY is a TAP-independent peptide and presented via HLA-A*02:01, which was not affected by the expression of EBV miRNAs (Fig. 3F). These experiments strongly supported the presence of additional immunoevasive mechanisms that affect recognition of the FLY epitope. To address the possibility that LMP2A/B gene expression may be regulated by EBV miRNAs, we evaluated the gene transcript levels by quantitative RT-PCR in infected B cells 15 d post infection and found LMP2A/B unaffected by viral miRNAs (Fig. 4D). In contrast, LMP1, a known target of EBV miRNAs (43, 44), was down-regulated as expected (Fig. 4D). Similarly, LMP2 protein levels did not depend on viral miRNAs (Fig. 4E), substantiating the conclusion that EBV miRNAs regulate the activation of LMP2A/B epitope-specific T-cell clones without affecting the viral source of the epitopes they recognize.
Fig. 4.
EBV miRNAs control recognition of diverse types of CD8+ T-cell epitopes. (A) Schematic overview of the experiments shown in B, C, F, and G of this figure. The presentation of viral epitopes from B cells infected with WT/B95-8 or ΔmiR EBV was analyzed with epitope-specific CD8+ T-cell clones or polyclonal lines. Equal numbers of B and T cells were cocultured for 16 h, and IFN-γ release was measured by ELISA at the indicated time points. (B and C) The presentation of two LMP2 epitopes by infected B cells was analyzed with CD8+ T-cell clones specific for the HLA-B*40:01–restricted IED epitope (B) or the HLA-A*02:01–restricted FLY (C) epitope. A representative time course experiment with mean values and SD of three replicates (Left in B and C) and the summary of all experiments performed 15 dpi with B cells from five different donors (Right in B and C) are shown. Results are normalized to values of ΔmiR EBV-infected cells. (D) Relative transcript levels of LMP1, LMP2, and EBNA1 were assessed by quantitative RT-PCR in B cells infected with WT/B95-8 or ΔmiR EBV at day 15 post infection. Transcript levels are normalized as in Fig. 3A. LMP1 is a known target of viral miRNAs (43, 44) and was used here as a positive control. Data are shown as mean values and SD of five donors. (E) Western blot analysis of EBNA1 and LMP2 in B cells infected with WT/B95-8 or ΔmiR EBV 15 dpi. Representative examples (Left) and protein levels relative to Tubulin (Right) are shown. The result from ΔmiR EBV-infected cells was set to 100%. Data are shown as mean values and SD of four different donors. (F) Recognition of the HLA-B*35:01–restricted EBNA1 epitope HPV presented by infected B cells as in B and C. (G) IL-12 neutralization reduces the activation of an epitope-specific CD8+ T-cell clone. Infected B cells (15 dpi) were cocultured as in C (Right) with the FLY-specific T-cell clone together with an anti-IL12B antibody or a control antibody of the same isotype (2.5 μg/mL). After 16 h, IFN-γ release was measured by ELISA. An overview of three experiments with different donors is shown. (H) Equal numbers of infected B cells and polyclonal EBV-specific CD8+ T cells were cocultured together with an anti-IL12B antibody or a control antibody of the same isotype (2.5 μg/mL). After 16 h, IFN-γ release was measured by ELISA. Results from two different donors are shown. *P < 0.05. **P < 0.01.
In contrast to LMP2A, EBNA1 transcripts appeared to be under the control of viral miRNAs (Fig. 4D) (35), also resulting in decreased levels of EBNA1 protein 15 d post infection (Fig. 4E). We therefore tested the recognition of the HPV epitope of EBNA1 presented by the HLA-B*35:01 allele in a time course experiment (Fig. 4F). After only 1 day post infection, the HPV-specific T-cell clone was clearly activated but only when challenged with ΔmiR EBV-infected B cells. Later on, B cells infected with either virus presented the HPV peptide, but ΔmiR EBV-infected B cells were preferentially recognized (Fig. 4F). Regulation of TAP (Fig. 3 A–C); lower surface levels of the presenting HLA-B allele, which might be down-regulated similar to other HLA-B allotypes (Fig. 3F); and reduced levels of EBNA1 gene expression (Fig. 4E) may all have contributed to this result.
The analysis of EBNA1 epitope presentation did not reveal why TAP-independent LMP2A-derived peptides presented via HLA-A*0201 were under the control of viral miRNAs (Fig. 4C). We speculated that other costimulatory molecules or proinflammatory cytokines (35) may be responsible for this TAP-independent immunoevasive function. In particular, IL-12, which contributes to the activation of effector T-cell functions (45), was a possible candidate because it is a direct and prominent target of at least five different EBV miRNAs 5 d post infection (35) and was also down-regulated at the transcript level 15 d post infection (Fig. 3A). To address this possibility, we neutralized IL-12 secreted from EBV-infected B cells with a suitable antibody and measured IFN-γ secretion by the FLY-specific T-cell clone (Fig. 4G). IL-12 neutralization dramatically reduced the activation of the T cells cocultured with ΔmiR EBV-infected cells. T-cell activation with WT/B95-8 EBV-infected target cells was also affected but to a lesser extent. Very similar results were observed with polyclonal EBV-specific CD8+ T cells cocultured with HLA-matched infected B cells (Fig. 4H), showing that miRNA-mediated regulation of IL-12 was globally decreasing recognition by CD8+ T cells. This effect, which was clearly evident for EBV-specific CD8+ T cells, was mild with EBV-specific CD4+ T cells (Fig. S3).
