Significance
Most male moths find their mates by following species-specific pheromones released by females. Despite the importance of pheromone communication in reproductive isolation, much is still unknown about its genetic basis. We investigated male responses in the two pheromone races of the European corn borer. A reasonable hypothesis, that males of the two races differ in the genes encoding the receptor proteins that respond to the two pheromone components, was previously rejected without a convincing alternative. We found that instead male choice was correlated with genes affecting growth and differentiation of the nerve cells that may contain these receptors. This unexpected finding resolves the dilemma and points to another layer of complexity in the evolution of sexual pheromone communication systems.
Keywords: Ostrinia nubilalis, Z chromosome, pheromone response, sexual communication, QTL analysis
Abstract
The sexual pheromone communication system of moths is a model system for studies of the evolution of reproductive isolation. Females emit a blend of volatile components that males detect at a distance. Species differences in female pheromone composition and male response directly reinforce reproductive isolation in nature, because even slight variations in the species-specific pheromone blend are usually rejected by the male. The mechanisms by which a new pheromone signal–response system could evolve are enigmatic, because any deviation from the optimally attractive blend should be selected against. Here we investigate the genetic mechanisms enabling a switch in male response. We used a quantitative trait locus-mapping approach to identify the genetic basis of male response in the two pheromone races of the European corn borer, Ostrinia nubilalis. Male response to a 99:1 vs. a 3:97 ratio of the E and Z isomers of the female pheromone is governed by a single, sex-linked locus. We found that the chromosomal region most tightly linked to this locus contains genes involved in neurogenesis but, in accordance with an earlier study, does not contain the odorant receptors expressed in the male antenna that detect the pheromone. This finding implies that differences in the development of neuronal pathways conveying information from the antenna, not differences in pheromone detection by the odorant receptors, are primarily responsible for the behavioral response differences among the males in this system. Comparison with other moth species reveals a previously unexplored mechanism by which male pheromone response can change in evolution.
Few communication systems in the natural world can rival the sensitivity and elegance of moth sexual pheromones, which reliably bring together males and females of the same species for reproduction (1). The signal is a volatile blend of fatty-acid–derived compounds with various modifications, synthesized by the specialized pheromone gland of the female and released into the air. The detection system is the male antenna, bearing sensillar hairs with specialized neurons that express pheromone receptors that are activated by the binding of individual pheromone components. Species-specific blends activate a behavioral response in which the male flies upwind, following the odor plume, and eventually finds the female (2). Subtle differences in the pheromone compositions of closely related species are sufficient to block this attraction and to maintain reproductive isolation in nature (3, 4). The manifold variety of pheromone systems has been suggested as one of the factors promoting the evolution of the high species diversity of Lepidoptera (4, 5).
However, the origin of such variety poses an evolutionary dilemma. In such a finely tuned communication system, any deviation by the female from the optimally attractive pheromone blend or any preference by the male for an atypical blend would decrease mating success and should be selected against, maintaining the status quo (6–9). There is empirical evidence for such selection against deviance in moths (4, 10, 11). A coordinated change of both signal and response would seem necessary to overcome this problem but is hard to imagine because genes affecting female pheromone production and male response are different and reside on separate chromosomes (12–17). Moreover, despite significant advances in the biochemistry of pheromone synthesis and the physiology of pheromone detection, the identity and mode of action of most genes that shape these traits are unknown. Knowledge of the underlying genetic basis is a requirement for any theory invoking genetic mechanisms for evolutionary change.
The European corn borer (ECB), Ostrinia nubilalis Hübner (Lepidoptera: Crambidae), is a well-studied model system of sexual communication in moths. This species consists of two strains, denoted “E” and “Z,” which use different ratios of the same two pheromone compounds, (E)-11-tetradecenyl acetate and (Z)-11-tetradecenyl acetate (henceforth “E11-14:OAc” and “Z11-14:OAc”). E-strain females produce E11-14:OAc and Z11-14:OAc in a ratio of 99:1, and Z-strain females produce these compounds in a nearly opposite ratio of 3:97 (18). E- and Z-strain males prefer the ratios produced by their respective females (19, 20). This preference leads to reproductive isolation (21), although hybrids are occasionally found in nature (22–25). In the laboratory, hybrids are readily formed by matings; hybrid females produce an intermediate compound ratio of 65:35 (22), whereas hybrid males show a response centered around a 50:50 ratio (18, 26).
The genes controlling these strain differences have been mapped to different chromosomes by interstrain crosses (13). The gene causing variation in female pheromone production is autosomal and encodes a fatty-acyl reductase (27, 28). The main gene controlling the male behavioral response difference, Resp, is sex-linked on the Z chromosome (13, 18) (males are ZZ and females ZW in the standard chromosomal nomenclature). The identities of the gene(s) controlling strain variation in male behavioral response in Ostrinia are still unknown.
So far, the most promising candidates for the control of male preference in moths are the genes encoding olfactory receptor proteins (ORs) that bind to specific pheromone components (29–33). In strong support of this idea, Gould et al. (14) showed that a cluster of OR genes expressed in the antenna is very tightly linked to an autosomal gene controlling the interspecific difference in male behavioral response among Heliothis virescens and Heliothis subflexa. Moreover, single-cell recordings showed that species-specific spike-amplitude responses to individual pheromone components were also linked to the OR cluster (14). In O. nubilalis, five of the seven pheromone ORs are located within a large cluster on the Z chromosome (34). The pheromone ORs show similar spatial antennal expression patterns in males of both strains (35), but OnubOR6, which responds to Z11-14:OAc (36), is expressed at a higher level in Z-strain males (35), and OnubOR4, which binds to E11-14:OAc (32), is expressed at a higher level in E-strain males; expression levels of both OnubOR6 and OnubOR4 are intermediate in hybrids (35). Three Z-linked genes—OnubOR4, OnubOR6, and OnubOR1—show more sequence differences among the E and Z strains than do the two autosomal ORs; however, linkage mapping of the OR cluster on the Z chromosome placed it ∼20 cM distant from the Resp gene (31). Although mapping of Resp was based on phenotype data from 78 males (13), it is difficult to judge the significance of this result, because the accuracy of the behavioral scoring is unknown. Furthermore, independent replicate backcrosses using the E and Z strains by another group showed a high variability in recombination rates among Z-linked genetic markers (37).
To identify the position of the Resp locus more precisely, we conducted a new quantitative trait locus (QTL) analysis, using 470 male progeny from a series of backcrosses. We confirmed that Resp is separated from the OR cluster (31) with a more precise estimate of 15 cM. Moreover, we identified several genes that are much more tightly linked to Resp and which point to a site of action in establishing the connections between olfactory sensory neurons (OSNs) and the brain or deeper within the brain itself. This finding opens up a set of candidate genes acting at a deeper mechanistic level that could be responsible for variations in male response.
Results
Crossing Design.
Hybrid crosses were set up between the Z and E strains, and resulting male progeny were backcrossed to Z-strain females for seven consecutive generations. In each generation we scored behavioral responses to Z, E, and hybrid pheromone blends using artificial lures for 50–150 males (fewer for the third and fourth backcrossed generations). In each generation, one of the scored males with a hybrid phenotype was chosen for backcrossing to a Z-strain female to produce the next generation. In total, 649 males were phenotyped, using the wind tunnel setup described in Methods. After the last generation of backcrossing, DNA was isolated from all 649 phenotyped males, and genotypes were scored at nine sex-linked markers (kettin, five ORs, ldh, bgi012356, and bgi03892). This preliminary scoring showed a loss of genetic polymorphism correlated with behavioral response in the last two backcross generations, probably because of the inadvertent selection of a homozygous male for continuation of the backcrossing. Therefore, only 470 males from the first five generations could be used for final mapping of 21 sex-linked genes and the QTL analysis. We also genotyped 143 females (∼20 per generation) to aid in map construction.
Behavioral Measurements.
The relative intensity of individual male behavioral responses to each of the pheromone blends—Z, hybrid (H), and E—was summarized using a quantitative score that assigned numerical weights to two key behaviors previously shown to be correlated with directed flight toward the pheromone source (38). These behaviors are wing-fanning, a rapid vibration of the wings while remaining in a stationary position, and extrusion of the hair pencil scales at the tip of the abdomen, which release the male pheromone during courtship at close range (Fig. 1). Wing-fanning could be strong and continuous or weak and intermittent (on–off), and hair pencil extrusion could be present or absent. The latency time, i.e., the number of seconds between presentation of the stimulus and the initiation of wing-fanning, was also factored in. An initial set of numerical weights for these behaviors was first chosen arbitrarily, such that the scored response to a given pheromone blend increased with the occurrence and intensity of wing-fanning, increased more with a shorter latency time, and increased even more with the extrusion of hair pencil scales. Results from the three lures were combined by subtracting the H and E lure responses from the Z lure response, so that Z-strain males would have the highest score, because these are male-informative backcross progeny from a pure Z-strain mother. Phenotypic scores then were calculated for each male (see Fig. S1A for details). After an initial QTL analysis, the parameter set of numerical weights was optimized using a discriminant function approach, to discriminate more precisely between the responses of Resp locus homozygotes vs. Resp locus heterozygotes (Fig. S1B).
