Significance
Pluripotent stem cells (PSCs) can exist in a naïve or primed pluripotency state. Naïve PSCs correspond to an earlier developmental state more closely related to cells from the preimplantation embryo and may have more robust developmental potential than primed PSCs. However, understanding the molecular mechanisms that regulate naïve PSC behaviors such as self-renewal, differentiation, or preservation of the naïve state is incomplete. Here, we report that Wnt/β-catenin signaling promotes the self-renewal of naïve human embryonic stem cells (hESCs). When grown in conditions that inhibit Wnt/β-catenin signaling, naïve hESCs remain undifferentiated but have a more primed-like protein expression profile. Our results suggest that Wnt/β-catenin signaling plays a critical role in regulating human naïve pluripotency.
Keywords: Wnt/β-catenin signaling, naïve pluripotency, naïve-to-primed transition, human embryonic stem cells, self-renewal
Abstract
In both mice and humans, pluripotent stem cells (PSCs) exist in at least two distinct states of pluripotency, known as the naïve and primed states. Our understanding of the intrinsic and extrinsic factors that enable PSCs to self-renew and to transition between different pluripotent states is important for understanding early development. In mouse embryonic stem cells (mESCs), Wnt proteins stimulate mESC self-renewal and support the naïve state. In human embryonic stem cells (hESCs), Wnt/β-catenin signaling is active in naïve-state hESCs and is reduced or absent in primed-state hESCs. However, the role of Wnt/β-catenin signaling in naïve hESCs remains largely unknown. Here, we demonstrate that inhibition of the secretion of Wnts or inhibition of the stabilization of β-catenin in naïve hESCs reduces cell proliferation and colony formation. Moreover, we show that addition of recombinant Wnt3a partially rescues cell proliferation in naïve hESCs caused by inhibition of Wnt secretion. Notably, inhibition of Wnt/β-catenin signaling in naïve hESCs did not cause differentiation. Instead, it induced primed hESC-like proteomic and metabolic profiles. Thus, our results suggest that naïve hESCs secrete Wnts that activate autocrine or paracrine Wnt/β-catenin signaling to promote efficient self-renewal and inhibit the transition to the primed state.
Pluripotent stem cells (PSCs) undergo unlimited self-renewal and can differentiate into cells representative of all three germ layers of the adult body (1). In both mice and humans, PSCs exist in at least two different states of pluripotency in vitro, commonly referred to as naïve and primed states. The different states of pluripotency likely reflect different stages of embryonic development in vivo (2–5). Understanding the molecular underpinnings of PSC behaviors in different pluripotency states and identifying the molecular factors that control transitions between these states may provide insight into early embryonic development and contribute to efforts designed to use PSCs and their derivatives for research and treatment of human diseases.
Much of our knowledge about the differences between naïve and primed states comes from research on mouse PSCs. Naïve mouse embryonic stem cells (mESCs) are derived from embryonic cells of the preimplantation blastocyst and have a broad and robust developmental capacity (6, 7). In contrast, primed PSCs, exemplified by mouse epiblast stem cells (mEpiSCs), display biases toward lineage-specific differentiation (8). For example, although mESCs can be used to generate a whole animal through tetraploid complementation when injected into a preimplantation blastocyst (9), mEpiSCs exhibit a poor capacity to contribute to such chimeras. Conventional human embryonic stem cells (hESCs) exist in the primed state and exhibit marked similarities in morphological, molecular, and epigenetic characteristics compared with mEpiSCs (10). More recently, naïve-state hESCs have been generated by toggling conventional hESCs back from the primed state (e.g., LIS1, WIN1, and H1-4iLIF lines) (3, 11–13) or by deriving new hESC lines from human six- to eight-cell embryos (e.g., ELF1 line) using naïve-state growth conditions (12). The molecular programs used to support naïve pluripotency in hESCs and the degree of conservation of the different states of pluripotency between human and mouse PSCs is an area of active investigation (14).
Wnt/β-catenin signaling is a key signaling pathway involved in embryonic development and is tightly regulated in PSCs (15–17). Wnt ligands are evolutionarily conserved secreted glycoproteins involved in autocrine and paracrine cell signaling. In the absence of Wnts, a “destruction complex” consisting of the cytosolic proteins glycogen synthase kinase 3 (GSK3), AXIN, adenomatous polyposis coli (APC) protein, and casein kinase 1α phosphorylates β-catenin (encoded by the CTNNB1 gene). This pool of phosphorylated β-catenin is ubiquitylated and thereby targeted to the proteasome for degradation. In the presence of Wnt ligands, binding of Wnts to a heteromeric receptor complex leads to inhibition of the destruction complex, thus enabling β-catenin to accumulate in the cytoplasm. β-catenin translocates to the nucleus, where it acts as a transcriptional coactivator for the T-cell factor (TCF) and lymphoid enhancing factor (LEF) family of DNA-binding transcription factors (18).
Wnt/β-catenin signaling is important in mESCs (19–22). Activation of Wnt/β-catenin signaling by recombinant Wnt3a or by inhibition of GSK3 synergizes with activation of JAK/STAT signaling by recombinant leukemia inhibitory factor (Lif) to promote self-renewal and inhibit spontaneous differentiation (19, 20). Dual inhibition of kinases GSK3 and MEK drives self-renewal and inhibits differentiation of naïve mESCs (2). Moreover, paracrine and autocrine Wnt signaling prevent naïve mESCs from converting into a primed mEpiSC-like state (22).
Whether Wnt/β-catenin signaling plays a similar role in naïve hESCs has not been fully investigated. Notably, naïve hESC lines generated without transgenes (3, 12, 23, 24), including ELF1 and H1-4iLIF hESCs, were derived and subsequently maintained in the presence of GSK3 inhibitors—a condition that may promote Wnt/β-catenin signaling. Recently, we reported that activation of a β-catenin–activated reporter (BAR) is increased when ELF1 hESCs are grown in naïve conditions compared with primed conditions (13). Moreover, the activity of BAR in naïve-state ELF1 hESCs is suppressed by inhibition of Wnt/β-catenin signaling (13), using the small molecules XAV939, which promotes degradation of β-catenin (25), or IWP2, which blocks the secretion of Wnt ligands (26), or by siRNA-mediated knockdown of CTNNB1 (13). We also found that the expression of genes involved in Wnt signaling pathways changes early during the naïve-to-primed transition in hESCs (13). Nevertheless, we did not establish a causal link between Wnt/β-catenin signaling and naïve hESC behaviors, such as self-renewal, differentiation, or preservation of the naïve state.
Here, we show that Wnt/β-catenin signaling promotes self-renewal of naïve hESCs but is dispensable for the maintenance of pluripotency marker expression. Moreover, inhibition of Wnt/β-catenin signaling in naïve hESCs induces a more primed-like global protein expression profile. Taken together, our results implicate Wnt/β-catenin signaling as a positive regulator of human naïve pluripotency.
Results
Naïve hESCs Display Active Wnt/β-Catenin Signaling.
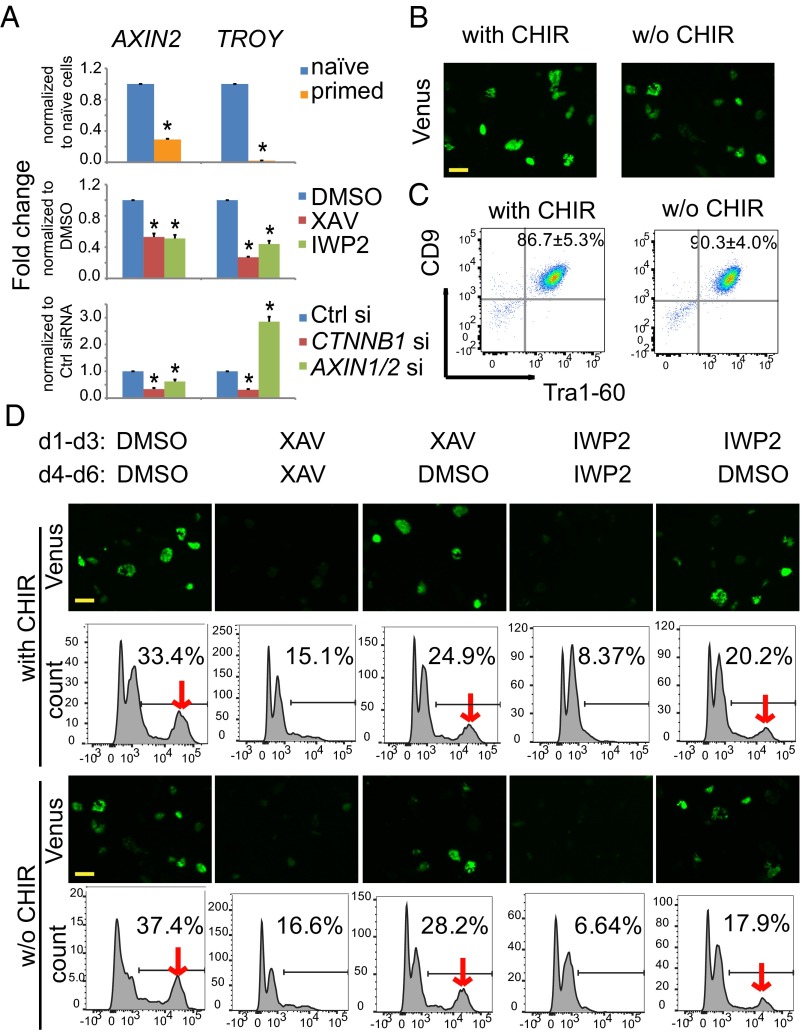
Wnt/β-catenin signaling plays distinct roles in naïve and primed PSCs (20, 27, 28), and we reported that BAR activity is greater in ELF1 hESCs grown in naïve conditions compared with those grown in primed conditions (13). To verify that changes in BAR activity reflect bona fide changes in β-catenin signaling activity in ELF1 hESCs, we compared the expression of mRNA transcripts of the Wnt/β-catenin target genes AXIN2 and TROY (TNFRSF19) using quantitative RT-PCR (qRT-PCR). The abundance of transcripts for both genes was greater in naïve ELF1 hESCs compared with that in primed ELF1 hESCs (Fig. 1A). In addition, inhibition of the Wnt/β-catenin pathway using XAV939, IWP2, or CTNNB1-siRNAs reduced the expression of AXIN2 and TROY (Fig. 1A), indicating that the greater BAR signal in naïve hESCs reflects increased signaling through the Wnt/β-catenin pathway. In contrast, inhibition of the destruction complex by siRNA-mediated knockdown of both AXIN1 and AXIN2 (hereafter referred to as AXIN1/2)—which promotes activation of the Wnt/β-catenin pathway (28)—enhanced both the BAR and TROY expression (Fig. 1A and Fig. S1 A and B). These experiments revealed that Wnt/β-catenin signaling activity is increased in naïve hESCs compared with the same hESC line cultured in the primed state.