Fig. S3.
IL-12 neutralization barely reduces the activation of EBV-specific CD4+ T cells. Equal numbers of infected B cells (5 dpi) were cocultured with polyclonal EBV-specific CD4+ T cells together with an anti–IL-12B antibody or a control antibody of the same isotype (2.5 μg/mL). After 16 h, IFN-γ release was measured by ELISA. One representative experiment out of four with different donors is shown.
Discussion
In this study, we show that EBV miRNAs inhibit surveillance of EBV by CD8+ T cells. Viral miRNAs reduce virus-specific proliferation, cytokine production, and killing of infected cells by CD8+ T cells with various EBV latent epitope specificities. We identified several mechanisms for this inhibition. First, miRNAs target TAP2 directly, down-regulate the entire TAP complex, and reduce HLA allotypes that preferentially present TAP-dependent epitopes. Second, miRNAs repress EBNA1, which limits the level of a protein but is essential during most forms of EBV latency. Third, miRNAs diminish IL-12 release by infected B cells, reducing the virus-specific activity of EBV-specific CD8+ T cells. Thus, EBV miRNAs limit surveillance by CD8+ T cells through multiple mechanisms, likely contributing to the maintenance of lifelong infection.
It is an attractive hypothesis (35) that T-cell immunoevasion in latency would be most economically achieved by miRNAs due to their nonantigenicity. This hypothesis is now fully substantiated by our present findings that EBV miRNAs interfere with several steps of antigen presentation preventing CD8+ T-cell recognition of latently infected B cells. These results are complementary to our previous findings documenting that EBV miRNAs regulate multiple pathways important for differentiation and activation of antiviral CD4+ T cells in the first days of infection (35). That study also provided some evidence that CD4+ T-cell recognition is also regulated later, as the structural protein gp350 could be detected by CD4+ T cells in cells 15 d post infection but only when miRNAs were absent (35). The mechanisms of regulation we identified in that context—that is, regulation of MHC II, lysosomal enzymes, and IL-12—are likely relevant in latency as well and may explain why EBNA-specific CD4+ T cells are generally impaired in recognizing LCLs (46). The present study focused on established latency, but because we observed a strong miRNA regulation of EBNA1 recognition by CD8+ T cells already on days 1, 2, and 3 after infection, the hypothesis that EBV miRNAs generally suppress CD8+ T-cell recognition already in the first days of infection during prelatency (13, 22, 47) deserves closer investigation in the future.
An overview of EBV miRNAs that directly target pathways involved in CD8+ and/or CD4+ T-cell recognition of infected B cells is provided in Table 2. As large subsets of viral miRNAs are expressed in all phases of EBV’s life cycle (48), it appears plausible that viral miRNAs inhibit these target molecules globally. For example, miRNAs miR-BART1, miR-BART2, and miR-BART22, which target IL-12 (35), are all highly expressed not only in latency III but also in EBV-infected germinal center B cells (latency II) and memory B cells (latency 0/I) from healthy donors as well as different types of EBV-associated cancer cells (34, 48, 49). Therefore, miRNA-mediated reduction of IL-12 could lead to decreased T-cell activation and recognition at different stages of infection and malignant disease. Among TAP-regulating miRNAs, miR-BHRF1–3 is predominantly expressed initially upon infection and in latency III in vitro (36), but miR-BART17 also shows expression in memory B cells and cancer cells (48), suggesting that EBV miRNA-mediated TAP regulation could likewise be important in vivo.
Table 2.
Direct targets of EBV miRNAs with immune functions
| Function | Gene (protein) | EBV miRNAs |
| Antigen processing | CTSB (Cathepsin B) | BHRF1–2 |
| BART2–5p | ||
| LGMN (AEP) | BART2–5p | |
| IFI30 (GILT) | BART1–5p | |
| BART1–3p | ||
| Peptide transport | TAP2 (TAP) | BHRF1–3 |
| BART17 | ||
| Cytokines | IL12B (IL-12p40)* | BHRF1–2 |
| BART1–3p | ||
| BART2–5p | ||
| BART10-3p | ||
| BART22 |
Component of IL-12 (p35/p40) and IL-23 (p19/p40).
Although EBV-specific immunity is likely to operate at different stages of latency and lytic replication to control viral infection, the question of interest is whether latency III, in its own right, is a target of EBV-specific immunosurveillance by T cells. For immunosuppressed patients, it appears clear that T-cell deficiency favors appearance of latency III malignancies (3), that adoptive T-cell therapy can prevent this (4, 5), and that T-cell therapy fails if the EBV strain in question does not express crucial CD8+ T-cell epitopes in a latency III protein (50). Regarding infection in immunocompetent carriers, there were early arguments against a T-cell surveillance of latency III (51), but later studies showed that latency III-associated CD8+ T-cell epitopes are in fact under a selective pressure that depends on the frequency of HLA class I allotypes in a population (52, 53). Cumulatively, these reports suggest that EBV-specific CD8+ T-cell surveillance of latency III is an important aspect of infection control in vivo.