Fig. 1.
Quantitative scoring of male behavioral response indicators.
Fig. S1.
Formula optimization for behavioral phenotype scoring. (A) Results for the starting parameter set. (B) Results for the optimized parameter set.
Linkage Maps.
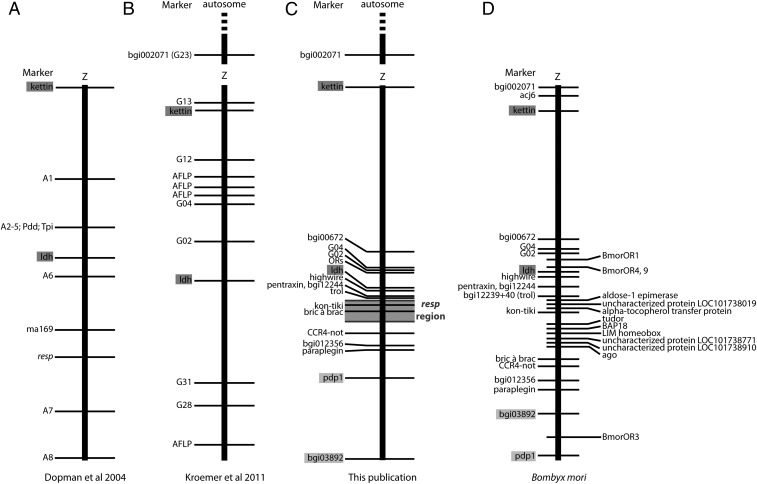
To examine the contribution of each chromosome to male behavioral response, a preliminary map using amplified fragment length polymorphism (AFLP) markers covering all 31 chromosomes was constructed, using data from the 120 progeny of the first backcross generation (Fig. S2). We then mapped an additional 21 genes on the Z chromosome using the progeny of all backcrosses. These genes were chosen based on their Z-chromosomal location in Bombyx mori, and their orthologs in O. nubilalis were PCR-amplified using degenerate primers designed from multispecies sequence alignments (Table S1). Overall, the gene order in O. nubilalis between kettin at one end and paraplegin near the other end was the same as in B. mori. However, we did not map any genes surrounding Tpi, where Wadsworth et al. (39) have detected a region of low recombination that probably corresponds to an inversion. Additionally, the order of the two end markers pdp1 and bgi03892 was inverted (Fig. 2). An inversion between bgi03892 and another marker was also detected in O. nubilalis by Kroemer et al. (Fig. 2) (37). Finally, we found that the Z-linked marker bgi002071 in B. mori was autosomal in O. nubilalis, as was also shown for marker G23 just above kettin by Kroemer et al. (37). Thus, the segment of the Z chromosome above kettin in B. mori appears to be autosomal in O. nubilalis and therefore segregates independently from the behavioral response trait.
Fig. S2.
Genome-wide QTL analysis with AFLPs to identify major linkage groups contributing to male behavioral response.
Table S1.
Primer sequences designed from multispecies sequence alignments
| Gene | Forward primer | Reverse primer |
| Tpi | TGTGAACACTCTGAAAAAGGGTCC | TCCCAGTTGCTGCCAATAGC |
| GCTATTGGCAGCAACTGGGA | ATGTCAACGAAYTCTGGCTTGAG | |
| kettin | TGAAATCCCGGAACCAGTAACA | CAAAACCTGAGTAAAAATGGGCTT |
| CCAGAAGGCATGAAGAAGATCCA | CAAGCTCCTATTTTCACTACTCACCTC | |
| bgi00672 | CTGAAAGAGTTCATCTGGAAGACG | CACTTTGTAGACGGCAGCCAC |
| G04 | CAGCTACGGAGCTCCGTCCGC | GTATACGAGGGTCTTCTCTTC |
| G02 | CCGAGCGTGCCGTTGC | GCTTCCGTGTGCTGGCG |
| GCTCTTCCTTTCTCCGTGTGC | GCTTCCGTGTGCTGGCG | |
| OnubOR4 | CTGTTCCTGCTGCATAACG | TTCGCAGTAGCATGAAGTAG |
| OnubOR5 | GTGCTGTTCCTGCTCTAC | CGTTCGCAAGAACATGAAG |
| OnubOR6 | CGATACGGACCTTTGACTATGATCG | AAGCAGTTCGCTGGTTGCTGTTG |
| ldh | ACCTCCACCCACGGCTACAT | TCTCTTTCCAGTTCTCGGGGTC |
| highwire | GGCCAGGCNATGGTNATHAAYGA | CTTCAGGGTGGGNGTNGGRAARAANGC |
| CATCAACGACGAGGTGGGCTTC | CAGGATGGAGTTGGTCAGCTCCAG | |
| pentraxin | ACCATGCTGCARCTNTAYCAYGTNGC | TCCTTCTCGGGRTTRTCNCKYTTRTC |
| ACCATGCTGCARCTNTAYCAYGTNGC | TGGCCGTTNACRTAYTCYTGYTG | |
| GCACACAAAGACCACAAACATCA | GACCGTTCACGTACTCTTGTTG | |
| bgi12244 | TTCCTGATCTCNCCNAARAARGARGC | CAGCTTCTGCCARTARAANGCYTCNAC |
| TAGGGCTCGCAGCTTATCACT | TGACTTATTTCGTTGACTAGATCG | |
| trol | CACAACGCCGGNGTNAAYATHAC | GGGGGGGCANGTRCAYTCYTCNAC |
| ATCACGGACTTCGTCATGGAGTC | TCTCTGTTGGTNAGNGGRCANGGRCA | |
| CACGGACTTCGTCATGGAG | TAGATGGGTCGCCGTAGGT | |
| kon-tiki | TGCGAGAAYATHACNGGNGTNATG | GATGAAGGGGGGNGANGCYTGDATYTC |
| CAGACTATTCATCGGGTCGAGT | TCCTGAGAAAACTGAGCGATCT | |
| bric-a-brac | TTCTGCCTGCGNTGGAAYAAYTAYCA | CTTCAAGTCGCAGTACTTTAT |
| ATGTGGCARAARTGYTGGAAYAC | GCGCTGGGGCCNAGCATRTTRTGNAC | |
| ATGTGGCAGAAGTGTTGGAACAC | CTGGGGCCGAGCATGTTGTGGAC | |
| CCR4-not | ATCTACCCCGAYGAYACNCARACNCC | TCCACCAGRTANAGRCANCGCCANGT |
| GGCGGCCCNAAYTCNCCNTTYGCNATG | CGCACGTGCTCRCARTAYTTRTGRTA | |
| CCTTACAACACAACACGAACG | GGTAGTCTTTCAACCGCGACT | |
| ATCGCCAACGCNACNAAYATHGAYAC | TCGCTGAAGTTNGCDATNCCYTTRTC | |
| GCACTGGACCGCTTCAAGTC | GTTCGATGGACGCTCTCTTC | |
| GCCGTGCTNGCNACNGARAAYATGGA | GGCTTGGGDATRAACATNCCNGCRTC | |
| GCCGTGCTNGCNACNGARAAYATGGA | ARGCACTCNAGNAGNGTRTGCATRTT | |
| AGAGCCAAGGGNCARACNCCNAAYATG | TGGGTTCTTGATGAGTTCGAT | |
| AGAGCCAAGGGNCARACNCCNAAYATG | CGATGAATGTGATGAGAAGTCC | |
| CGACTTCGACTATGAGGGAAG | GAGGCAGCACGAGAAATAGTG | |
| On-bgi012356 | TGAGCATATCGTTCCCGACTG | TTCAAGACCAAGAGGGTGCTTC |
| paraplegin | GGCCCCCCCGGNTGYGGNAARAC | CGGCTCTTCATNCCRTCCATYTCNAC |
| CGCCCTGCATCATCTACATA | CGCGTACGATGGTCTGTTT | |
| CATCTACATAGACGAGATGGATGC | GTACGATGGTCTGTTTGTACCG | |
| CCCGGCMGNTTCGACMGNCACATCYT | TGTGATCTGTGAGCTGAGTTTGA | |
| bgi03892 | TCGTACCTCCCGTCGCAAAC | TGATGGCTTCCTCCATTTCCTT |
| TGTTGACGCTGGAACTCCTCC | CTTCTAAGGCTAGGTCGTCGATCTC | |
| pdp1 | GAGGAGCTGAARCCNCARCCNATGAT | TCTCAGGGCGATYTGRTTYTCYTTCAT |
| TAAACGCCGAGGAAGTAGTCG | TCCGATAGATTTGTGAACCTTCC |
Fig. 2.