Fig. 1.
Naïve hESCs have active endogenous Wnt/β-catenin signaling independent of the presence of CHIR. (A) qRT-PCR results of AXIN2 and TROY expression in naïve (2iLIF) or primed (AF) ELF1 hESCs (Top); naïve ELF1 hESCs cultured in media with vehicle control (DMSO), XAV939 (XAV, 5 µM), or IWP2 (2 µM) for 3 d (Middle); or naïve ELF1 hESCs in media with 2iLIF for 3 d after transfection with negative control (Ctrl) siRNAs, CTNNB1 siRNAs, and AXIN1/2 siRNAs (Bottom). Cells in all conditions were cultured on Matrigel with MEF-CM for 3 d (mean ± SEM of n = 3 biological replicates; *P < 0.05 by t test). (B) Representative fluorescence images of BAR-Venus expression of ELF1-BAR hESCs grown on MEFs in media with CHIR or without CHIR (w/o CHIR) for three passages. (Scale bar, 100 µm.) (C) Representative plots of flow cytometry for Tra1-60 and CD9 expression in ELF1 hESCs cultured in media with or w/o CHIR for three passages (last passage on Matrigel) (mean ± SEM of n = 3 biological replicates; *P < 0.05 by t test). (D) Representative images and flow cytometry histograms of BAR-Venus expression in naïve ELF1-BAR hESCs cultured on MEFs in media with or w/o CHIR and with the indicated small molecules for 3 d. At the end of day 3, cells were lifted and passaged into new wells with MEFs and cultured with the indicated small molecules from days 4–6 (single data point representative of n = 2 biological replicates). (Scale bar, 100 µm.)
Fig. S1.
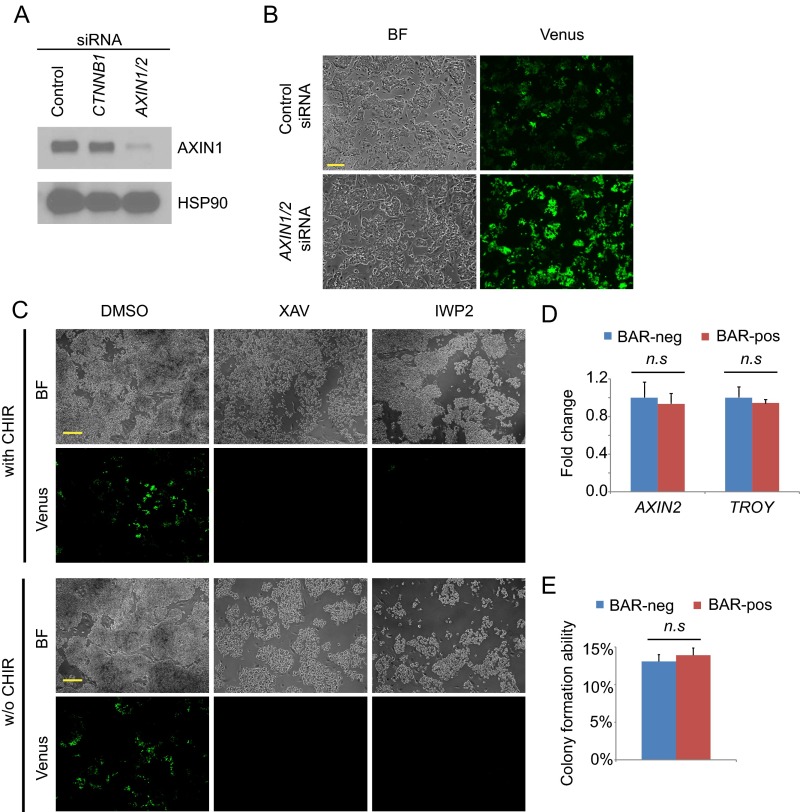
(A) Western blot of AXIN1 after naïve ELF1 hESCs were treated with the indicated small molecules and cultured on Matrigel for 3 d. HSP90 was used as loading control. (B) Representative bright field (BF) and fluorescent (Venus) images of naïve ELF1-BAR hESCs plated on Matrigel and transfected with control (Ctrl) siRNA and AXIN1/2 siRNA, respectively, for 3 d. (Scale bar, 100 µm.) (C) Representative fluorescent images of BAR expression in naïve ELF1-BAR hESCs on Matrigel and treated with the indicated small molecules for 3 d with CHIR (Top) or without (w/o) CHIR (Bottom). (Scale bar, 100µm). (D) qRT-PCR analysis of AXIN2 and TROY expression in FACS-sorted BAR-negative (BAR-neg) and BAR-positive (BAR-pos) ELF1-BAR hESCs. All of the cells were FACS-sorted and then cultured on Matrigel for 3 d. Fold changes were normalized to the value in BAR-neg cells (mean + SEM of n = 3 biological replicates; n.s., not significant). (E) Colony formation ability of BAR-neg and BAR-pos cells, respectively (mean + SEM of n = 3 biological replicates; n.s., not significant).
We found that the BAR reporter signal was heterogeneously expressed among naïve ELF1 hESCs. However, FACS-sorted BAR-positive and BAR-negative cells had comparable amounts of AXIN2 and TROY expression (Fig. S1D) as well as a similar ability to form pluripotent colonies (Fig. S1E). Because epigenetic silencing of reporter transgenes occurs in ESCs (29, 30), these results suggest that the BAR-negative population was likely a result of reporter silencing.
The GSK3β inhibitor CHIR99021 (CHIR) is included in the culture media of all reported naïve hESCs at concentrations ranging from 0.3 to 3 µM (11, 12, 31). For example, naïve ELF1 hESCs are grown in media with 1.5 µM CHIR, 1 µM MEK inhibitor PD0325901, 10 ng/mL Lif, 10 ng/mL IGF1, and 10 ng/mL FGF2 (termed 2iLIF). To test if activation of Wnt/β-catenin signaling in naïve ELF1 hESCs was due to the presence of CHIR, we cultured ELF1 hESCs in 2iLIF without CHIR (termed 1iLIF). We found that the activation of BAR was comparable in ELF1 hESCs for at least three passages when grown in either 2iLIF or 1iLIF (Fig. 1B), suggesting that endogenous Wnt activation may be present in naïve hESCs. Moreover, ELF1 hESCs grown in 2iLIF or 1iLIF had a comparably high percentage of cells expressing the pluripotency markers Tra1-60 and CD9 (Fig. 1C). Incubation with XAV939 or IWP2 for 3 d reduced BAR activity in ELF1 hESCs grown in 2iLIF or 1iLIF, and BAR activity recovered when Wnt inhibitors were subsequently withdrawn (Fig. 1D). Because IWP2 blocks the secretion of Wnts (26), this result indicates that in naïve growth conditions ELF1 hESCs and/or the mouse embryo fibroblast (MEF) feeder cells secrete Wnt ligands that activate autocrine and/or paracrine β-catenin signaling in the absence of exogenous stimuli, such as CHIR.
To test if autocrine Wnt signaling induces BAR activity in naïve ELF1 hESCs, we cultured cells in the absence of MEFs on Matrigel in MEF-conditioned media (MEF-CM). Consistent with our findings in naïve ELF1 hESCs grown on MEFs, BAR activity was comparable in ELF hESCs grown on Matrigel with either 2iLIF or 1iLIF (Fig. S1C). Moreover, adding XAV939 or IWP2 to naïve ELF1 hESCs reduced BAR activity in the absence of MEFs (Fig. S1C), supporting the conclusion that ELF1 hESCs secrete canonical Wnt ligands to activate the endogenous Wnt/β-catenin signaling through an autocrine and/or paracrine mechanism.
Wnt/β-Catenin Signaling Is Not Required for the Expression of Pluripotency Markers in Naïve hESCs.
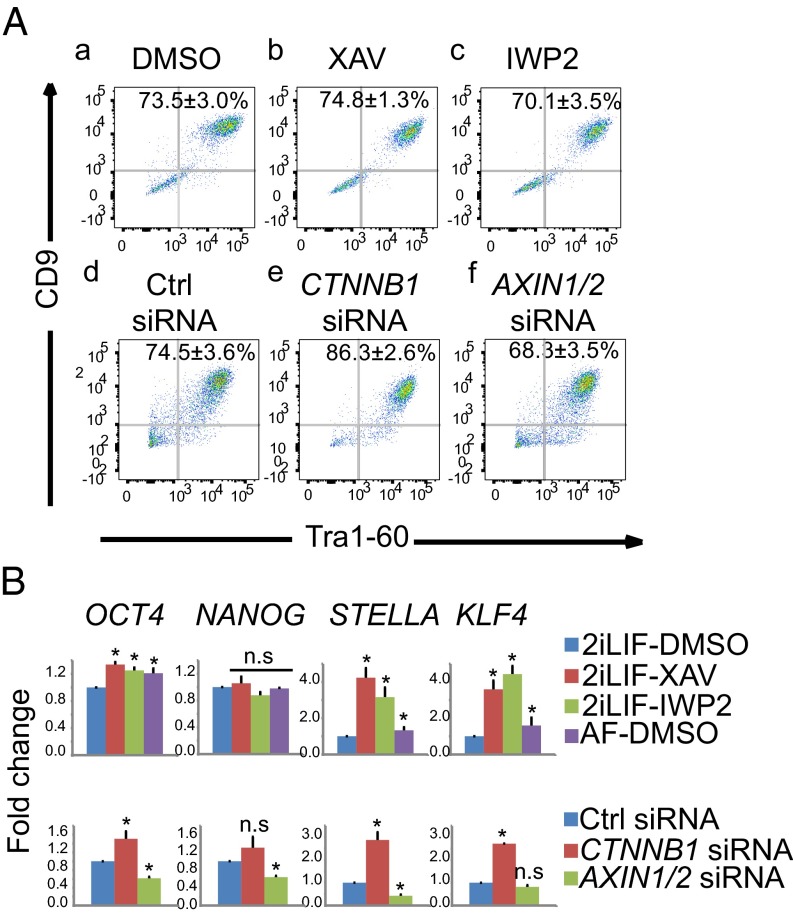
The presence of endogenous Wnt/β-catenin signaling in naïve ELF1 hESCs suggests a potential functional role for Wnt signaling in these cells. Thus, to explore this hypothesis, we first tested whether Wnt/β-catenin signaling was required for maintenance of pluripotency. However, incubation with XAV939 or IWP2 for 3 d did not affect the percent of Tra1-60/CD9 double-positive ELF1 hESCs grown on Matrigel with MEF-CM supplemented with 2iLIF (Fig. 2 A, a–c). Moreover, siRNA-mediated knockdown of β-catenin did not significantly affect the percent of Tra1-60/CD9 double-positive ELF1 hESCs, compared with the negative control siRNAs. Similarly, siRNA-mediated knockdown of AXIN1/2 siRNAs had no significant effect on the percent of Tra1-60/CD9 double-positive ELF1 hESCs (Fig. 2 A, d–f).