In this work, we have analyzed expansion of EBV-specific T cells, cytokine secretion, and cytolysis to study the interference of EBV miRNAs with CD8+ T-cell functions. We found such interference for T cells specific for five out of five epitopes from three different antigens (LMP2A/B, EBNA1, and EBNA3A). Because this collection contained epitopes with different HLA class I restrictions, derived from different categories of antigen (nuclear vs. transmembrane proteins) and with different processing requirements (TAP-dependent or -independent, proteasome- or immunoproteasome-dependent), our data indicate that the cumulative functional impact of miRNAs allows latently infected B cells to hide from CD8+ T cells in general. Our findings with polyclonal CD8+ T cells of complex composition corroborated our view.
miRNAs mediate their effects through direct binding to their target transcripts such as TAP2, for example (Fig. 3C). Because the recognition of TAP-independent epitopes is also inhibited by miRNAs, other mechanisms must contribute to the reduced presentation of the TAP-independent LMP2 epitopes CLG (Fig. 2G) and FLY (Fig. 4). Only a minority of all CD8+ T-cell epitopes (including those of viral origin) are expected to be TAP-independent (42), and the importance of TAP is reflected by the many herpesviruses that have evolved their own TAP-inhibitory proteins (54, 55). A broad impact of TAP regulation on the immunological status in latency is also suggested by our observation that viral miRNAs do not affect global levels of HLA-A2, which is capable of presenting highly hydrophobic peptides that are more likely to be TAP-independent (39, 56), but do reduce levels of the HLA class I allotypes tested (HLA-B7, B8, and B40). These and most HLA class I allotypes are less likely to present TAP-independent epitopes, because they require the presence of polar or charged anchor residues in the peptide (57, 58).
Another effect of EBV miRNAs, suppression of IL-12, seems to act globally on the function of antigen-specific T cells. Although IL-12 was originally identified as a product of EBV-infected LCLs (59), its role in EBV-specific CD8+ T-cell immunity has remained obscure. In addition to its well-known function in promoting Th1 differentiation (60), IL-12 was shown in mouse and human studies to promote CD8+ T-cell functions such as proliferation, cytolysis, and IFN-γ production (60, 61) through STAT4 signaling, up-regulating T-bet, and increasing IL-2 sensitivity (62, 63). In our experiments, blockade of IL-12 fully reverted the effect of the miRNA deletion for polyclonal EBV-specific CD8+ T cells (Fig. 4 G and H) but unexpectedly had only a minor effect on polyclonal EBV-specific CD4+ T cells (Fig. S3). The reason for this difference is not clear yet, but one possibility may be a differential requirement for costimulatory signals (64). However, as we have shown (35), EBV miRNAs affect CD4+ T-cell responses through IL-12 regulation already at the level of differentiation from naive T cells and thus act on both major classes of T cells. Because we found IL-12 to be the gene product most strongly down-regulated by EBV miRNAs, control of this cytokine appears to be central to the maintenance of EBV infection.
Materials and Methods
Patient Samples.
PBMCs and surgically removed adenoid biopsies were obtained from volunteer blood donors and patients from the Department of Otorhinolaryngology of the Universitätsklinikum der Ludwig-Maximilians-Universität München, respectively. The local ethics committee (Ethikkommission bei der Ludwig-Maximilians-Universität München) approved the use of this human material. Informed consent was not required because the biopsies originated from disposed tissues from anonymous donors who underwent routine surgery.
Human Primary Cells, Cell Lines, and Cell Culture.
Human primary B and T cells were prepared from adenoidal mononuclear cells (MNCs) or PBMCs as described (35). The EBV-positive Burkitt’s lymphoma cell line Raji, HEK293-based EBV producer cell lines, infected human primary B cells, LCLs, and isolated T cells were cultivated as described in SI Materials and Methods.
Preparation of EBV Stocks and Infection of Human Primary B Cells.
Stocks of recombinant EBV strains were essentially prepared and quantitated as described (65). Details can be found in SI Materials and Methods.
In Vitro Model of EBV Infection (B-Cell Outgrowth Assay).
B cells (CD19+) were isolated from PBMCs of EBV-positive donors and infected with WT/B95-8 or ΔmiR EBV strains. After 12 h, the infected B cells were extensively washed to remove free virions. CD8+ T cells isolated from the same donors were cocultured at B:T cell ratios ranging from 8:1–1:2 (seeding 32,000 B cells per well). B cells cultivated without T cells served as control. Cells were refed weekly. After 4 wk, the cultures were analyzed for viable cells in MTT assays as previously described (22).
Establishment of EBV-Specific Effector T Cells and T-Cell Clones.