Comparison of linkage maps of O. nubilalis and linkage and physical maps for B. mori. Genes common to all maps are shown in dark gray, and gene inversions are shown in light gray. (A–C) Compare our results with previously published results on O. nubilalis. (A) Adapted from Dopman et al. (13). (B) Adapted from Kroemer et al. (37). (C) Our results. (D) B. mori sex chromosome gene linkage. Z is the sex chromosome. Thick lines represent the chromosomes, and the dashed line shows the direction in which the chromosome continues.
QTL Analyses.
A QTL analysis using 198 AFLP markers that were mapped to the 31 chromosomes spread over 45 linkage groups showed that the Z chromosome was the only one with a significant effect on the male behavioral response, (Fig. S2). This result is in agreement with the findings of Dopman et al. (13), who used a qualitative flight assay to map the major Resp gene to the Z chromosome in a backcross to the E strain. Our quantitative measurement of male behavior additionally permits an estimate of the contribution of minor genes. In comparison with the Z chromosome with a significant logarithm of odds (LOD) score of 5.0, explaining 25% of the variance in behavioral score, the chromosome with the next largest effect is AFLP linkage group 31, explaining 11% of the variance, but this effect was below the P < 0.05 significance threshold (Fig. S2). No other linkage group has an LOD score higher than 1.5 (Fig. S2).
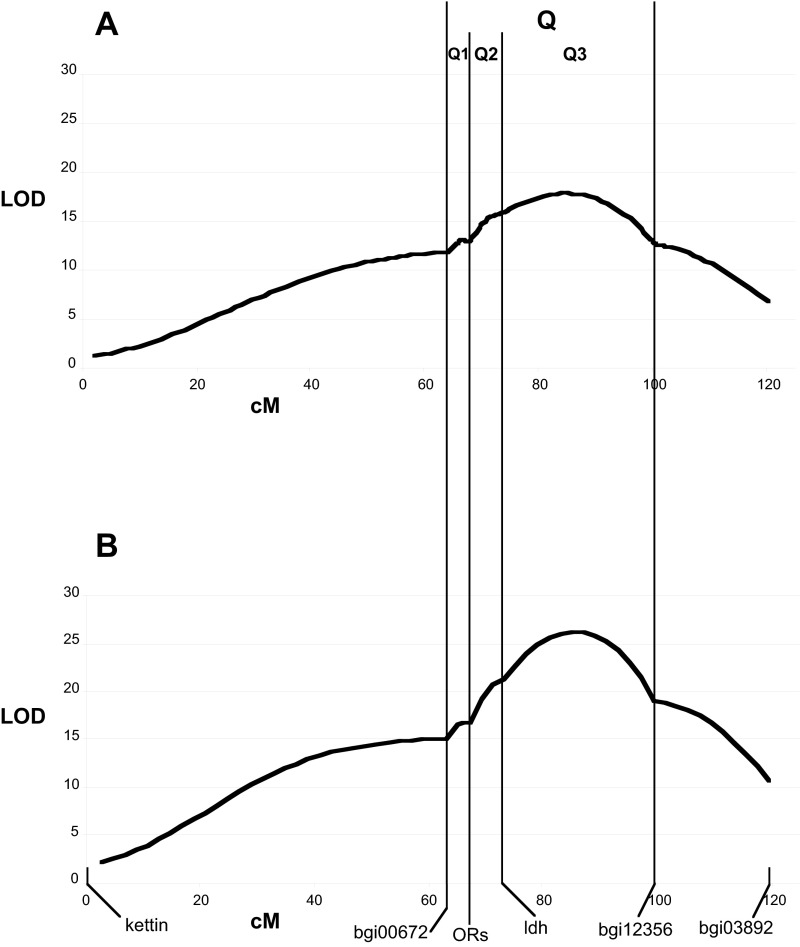
We then conducted a more extensive QTL analysis using 12 genes on the Z chromosome, using data from all 470 phenotyped and genotyped males. Interval mapping produced a single broad peak of the log-likelihood function at position 84.2 cM with an LOD score of 8.7 covering an interval of 36 cM, containing one or more QTLs for the behavioral response. The OR cluster occurred near one edge of this broad interval (Fig. S3A). We used this large interval to optimize the behavioral scoring function (SI Materials and Methods), which increased the LOD score at the peak to 27.8 cM and narrowed its width to 12 cM (Fig. S3B). Finally, for the final QTL analysis we added genotype scores for an additional 12 markers, 6 of which were located within the 12-cM interval (Fig. 3).
Fig. S3.
LOD scores for the starting and optimized parameter sets. (A) The starting parameter set. (B) The optimized parameter set.
Fig. 3.
Final QTL analysis with 21 sex-linked markers and optimized behavioral scores. The male response locus (Resp) is predicted to be inside the gray zone. Every locus outside this zone is at least 100 times less likely to be the Resp locus. The distance between the different loci is measured in centimorgans, and the LOD scores obtained for the ORs and the second peak are shown in boldface type.
The resulting log-likelihood function showed a single narrow peak at position 85.3 cM near the marker kon-tiki with an LOD score of 29.6. The 2-LOD confidence interval in which the LOD score exceeds 27.6 is 8 cM wide and covers positions 83 cM to 91 cM. The likelihood that Resp occurs at the peak is more than 100 times greater than the likelihood that it occurs outside the 2-LOD confidence interval. The log-likelihood function also showed a second peak at 108 cM with a maximum LOD score of 20.5, flanked by two markers (paraplegin and pdp1). This peak is approximately 107 times less likely to be the Resp locus than any position within the 2-LOD confidence interval, but it could represent a second QTL with a smaller effect on the behavioral score. To examine this possibility, we conducted fixed-effects two-way ANOVA using genotypes at the markers nearest the major and minor peaks as factors (Fig. S4). The major peak effect was highly significant (P < 0.0001); the minor peak effect and interaction effects were not (P = 0.46 and P = 0.37, respectively for the marker to the left of the minor peak and P = 0.49 and P = 0.18 for the marker to the right of the minor peak), indicating that any possible contribution of the minor peak effect and interaction effect to the overall variance of the behavioral score is less than 10% of the contribution of the major QTL.
Fig. S4.
Two-way fixed-effects ANOVAs on the preference score comparing the marker closest to the main QTL peak (kon-tiki) with two markers flanking the minor QTL peak (paraplegin and pdp1). Preference is strongly associated with the kon-tiki phenotype but not with the genotypes of the other markers or their interactions.
Evaluation of Candidate Genes for the Behavioral Response.
We next evaluated the OR cluster as a candidate for the Resp locus. The cluster maps to position 72 cM; in scoring OnubOR5, OnubOR4, and OnubOR6, we did not detect any crossovers within the cluster (Figs. 2 and 3). The OR genes occur at 85.3 cM, 13.3 cM away from the larger QTL peak, somewhat closer than the distance of 20 cM found by Lassance et al. (31) between the ORs and the Resp locus. The OR cluster is even further away (36 cM) from the smaller peak at 108 cM. The LOD score of the OR cluster is 20.5, making it very unlikely to be the Resp locus. The peak at 85.3 cM is 109 times more likely, and any location within the 2-LOD confidence interval is at least 107 times more likely to be the Resp locus than is the OR cluster. The slight hump in the LOD score at the OR cluster (Fig. 3) might indicate a smaller contribution to the behavioral response. To test the possibility that the ORs represent a third, minor QTL, we conducted a two-way ANOVA using the marker genotypes at the QTL peak and the ORs as factors (Fig. S5). As before, the main effect of the peak was highly significant (P < 0.0001), but neither the main effect of the OR genotype (P = 0.84) nor the interaction effect (P = 0.13) was significant. Moreover, the nonsignificant effect of the OR genotype depended on the genotype of the markers near the peak. Thus, sequence variation at the OR cluster makes no consistent contribution to male pheromone preference.
Fig. S5.
Two-way fixed-effects ANOVA on the preference score comparing the marker closest to the major QTL peak (kon-tiki) with the cluster of OR genes. Preference is strongly affected by the kon-tiki genotype but not by the OR genotype or interaction.
The 2-LOD confidence interval around the major peak contains two of the marker genes, kon-tiki and bric-a-brac (bab). Comparing our O. nubilalis map with the B. mori and Danaus plexippus genomes, a total of seven genes have been mapped between ldh and bgi012356 (Fig. 3). Interestingly, six of these genes—highwire, pentraxin, trol, kon-tiki, bab, and CCR4-not—seem to be involved in neurogenesis (Table 1). These genes seem to control neuronal axonal growth and orientation processes specifically in insects and other animals (see references in Table 1). Furthermore, the genome comparisons to identify other candidates for Resp showed a total of 12 genes (including kon-tiki and bab) located within the Resp region. Eight have one or more identifiable protein domains, six have homologs in Drosophila with some functional information, and three are not similar to any protein of known function. Although none has an obvious connection to pheromone sensory physiology, several are implicated in neurogenesis; we discuss these possible roles below.
Table 1.