Fig. 2.
Wnt/β-catenin signaling is dispensable for maintaining pluripotency in ELF1 hESCs. (A) Representative plots of flow cytometry for Tra1-60 and CD9 in ELF1 hESCs grown on Matrigel and with the indicated small molecules for 3 d or after transfection with the indicated siRNAs for 3 d (mean ± SEM of n = 3 biological replicates; *P < 0.05 by t test). (B) Results of qRT-PCR–based analysis of gene expression in primed ELF1 hESCs (AF-DMSO) or naïve ELF1 hESCs cultured with indicated small molecules or transfected with siRNAs as described for A. Fold changes were normalized to the value of 2iLIF-DMSO (Top) or Ctrl-siRNA (Bottom) (mean + SEM of n = 3 biological replicates; *P < 0.05 by t test; n.s., not significant).
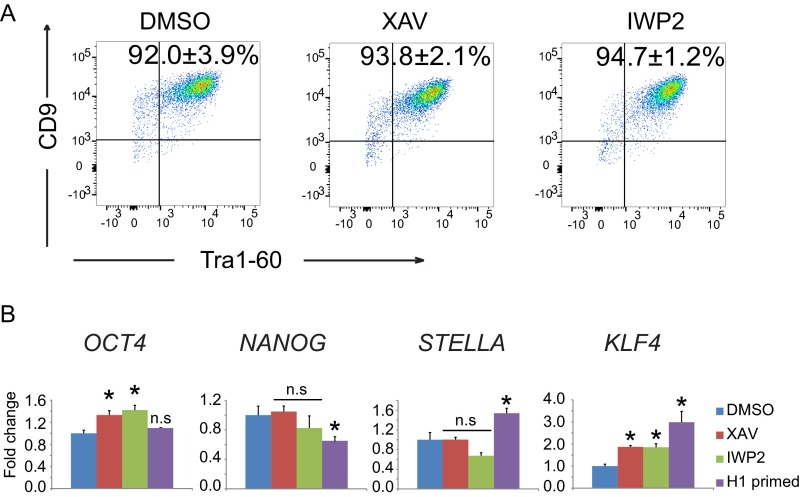
To evaluate the possibility of cell line-specific effects, we tested the role of Wnt/β-catenin signaling in the maintenance of pluripotency in an additional naïve hESC line. The conventional primed hESC line H1 was toggled back to the naïve state and is maintained in 2iLIF supplemented with a JNK inhibitor (SP600125) and a p38 MAPK inhibitor (BIRB796) (collectively termed 4iLIF) to create H1-4iLIF hESCs (12, 13). Incubation of XAV939 or IWP2 did not affect the proportion of Tra1-60/CD9 double-positive H1-4iLIF hESCs (Fig. S2A).
Fig. S2.
Wnt/β-catenin signaling is dispensable for maintaining pluoripotency in H1-4iLIF hESCs. (A) Representative plots of flow cytometry for Tra1-60 and CD9 in H1-4iLIF hESCs grown on Matrigel and with the indicated small molecules for 3 d (mean ± SEM of n = 3 biological replicates). (B) Results of qRT-PCR–based analysis of gene expression in primed H1 hESCs or naïve H1-4iLIF hESCs cultured with the indicated small molecules as described for A. Fold changes were normalized to the value in DMSO-treated samples (mean + SEM of n = 3 biological replicates; *P < 0.05 by t test; n.s., not significant).
To independently assess the role of Wnt/β-catenin signaling in pluripotency of naïve hESCs, we analyzed the expression of a panel of pluripotency-associated genes (OCT4, NANOG, STELLA, and KLF4) in ELF1 and H1-4iLIF hESCs. In ELF1 hESCs grown in 2iLIF, inhibition of Wnt/β-catenin signaling by XAV939, IWP2, or CTNNB1-siRNAs increased the abundance of transcripts for OCT4, STELLA, and KLF4 but not NANOG (Fig. 2B). In H1-4iLIF hESCs, incubation with Wnt inhibitors increased expression of OCT4 and KLF4 but did not affect expression of NANOG and STELLA (Fig. S2B). In contrast, activation of Wnt/β-catenin signaling by AXIN1/2-siRNA transfection modestly decreased the expression of OCT4, STELLA, and NANOG compared with control siRNA transfection in naïve ELF1 hESCs (Fig. 2B).
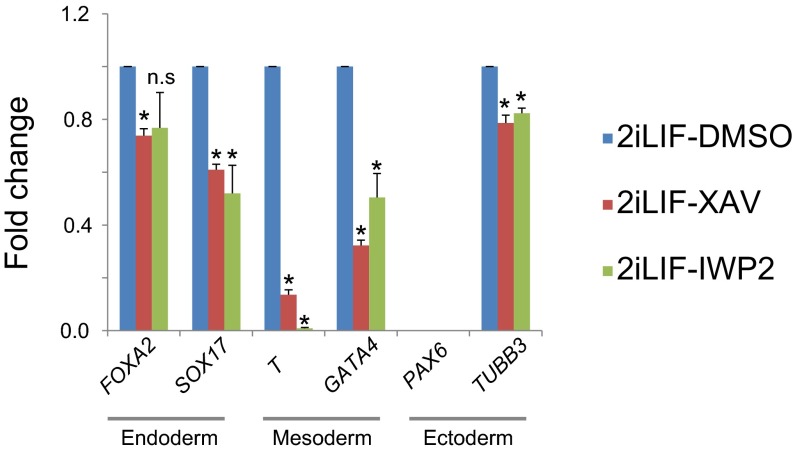
We also tested whether inhibition of Wnt signaling increases expression of markers of lineage differentiation in ELF1 hESCs grown in naïve conditions. We analyzed the expression of genes associated with early stages of differentiation, including markers for endoderm (FOXA2, SOX17), mesoderm (T, GATA4), and ectoderm (PAX6, TUBB3) lineages. We found that incubation with XAV939 or IWP2 did not increase the expression of genes from all three lineages (PAX6 expression was below detection threshold) (Fig. S3).
Fig. S3.
Inhibition of Wnt/β-catenin signaling reduces the gene expression of early differentiation markers. Shown are results of qRT-PCR–based analysis of gene expression in naïve ELF1 hESCs cultured on Matrigel with the indicated small molecules for 3 d. Fold changes were normalized to the value in DMSO-treated samples (mean + SEM of n = 3 biological replicates; *P < 0.05 by t test; n.s., not significant; expression of PAX6 is under the detection threshold).
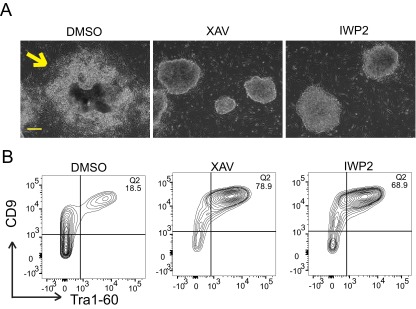
Finally, we tested whether sustained inhibition of Wnt/β-catenin signaling affects spontaneous differentiation of ELF1 hESCs. ELF1 cells are passaged at a split ratio of 1:10 every 3–4 d for optimal self-renewal of undifferentiated cells. If ELF1 hESCs are not passaged or the density becomes too high, even if they were maintained in 2iLIF, cells spontaneously differentiate rapidly, which is indicated by overt morphological changes in colonies such as uneven edges and loss of Tra1-60/CD9 expression. We grew ELF1 hESCs in 2iLIF on MEFs with or without XAV939 or IWP2 for 7 d with daily media changes. Whereas ELF1 hESCs incubated with the vehicle control (DMSO) underwent dramatic spontaneous differentiation (Fig. S4A), colonies from ELF1 hESCs incubated with XAV939 or IWP2 exhibited mostly undifferentiated morphology with well-defined smooth edges and had a significantly higher percent of Tra1-60/CD9 double-positive cells (Fig. S4 A and B).
Fig. S4.
Inhibition of Wnt/β-catenin signaling does not lead to spontaneous differentiation in ELF1 hESCs. (A) Representative bright field images of ELF1 hESCs after they were cultured on MEF for 7 d in the indicated small molecules. Yellow arrow indicates spontaneous differentiation. (Scale bar, 100 µm.) (B) Representative flow cytometry plots of the Tra1-60+/CD9+ population of ELF1 hESCs treated as described in A of two biological replicates.
Collectively, these results revealed that inhibiting Wnt/β-catenin signaling in hESCs grown in naïve conditions does not decrease pluripotency marker expressions or induce differentiation.
Wnt/β-Catenin Signaling Is Required for the Self-Renewal of Naïve hESCs.
Self-renewal of PSCs enables cells to proliferate while remaining pluripotent. We found that Wnt/β-catenin signaling was not required to maintain pluripotency marker expression (Fig. 2); thus, we tested whether Wnt/β-catenin signaling plays a role in the proliferation of naïve hESCs.