EBV-specific CD8+ T-cell clones were established by limiting dilution from polyclonal T-cell lines that were generated by stimulating PBMCs with LCLs infected with WT/B95-8 or from specific T cells directly obtained from peripheral blood cells by peptide stimulation, IFN-γ capture (Miltenyi Biotec), and magnetic isolation (66, 67). Likewise, EBV-specific CD4+ T cells were generated by repetitive stimulation of sorted CD4+ T cells with autologous LCLs infected with WT/B95-8 EBV as described previously (66).
EBV-Specific T-Cell Recognition.
EBV-specific effector T-cell activities were measured with IFN-γ ELISA and calcein release assays. For IFN-γ detection from T cells, effector and target cells were cocultured at a 1:1 ratio (5 × 104 cell per well) for 16 h in a 96-well plate (V bottom). IFN-γ levels were detected with ELISA following the manufacturer’s protocol (Mabtech). IFN-γ concentrations below 16 pg/mL were regarded negative. Neutralization of IL-12 was performed with an antibody (2.5 μg/mL), which was added directly to the coculture and is directed against the p40 subunit of IL-12 (C8.6; BioLegend). An analogous isotype control antibody (MOPC-21; BioLegend) was used as a control.
T-Cell Cytotoxicity Assays.
EBV-infected B cells were purified by Ficoll-Hypaque (PAN-Biotech) gradient centrifugation, and 5 × 105 target cells were labeled with calcein (Invitrogen) at 0.5 µg/mL. After three washing steps with PBS, target and effector cells were cocultured in V bottom 96-well plates with different ratios in RPMI without Phenol Red (PAN-Biotech). After 4 h of coculture, fluorescence intensities in supernatants were measured by the Infinite F200 PRO fluorometer (Tecan). As controls, spontaneous calcein release of target cells cultivated without effector cells and cells lysed with 0.5% Triton-X100 (Carl Roth) were used to define the levels of no and fully lysed target cells, respectively.
T-Cell Expansion Assay.
CD8+ T cells were isolated from PBMCs of EBV-positive donors. We stimulated 1 × 106 CD8+ T cells with 1 × 105 autologous irradiated B cells (infected for 15 d) and 20 U/mL IL-2. Cells were restimulated every 10 d. At 10 and 20 d after the first stimulation, T cells were stained with unlabeled HLA/peptide pentamers (Proimmune) for 20 min at 37 °C. Counterstaining was done with CD8 and CD3-specific antibodies and Pro5 fluorotag (Proimmune) on ice for 30 min. T-cell numbers were determined using calibrated APC-beads as volume standard by flow cytometry (68).
Luciferase Reporter Assays.
Details of the reporter plasmids and the technical aspects of the dual luciferase reporter assays can be found in SI Materials and Methods.
Quantitative RT-PCR.
Isolation of RNAs and their analyses by PCR are described in SI Materials and Methods.
Western Blotting.
Cell lysis and antibodies used to detect viral and cellular proteins of interest can be found in SI Materials and Methods.
Flow Cytometry and Antibodies.
Techniques and antibodies used to detect various surface molecules are described in detail in SI Materials and Methods.
Statistical Analysis.
We used Prism 6.0 software (GraphPad) for statistical analysis, and the two-tailed ratio t test was applied unless otherwise mentioned.
SI Materials and Methods
Cell Lines and Cell Culture.
The EBV-positive Burkitt’s lymphoma cell line Raji, HEK293-based EBV producer cell lines (36), infected human primary B cells, and isolated T cells were maintained in RPMI medium 1640 (Life Technologies). HEK293T cells were maintained in DMEM. All media were supplemented with 10% (vol/vol) FBS (Life Technologies), penicillin (100 U/mL; Life Technologies), and streptomycin (100 mg/mL; Life Technologies). Cells were cultivated at 37 °C in a 5% (vol/vol) CO2 incubator.
Separation of Human Primary Lymphocytes.
Human primary B and T cells were prepared from adenoidal MNCs or PBMCs by Ficoll-Hypaque gradient centrifugation. B cells and CD8+ T cells were isolated using MACS separation columns (Miltenyi Biotec) with CD19 or CD8 MicroBeads, respectively.
Preparation of EBV Stocks.
The recombinant EBV genome designated WT/B95-8 in this study is identical to plasmid 2089, which contains the complete EBV strain B95-8 genome, the F factor origin of replication, the chloramphenicol resistance gene, the gene for the green fluorescent protein (GFP) under the control of the cytomegalovirus (CMV) promoter, and the hygromycin resistance gene as a selectable marker in eukaryotic cells (69). Inactivation of all miRNA genes from this construct resulted in plasmid 4027, also called ΔmirALL or ΔmiR (36). Infectious virus was produced by lytic induction of producer cell lines stably carrying these recombinant EBV genomes in episomal form.
To induce EBV’s lytic cycle in the ΔmiR (4027) or WT/B95-8 (2089) HEK 293 producer cell lines, plasmids coding for BZLF1 and BALF4 were transiently transfected. Supernatants were collected after 3 d. Virus stocks were titrated using Raji cells as described in detail recently (65). Isolated primary B cells were infected with the virus stocks at a multiplicity of infection of 0.1 GRU (Green Raji Units, see ref. 65). Eighteen hours later, the infected B cells were cultivated in fresh medium at an initial density of 5 × 105 cells per milliliter.
