Significance
In in vitro fertilization (IVF), current methods of diagnosing chromosome abnormality and screening for viability of transfer require biopsy of embryos, which affects embryo quality, awaits long-term biosafety test, and requires specialized skills. We demonstrate the principle of noninvasive chromosome screening (NICS), which is based on sequencing the genomic DNA secreted into the culture medium from the embryo, avoiding the need for embryo biopsy and substantially increasing the safety. By characterizing its precision and demonstrating successful live births, we validate that NICS offers the potential of significantly improving the clinical outcome of IVF.
Keywords: chromosomal abnormalities, PGS, IVF, WGA, MALBAC
Abstract
Preimplantation genetic screening (PGS) is widely used to select in vitro-fertilized embryos free of chromosomal abnormalities and to improve the clinical outcome of in vitro fertilization (IVF). A disadvantage of PGS is that it requires biopsy of the preimplantation human embryo, which can limit the clinical applicability of PGS due to the invasiveness and complexity of the process. Here, we present and validate a noninvasive chromosome screening (NICS) method based on sequencing the genomic DNA secreted into the culture medium from the human blastocyst. By using multiple annealing and looping-based amplification cycles (MALBAC) for whole-genome amplification (WGA), we performed next-generation sequencing (NGS) on the spent culture medium used to culture human blastocysts (n = 42) and obtained the ploidy information of all 24 chromosomes. We validated these results by comparing each with their corresponding whole donated embryo and obtained a high correlation for identification of chromosomal abnormalities (sensitivity, 0.882, and specificity, 0.840). With this validated NICS method, we performed chromosome screening on IVF embryos from seven couples with balanced translocation, azoospermia, or recurrent pregnancy loss. Six of them achieved successful clinical pregnancies, and five have already achieved healthy live births thus far. The NICS method avoids the need for embryo biopsy and therefore substantially increases the safety of its use. The method has the potential of much wider chromosome screening applicability in clinical IVF, due to its high accuracy and noninvasiveness.
Human embryos are prone to chromosomal abnormalities, mainly due to age-dependent chromosome segregation errors during meiosis I (1). Chromosomal abnormalities could cause early pregnancy loss or severe chromosomal diseases such as Down and Patau syndrome among many others (2, 3). The occurrence of chromosomal abnormalities is substantially higher in patients of advanced maternal age, patients with recurrent pregnancy loss, or those who carry chromosomal aberrations such as translocations, all of which result in poor clinical outcome for reproduction.
Chromosomal abnormalities can be prevented in in vitro fertilization (IVF) by performing preimplantation genetic screening (PGS) of all 24 chromosomes. There are various PGS methods for comprehensive chromosome screening currently in clinical use, including comparative genomic hybridization (array-CGH) (4, 5), single-nucleotide polymorphism (SNP) arrays (6–9), multiplex quantitative PCR (10), and next-generation sequencing (NGS) (11, 12). Multiple clinical trials have confirmed the clinical efficacy of PGS, including increasing implantation and clinical pregnancy rates, as well as decreasing miscarriage rates (13–16). However, the applicability of PGS has been limited for a number of reasons: (i) PGS requires invasive embryo biopsy, which has been shown to decrease embryo quality after cleavage-stage biopsy (17); (ii) long-term biosafety of embryo biopsy in humans has not been evaluated, whereas animal studies have shown negative influences on neural and adrenal development (18–20); (iii) it involves technical expertise, requiring special training and experienced embryologists to perform the biopsy, which significantly increase the overall costs of clinical PGS cycles. Therefore, a noninvasive and easy-to-perform screening tool would greatly facilitate the widespread performing of chromosome screening before embryo implantation, thereby improving success rates.
Efforts have been made to develop noninvasive approaches for PGS (21, 22). Palini et al. (23) reported the observation of the existence of DNA in the blastocoele fluid, Gianaroli et al. (24) performed a pilot study on chromosome screening using blastocentesis, and Stigliani et al. (25, 26) observed genomic and mitochondria DNA contents in the culture medium, which were correlated with embryo quality. Wu et al. (27) reported the PCR detection of the secreted genomic DNA in the culture medium for preimplantation genetic diagnosis (PGD) of α-thalassemia cases. Although the mechanism of secretion is not known, it is most likely that the DNA in the media results from the apoptotic cells of the growing embryo. The genome-wide analysis of the secreted DNA in the culture medium has not been previously reported, and would provide the basis for PGS, assuming that the detected DNA arises from cells of the growing embryo. From the perspective of noninvasiveness and ease in handling the embryos, blastocyst culture medium would be an ideal source for chromosome screening. However, such an approach would require high sensitivity and reproducibility of the whole-genome amplification (WGA) of the DNA from the blastocyst culture medium. To the best of our knowledge, no previous studies have reported validation studies using blastocyst culture medium for noninvasive comprehensive screening of chromosomes to improve the clinical outcome of in vitro fertilization (IVF)/intracytoplasmic sperm injection (ICSI) patients.