Description of genes near the resp locus region
| Resp region | Full name (abbreviation) | Homologs/ synonyms | BLASTp best hit | Function | Expression | Ref. or source | |
| Accession number | Organism | ||||||
| Out | Highwire (hiw)* | PHR protein family: | AF262977 | * | Ubiquitin ligase, negative regulator of synaptic growth | Muscle neurons; CNS; retinal cone | (75–78) |
| Myc (PAM)† | Presynaptically: synaptic growth and axon guidance | ||||||
| Rpm-1 gene‡ | Postsynaptically: endocytosis of glutamate receptors | ||||||
| Esrom§ | |||||||
| Out | Pentraxin (pent)* | Lectins | U18772 | § | Innate immunity | Sensory neurons | (79) |
| Synapse formation and remodeling, neural crest cell migration | Brain | ||||||
| Out | terribly reduced optic lobes (trol)* | Heparan sulfate | XP_014364776.1 | ¶ | Activates neuroblasts’ adhesion, growth migration and differentiation | Larval brain, imaginal discs, fat body, muscles; adult gonads | (43, 80, 81) |
| Proteoglycan¶ | Signal transduction | ||||||
| Perlecan†,‡ | Regulates neuromuscular junction | ||||||
| In | Aldose-1 epimerase# | Galactose mutarotase† | XP_004933180.1 | # | Cytoplasm; testis | (82) | |
| In | LOC101738019# | KGM_08728|| | XP_004933181.1 | # | Uncharacterized protein | unknown | GenBank |
| In | Cralbp* | KGM_088417 | XP_013184856.1 | ** | Cellular retinaldehyde binding protein | Fat body, gut: hemocytes | (83) |
| α-Tocopherol transfer protein | α-Tocopherol transfer protein | ||||||
| In | kon-tiki (kon)* | NG2 proteoglycans: | XP_013184902.1 | ** | Muscle development, i.e., filopodium assembly and orientation targeting skeletal positions, directed myotube migration | Myotube tips: Embryonic founder; Myoblasts: CNS | (51, 52) |
| Chondroitin sulfate proteoglycan** | Target-derived signal initiating stable connection | ||||||
| Perdido* | Neurogenesis i.e., cell migration and differentiation | ||||||
| NG2/MCSP†,§ | |||||||
| In | Tudor domain* | RING finger protein 17 | XP_013140855.1 | †† | Germline development | (53) | |
| In | BAP18¶ | SANT domain | XP_004933184.1 | # | Chromatin modification | (54) | |
| DNA binding | |||||||
| In | LIM homeobox | Transcription factor | XP_013184855.1 | ** | Tissue patterning and differentiation | (55) | |
| Neuronal patterning | |||||||
| In | bgi12353A | LOC101738771# | EHJ68261.1 | || | Uncharacterized protein | Testis | GenBank |
| In | bgi12353B | LOC101738910# | XP_013184860.1 | ** | Uncharacterized protein | Testis | GenBank |
| KGM_07611¶ | |||||||
| In | Archipelago (ago) | F-box protein | XP_013184917.1 | ** | Cyclin binding | Imaginal tissues: eye; imaginal disk: photoreceptor cell | (56, 57) |
| Ubiquitin–protein transferase activity: negative regulator of cell growth, including axon guidance | |||||||
| In | Bric à brac (bab)* | BTB POZ domain | XP_014364829.1 | ¶ | Leg and antenna segmentation | Leg and antenna imaginal disks; ovaries | (58–60) |
| Eye–antennal disk morphogenesis and imaginal disk-derived leg morphogenesis | |||||||
| Sex differentiation, i.e., abdominal color sexual dimorphism in D. melanogaster | |||||||
| Female gonad development | |||||||
| Out | CCR4-NOT* | XP_013184897.1 | ** | Regulation of synaptic growth of the neuromuscular junction | (61, 62) | ||
| Inhibition of miRNA degradation | |||||||
Drosophila melanogaster.
Homo sapiens.
Caenorhabditis elegans.
Mus musculus.
Papilio machaon.
Bombyx mori.
Danaus plexippus.
Amyelois transitella.
Papilio polytes.
SI Materials and Methods
Strategies for QTL Analysis of the Male Response.
For the purpose of fine-scale genetic mapping, there are disadvantages in using any male response as a dichotomous trait. A single error in scoring that trait will produce a spurious recombinant in the linkage map, resulting in an inaccurate location of the Resp locus. Multiple errors in scoring the trait will inflate the estimated recombination rate in the region of the Resp locus, increase the inaccuracy of its location, and tend to bias the location of the Resp gene toward the end of a chromosome. When the behavioral trait is assumed to be scored without error, there is no way to estimate statistically the effects of possible scoring errors. The likelihoods of different gene orders can be compared, but the spurious recombinants will decrease the likelihood of the most likely order and will inflate the estimate of recombination rates. Therefore, we took great care to devise a quantitative score and used a discriminant function approach to discriminate more precisely between homozygote and heterozygote responses, as explained in detail below. Two behavioral characters were measured as a response to each of the pheromone blends (Z, H, and E): wing-fanning and hair pencil extrusion. For each blend, a behavioral score was calculated by weighing the behaviors as shown in Fig. S1. Each male was held within a tube with screening at both ends. The pheromone plume could pass through the tube, and the male could be observed at close range. This method allowed us to test many males in succession, greatly increasing the sample size in each generation. If males had been allowed to fly to the lure, recapture of the male within the wind tunnel would have been required, possibly resulting in damage and likely influencing the response to the next blend to be tested.
Optimization of Preference Score.
The LOD function resulting from the preliminary QTL analysis exhibited a broad peak at 84.2 cM in the interval Q between bgi00672 and bgi12356, with a maximum LOD score of 17.7, explaining 22% of the phenotypic variance in the behavioral score (Fig. S3A). We used this entire interval Q to optimize the weightings in the behavioral score. We identified four groups: 149 males homozygous for the Z allele at all marker loci in the interval (group A); 149 heterozygous males with one Z allele and one E allele at all marker loci in interval Q (group B); 73 males with at least one crossover in the interval, producing some homozygous and some heterozygous markers within interval Q (group C); and 99 males with missing data, such that none of the other conditions could be established with complete certainly (group D).
Unless there is a double crossover within one of the subintervals Q1, Q2, or Q3 (Fig. S3), group A males are homozygous for the entire interval Q of the Z-strain chromosome and therefore must be homozygous for the Z allele of the Resp locus. Double crossovers within one of the three subintervals would not be experimentally detectable, unless additional markers were scored within them, but we can estimate the probability of a double crossover as ≤0.05 (see below); thus overall at least 95% of group A males are expected not to show a double crossover and thus to be homozygous for the Z allele of the Resp locus. By the same reasoning, at least 95% of group B males are expected to be heterozygotes, with one Z allele and one E allele at the Resp locus. Therefore, independent of the actual position of the Resp gene within interval Q, we can take A and B as representative groups of Resp ZZ homozygotes and ZE heterozygotes, respectively, and can use these two groups in a discriminant function analysis. This analysis determines which combination of parameters in the formula for the behavioral score best discriminates between the two genotypes at the Resp locus.
The goal of discriminant analysis is to find the set of predictor variables that best discriminates between two types of observations described by a known class variable. Here the class variable is the genotype at the Resp locus (assumed to be known for groups A and B), and the predictor variables are the parameters used to calculate the behavioral score. We ask the following: Is the way we initially chose to measure the phenotypic score the best way to characterize the behavioral differences between the genotypes, and, if not, can we find a better way? Although classical discriminant function analysis assumes that the predictor variables are normally distributed, we make no such assumptions because we return to the original concept of the discriminant rule as defined by Fisher (84), namely to maximize the between-groups sum of squares, SSbetween, relative to the within-groups sum of squares, SSwithin. SSbetween is equivalent to the squared difference of the behavioral score means of groups A and B, and SSwithin is related to the weighted average of the variance of the behavioral score within the two groups. We searched the space of parameters used to define the behavioral score over a regular grid and calculated the SSbetween/SSwithin ratio (hereafter the “SS ratio”) for each parameter set. We defined the optimized parameter set (Par13.3) as that which produced the largest value of this ratio. We found that the optimized parameter set increased the SS ratio from 84.2 to 137.5. QTL analysis using the same markers with the new behavioral score calculated for all the males (groups A–D) shifted the LOD peak 2 cM to the right, to 86.2 cM, increased the maximum LOD value from 17.7 to 26.1, and increased the fraction of phenotypic variance explained from 22 to 31%. We subsequently used the optimized parameter set to calculate the behavioral scores used for the final QTL analysis using more markers within interval Q.
We estimated the probability of an undetected double crossover within intervals Q1, Q2, or Q3 by taking each interval length (3.9, 5.9, or 26.3 cM), converting them to crossover probabilities (0.037, 0.055, or 0.204), squaring these, and adding them to produce a sum of 0.046 ∼5%. Squaring the crossover probabilities assumes no interference; typically there is interference, and the frequency of double crossovers will be much smaller than 5%. Double crossovers within interval Q involving single crossovers in Q1 and Q2, or in Q1 and Q3, or in Q2 and Q3 will be more common, but these are detectable, and such males are classified into group C. The occurrence of undetected double crossovers in interval Q will affect only the assumption that group A males have no E allele at the Resp locus and that group B males have no Z allele at the Resp locus. If these assumptions are violated up to 5% of the time, the discriminant analysis will be only slightly less discriminating than optimal. We have seen that, even when suboptimal, the discriminant analysis approach greatly improved the peak LOD score and the fraction of variance explained.