Luciferase Reporter Assays.
The 3′UTRs of TAP2 (ENST00000374897), TAP1 (ENST00000354258), IPO7 (ENST00000379719), and HLA-B7/B8 (ENST00000412585/ENST00000425848) were cloned downstream of Renilla luciferase (Rluc) into the expression plasmid psiCHECK-2 (Promega). The pCDH vectors expressing single viral miRNA were used as previously described (35). The psiCHECK-2 reporter and pCDH-EF1-MCS plasmid DNAs (System Biosciences) with a viral miRNA of interest were cotransfected into HEK293T cells using Metafectene Pro (Biontex). miR-BHRF1–3 was expressed from a modified pLSP plasmid vector (70). It was digested with BamHI and EcoRI and ligated with miR-BHRF1–3 sequences obtained from p2089 (36). The resulting pLSP-BHRF1–3 plasmid was digested with SfiI and XbaI, and the Cerulean gene was inserted as a phenotypic marker. Twenty-four hours after DNA transfection, we measured luciferase activities with the Dual-Luciferase Assay Kit (Promega) and the Orion II Microplate Luminometer (Titertek-Berthold). The activity of Rluc was normalized to the activity of Firefly luciferase (Fluc) encoded by the psiCHECK-2 reporter. Site-specific mutagenesis was performed as previously described (35). We performed in silico prediction of EBV miRNA binding sites on 3′UTRs with TargetScan (www.targetscan.org) (41).
Quantitative RT-PCR.
RNA was isolated using the Direct-zol RNA MiniPrep columns (Zymo Research). RNA was treated with DNase I (Thermo Fisher Scientific) and reverse transcribed with SuperScript III Reverse Transcriptase (Thermo Fisher Scientific), and quantitative PCR was performed using the LightCycler 480 SYBR Green I Mix (Roche) and the LightCycler 480 Instrument II (Roche) according to the manufacturer’s instructions. The following primers were used:
HPRT1 for 5′-tgaccttgatttattttgcatacc-3′ and rev 5′-cgagcaagacgttcagtcct-3′,
HMBS for 5′-ctgaaagggccttcctgag-3′ and rev 5′-cagactcctccagtcaggtaca-3′,
GUSB for 5′-cgccctgcctatctgtattcattggaggtg-3′ and rev 5′-gatgaggaactcttggtgacagcc-3′,
IPO7 for 5′-tcgccattgtattcgagaaa-3′ and rev 5′-gaatgcatgtagtaagctgtaccc-3′,
IL12B for 5′-ccctgacattctgcgttca-3′ and rev 5′-aggtcttgtccgtgaagactcta-3′,
TAP1 for 5′-agtgccctggatgcaaac-3′ and rev 5′-agaaagaggatgtggtcacg-3′,
TAP2 for 5′-tgcgggacagaaacaacgtc-3′ and rev 5′-agcctgtgagcaatcaccag-3′,
EBNA1 for 5′-aagcatcgtggtcaaggagg-3′ and rev 5′-gcgacccaagttccttcgtc-3′,
LMP1 for 5′-aggctaggaagaaggccaaa-3′ and rev 5′-ctgttcatcttcgggtgctt-3′.3gtt
LMP2 for 5′-atcgctggtggcagtatttt-3′ and rev 5′-gagtatgccagcgacaatca-3′.
Western Blotting.
We lysed cells with RIPA buffer [50 mM Tris·HCl (pH 8), 150 mM NaCl, 0.1% SDS, 1% Nonidet P-40, 0.5% DOC] and boiled the extracts with Laemmli buffer. Proteins were separated on 10% (vol/vol) SDS/PAGE gels (Carl Roth) and transferred to nitrocellulose membranes (GE Healthcare Life Science) using the Mini-PROTEAN Tetra Cell apparatus (Bio-Rad). Membranes were blocked for 30 min with Roti-Block (Carl Roth) followed by antibody incubation. Secondary antibodies conjugated with horseradish peroxidase were used (Cell Signaling) and exposed to CEA films (Agfa HealthCare). Protein levels were quantified with the software ImageJ. The following primary antibodies reactive to human proteins were used: anti-human Tubulin (B-5–1-2; Santa Cruz), anti-human Actin (AC-74; Sigma), anti-human IPO7 (ab88339; Abcam), anti-human TAP1 (1.28; Acris), and anti-human TAP2 (2.17, Acris). Elisabeth Kremmer, Helmholtz Zentrum München, German Research Center for Environmental Health, Institute of Molecular Immunology, Munich, provided the antibodies specific for the EBV proteins LMP2, LMP1, and EBNA1.
Flow Cytometry and Antibodies.