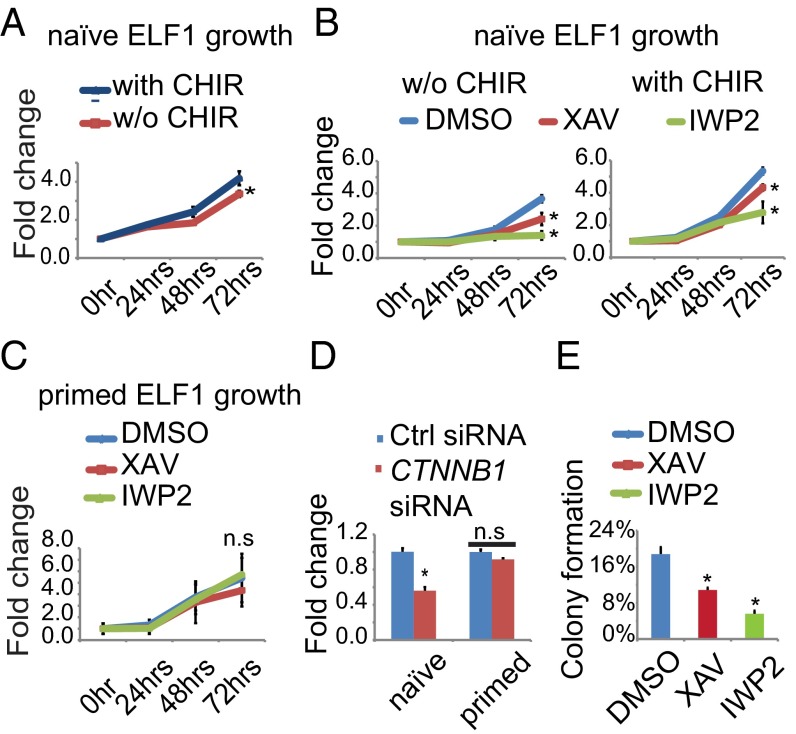
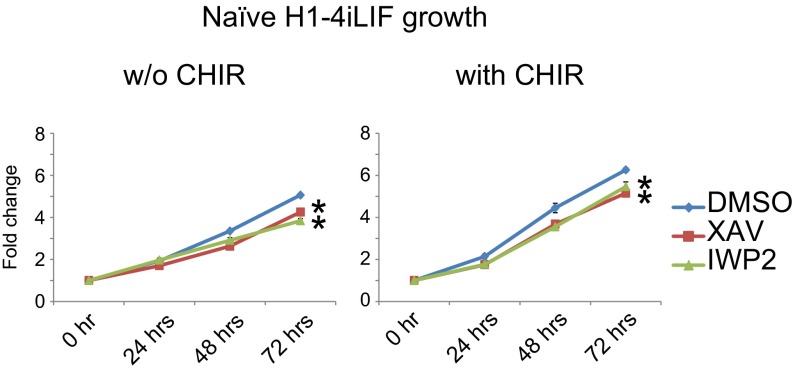
We counted the number of ELF1 hESCs grown in naïve conditions in the presence (2iLIF) or absence (1iLIF) of CHIR. ELF1 hESCs previously maintained on MEFs were subcultured on Matrigel with MEF-CM for the duration of the experiment to minimize the contribution of MEFs to the cell counts. Removing CHIR from the culture media reduced the number of ELF1 hESCs after as little as 3 d (Fig. 3A). Moreover, incubation with XAV939 or IWP2 reduced the number of ELF1 hESCs grown in 1iLIF or 2iLIF (Fig. 3B). Similarly, we observed that incubation with Wnt inhibitors decreased the number of H1-4iLIF hESCs grown with or without CHIR (Fig. S5).
Fig. 3.
Wnt/β-catenin activity is important for efficient self-renewal of naïve hESCs. (A–C) Graphs of the relative number of naïve (A and B) or primed (C) ELF1 hESCs grown on Matrigel with the indicated small molecules. Cells were allowed to attach for 16–20 h before the addition of DMSO, XAV, or IWP2. The media was changed daily. Cell number was measured using the CyQUANT assay at specified times. Fold changes were calculated relative to the cell number at the time the small molecules were first added (defined as 0 h). (D) Graph of the relative number of naïve or primed ELF1 hESCs transfected with the indicated siRNAs and grown on Matrigel for 3 d. Cell number was measured using CyQUANT assay and normalized to the value for Ctrl siRNA-transfected cells. (E) Graph of the relative colony formation potential of naïve ELF1 hESCs. Cells were cultured on Matrigel and with the indicated small molecules for 3 d, then lifted and plated onto MEFs at clonal density (250 cells per cm2), and cultured with the indicated small molecules for 3 more days. Colonies were subsequently stained for AP activity, and the number of AP-positive colonies was manually counted. The percentage is calculated as the number of colonies divided by the number of cells seeded (all of the data are means, and error bars represent S.E.M of n = 3 biological replicates; *P < 0.05 by t test; n.s., not significant).
Fig. S5.
Wnt/β-catenin activity is important for the self-renewal of naïve H1-4iLIF hESCs. Shown are graphs of the relative number of H1-4iLIF hESCs grown on Matrigel with or without (w/o) CHIR and with the indicated small molecules. Cells were allowed to attach for 16–20 h before the addition of DMSO, XAV, or IWP2. The media was changed daily. Cell number was measured using the CyQUANT assay at specified times. Fold changes were calculated relative to the cell number at the time the small molecules were first added (defined as 0 h) (mean ± SEM of n = 3 biological replicates; *P < 0.05 by t test).
We also tested if the reduction in cell numbers due to inhibition of Wnt/β-catenin signaling was unique to naïve-state hESCs. ELF1 hESCs can be toggled to the primed state by switching to culture media containing Activin A and FGF2 (termed AF conditions) (12). In contrast to ELF1 hESCs grown in 2iLIF, the number of ELF1 hESCs grown in AF was not affected by XAV939 or IWP2 (Fig. 3C). In addition, siRNA-mediated knockdown of CTNNB1 decreased the number of ELF1 hESCs grown in 2iLIF but not AF conditions (Fig. 3D).
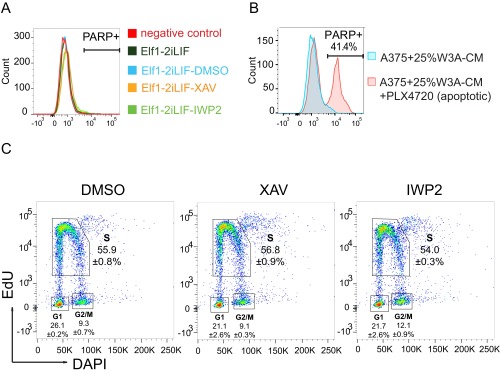
Reductions in cell number over time in culture could reflect increased cell death and/or decreased cell proliferation. However, the number of ELF1 hESCs positive for cleaved PARP, a marker for caspase-induced apoptosis, was not increased by incubation with XAV939 or IWP2 (Fig. S6 A and B). Moreover, cell-cycle analysis revealed that the proportion of ELF1 hESCs in G1, S, and G2/M phases was not affected upon Wnt inhibition (Fig. S6C). These data suggest that inhibition of Wnt/β-catenin signaling does not induce apoptosis nor affect any specific stages of the cell cycle but likely prolongs the overall cell-cycle transit time and thereby decreases the rate of cell proliferation in naïve ELF1 hESCs.
Fig. S6.
Wnt inhibition does not induce apoptosis or affect specific stages of the cell cycle in naïve ELF1 hESCs. (A) Representative flow cytometry histogram of the PARP+ population in naïve ELF1 hESCs grown on Matrigel and with the indicated small molecules for 3 d. (B) Flow cytometry histogram of the PARP+ population in A375 cells cultured with 25% (vol/vol) Wnt3a-CM (W3A-CM). Addition of PLX4720 was used as a positive control for cleaved PARP in the same experiment. (C) Representative flow cytometry plots of the EdU assay for cell-cycle analysis of ELF1 hESCs grown on Matrigel with the indicated small molecules for 3 d (mean ± SEM of n = 2 biological replicates).
Colony formation from single cells (clonogenicity, or also called colony forming ability) is commonly used as a measure of PSC self-renewal (22). We performed colony formation assays using ELF1 hESCs and stained colonies for alkaline phosphatase (AP) activity, which marks undifferentiated PSCs (32). We found that inhibition of Wnt signaling by XAV939 or IWP2 reduced the number of AP-positive colonies formed from ELF1 hESCs grown in 2iLIF (Fig. 3E).
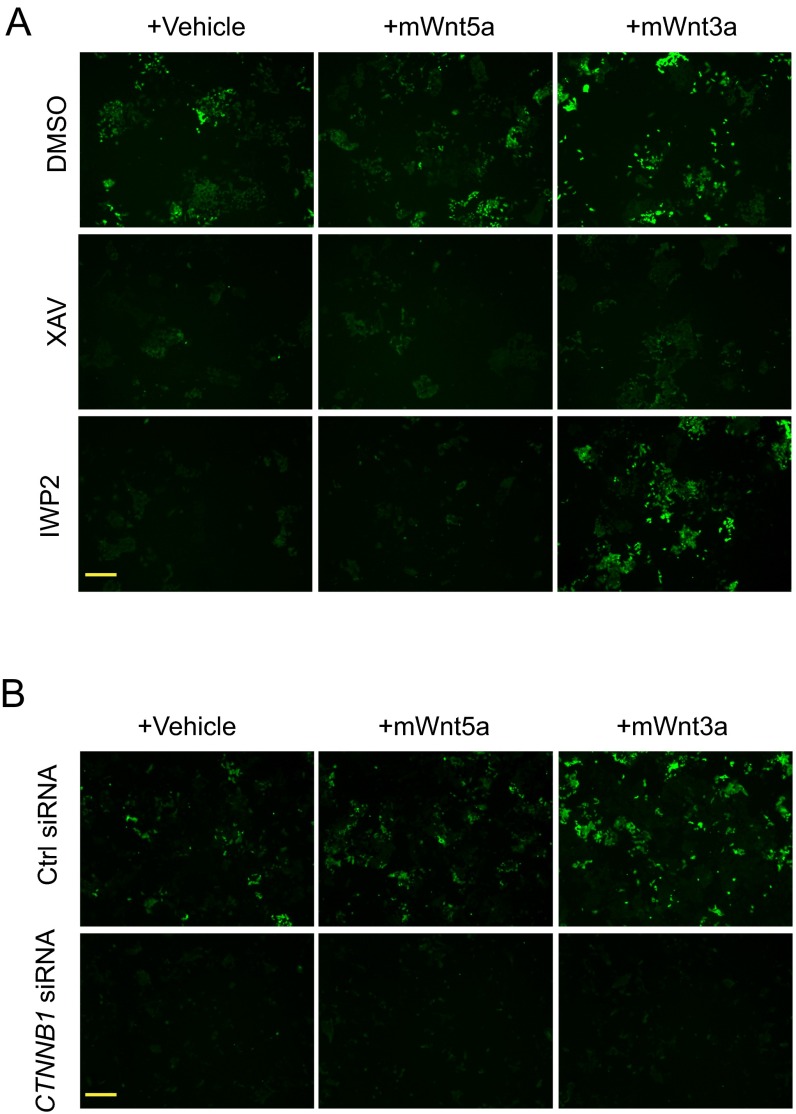
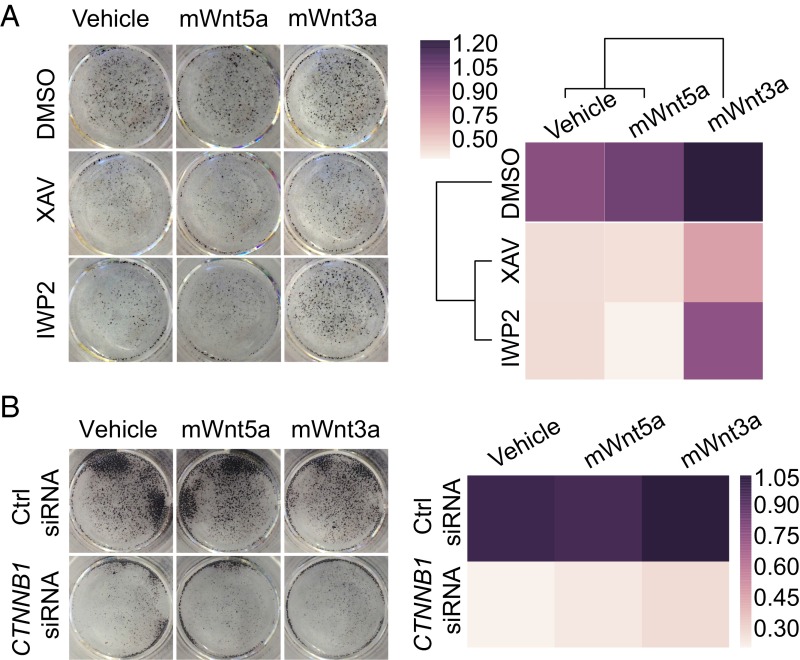
If the inhibition of cell proliferation in naïve hESCs by IWP2 is due to inhibition of endogenous Wnt/β-catenin signaling, then restoring pathway activation should increase cell numbers in the presence of IWP2. To test this hypothesis, we compared the effects of recombinant mouse Wnt3a protein (mWnt3a), which activates canonical Wnt signaling (33), and recombinant mouse Wnt5A protein (mWnt5A), which generally does not activate Wnt/β-catenin signaling (34). We verified that mWnt3a, but not mWnt5A, increased BAR activity in ELF1 hESCs grown in 2iLIF in the presence of IWP2, which blocks Wnt secretion, but not XAV939, which acts downstream of Wnt receptor activation (25) (Fig. S7A). Moreover, similar to our observations using colony formation assays, incubation with XAV939 or IWP2 reduced the number of AP-positive ELF1 hESCs grown in 2iLIF in a nonclonogenic format (Fig. 4A). Importantly, mWnt3A markedly increased the number of AP-positive naïve ELF1 hESCs in the presence of IWP2 (Fig. 4A). Moreover, mWnt3A slightly increased the number of AP-positive ELF1 hESCs in the presence of XAV939 (Fig. 4A), an effect that might be due to incomplete inhibition of Wnt/β-catenin signaling by XAV939 or to β-catenin–independent effects of mWnt3A (35, 36). Exogenous mWnt3a did not increase the number of AP-positive ELF1 hESCs transfected with CTNNB1 siRNAs (Fig. 4B and Fig. S7B). Thus, ligand-mediated Wnt/β-catenin signaling is critical for the self-renewal of naïve ELF1 hESCs.