Our subsequent analysis (see below) eliminated the OR cluster from serious consideration as a candidate for the Resp locus. However, the OR cluster had been included in the interval Q along with the Resp locus, so no bias against the OR cluster was introduced by excluding it from the discriminant analysis.
Evaluation of Peaks in the Likelihood Function.
In addition to the major peak, the log-likelihood function resulting from the final QTL analysis showed a second peak at 108 cM with a maximum LOD score of 20.5, flanked by the two markers paraplegin and pdp1. This peak is approximately 107 times less likely to be the Resp locus than is any position within the 2-LOD confidence interval, but it could represent a second QTL with a smaller effect. To examine this possibility, we conducted two-way fixed-effects ANOVA, using the genotype at kon-tiki (the marker nearest the major peak in the largest number of males genotyped) as one factor and the genotype at paraplegin or pdp1 as the other factor. The dependent variable was the behavioral score for each individual. In each ANOVA, kon-tiki had a highly significant main effect on the behavioral score (P < 0.0001), but the main effect of the other marker and the interaction effect were not significant (Fig. S4). Thus, when the large effect of the main peak is accounted for, the overall contributions of markers near the minor peak are not significant. The presence of a peak rising above two nonsignificant markers may be caused by the contrasting marker effects within a given genotype class for kon-tiki (Fig. S4). For kon-tiki EZ heterozygotes the pdp1 ZZ homozygote has a higher average score than the pdp1 EZ heterozygote, but for kon-tiki ZZ homozygotes the pdp1 ZZ and EZ scores are the same. For kon-tiki EZ heterozygotes the paraplegin ZZ and EZ are the same, but for kon-tiki ZZ homozygotes paraplegin ZZ is higher than paraplegin EZ. Therefore, possessing two copies of the Z allele at markers around the minor peak increases the tendency to prefer the Z blend in some but not all genetic backgrounds. If there is a second minor QTL under this peak, its effect depends on the genotype at the major QTL, and its contribution to the overall variance of the behavioral score is less than 10% of the contribution of the major QTL.
Evaluation of ORs as Candidate Genes.
To test the possibility that the ORs represent a third, minor QTL, we conducted a two-way, fixed-effect ANOVA using the genotypes at kon-tiki and the ORs as factors (Fig. S5). The dependent variable was the behavioral score for each individual. As before, the main effect of kon-tiki was highly significant (P < 0.0001), but the main effect of the OR genotype and the interaction effect were both nonsignificant. The OR genotype had contrasting effects on the behavioral score, depending on the kon-tiki genotype. Within kon-tiki EZ genotypes, OR ZZ homozygotes had higher scores than OR ZE heterozygotes, but for kon-tiki ZZ homozygotes, OR ZZ homozygotes had lower scores than OR EZ heterozygotes (Fig. S5). Thus, sequence variation at the OR cluster makes no consistent contribution to male pheromone preference.
Discussion
By fine-scale mapping of the male response in O. nubilalis, we found candidate genes that had not been implicated in previous studies of male pheromone perception in moths. Moreover, confirming previous work (31), we ruled out variation in other candidate genes, i.e., the ORs that detect pheromone components and have been implicated in studies of other species. We will explain the justification for our approach in analyzing behavior, point out some aspects of our mapping methods, reconsider the role of the pheromone receptors, and discuss the possible roles of genes in the chromosomal region we have identified.
In mapping the genes underlying the male behavioral response, we deliberately chose to measure traits different from but correlated with the final, evolutionarily relevant outcome: a mating with a female of the same strain. This outcome has traditionally been measured by observing male behavior and flight toward a female or an artificial pheromone dispenser at the upwind end of a wind tunnel. The ultimate criterion of response is considered to be touching and attempting to copulate with the pheromone source, and this criterion has been used as a qualitative, dichotomous trait in previous mapping studies (13, 18). This approach has been justified by the observation that E-strain males may exhibit earlier signs of attention to the Z lure and the opposite applies to Z-strain males; such signs are unreliable predictors of the final outcome (38). However, recent studies have shown that early responses by Z-strain males can lock them in to subsequent behaviors that are maintained even if the pheromone lure is experimentally switched from Z to E while the moth is in midflight (40). Thus, variation in the early responses also may be a relevant indicator of genetic differences between the Z and E strains. Therefore, we scored male response as a quantitative trait by observing the preflight responses of males to different pheromone blends. This reduction in individual assay time greatly increased the number of males that could be scored against all three pheromone blends over the generations. Even though 68% of the variance in this quantitative trait was nongenetic or was caused by minor genetic factors, QTL analysis could define an interval on the Z chromosome that accounted for 32% of the variance. Our methods enabled the quantitative assessment of previously suspected candidate genes and identified candidates that had not previously been considered; this assessment would have not been possible if the response had been scored as a dichotomous trait. Moreover, our method avoids inaccuracies in fine-scale genetic mapping caused by errors in scoring male response as a dichotomous trait, which are inevitably confounded with recombination (see the discussion of mapping strategies in SI Materials and Methods).
The exclusion of the cluster of ORs from the QTL region means that sequence variation in those genes, whether in the coding sequence or in noncoding regions that affect their expression patterns, cannot be directly responsible for the difference in male response. However the possibility remains that differences in OR expression or function may be controlled by a trans-acting transcription factor located within the QTL region. We previously found no qualitative difference in OR expression: Both strains have the same ORs, localized at the same positions on the antenna, with the same number of sensilla and neurons housing them (35). However, we did find quantitative differences in the expression of OnubOR4 and OnubOR6 (35). These differences could be caused by a trans-acting transcription factor or could be affected by changes in neuron size and number. Nevertheless, the genetic factor underlying male response in the Ostrinia pheromone strains must be different from that of H. virescens and H. subflexa, in which the male behavioral responses and most of the neurophysiological responses to pheromone components cosegregated with the autosomal OR cluster (14).
The candidate genes in the Resp QTL region (Table 1) suggest that other factors affect the structure, function, or connectivity of the OSNs or other neurons in the antennal lobe or elsewhere in the brain. Three candidates are similar to genes of unknown function in the sequenced genomes of other lepidopteran species (Table 1). Two other candidates could potentially play a role in gene regulation: One has a Tudor domain also found in proteins that bind to methylated histones (41), and the other has a SANT (switching-defective protein 3, adaptor 2, nuclear receptor corepressor, and transcription factor IIIB) domain involved in histone modification (42). Two candidates, aldose epimerase and a lipid-binding α-tocopherol transfer protein, have domains not necessarily specific to the nervous system. Four genes within the QTL region and two just outside it are similar to genes with roles in neurogenesis. Of the latter, trol (terribly reduced optic lobes, also called perlecan) is a large multidomain heparan sulfate proteoglycan in the extracellular matrix with Laminin G domains that binds to and stores other signaling molecules controlling neuroblast proliferation (43). On the other side of the QTL region is CCR4-Not, coding for the catalytic subunit of the deadenylase complex (44, 45), which interacts with the conserved P body component HPat/Pat1 to regulate synaptic terminal growth in the Drosophila neuromuscular junction (46).
We consider that four genes mapping within the QTL region are most likely to play a relevant role in neurogenesis. In Drosophila melanogaster, kon-tiki encodes a transmembrane protein that promotes cell migration in muscle development (47, 48). Similar mechanisms have been proposed to generate directed migration and target recognition of myotubes as well as neuronal axon and dendrite growth toward their synaptic partners (49). The second likely candidate is a gene with LIM and homeobox domains characteristic of many transcription factors that has also been described as a neural-patterning gene in several organisms [i.e., Drosophila, mice, and Caenorhabditis elegans (50)]. The third likely candidate is archipelago (ago), which encodes an F-box protein and part of a ubiquitin ligase complex that interacts with the Notch signaling pathway and suppresses tissue growth in flies and tumor development in vertebrates (51, 52). Mutations in the fourth likely candidate, bab, have many consequences, including disordered arrangements of eggs in the ovaries in D. melanogaster (53). In both antennae and tarsi, bab causes distal segment fusion (54). The protein encoded by bab possesses a BTB domain also found in the Drosophila genes tramtrack and Broad-Complex (55). Moreover, bab was recently found to interact with other transcription factors in a network that patterns the developing olfactory tissue in D. melanogaster (56).