Stained cell suspensions were measured with the LSRFortessa or FACSCanto (BD Biosciences) flow cytometers and the FACSDiva software (BD Biosciences). Acquired data were analyzed with FlowJo software Ver. 9.8 (FlowJo). The following human-specific antibodies were used:
anti–HLA-ABC APC (W6/32, IgG2a; BioLegend),
anti–HLA-Bw6 PE (REA143, IgG1; Miltenyi Biotec),
anti–HLA-A2 PE (BB7.2, IgG2b; BioLegend),
anti–HLA-B7 PE (BB7.2, IgG2b; Santa Cruz),
anti–HLA-B8 unlabeled mAb (8.L.215; USbiological),
anti-mouse IgG PE (poly4053; BioLegend),
isotype IgG1 PE (MOPC-21; BioLegend),
isotype IgG2b PE (MPC-11; BioLegend),
isotype IgG2a APC (MOPC-173; BioLegend),
anti-CD8 Pacific Blue (RPA-T8; BioLegend),
anti-CD8 PerCP/Cy5.5 (RPA-T8; BioLegend),
anti-CD4 PE (RPA-T4; BioLegend),
anti-CD3 APC (HIT3a; BioLegend),
anti-CD3 APC/Cy7 (HIT3a; BioLegend),
anti-CD19 FITC (HIB19; BioLegend),
HLA-A*0201/CLG pentamer (CLGGLLTMV, LMP2; Proimmune),
HLA-B*0702/RPP pentamer (RPPIFIRRL, EBNA3A; Proimmune), and
HLA-B*0801/QAK pentamer (QAKWRLQTL, EBNA3A; Proimmune).
Acknowledgments
We thank Elisabeth Kremmer and Dagmar Pich for monoclonal antibodies and valuable experimental advice, respectively. This work was financially supported by grants of the Deutsche Forschungsgemeinschaft (SFB1054/TP B05, SFB1064/TP A13, SFB-TR36/TP A04), Deutsche Krebshilfe (107277 and 109661), National Cancer Institute (CA70723 and CA022443), and personal grants to T.T. from Deutscher Akademischer Austauschdienst (DAAD, Studienstipendien für ausländische Graduierte aller wissenschaftlichen Fächer) and to M.B. from the European Molecular Biology Organization (EMBO).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605884113/-/DCSupplemental.
References
- 1.van Baarle D, et al. Dysfunctional Epstein-Barr virus (EBV)-specific CD8(+) T lymphocytes and increased EBV load in HIV-1 infected individuals progressing to AIDS-related non-Hodgkin lymphoma. Blood. 2001;98(1):146–155. doi: 10.1182/blood.v98.1.146. [DOI] [PubMed] [Google Scholar]
- 2.Smets F, et al. Ratio between Epstein-Barr viral load and anti-Epstein-Barr virus specific T-cell response as a predictive marker of posttransplant lymphoproliferative disease. Transplantation. 2002;73(10):1603–1610. doi: 10.1097/00007890-200205270-00014. [DOI] [PubMed] [Google Scholar]
- 3.Landgren O, et al. Risk factors for lymphoproliferative disorders after allogeneic hematopoietic cell transplantation. Blood. 2009;113(20):4992–5001. doi: 10.1182/blood-2008-09-178046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heslop HE, et al. Long-term outcome of EBV-specific T-cell infusions to prevent or treat EBV-related lymphoproliferative disease in transplant recipients. Blood. 2010;115(5):925–935. doi: 10.1182/blood-2009-08-239186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moosmann A, et al. Effective and long-term control of EBV PTLD after transfer of peptide-selected T cells. Blood. 2010;115(14):2960–2970. doi: 10.1182/blood-2009-08-236356. [DOI] [PubMed] [Google Scholar]
- 6.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: Lessons from Epstein-Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 7.Bihl F, et al. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol. 2006;176(7):4094–4101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 8.Kalla M, Göbel C, Hammerschmidt W. The lytic phase of Epstein-Barr virus requires a viral genome with 5-methylcytosine residues in CpG sites. J Virol. 2012;86(1):447–458. doi: 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med. 2004;350(13):1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- 10.Rowe M, Lear AL, Croom-Carter D, Davies AH, Rickinson AB. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J Virol. 1992;66(1):122–131. doi: 10.1128/jvi.66.1.122-131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longnecker RM, Kieff E, Cohen JI. 2013. in Fields Virology, eds Knipe DM, et al. (Wolters Kluver, Philadelphia), Vol 2, pp 1898–1959.