Fig. S7.
Effects of Wnt inhibitors and recombinant Wnt ligands. Shown are representative fluorescent images of BAR expression in ELF1-BAR hESCs cultured on Matrigel and (A) treated as in Fig. 4A or (B) treated as in Fig. 4B (n = 3 biological replicates). (Scale bar, 100 µm.)
Fig. 4.
Exogenous activation of Wnt/β-catenin signaling by recombinant Wnt3a partially restores self-renewal in naïve hESCs incubated with Wnt inhibitors. (A and B) Representative images of naïve ELF1 hESCs grown on Matrigel for 3 d (A) in the presence of the indicated small molecules or (B) transfected with the indicated siRNAs. Cells were plated 20 h before the addition of small molecules and vehicle control (1% CHAPS) or murine recombinant Wnt3a (20 ng/mL) or Wnt5a (20 ng/mL). The media was changed daily. Colonies were subsequently stained for AP activity, and the amount of AP-positive area was measured using ImageJ and normalized to the value of the DMSO-Vehicle (in A) or Ctrl-Vehicle (in B) (normalized fold change). The heat maps represent the average normalized fold change in AP-positive area across n = 3 biological replicates. The heat map in A was organized by unsupervised hierarchical clustering.
Inhibition of Wnt/β-Catenin Signaling Induces Naïve hESCs to Transition Toward a Primed-Like State.
Upon Wnt inhibition in naïve hESCs, cells exhibit continued expression of stem cell markers and lack of expression of differentiation markers, a slower rate of proliferation, as well as reduced clonogenic self-renewal. These differences are reminiscent of the molecular and cellular changes that occur during the naïve-to-primed state transition; therefore, we asked whether Wnt inhibition induced naïve hESCs to a more primed-like state.
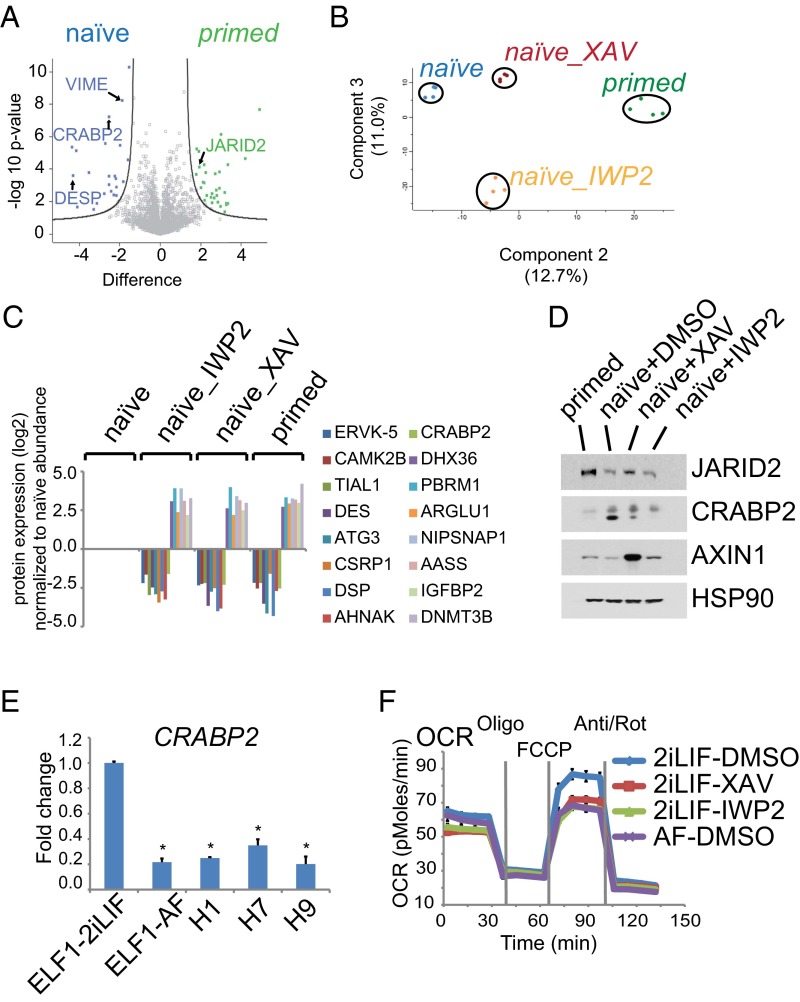
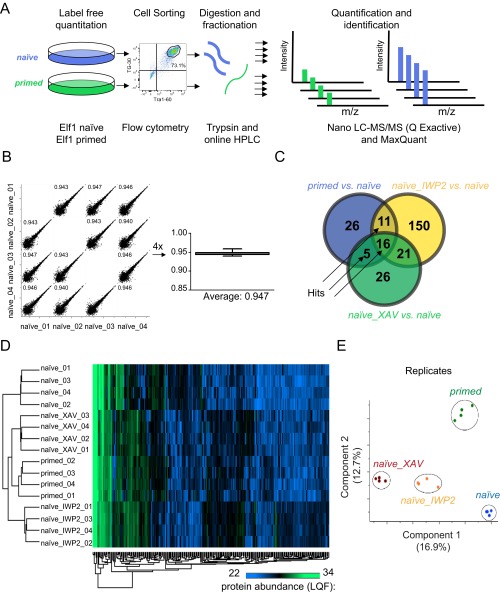
To address this question, we characterized the proteomic changes induced by inhibition of Wnt/β-catenin signaling in ELF1 hESCs grown in naïve (2iLIF) conditions and compared those to the molecular differences between ELF1 hESCs grown in naïve or primed (AF) conditions. We assayed the protein expression profile of sorted Tra1-60/CD9 double-positive ELF1 hESCs using mass spectrometry (Fig. 5A and Fig. S8A). We identified a total of 2,930 proteins from primed ELF1 hESCs and naïve ELF1 hESCs treated with DMSO, XAV939, or IWP2 for 7 d (Dataset S1). Quantification of relative protein abundances was reproducible across two biological replicates (each with two technical replicates) (Fig. S8B). Among these proteins, we found 58 proteins that were differentially expressed between naïve and primed ELF1 hESCs (Dataset S2). We also found 198 or 68 proteins that were differentially expressed between naïve ELF1 hESCs treated with DMSO and IWP2 or XAV939, respectively (Fig. S8C and Datasets S3 and S4). Principle components analysis of these differentially expressed proteins revealed that the protein profiles of naïve ELF1 hESCs with inhibition of Wnt signaling clustered closer to the protein profiles of primed rather than naïve ELF1 hESCs (Fig. 5B and Fig. S8 D and E). Moreover, among the differentially expressed proteins in naïve and primed ELF1 hESCs, 16 proteins had expression changes in naïve ELF1 hESCs incubated with XAV939 and IWP2 (Fig. S8C and Dataset S5). Interestingly, inhibition of Wnt signaling in naïve ELF1 hESCs led to protein expression patterns that mimicked the expression patterns of primed ELF1 hESCs (Fig. 5C), consistent with the principle component analysis.
Fig. 5.
Inhibition of Wnt signaling in naïve ELF1 hESCs promotes primed-like protein expression and metabolic profiles. (A) Volcano plot of differentially expressed proteins in primed and naïve ELF1 hESCs. Proteins were quantified by nano liquid chromatography tandem mass spectrometry (nano-LC-MS/MS). The values on the x axis are the average difference (primed-naïve) of log2-transformed relative protein abundances for n = 2 biological replicates. (B) Principal component (PC) analysis of MS-based global protein expression profiles in ELF1 hESCs. (C) Relative expression of proteins that were differentially expressed in naïve ELF1 hESCs compared with primed ELF1 hESCs and naïve ELF1 hESCs incubated with XAV939 or IWP2. The data were quantified by MS as in A and normalized to the value in naïve ELF1 hESCs. (D) Western blot validation of MS results for JARID2 and CRABP2 in ELF1 hESCs. AXIN1 is stabilized by XAV939. HSP90 was used as a loading control. The blot is representative of n = 2 biological replicates. (E) Results of qRT-PCR–based analysis of CRABP2 expression in naïve (ELF1-2iLIF) and primed (ELF1-AF, H1, H7, and H9) hESC lines (mean + SEM of n = 3 biological replicates normalized to the value in ELF1-2iLIF hESCs; *P < 0.05 by t test). (F) Plot of OCR measured using the Seahorse assay in naïve (2iLIF) or primed (AF) ELF1 hESCs grown on Matrigel with the indicated small molecules for 3 d. The data are the mean ± SEM of n = 5 technical replicates. The plot is representative of n = 3 biological replicates. Ant/Rot, Antimycin and Rotenone; FCCP, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone; Oligo, oligomycin.