In addition to differing in their behavioral response to female pheromones, E- and Z-strain males also differ in the connections between the antennal OSNs and the antennal lobe of the brain. In the Z strain, the OSNs that respond to Z11-14:OAc project their axons onto the large, medial glomerulus of the antennal lobe, and the OSNs responding to E11-14:OAc project onto the smaller, lateral glomerulus (57). In the E strain, these connections are reversed (57). In F1 hybrids, the connections are similar to those in the E strain, but the lateral and medial glomeruli are more similar in size, and glomerular size appears to be Z-linked (58). A Z-linked mutant in B. mori shows a similar rewiring of neuronal connections. In wild-type B. mori males, OSNs expressing the bombykol receptor BmOR1 target their axons to the larger toroid glomerulus, whereas OSNs expressing the bombykal receptor BmOR3 send their axons to the smaller cumulus (59, 60). In null mutants of the Z-linked acj6 gene, BmOR1-expressing OSNs are rare or absent, and BmOR3-expressing OSNs project instead to the toroid (61). Mutant males are attracted not by the main pheromone component bombykol but instead by the minor component bombykal. We have confirmed that acj6 is also Z-linked in O. nubilalis, but it segregates independently from the QTL interval. Furthermore, acj6 maps distal to kettin on the Z chromosome of B. mori, far from the QTL interval (Fig. 2). Thus, despite intriguing similarities in the phenotype caused by mutation of acj6 in Bombyx, acj6 does not seem to be responsible for male response in O. nubilalis.
Nevertheless, it is possible that one of the genes in the QTL interval could have a similar effect on axonal targeting. In Drosophila (unlike mammals) the expression of a given OR in a sensory neuron is not directly involved in glomerular targeting (62). Instead, upstream-acting transcription factors involved in neuronal differentiation separately specify both the downstream expression of a particular OR (63) and the axonal guidance of that neuron to its cognate glomerulus in the brain. In O. nubilalis, both strains possess the same cluster of ORs for pheromone detection. One of the newly identified candidate genes could control the expression of specific ORs in different OSNs [as inferred by Koutroumpa et al. (35)] or the targeting of these OSNs to the antennal lobe, or both processes. A third possibility is a change in the antennal lobe itself, governing which OSNs will connect to which glomeruli. Such a change is suggested by the results of transplanting antennae of E-strain males onto Z-strain males: The chimeric males responded only to the Z blend (64). In this example, the Z genotype of the recipient bearing the antennal lobe had a greater influence than the E genotype of the OSNs in the transplanted antennae.
One other extensively studied trait is the spike amplitude of OSNs responding to the E or Z component in the two strains. In E-strain males, the E-responding OSN has bigger spike amplitude than the Z-responding OSN. In Z-strain males, the magnitudes of the spike amplitudes are reversed, and in hybrids they are more nearly equal. Spike amplitude is correlated with the diameter of the dendrite projecting into the sensillar lymph (65). Early studies found that spike amplitude was autosomally inherited (66), not sex-linked (18), and was not on the autosome controlling female pheromone production (12). Later studies found that spike amplitude was not correlated with male behavioral response (67) but also may have a minor Z-linked component (15). There is a strain difference in electroantennogram responses that also appears to be Z-linked (58). Further discussion on spike amplitude measurements and the interpretation of these results in the light of sex-linkage can be found in Koutroumpa et al. (35). We did not score spike amplitudes or whole-antennal responses in this study because of the prohibitive workload of neurophysiological preparations of each male after the behavioral assays, but further mapping of these traits would be worthwhile.
In conclusion, our QTL-mapping approach has revealed several genes involved in neurogenesis that may account for the differences in male behavioral response between pheromone strains in O. nubilalis. In addition to the previously established importance of changes in the ORs, as supported by phylogenetic analysis, functional expression, and genetics, other changes can shift preference without any change in the ORs themselves. Further advances will result from the application of QTL mapping to other recently diverged species pairs or species with pheromone races, as well as from functional studies of the candidate genes we have identified.
Materials and Methods
Insects.
Laboratory colonies of ECB Z and E strains were used. The Z-strain colony derived from cornfield-collected adults in Kéty town, county of Tolna, Hungary in 2004. The E-strain colony was established from larvae extracted from maize stems collected by Magda Rak-Cizej of the Agriculture and Forestry Institute, Novo Mesto, Slovenia. The purity of the strains was monitored by GC analysis of female pheromone production following the protocol of Kárpáti et al. (68), and male response was evaluated with an electroantennogram (58). All insects were reared on a semiartificial diet (69) until pupation. Adults were fed a 5% honey–water solution throughout their adult lives. All animals were kept at 25 °C, relative humidity 70% under an 18-h/6-h light/dark photoperiod. The day of emergence was considered day 0.
Backcrosses.
All crosses were single-pair crosses. Because there is crossing-over in male Lepidoptera (70), we conducted male-informative backcrosses to generate recombination in the Z chromosome. Single-pair matings were set up hybridizing pure Z-strain females with E-strain males (ZE hybrid) and E-strain females with Z-strain males (EZ hybrid). The F1 hybrid males were backcrossed to pure Z-strain females. We selected one out of 30 hybrid families, namely family EZ2, because it was highly fecund. Sons of the EZ2 family that showed hybrid behavior in the wind tunnel bioassays were backcrossed with pure Z-strain females, giving rise to the first backcross generation (BC1: ZEZ2). Two BC1 families were continued to second-generation backcrosses, i.e., ZEZ2-4 and ZEZ2-7 families. Because only the ZEZ2-7 family generated enough offspring, we continued the backcrossing procedure with this family for six additional generations (BC2–BC7). One family for each of the subsequent BC2–BC7 families was phenotyped and genotyped.
Wind Tunnel Phenotyping.
All 649 backcross males were tested separately in the wind tunnel for response to each of the Z, E, and H blends. Our wind-tunnel assays were based on a series of behavioral male responses to each of the pheromone blends, i.e., resting, wing fanning (on/off), strong wing fanning, and hair pencil extrusion. Each category was given a score (Fig. 1 and Fig. S1), so that we had a weighted and quantitative measurement for each male.
The males were 1–2 d old and were sexually and olfactorily virgin. All males were behaviorally tested at the second hour of their scotophase, with 70% humidity and optimal temperature of 19 °C. Each male was kept in a cylinder 2 cm in diameter with gauze on both ends so that the air plume with the pheromone could pass through the tube. With this setup we were able to keep the males captured during the assay, avoiding the need to recapture them (and possibly damage them) at the end of the experiment. We used the same males for subsequent matings.
The three pheromone blends were ordered from Pherobank and were separately presented to each of the males for 1 min maximally. During one experimental day, all males were tested first with the Z blend, followed successively by the H and E blends, with an interval of 30 min to ventilate the wind tunnel between the tests. After a male showed the hair pencil extrusion response or after 1 min, the male was removed from the wind tunnel. The latency of each reaction to the blend was recorded. If the behavioral response was ambiguous, the male was retested on the following day.
Mapping and Genotyping.
We mapped the chromosomes using AFLP markers, using a protocol adapted from ref. 71. On polyacrylamide gels, the markers scored were present in the pure E-strain female, the F1 hybrid male, and the heterozygous backcross males and were absent in the pure Z-strain male, the F1 Z-strain female, and in the homozygous backcross offspring. The opposite pattern was scored as well. We scored 198 AFLP markers from 47 primer combinations in 123 samples (parents and F1 and BC1 males and females). Using MapMaker 3.0 (72) we grouped 180 of these markers into 45 linkage groups, with at least two markers from different primer pairs. The number of markers for the 35 linkage groups varied from 2 to 11; the average number was 5.14. For fine-scale mapping on the Z chromosome, we used intron-size polymorphisms or SNP variation in 21 Z-linked genes that were scored in all backcross males; then recombination rates were determined in MapMaker as well. We verified that all polymorphic differences used in the mapping also differed among wild-type Z and E males in the laboratory strains.
QTL Analysis.
With the phenotypic scores for all male behaviors, we first determined which linkage groups explained a significant portion of the variation, using R-studio using t test-based marker regression as implemented in R/qtl (version 0.98.490). We established significance thresholds for LOD scores empirically by permutation tests using 10,000 permutations. After finding a significant LOD score for only the Z chromosome, we conducted QTL analysis with the 21 Z-linked genes using MAPMAKER/QTL 1.1 (73, 74). We used the gene order determined by MapMaker 3.0, with a backcross design. We extracted the numerical values of the LOD scores from the program output and plotted them in Excel.
Acknowledgments
The behavioral investigations were supported by Grant Vetenskapsrådet (the Swedish Research Council), Project 621-2014-4816 (to T.D.) and by a Linnaeus Chemical Ecology, Ethology, and Evolution Initiative (ICE3) grant [The Swedish Research Council for Sustainable Development (FORMAS)/Swedish University of Agricultural Sciences] (to F.A.K. and T.D.). The molecular work and mapping analysis were supported by the Max Planck Institute for Chemical Ecology (D.G.H., A.T.G., and F.A.K.). F.A.K. received further funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie Grant Agreement 661322.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610515113/-/DCSupplemental.