- 12.Joseph AM, Babcock GJ, Thorley-Lawson DA. Cells expressing the Epstein-Barr virus growth program are present in and restricted to the naive B-cell subset of healthy tonsils. J Virol. 2000;74(21):9964–9971. doi: 10.1128/jvi.74.21.9964-9971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerschmidt W. The epigenetic life cycle of Epstein-Barr virus. Curr Top Microbiol Immunol. 2015;390(Pt 1):103–117. doi: 10.1007/978-3-319-22822-8_6. [DOI] [PubMed] [Google Scholar]
- 14.Hislop AD, et al. A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates. J Exp Med. 2007;204(8):1863–1873. doi: 10.1084/jem.20070256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horst D, et al. Specific targeting of the EBV lytic phase protein BNLF2a to the transporter associated with antigen processing results in impairment of HLA class I-restricted antigen presentation. J Immunol. 2009;182(4):2313–2324. doi: 10.4049/jimmunol.0803218. [DOI] [PubMed] [Google Scholar]
- 16.Zuo J, et al. The Epstein-Barr virus G-protein-coupled receptor contributes to immune evasion by targeting MHC class I molecules for degradation. PLoS Pathog. 2009;5(1):e1000255. doi: 10.1371/journal.ppat.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuo J, et al. The Epstein-Barr virus-encoded BILF1 protein modulates immune recognition of endogenously processed antigen by targeting major histocompatibility complex class I molecules trafficking on both the exocytic and endocytic pathways. J Virol. 2011;85(4):1604–1614. doi: 10.1128/JVI.01608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe M, et al. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci USA. 2007;104(9):3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo J, et al. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J Virol. 2008;82(5):2385–2393. doi: 10.1128/JVI.01946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinn LL, et al. The missing link in Epstein-Barr Virus immune evasion: The BDLF3 gene induces ubiquitination and downregulation of major histocompatibility complex class I (MHC-I) and MHC-II. J Virol. 2015;90(1):356–367. doi: 10.1128/JVI.02183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croft NP, et al. Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle. PLoS Pathog. 2009;5(6):e1000490. doi: 10.1371/journal.ppat.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jochum S, Moosmann A, Lang S, Hammerschmidt W, Zeidler R. The EBV immunoevasins vIL-10 and BNLF2a protect newly infected B cells from immune recognition and elimination. PLoS Pathog. 2012;8(5):e1002704. doi: 10.1371/journal.ppat.1002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cassell DJ, Schwartz RH. A quantitative analysis of antigen-presenting cell function: Activated B cells stimulate naive CD4 T cells but are inferior to dendritic cells in providing costimulation. J Exp Med. 1994;180(5):1829–1840. doi: 10.1084/jem.180.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiesner M, et al. Conditional immortalization of human B cells by CD40 ligation. PLoS One. 2008;3(1):e1464. doi: 10.1371/journal.pone.0001464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowe M, et al. Epstein-Barr virus (EBV)-associated lymphoproliferative disease in the SCID mouse model: Implications for the pathogenesis of EBV-positive lymphomas in man. J Exp Med. 1991;173(1):147–158. doi: 10.1084/jem.173.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lautscham G, et al. Identification of a TAP-independent, immunoproteasome-dependent CD8+ T-cell epitope in Epstein-Barr virus latent membrane protein 2. J Virol. 2003;77(4):2757–2761. doi: 10.1128/JVI.77.4.2757-2761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill AB, et al. Class I major histocompatibility complex-restricted cytotoxic T lymphocytes specific for Epstein-Barr virus (EBV)-transformed B lymphoblastoid cell lines against which they were raised. J Exp Med. 1995;181(6):2221–2228. doi: 10.1084/jem.181.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 29.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2(4):269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 30.Cai X, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2(3):e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kincaid RP, Sullivan CS. Virus-encoded microRNAs: An overview and a look to the future. PLoS Pathog. 2012;8(12):e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haneklaus M, et al. Cutting edge: MiR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J Immunol. 2012;189(8):3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 33.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5(4):376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Xia T, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68(5):1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagawa T, et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T-cell responses targeting IL-12 and peptide processing. J Exp Med. 2016 doi: 10.1084/jem.20160248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seto E, et al. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6(8):e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feederle R, et al. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011;7(2):e1001294. doi: 10.1371/journal.ppat.1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feederle R, et al. The members of an Epstein-Barr virus microRNA cluster cooperate to transform B lymphocytes. J Virol. 2011;85(19):9801–9810. doi: 10.1128/JVI.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lautscham G, et al. Processing of a multiple membrane spanning Epstein-Barr virus protein for CD8(+) T cell recognition reveals a proteasome-dependent, transporter associated with antigen processing-independent pathway. J Exp Med. 2001;194(8):1053–1068. doi: 10.1084/jem.194.8.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dölken L, et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. 2010;7(4):324–334. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Garcia DM, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18(10):1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Val M, Lázaro S, Ramos M, Antón LC. Are membrane proteins favored over cytosolic proteins in TAP-independent processing pathways? Mol Immunol. 2013;55(2):117–119. doi: 10.1016/j.molimm.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 43.Lo AK, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA. 2007;104(41):16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley KJ, et al. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31(9):2207–2221. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gately MK, Wolitzky AG, Quinn PM, Chizzonite R. Regulation of human cytolytic lymphocyte responses by interleukin-12. Cell Immunol. 1992;143(1):127–142. doi: 10.1016/0008-8749(92)90011-d. [DOI] [PubMed] [Google Scholar]