Fig. S8.
Proteomic analysis of ELF1 hESCs. (A) Example experimental workflow used to identify differentially regulated protein expression in different samples. (B) Label-free quantification of protein expression is reproducible (average Pearson’s correlation of 0.947; n = 2 biological replicates, each with two technical replicates). (C) Statistics of the ELF1 proteomic analysis. Hits: Wnt/β-catenin signaling regulated naïve or primed ELF1 enriched proteins (n = 2 biological replicates, each with two technical replicates). (D) Unsupervised hierarchical clustering of protein expression profiles in naïve, primed, and naïve cells treated with XAV939 or IWP2 (relative protein expression: green, high; black, medium; blue, low). (E) Principle component analysis using component 1 and component 2. Blue, ELF1 naïve; green, ELF1 primed; yellow, ELF1 naïve cells treated with IWP2 (naïve_IWP2); brown, ELF1 naïve cells treated with XAV939 (naïve_XAV).
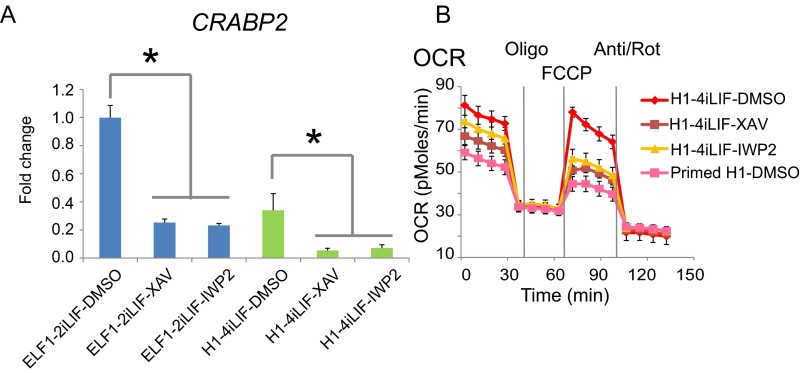
We independently tested the mass spectrometry-based results using additional assays. Western blot for two of the differentially expressed proteins, CRABP2 and JARID2, confirmed the changes in protein expression among primed ELF1 hESCs and naïve ELF1 hESCs incubated with DMSO, XAV939, or IWP2 (Fig. 5D). Moreover, analysis of gene expression by qRT-PCR showed that CRABP2 mRNA expression was higher in naïve ELF1 hESCs compared with that in multiple primed-state hESC lines (Fig. 5E). Furthermore, Wnt inhibition reduced CRABP2 expression in both naïve ELF1 and H1-4iLIF hESCs (Fig. S9A). Thus, inhibition of Wnt signaling in naïve hESCs promotes a more primed-like protein expression profile.
Fig. S9.
Wnt inhibition leads to primed-like changes in naïve hESCs. (A) Results of qRT-PCR–based analysis of CRABP2 expression in ELF1-2iLIF and H1-4iLIF hESCs grown on Matrigel and treated with the indicated small molecules for 3 d. Fold changes were normalized to the value in ELF1-2iLIF-DMSO. For ELF1-2iLIF hESCs, data represent mean + SEM of n = 2 biological replicates. For H1-4iLIF hESCs, data represent mean + SEM of n = 3 biological replicates (*P < 0.05 by t test). (B) Plot of OCR measured using the Seahorse assay in H1-4iLIF hESCs grown on Matrigel with the indicated small molecules for 3 d. The data are the mean ± SEM of n = 5 technical replicates. The plot is representative of n = 3 biological replicates. Ant/Rot, Antimycin and Rotenone; FCCP, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone; Oligo, oligomycin.
We previously reported that both mouse and human naïve and primed PSCs have differences in mitochondrial respiration. Specifically, naïve PSCs exhibit a greater maximal mitochondrial activity, measured by the increase of oxygen consumption rate (OCR) due to 4-(trifluoromethoxy)phenylhydrazone (FCCP), than primed PSCs (13, 37). Thus, we tested if inhibition of Wnt signaling affects the OCR of naïve ELF1 and H1-4iLIF hESCs. We found that in both cell lines, XAV939 or IWP2 decreased the maximal OCR change, causing the mitochondrial respiration to resemble a more primed hESC-like state (Fig. 5F and Fig. S9B). Therefore, inhibition of Wnt/β-catenin signaling may facilitate the ability of naïve hESCs to transition to a more metabolically primed hESC-like state.
Discussion
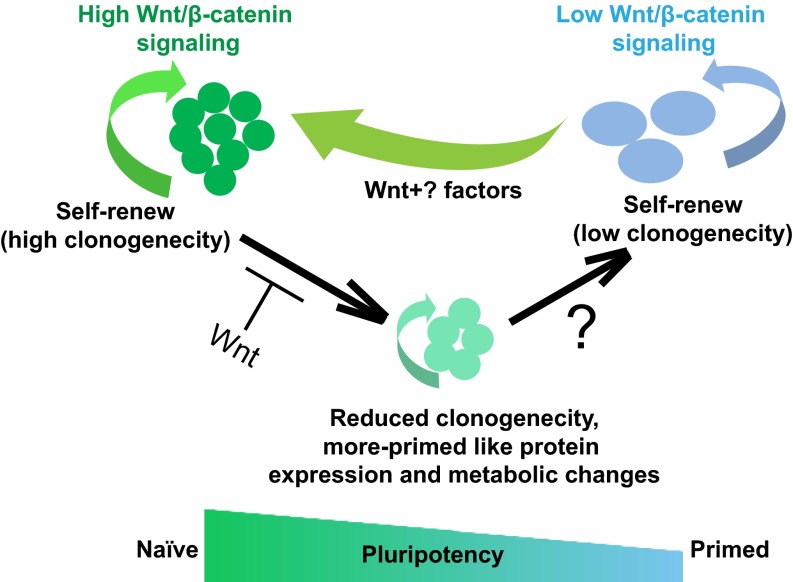
We are interested in understanding the molecular mechanisms of pluripotency and self-renewal in naïve hESCs. In this study, we showed that Wnt/β-catenin signaling is an important factor during self-renewal of naïve hESCs but is not required for the expression of pluripotency markers. Furthermore, we found that inhibition of Wnt/β-catenin signaling induces protein and metabolic changes associated with the naïve-to-primed state transition of hESCs. A model summarizing these findings is included in Fig. S10.
Fig. S10.
A model depicting the role of Wnt/β-catenin signaling in naïve-state hESCs. Cells remain pluripotent following inhibition of Wnt signaling but have decreased self-renewal ability and transitioned to a more primed-like intermediate state.
We found that naïve hESCs cultured on MEFs have increased activity of Wnt/β-catenin signaling compared with primed hESCs and, interestingly, that this signaling remained active after removal of CHIR (a potential source of exogenous Wnt signaling activation) from the media, suggesting that naïve hESCs and/or MEFs secrete Wnt ligands to maintain the endogenous Wnt/β-catenin signaling activity. Importantly, we also found that adding the small molecule IWP2, which blocks lipid modification of Wnts required for secretion, inhibited Wnt/β-catenin activity in the presence or absence of CHIR in naïve ELF1 hESCs grown without MEFs, indicating that Wnt ligands are secreted by hESCs rather than or in addition to supporting feeder cells. Previous reports have shown that mESCs and MEFs secreted multiple Wnt ligands (19, 22), and RNA-seq analysis suggests that the expression of mRNAs encoding Wnts are increased in naïve hESCs compared with primed hESCs (13). Our results provide clear evidence that naïve hESCs have autocrine canonical Wnt ligands that support activation of β-catenin, suggesting that autocrine Wnt/β-catenin signaling may be a conserved feature between naïve-state hESCs and mESCs. It will be important in future experiments to address the particular spectrum of secreted Wnt ligands produced by naïve hESCs, as different Wnts can alternatively activate both β-catenin–dependent and β-catenin–independent signaling pathways (38).
Stem cell self-renewal is defined as proliferation of pluripotent cells. We found that endogenous Wnt/β-catenin signaling in naïve hESCs stimulates the self-renewal by promoting proliferation and is not required for maintaining pluripotency. This finding revealed a conserved role of Wnt/β-catenin signaling in promoting self-renewal in both naïve mouse and human PSCs. Importantly, our data imply that Wnt/β-catenin signaling increases the overall cell-cycle transit time during proliferation, which is a characteristic that differs between naïve and primed hESCs (12). Therefore, our finding suggests potential directions for future work that may shed light on the establishment of naïve pluripotency, such as studying molecular networks associated with Wnt signaling that regulate the cell cycle in naïve hESCs.
We also found that stimulation with recombinant Wnt3a, but not Wnt5a, efficiently rescued the defects of naïve ELF1 hESC self-renewal with IWP2 treatment, but not in cells lacking β-catenin. This implies that canonical Wnt ligands are sufficient to partially recover self-renewal, and noncanonical Wnt ligands—if also being secreted by naïve hESCs—are likely not major players in regulating naïve cell self-renewal.
In mouse PSCs, cells in different pluripotent states possess distinctive gene expression profiles (39). For example, STELLA and KLF4 are associated with the naïve pluripotency network in mouse cells (40, 41), their expression is greater in mESCs relative to mEpiSCs (22, 41), and they are used as markers of the naïve state in mouse PSCs (7, 24). However, we found that STELLA and KLF4 expression was not increased in naïve relative to primed hESCs (Fig. 2B and Fig. S2). This result highlights the complexity of naïve-state pluripotency in mammalian PSCs. Despite some conserved features between naïve mESCs and hESCs (e.g., morphology, Lif dependency, increased mitochondria respiration), it is likely that naïve hESCs possess distinct transcription factor circuitries to maintain the naïve-state pluripotency. We identified differentially expressed proteins that may serve as markers of human naïve and primed states, such as CRABP2 and JARID2. Intriguingly, CRABP2 was expressed at a lower level across multiple primed hESC lines compared with that of the naïve ELF1 cells, and CRABP2 protein expression was reduced by inhibition of Wnt/β-catenin signaling. Thus, Wnt/β-catenin–mediated regulation of CRABP2 expression and the role of this and other proteins identified in our study merit further investigation.