References
- 1.Cardé RT, Baker TC. Sexual communication with pheromones. In: Bell WJ, Cardé RT, editors. Chemical Ecology of Insects. Chapman and Hall; New York: 1984. pp. 355–383. [Google Scholar]
- 2.Cardé RT, Willis MA. Navigational strategies used by insects to find distant, wind-borne sources of odor. J Chem Ecol. 2008;34(7):854–866. doi: 10.1007/s10886-008-9484-5. [DOI] [PubMed] [Google Scholar]
- 3.Groot AT, Dekker T, Heckel DG. The genetic basis of pheromone evolution in moths. Annu Rev Entomol. 2016;61:99–117. doi: 10.1146/annurev-ento-010715-023638. [DOI] [PubMed] [Google Scholar]
- 4.Löfstedt C. Moth pheromone genetics and evolution. Philos Trans R Soc Lond B Biol Sci. 1993;340(1292):167–177. [Google Scholar]
- 5.Johansson BG, Jones TM. The role of chemical communication in mate choice. Biol Rev Camb Philos Soc. 2007;82(2):265–289. doi: 10.1111/j.1469-185X.2007.00009.x. [DOI] [PubMed] [Google Scholar]
- 6.Brooks R, et al. Experimental evidence for multivariate stabilizing sexual selection. Evolution. 2005;59(4):871–880. [PubMed] [Google Scholar]
- 7.Kirkpatrick M, Nuismer SL. Sexual selection can constrain sympatric speciation. Proc Biol Sci. 2004;271(1540):687–693. doi: 10.1098/rspb.2003.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne JA, Orr HA. Speciation. Sinauer Associates, Inc.; Sunderland, MA: 2004. pp. 1–545. [Google Scholar]
- 9.Ritchie MG. Sexual selection and speciation. Annu Rev Ecol Evol Syst. 2007;38(7):79–102. [Google Scholar]
- 10.Butlin RK, Trickett AJ. Can Population Genetic Simulations Help to Interpret Pheromone Evolution? Chapman & Hall; New York: 1997. pp. 548–562. [Google Scholar]
- 11.Groot AT, et al. Experimental evidence for interspecific directional selection on moth pheromone communication. Proc Natl Acad Sci USA. 2006;103(15):5858–5863. doi: 10.1073/pnas.0508609103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Löfstedt C, Hansson BS, Roelofs W, Bengtsson BO. No linkage between genes controlling female pheromone production and male pheromone response in the European corn borer, Ostrinia nubilalis Hübner (Lepidoptera; Pyralidae) Genetics. 1989;123(3):553–556. doi: 10.1093/genetics/123.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dopman EB, Bogdanowicz SM, Harrison RG. Genetic mapping of sexual isolation between E and Z pheromone strains of the european corn Borer (Ostrinia nubilalis) Genetics. 2004;167(1):301–309. doi: 10.1534/genetics.167.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould F, et al. Sexual isolation of male moths explained by a single pheromone response QTL containing four receptor genes. Proc Natl Acad Sci USA. 2010;107(19):8660–8665. doi: 10.1073/pnas.0910945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsson SB, et al. Ostrinia revisited: Evidence for sex linkage in European Corn Borer Ostrinia nubilalis (Hubner) pheromone reception. BMC Evol Biol. 2010;10:285. doi: 10.1186/1471-2148-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lassance JM. Journey in the Ostrinia world: From pest to model in chemical ecology. J Chem Ecol. 2010;36(10):1155–1169. doi: 10.1007/s10886-010-9856-5. [DOI] [PubMed] [Google Scholar]
- 17.Heckel DG. Smells like a new species: Gene duplication at the periphery. Proc Natl Acad Sci USA. 2010;107(21):9481–9482. doi: 10.1073/pnas.1004511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roelofs W, et al. Sex pheromone production and perception in European corn borer moths is determined by both autosomal and sex-linked genes. Proc Natl Acad Sci USA. 1987;84(21):7585–7589. doi: 10.1073/pnas.84.21.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardé RT, Kochansky J, Stimmel JF, Wheeler AG, Roelofs WL. Sex-pheromone of European corn-borer (Ostrinia-nubilalis) - cis-responding and trans-responding males in Pennsylvania. Environ Entomol. 1975;4(3):413–414. [Google Scholar]
- 20.Klun JA. Isolation of a sex pheromone of the European corn borer. J Econ Entomol. 1968;61(2):484–487. [Google Scholar]
- 21.Malausa T, et al. Assortative mating in sympatric host races of the European corn borer. Science. 2005;308(5719):258–260. doi: 10.1126/science.1107577. [DOI] [PubMed] [Google Scholar]
- 22.Klun JA, Maini S. Genetic basis of an insect chemical communication system: The European corn borer Lepidoptera, Pyralidae. Environ Entomol. 1979;8(3):423–426. [Google Scholar]
- 23.Buechi R, Priesner E, Brunetti R. Das sympatrische vorkommen von zwei pheromonstammen des Maiszunslers, Ostrinia nubilalis Hbn., in der Sudschweiz. Mitt Schweiz Entomol Ges. 1982;55(1-2):33–53. [Google Scholar]
- 24.Roelofs WL, Du JW, Tang XH, Robbins PS, Eckenrode CJ. Three European corn borer populations in New York based on sex pheromones and voltinism. J Chem Ecol. 1985;11(7):829–836. doi: 10.1007/BF01012071. [DOI] [PubMed] [Google Scholar]
- 25.Bengtsson BO, Löfstedt C. No evidence for selection in a pheromonally polymorphic moth population. Am Nat. 1990;136(5):722–726. [Google Scholar]
- 26.Glover TJ, Campbell MG, Linn CE, Roelofs WL. Unique sex-chromosome mediated behavioral-response specificity of hybrid male European corn-borer moths. Experientia. 1991;47(9):980–984. [Google Scholar]
- 27.Lassance JM, et al. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature. 2010;466(7305):486–489. doi: 10.1038/nature09058. [DOI] [PubMed] [Google Scholar]
- 28.Lassance JM, et al. Functional consequences of sequence variation in the pheromone biosynthetic gene pgFAR for Ostrinia moths. Proc Natl Acad Sci USA. 2013;110(10):3967–3972. doi: 10.1073/pnas.1208706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carraher C, Authier A, Steinwender B, Newcomb RD. Sequence comparisons of odorant receptors among tortricid moths reveal different rates of molecular evolution among family members. PLoS One. 2012;7(6):e38391. doi: 10.1371/journal.pone.0038391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krieger J, et al. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens) Proc Natl Acad Sci USA. 2004;101(32):11845–11850. doi: 10.1073/pnas.0403052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lassance J-M, Bogdanowicz SM, Wanner KW, Löfstedt C, Harrison RG. Gene genealogies reveal differentiation at sex pheromone olfactory receptor loci in pheromone strains of the European corn borer, Ostrinia nubilalis. Evolution. 2011;65(6):1583–1593. doi: 10.1111/j.1558-5646.2011.01239.x. [DOI] [PubMed] [Google Scholar]
- 32.Leary GP, et al. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci USA. 2012;109(35):14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinwender B, Thrimawithana AH, Crowhurst RN, Newcomb RD. Pheromone receptor evolution in the cryptic leafroller species, Ctenopseustis obliquana and C. herana. J Mol Evol. 2015;80(1):42–56. doi: 10.1007/s00239-014-9650-z. [DOI] [PubMed] [Google Scholar]
- 34.Yasukochi Y, Miura N, Nakano R, Sahara K, Ishikawa Y. Sex-linked pheromone receptor genes of the European corn borer, Ostrinia nubilalis, are in tandem arrays. PLoS One. 2011;6(4):e18843. doi: 10.1371/journal.pone.0018843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koutroumpa FA, et al. Shifts in sensory neuron identity parallel differences in pheromone preference in the European corn borer. Front Ecol Evol. 2014;2(65):1–12. [Google Scholar]
- 36.Wanner KW, et al. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS One. 2010;5(1):e8685. doi: 10.1371/journal.pone.0008685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroemer JA, et al. A rearrangement of the Z chromosome topology influences the sex-linked gene display in the European corn borer, Ostrinia nubilalis. Mol Genet Genomics. 2011;286(1):37–56. doi: 10.1007/s00438-011-0624-1. [DOI] [PubMed] [Google Scholar]
- 38.Glover TJ, Tang XH, Roelofs WL. Sex pheromone blend discrimination by male moths fromE andZ strains of European corn borer. J Chem Ecol. 1987;13(1):143–151. doi: 10.1007/BF01020358. [DOI] [PubMed] [Google Scholar]
- 39.Wadsworth CB, Li X, Dopman EB. A recombination suppressor contributes to ecological speciation in OSTRINIA moths. Heredity (Edinb) 2015;114(6):593–600. doi: 10.1038/hdy.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kárpáti Z, Tasin M, Cardé RT, Dekker T. Early quality assessment lessens pheromone specificity in a moth. Proc Natl Acad Sci USA. 2013;110(18):7377–7382. doi: 10.1073/pnas.1216145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312(5774):748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 42.Badri KR, Zhou Y, Dhru U, Aramgam S, Schuger L. Effects of the SANT domain of tension-induced/inhibited proteins (TIPs), novel partners of the histone acetyltransferase p300, on p300 activity and TIP-6-induced adipogenesis. Mol Cell Biol. 2008;28(20):6358–6372. doi: 10.1128/MCB.00333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voigt A, Pflanz R, Schäfer U, Jäckle H. Perlecan participates in proliferation activation of quiescent Drosophila neuroblasts. Dev Dyn. 2002;224(4):403–412. doi: 10.1002/dvdy.10120. [DOI] [PubMed] [Google Scholar]