- 46.Long HM, et al. CD4+ T-cell responses to Epstein-Barr virus (EBV) latent-cycle antigens and the recognition of EBV-transformed lymphoblastoid cell lines. J Virol. 2005;79(8):4896–4907. doi: 10.1128/JVI.79.8.4896-4907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jochum S, Ruiss R, Moosmann A, Hammerschmidt W, Zeidler R. RNAs in Epstein-Barr virions control early steps of infection. Proc Natl Acad Sci USA. 2012;109(21):E1396–E1404. doi: 10.1073/pnas.1115906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu J, et al. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011;7(8):e1002193. doi: 10.1371/journal.ppat.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim DN, et al. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J Virol. 2007;81(2):1033–1036. doi: 10.1128/JVI.02271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gottschalk S, et al. An Epstein-Barr virus deletion mutant associated with fatal lymphoproliferative disease unresponsive to therapy with virus-specific CTLs. Blood. 2001;97(4):835–843. doi: 10.1182/blood.v97.4.835. [DOI] [PubMed] [Google Scholar]
- 51.Khanna R, et al. Evolutionary dynamics of genetic variation in Epstein-Barr virus isolates of diverse geographical origins: Evidence for immune pressure-independent genetic drift. J Virol. 1997;71(11):8340–8346. doi: 10.1128/jvi.71.11.8340-8346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Midgley RS, Bell AI, McGeoch DJ, Rickinson AB. Latent gene sequencing reveals familial relationships among Chinese Epstein-Barr virus strains and evidence for positive selection of A11 epitope changes. J Virol. 2003;77(21):11517–11530. doi: 10.1128/JVI.77.21.11517-11530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Midgley RS, et al. HLA-A11-restricted epitope polymorphism among Epstein-Barr virus strains in the highly HLA-A11-positive Chinese population: Incidence and immunogenicity of variant epitope sequences. J Virol. 2003;77(21):11507–11516. doi: 10.1128/JVI.77.21.11507-11516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ressing ME, Luteijn RD, Horst D, Wiertz EJ. Viral interference with antigen presentation: Trapping TAP. Mol Immunol. 2013;55(2):139–142. doi: 10.1016/j.molimm.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Verweij MC, et al. Viral inhibition of the transporter associated with antigen processing (TAP): A striking example of functional convergent evolution. PLoS Pathog. 2015;11(4):e1004743. doi: 10.1371/journal.ppat.1004743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinzierl AO, et al. Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur J Immunol. 2008;38(6):1503–1510. doi: 10.1002/eji.200838136. [DOI] [PubMed] [Google Scholar]
- 57.Sutton J, et al. A sequence pattern for peptides presented to cytotoxic T lymphocytes by HLA B8 revealed by analysis of epitopes and eluted peptides. Eur J Immunol. 1993;23(2):447–453. doi: 10.1002/eji.1830230222. [DOI] [PubMed] [Google Scholar]
- 58.Falk K, et al. Peptide motifs of HLA-B58, B60, B61, and B62 molecules. Immunogenetics. 1995;41(2-3):165–168. doi: 10.1007/BF00182333. [DOI] [PubMed] [Google Scholar]
- 59.Kobayashi M, et al. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J Exp Med. 1989;170(3):827–845. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manetti R, et al. Interleukin 12 induces stable priming for interferon gamma (IFN-gamma) production during differentiation of human T helper (Th) cells and transient IFN-gamma production in established Th2 cell clones. J Exp Med. 1994;179(4):1273–1283. doi: 10.1084/jem.179.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valiante NM, Rengaraju M, Trinchieri G. Role of the production of natural killer cell stimulatory factor (NKSF/IL-12) in the ability of B cell lines to stimulate T and NK cell proliferation. Cell Immunol. 1992;145(1):187–198. doi: 10.1016/0008-8749(92)90322-g. [DOI] [PubMed] [Google Scholar]
- 62.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12(11):749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Starbeck-Miller GR, Xue HH, Harty JT. IL-12 and type I interferon prolong the division of activated CD8 T cells by maintaining high-affinity IL-2 signaling in vivo. J Exp Med. 2014;211(1):105–120. doi: 10.1084/jem.20130901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elloso MM, Scott P. Differential requirement of CD28 for IL-12 receptor expression and function in CD4(+) and CD8(+) T cells. Eur J Immunol. 2001;31(2):384–395. doi: 10.1002/1521-4141(200102)31:2<384::aid-immu384>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 65.Steinbrück L, et al. K1 and K15 of Kaposi’s sarcoma-associated herpesvirus are partial functional homologues of latent membrane protein 2A of Epstein-Barr virus. J Virol. 2015;89(14):7248–7261. doi: 10.1128/JVI.00839-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adhikary D, et al. Immunodominance of lytic cycle antigens in Epstein-Barr virus-specific CD4+ T cell preparations for therapy. PLoS One. 2007;2(7):e583. doi: 10.1371/journal.pone.0000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rancan C, Schirrmann L, Hüls C, Zeidler R, Moosmann A. Latent membrane protein LMP2A impairs recognition of EBV-infected cells by CD8+ T cells. PLoS Pathog. 2015;11(6):e1004906. doi: 10.1371/journal.ppat.1004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iskra S, Kalla M, Delecluse HJ, Hammerschmidt W, Moosmann A. Toll-like receptor agonists synergistically increase proliferation and activation of B cells by Epstein-Barr virus. J Virol. 2010;84(7):3612–3623. doi: 10.1128/JVI.01400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95(14):8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37(6):e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]