Homogenous primed-state mEpiSC lines can be more efficiently established in the presence of Wnt inhibitors, such as IWP2 (42) and XAV939 (43). Our mass spectrometry-based analysis of protein expression profiles of naïve ELF1 hESCs with inhibition of Wnt/β-catenin signaling also suggests that blocking of Wnt/β-catenin signaling could lead naïve hESCs to a more primed-like state (Fig. 5B). Moreover, profiling of differences in ATP metabolism also supports the conclusion that inhibition of Wnt/β-catenin signaling in naïve hESCs causes a primed-state cell behavior (Fig. 5F). Together these results indicate that Wnt/β-catenin signaling may be an important player during naïve-to-primed transition in human PSCs. Notably, Wnt inhibition alone was not sufficient to complete the naïve-to-primed transition. Naïve ELF1 hESCs incubated with Wnt inhibitors exhibited some protein expression patterns that were primed-like and others that were distinct from both naïve and primed hESCs. Thus, other pathways in addition to the Wnt/β-catenin signaling pathway are likely necessary to toggle hESCs between naïve and primed states in a multipathway/multistep manner.
Materials and Methods
Cell Culture.
Naïve ELF1 hESCs were cultured in hESC Knockout Serum Replacement (KSR) medium [DMEM/F12 with 20% (vol/vol) KSR, 0.1 mM nonessential amino acid (NEAA) solution, 2.0 mM l-Glutamine, 1.0 mM sodium pyruvate, 1% Penicillin–Streptomycin solution, 0.1 mM β-ME] with freshly supplemented LIF (10 ng/mL, Invitrogen), PD 0325901 (1 µM, Tocris), CHIR99021(1.5 µM, AxonMedChem), IGF(10 ng/mL, Peprotech), and FGF2 (10 ng/mL, Invitrogen). Naïve H1-4iLIF hESCs were cultured in KSR medium and supplemented with 1 µM MEK inhibitor PD0325901, 1 µM GSK3 inhibitor CHIR99021, 5 µM JNK inhibitor SP600125, 2 µM p38 inhibitor BIRB796, 10 ng/mL human LIF, 5 ng/mL IGF, and 10 ng/mL bFGF. Medium was changed every other day. Naïve cultures were passaged with TrypLE Express (Invitrogen) every 3 or 4 d. Primed ELF1 hESCs were cultured in KSR medium freshly supplemented with FGF2 (8 ng/mL) and Activin A (10 ng/mL) with media changed every day. Primed H1 hESCs were cultured in KSR medium freshly supplemented with FGF2 (8 ng/mL) with media changed every day. Primed cultures were passaged with dispase every 7 d. Both naïve and primed hESCs were grown on irradiated MEF feeders (except where indicated) in 5% (vol/vol) O2 hypoxia condition. In some experiments, where indicated cells were grown on Matrigel (coating concentration 0.035 g/cm2, BD #354234) in MEF-CM with the supplements described above. Production of MEF-CM and generation of naïve and primed BAR ELF1 hESCs was previously described in Sperber et al. (13). Unless specified, naïve ELF1 and H1-4iLIF were cultured in media with all of the supplements including CHIR during experiments.
Antibody Labeling and Flow Cytometry.
Naïve ELF1 or H1-4iLIF hESCs were harvested as single cells via TrypLE Express and counted using a NucleoCounter (New Brunswick Scientific). After one wash with DPBS, live unfixed cells (200,000–500,000 cells per tube) were pelleted at 300 RCF (relative centrifugal force) and then resuspended to be incubated with 100 µL diluted primary antibodies Tra1-60 (clone Tra-1–60, 1:100, Millipore MAB4360) and CD9 (clone TG30, 1:100, Millipore MAB4427) at 4 °C for 30 min in blocking buffer (KSR). Cells were then washed with PBS once and incubated with isotype-specific secondary antibodies goat anti-mouse IgM-Alexa 488 (1:1,000) and goat anti-mouse IgG2a-Alexa 647 (1:1,000) (all secondary antibodies were from Invitrogen) at 4 °C for 10 min. After secondary incubation, cells were washed with PBS one time and resuspended in PBS and transferred through a cell strainer before analyzing on a BD FACS Canto II. Results were analyzed using FlowJo software.
Apoptosis Assay.
Naïve ELF1 hESCs were harvested as single cells via TrypLE Express and the culture media right before the assays were collected and pooled with cell suspension. All of the live and dead cells were pelleted at 300 RCF for 3 min at 4 °C. Cells were fixed with 4% (wt/vol) PFA for 10 min at 37 °C and then chilled on ice for 1 min. Pelleted cells were resuspended in cold 90% (vol/vol) methanol and incubated on ice for 30 min. Cells were washed with blocking buffer (1×PBS +0.5% BSA) two times and then blocked for 10 min at room temperature (RT). Cells were incubated with cleaved PARP antibody (1:50, Cell Signaling #6987S) for 60 min at RT in the dark. A375 cells treated overnight with 25% (vol/vol) Wnt3a-CM plus 2 µM PLX4720 (Symansis, cat. no. SY-PLX4720) were used as positive control (44). After secondary incubation, cells were washed with PBS one time and resuspended in PBS and transferred through a cell strainer before analyzing on a BD FACS Canto II. Results were analyzed using FlowJo software.
EdU Assay for Cell-Cycle Analysis.
Naïve ELF1 hESCs were plated onto Matrigel-coated tissue culture plates and treated as desired and cultured for 3 d. Two hours before the assay started, EdU (10 mM stock, 1:1,000) was added to the culture media and incubated. After the incubation, cells were harvested as single cells via TrypLE Express and were fixed with 4% PFA for 15 min at RT and then washed with PBS + 5% (vol/vol) FBS once. Cells were then permeabilized with 0.1% Triton-100 for 5 min at RT and washed with PBS + 5% FBS twice. Next, the Click-iT mixture was prepared following the manufacturer’s instruction (Molecular Probes cat. no. C10340), and the cells were incubated with the freshly made Click-iT mixture solution for 30 min in the dark at RT. After incubation, the cells were washed once with PBS + 5% FBS, and DAPI (1 µg/mL) was added and incubated for 15 min in the dark at RT. The cells were washed again with PBS + 5% FBS and resuspended in PBS and transferred through a cell strainer for flow analysis on a BD FACS Canto II. Results were analyzed using FlowJo software.
siRNA Transfection.
Naïve ELF1 hESCs were lifted with TrypLE Express as single cells and counted on a NucleoCounter. The required number of cells were used to mix with the transfection reagent and then seeded onto Matrigel-coated plates in MEF-CM supplemented with 10 µM Y-27632 and 1 µM Thiazovivin (Tocris). Typically, for one well in a 12-well plate (4 cm2 surface area), we seeded 100,000 cells and used 3 µL RNAiMAX (Invitrogen) plus 0.5 µL of 20 µM siRNA stock to make the transfection reagent mix according to the manufacturer’s instructions. Cells were allowed to attach overnight and then received daily media changes for the duration of the experiment. Cells were harvested for analysis at 72 h after transfection. Silencer Select siRNAs were purchased from Invitrogen: negative control (4390843), AXIN1 (s15814), AXIN2 (s15818), and CTNNB1 (s437).
Recombinant Wnt3a, Wnt5a, and Inhibitors.
Recombinant Wnt3a (Peprotech #315–20) and Wnt5a (R&D Systems #645-WN-010/CF) were reconstituted at 100 µg/mL in a final concentration of 1% Chaps in PBS, and bioactivity was confirmed for each lot in 293T-BAR or A375-BAR cells. XAV939 (Tocris), IWP2 (Tocris), and CHIR99021 (AxonMedChem) were reconstituted in DMSO. All of the experiments used 5 µM XAV939 and 2 µM IWP2.
AP Assay.
Black Alkaline Phosphatase Substrate Kit II (Vector Labortories, SK-5200) was used. Cells were washed briefly with PBS and fixed with 70% (vol/vol) EtOH for 10 min at RT. Substrate solution was prepared by mixing 4.5 mL water, 0.5 mL 1 M Tris (pH 9.5), two drops of reagent 1, two drops of reagent 2, and two drops of reagent 3 (volume was scaled as needed). EtOH was aspirated, and the cells were rinsed once with water. Approximately 0.8 mL of substrate solution was added per well of a 12-well plate and incubated at RT for 30 min. Cells were washed once with water and imaged using iphone4S.
Cell Proliferation Assay (CyQUANT Assay) for Measuring Cell Numbers.
Naïve ELF1 or H1-4iLIF cells grown on MEFs were lifted with TrypLE Express and subcultured on Matrigel in MEF-CM. Cells were plated at a density of 3,750 cells per cm2 in 12-well plates and left to attach overnight (16–20 h) before addition of DMSO, XAV939, or IWP2 for 3 d. After the duration of desired treatments (24, 48, and 72 h), media were withdrawn and the respective 12-well plates were put in –80 °C until all samples were collected and the assays were performed. The CyQUANT assays were performed according to the manufacturer’s instructions (ThermoScientific cat. no. C7026).
Colony Formation Assay.
Naïve ELF1 cells were lifted with TrypLE Express and plated onto Matrigel-coated tissue culture plates and were cultured in MEF-CM supplemented with DMSO, XAV939, or IWP2 for 3 d. In the case of FACS-sorted BAR-ELF1-2iLIF cells, BAR-positive and BAR-negative cells were plated onto Matrigel-coated tissue culture plates in MEF-CM for 3 d before proceeding to the next step. After 3 d, cells were then lifted by TrypLE Express and counted with NucleoCounter, and the same number of cells from each condition was plated in 12-well plates on MEF feeders with a plating density of 250 cells per cm2. Cells were cultured in the same media as they were on Matrigel for 3 more days and then stained with AP. Colonies were counted manually.
RNA isolation, cDNA Synthesis, and qRT-PCR.
Cells cultured on MEFs were lifted with TrypLE Express and transferred onto Matrigel and cultured for 3 d while being treated with Wnt inhibitors or siRNAs as indicated. Total RNA was extracted using GeneJet RNA purification kit (Thermo Scientific K0732) according to the manufacturer’s protocol. We used 2.0 µg RNA to generate cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific K1622) per 10 µL reaction. The reaction product was then diluted 10 times, and qRT-PCR was carried out using 2 µL of diluted cDNA (per 10 µL qRT-PCR, in duplicates for each run). qRT-PCR was performed using Applied Biosystems SYBR Green-based detection (Applied Biosystems) according to the manufacturer’s protocol on a Roche Lightcycler 480 instrument. All primer pairs were validated for not yielding a signal from MEF cDNA in the total cDNA samples. Transcript copy numbers were normalized to GAPDH for each sample, and fold expression over control was calculated for each gene of interest. Primer sequences are as follows (all are 5′–3′): GAPDH (forward: TGAAGGTCGGAGTCAACGGA; reverse: CCATTGATGACAAGCTTCCCG), AXIN2 (forward: GCGATCCTGTTAATCCTTATCAC; reverse: AATTCCATCTACACTGCTGTC), TROY (forward: GGAGTTGTCTAAGGAATGTG; reverse: GCTGAACAATTTGCCTTCTG), OCT4 (forward: GGGAAGGTATTCAGCCAAACG; reverse: GGTTCGCTTTCTCTTTCGGG), NANOG (forward: AGAAGGCCTCAGCACCTAC; reverse: GGCCTGATTGTTCCAGGATT), STELLA (forward: GTTACTGGGCGGAGTTCGTA; reverse: TGAAGTGGCTTGGTGTCTTG), KLF4 (forward: GATGGGGTCTGTGACTGGAT; reverse: CCCCCAACTCACGGATATAA), FOXA2 (forward: GACAAGTGAGAGAGCAAGTG; reverse: ACAGTAGTGGAAACCGGAG), SOX17 (forward: CCTGGGTTTTTGTTGTTGCT; reverse: GCTGTTTTGGGACACATTCA), T (forward: CTCCTTCAGCAAAGTCAAGC; reverse: TTAAGAGCTGTGATCTCCTCG), GATA4 (forward: CCAATCTCGATATGTTTGACGA; reverse: TTGATGCCGTTCATCTTGTG), PAX6 (forward: CTTCACCATGGCAAATAACC; reverse: GAAATGAGTCCTGTTGAAGTG), and TUBB3 (forward: CGGTGGTGGAGCCCTACAAC; reverse: AGGTGGTGACTCCGCTCAT).