- 44.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23(14):2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villanyi Z, Collart MA. Ccr4-Not is at the core of the eukaryotic gene expression circuitry. Biochem Soc Trans. 2015;43(6):1253–1258. doi: 10.1042/BST20150167. [DOI] [PubMed] [Google Scholar]
- 46.Pradhan SJ, et al. The conserved P body component HPat/Pat1 negatively regulates synaptic terminal growth at the larval Drosophila neuromuscular junction. J Cell Sci. 2012;125(Pt 24):6105–6116. doi: 10.1242/jcs.113043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Estrada B, Gisselbrecht SS, Michelson AM. The transmembrane protein Perdido interacts with Grip and integrins to mediate myotube projection and attachment in the Drosophila embryo. Development. 2007;134(24):4469–4478. doi: 10.1242/dev.014027. [DOI] [PubMed] [Google Scholar]
- 48.Schnorrer F, Kalchhauser I, Dickson BJ. The transmembrane protein Kon-tiki couples to Dgrip to mediate myotube targeting in Drosophila. Dev Cell. 2007;12(5):751–766. doi: 10.1016/j.devcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 49.Schnorrer F, Dickson BJ. Muscle building; mechanisms of myotube guidance and attachment site selection. Dev Cell. 2004;7(1):9–20. doi: 10.1016/j.devcel.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16(2):75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 51.Moberg KH, Mukherjee A, Veraksa A, Artavanis-Tsakonas S, Hariharan IK. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr Biol. 2004;14(11):965–974. doi: 10.1016/j.cub.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 52.Nicholson SC, Nicolay BN, Frolov MV, Moberg KH. Notch-dependent expression of the archipelago ubiquitin ligase subunit in the Drosophila eye. Development. 2011;138(2):251–260. doi: 10.1242/dev.054429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Godt D, Laski FA. Mechanisms of cell rearrangement and cell recruitment in Drosophila ovary morphogenesis and the requirement of bric à brac. Development. 1995;121(1):173–187. doi: 10.1242/dev.121.1.173. [DOI] [PubMed] [Google Scholar]
- 54.Chu J, Dong PDS, Panganiban G. Limb type-specific regulation of bric a brac contributes to morphological diversity. Development. 2002;129(3):695–704. doi: 10.1242/dev.129.3.695. [DOI] [PubMed] [Google Scholar]
- 55.Godt D, Couderc JL, Cramton SE, Laski FA. Pattern formation in the limbs of Drosophila: bric à brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development. 1993;119(3):799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- 56.Li Q, et al. A functionally conserved gene regulatory network module governing olfactory neuron diversity. PLoS Genet. 2016;12(1):e1005780. doi: 10.1371/journal.pgen.1005780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kárpáti Z, Dekker T, Hansson BS. Reversed functional topology in the antennal lobe of the male European corn borer. J Exp Biol. 2008;211(Pt 17):2841–2848. doi: 10.1242/jeb.017319. [DOI] [PubMed] [Google Scholar]
- 58.Kárpáti Z, Olsson S, Hansson BS, Dekker T. Inheritance of central neuroanatomy and physiology related to pheromone preference in the male European corn borer. BMC Evol Biol. 2010;10:286. doi: 10.1186/1471-2148-10-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307(5715):1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- 60.Sakurai T, et al. A single sex pheromone receptor determines chemical response specificity of sexual behavior in the silkmoth Bombyx mori. PLoS Genet. 2011;7(6):e1002115–e1002115. doi: 10.1371/journal.pgen.1002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujii T, et al. Sex-linked transcription factor involved in a shift of sex-pheromone preference in the silkmoth Bombyx mori. Proc Natl Acad Sci USA. 2011;108(44):18038–18043. doi: 10.1073/pnas.1107282108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37(5):827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- 63.Jafari S, et al. Combinatorial activation and repression by seven transcription factors specify Drosophila odorant receptor expression. PLoS Biol. 2012;10(3):e1001280–e1001280. doi: 10.1371/journal.pbio.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Linn C, Poole K, Zhang A, Roelofs W. Pheromone-blend discrimination by European corn borer moths with inter-race and inter-sex antennal transplants. J Comp Physiol A. 1999;184(3):273–278. [Google Scholar]
- 65.Hansson BS, Hallberg E, Löfstedt C, Steinbrecht RA. Correlation between dendrite diameter and action potential amplitude in sex pheromone specific receptor neurons in male Ostrinia nubilalis (Lepidoptera: Pyralidae) Tissue Cell. 1994;26(4):503–512. doi: 10.1016/0040-8166(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 66.Hansson BS, Löfstedt C, Roelofs WL. Inheritance of olfactory response to sex pheromone components in Ostrinia nubilalis. Naturwissenschaften. 1987;74(10):497–499. [Google Scholar]
- 67.Cosse AA, et al. Pheromone behavioral-responses in unusual male European corn-borer hybrid progeny not correlated to electrophysiological phenotypes of their pheromone-specific antennal neurons. Experientia. 1995;51(8):809–816. [Google Scholar]
- 68.Kárpáti Z, Molnar B, Szoecs G. Pheromone titer and mating frequency of E- and Z-strains of the European corn borer, Ostrinia nubilalis: Fluctuation during scotophase and age dependence. Acta Phytopathol Entomol Hung. 2007;42(2):331–341. [Google Scholar]
- 69.Mani E, Riggenbach W, Mendik M. Rearing of the codling moth Laspeyresia pomonella on artificial diet 1968-1978. Mitt Schweiz Entomol Ges. 1978;51(4):315–326. [Google Scholar]
- 70.Heckel DG. Comparative genetic linkage mapping in insects. Annu Rev Entomol. 1993;38:381–408. [Google Scholar]
- 71.Sheck AL, et al. Genetics of sex pheromone blend differences between Heliothis virescens and Heliothis subflexa: A chromosome mapping approach. J Evol Biol. 2006;19(2):600–617. doi: 10.1111/j.1420-9101.2005.00999.x. [DOI] [PubMed] [Google Scholar]
- 72.Lander ES, et al. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1(2):174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 73.Lincoln S, Daly M, Lander E. Whitehead Institute Technical Report. Whitehead Institute for Biomedical Research; Cambridge, MA: 1993. Mapping genes controlling quantitative traits using MAPMAKER/QTL 1.1: A tutorial and reference manual, 2nd edition. [Google Scholar]
- 74.Paterson AH, et al. Mendelian factors underlying quantitative traits in tomato: Comparison across species, generations, and environments. Genetics. 1991;127(1):181–197. doi: 10.1093/genetics/127.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han S, et al. The E3 ubiquitin ligase protein associated with Myc (Pam) regulates mammalian/mechanistic target of rapamycin complex 1 (mTORC1) signaling in vivo through N- and C-terminal domains. J Biol Chem. 2012;287(36):30063–30072. doi: 10.1074/jbc.M112.353987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Guyader S, Maier J, Jesuthasan S. Esrom, an ortholog of PAM (protein associated with c-myc), regulates pteridine synthesis in the zebrafish. Dev Biol. 2005;277(2):378–386. doi: 10.1016/j.ydbio.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 77.Murthy V, et al. Pam and its ortholog highwire interact with and may negatively regulate the TSC1 center dot TSC2 complex. J Biol Chem. 2004;279(2):1351–1358. doi: 10.1074/jbc.M310208200. [DOI] [PubMed] [Google Scholar]
- 78.Wan HI, et al. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26(2):313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 79.Sia GM, et al. Interaction of the N-terminal domain of the AMPA receptor GIuR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55(1):87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 80.Datta S, Kankel DR. L(1) trol and L(1)Devl, loci affecting the development of the adult central-nervous system in Drosophila melanogaster. Genetics. 1992;130(3):523–537. doi: 10.1093/genetics/130.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park Y, et al. Drosophila Perlecan modulates FGF and Hedgehog signals to activate neural stem cell division. Dev Biol. 2003;253(2):247–257. doi: 10.1016/s0012-1606(02)00019-2. [DOI] [PubMed] [Google Scholar]
- 82.Thoden JB, Timson DJ, Reece RJ, Holden HM. Molecular structure of human galactose mutarotase. J Biol Chem. 2004;279(22):23431–23437. doi: 10.1074/jbc.M402347200. [DOI] [PubMed] [Google Scholar]
- 83.Werner T, et al. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97(25):13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fisher RA. The use of multiple measurements in taxonomic problems. Ann Eugen. 1936;7:179–188. [Google Scholar]