Western Blots.
Cells were lysed on ice in 1× RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 0.2% Deoxycholate) freshly supplemented with protease and phosphatase inhibitor mixtures (Roche). Lysates were homogenized by ultrasound and centrifuged on maximum speed in a table-top centrifuge (Eppendorf 5415R) for 10 min at 4 °C, and the supernatants were recovered. Cleared lysates were normalized by protein concentration as determined by BCA assay (Pierce) before diluting with sample buffer + 20 mM DTT. We loaded 5–10 µg protein per lane and transferred them onto nitrocellulose membranes. Blots were blocked in TBST (Tris-buffered saline, 0.1% Tween 20) with BSA (3% wt/vol) for 1 h at RT and then incubated with primary antibodies at 4 °C overnight. Blots were washed with TBST, and species-specific HRP-conjugated secondary antibodies were used, followed by ECL-based detection (Pierce). The primary antibodies used were as follows: rabbit anti-human Axin1 (1:1,000, Cell Signaling #2087), rat anti-human HSP90 (1:20,000, clone 16F1, Abcam #ab13494), rabbit anti-human CRABP2 [1:1,000, clone EPR14256(B), Abcam #ab181255], and rabbit anti-human JARID2 (1:1,000, D6M9X, Cell Signaling #13594).
Metabolic Assay (Seahorse Assay).
Naïve ELF1 or H1-4iLIF hESCs were seeded onto XF96 polystyrene Seahorse microplates (V3-PS, Seahorse Bioscience part 101085–004) precoated with Matrigel at a density of 25 × 103 cells per well. At 16–20 h after plating, the culture media was exchanged for fresh MEF-CM supplemented with small molecules, and the cells were cultured for another 72 h. One hour before the assay, culture media was exchanged for Seahorse XF Base Medium (cat. no. 102353–100, Seahorse Bioscience) supplemented with sodium pyruvate (Gibco, 1 mM) and with 25 mM glucose (for the MitoStress assay). Substrates and selective inhibitors were injected during the measurements to achieve final concentrations of FCCP (1 μM), oligomycin (2.5 μM), antimycin (2.5 μM), and rotenone (2.5 μM). The OCR values were normalized to the number of cells present in each well, quantified by the Hoechst staining (HO33342, Sigma-Aldrich). Changes in OCR in response to the addition of substrates and inhibitors were defined as the maximal change after the FCCP injection compared with the last OCR value before the injection.
Proteomics.
Cells were stained with Tra1-60/CD9 as described in Antibody Labeling and Flow Cytometry, and the Tra1-60/CD9 double-positive population was FACS sorted and analyzed as previously described (13). For mass spectrometry analysis, cells were lysed in 1 M urea and 50 mM ammonium bicarbonate (pH 7.8) at 50 °C for 20 min. Normalized quantities of protein were reduced with 2 mM DTT, alkylated with 15 mM iodoacetamide, and digested overnight with trypsin. The resulting peptides were desalted on Waters Sep-Pak C18 cartridges. Peptides were separated using a heated 50 °C, 30-cm C18 column in a 180-min gradient [1–45% (vol/vol) acetonitrile with 0.1% (vol/vol) formic acid]. Peptides were measured on a Thermo Scientific Q Exactive (QE) operated in data-dependent mode with the following settings: 70,000 resolution, 400–1,600 m/z full scan, Top 10, and an 1.8 m/z isolation window. Identification and label-free quantification of peptides was done with MaxQuant 1.5 using a 1% false discovery rate against the human dataset downloaded from Uniprot on October 11, 2013. Peptides were searched using a 5 ppm mass error and a match between-run window of 2 min. Proteins that were significantly regulated between conditions were identified using a permutation-based t test (false discovery rate 5%) in Perseus 1.4.1.3.
Supplementary Material
Acknowledgments
We thank Dr. Carol B. Ware for providing the ELF1 hESC cell lines and offering valuable intellectual input during the manuscript preparation. This work was supported in part by University of Washington’s Proteomics Resource Grant UWPR95794 and National Institutes of Health Grants U01 HL100395 and P01 GM081619. R.T.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613849113/-/DCSupplemental.
References
- 1.Medvedev SP, Shevchenko AI, Zakian SM. Molecular basis of mammalian embryonic stem cell pluripotency and self-renewal. Acta Naturae. 2010;2(6):30–46. [PMC free article] [PubMed] [Google Scholar]
- 2.Ying Q-L, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453(7194):519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504(7479):282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 4.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 5.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 6.Ware CB. Naive embryonic stem cells: The future of stem cell research? Regen Med. 2014;9(4):401–403. doi: 10.2217/rme.14.31. [DOI] [PubMed] [Google Scholar]
- 7.Welling M, Geijsen N. Uncovering the true identity of naïve pluripotent stem cells. Trends Cell Biol. 2013;23(9):442–448. doi: 10.1016/j.tcb.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Bernemann C, et al. Distinct developmental ground states of epiblast stem cell lines determine different pluripotency features. Stem Cells. 2011;29(10):1496–1503. doi: 10.1002/stem.709. [DOI] [PubMed] [Google Scholar]
- 9.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90(18):8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassani S, Scho HR. Signaling roadmap modulating naive and primed pluripotency. Stem Cells Dev. 2014;23(3):193–208. doi: 10.1089/scd.2013.0368. [DOI] [PubMed] [Google Scholar]
- 11.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15(4):471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ware CB, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(12):4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sperber H, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol. 2015;17(12):1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidson KC, Mason EA, Pera MF. The pluripotent state in mouse and human. Development. 2015;142(18):3090–3099. doi: 10.1242/dev.116061. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka SS, Kojima Y, Yamaguchi YL, Nishinakamura R, Tam PPL. Impact of WNT signaling on tissue lineage differentiation in the early mouse embryo. Dev Growth Differ. 2011;53(7):843–856. doi: 10.1111/j.1440-169X.2011.01292.x. [DOI] [PubMed] [Google Scholar]
- 16.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31(12):2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nusse R, Varmus H. Three decades of Wnts: A personal perspective on how a scientific field developed. EMBO J. 2012;31(12):2670–2684. doi: 10.1038/emboj.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon RT. Wnt/beta-catenin pathway. Sci STKE. 2005;2005(271):cm1. doi: 10.1126/stke.2712005cm1. [DOI] [PubMed] [Google Scholar]
- 19.Hao J, Li T-G, Qi X, Zhao D-F, Zhao G-Q. WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev Biol. 2006;290(1):81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Biophys Res Commun. 2006;343(1):159–166. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- 21.Lyashenko N, et al. Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol. 2011;13(7):753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ten Berge D, et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13(9):1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan Y, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13(6):663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Takashima Y, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158(6):1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurek D, et al. Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Rep. 2015;4(1):114–128. doi: 10.1016/j.stemcr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson KC, et al. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc Natl Acad Sci USA. 2012;109(12):4485–4490. doi: 10.1073/pnas.1118777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart R, et al. Silencing of the expression of pluripotent driven-reporter genes stably transfected into human pluripotent cells. Regen Med. 2008;3(4):505–522. doi: 10.2217/17460751.3.4.505. [DOI] [PubMed] [Google Scholar]
- 30.Rival-Gervier S, et al. Kinetics and epigenetics of retroviral silencing in mouse embryonic stem cells defined by deletion of the D4Z4 element. Mol Ther. 2013;21(8):1536–1550. doi: 10.1038/mt.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duggal G, et al. Alternative routes to induce naïve pluripotency in human embryonic stem cells. Stem Cells. 2015;33(9):2686–2698. doi: 10.1002/stem.2071. [DOI] [PubMed] [Google Scholar]
- 32.Dravid G, et al. Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23(10):1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- 33.Willert K, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423(6938):448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 34.Oishi I, et al. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a / JNK signalling pathway. Genes Cells. 2003;8(7):645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- 35.Orme MH, Giannini AL, Vivanco MD, Kypta RM. Glycogen synthase kinase-3 and Axin function in a beta-catenin-independent pathway that regulates neurite outgrowth in neuroblastoma cells. Mol Cell Neurosci. 2003;24(3):673–686. doi: 10.1016/s1044-7431(03)00229-x. [DOI] [PubMed] [Google Scholar]
- 36.Carthy JM, Engström U, Heldin CH, Moustakas A. Commercially available preparations of recombinant Wnt3a contain non-Wnt related activities which may activate TGF-β signaling. J Cell Biochem. 2016;117(4):938–945. doi: 10.1002/jcb.25378. [DOI] [PubMed] [Google Scholar]
- 37.Zhou W, et al. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31(9):2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: Diseases and therapies. Nat Rev Genet. 2004;5(9):691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 39.Merrill BJ. Wnt pathway regulation of embryonic stem cell self-renewal. Cold Spring Harb Perspect Biol. 2012;4(9):a007971–a007971. doi: 10.1101/cshperspect.a007971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayashi K, Lopes SMCdeS, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3(4):391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo G, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136(7):1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimoto M, et al. A simple and robust method for establishing homogeneous mouse epiblast stem cell lines by wnt inhibition. Stem Cell Rep. 2015;4(4):744–757. doi: 10.1016/j.stemcr.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sumi T, Oki S, Kitajima K, Meno C. Epiblast ground state is controlled by canonical Wnt/β-catenin signaling in the postimplantation mouse embryo and epiblast stem cells. PLoS One. 2013;8(5):e63378. doi: 10.1371/journal.pone.0063378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biechele TL, et al. Wnt/β-catenin signaling and AXIN1 regulate apoptosis triggered by inhibition of the mutant kinase BRAFV600E in human melanoma. Sci Signal. 2012;5(206):ra3. doi: 10.1126/scisignal.2002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.