In this study, we performed noninvasive chromosome screening (NICS) on spent culture medium samples used for growing human embryos from days 3–5 (D3–D5). The NICS assay was validated by comparing results obtained from the culture medium with the chromosome ploidy information obtained directly from the corresponding D5 whole embryos. We then calculated the sensitivity and specificity of the NICS assay in screening chromosomal abnormalities. After the NICS assay had been validated, we performed this assay on seven couples with balanced chromosomal translocation or recurrent pregnancy loss to select embryos with normal chromosomal ploidy. Six of the seven couples obtained successful pregnancies, and five have already achieved healthy live births.
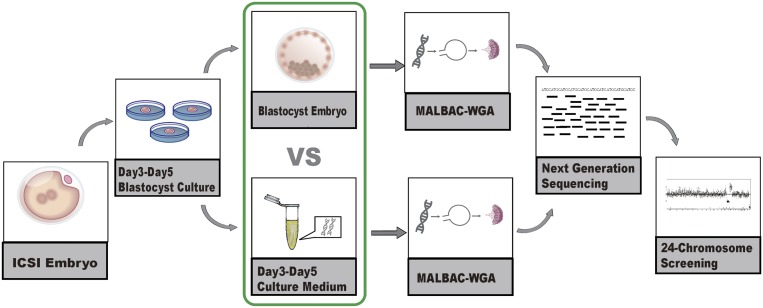
In brief, we performed validation experiments on 42 embryos. The donated and institutional review board (IRB)-approved embryos were created using ICSI and were vitrified on D3 and subsequently warmed and placed in blastocyst culture medium. We collected the spent culture media samples on D5 and performed WGA and sequencing by multiple annealing and looping-based amplification cycles (MALBAC)–NGS (28) (Fig. 1). The MALBAC-NGS protocol has been previously validated in performing PGS with cleavage-stage and blastocyst-stage biopsies (29–31), and is increasingly used for single-gene PGD combined with chromosomal PGS (30, 32). Similarly, we performed MALBAC-NGS on all 24 chromosomes from the corresponding D5 whole embryos, which we used as the gold standard to evaluate the chromosome screening results from the culture media.
Fig. 1.
Diagram of the validation procedure of the noninvasive chromosome screening (NICS) assay. Briefly, D3 embryos achieved via intracytoplasmic sperm injection (ICSI) were placed in blastocyst culture medium. Both the D3–D5 culture medium and the corresponding whole embryos were collected and used for WGA by multiple annealing and looping-based amplification cycles (MALBAC). Whole-genome–amplified products from both the D3–D5 culture medium and the embryo were sequenced using an Illumina HiSeq 2500 platform. The chromosome ploidy information was obtained from both the culture medium and the corresponding embryo.
Results
NICS Using Blastocyst Culture Medium.
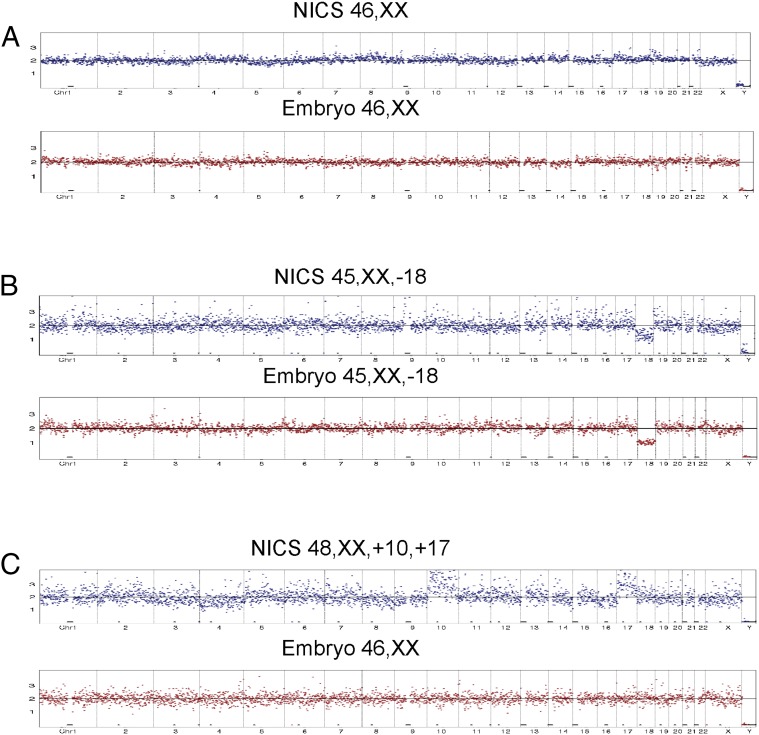
We sequenced ∼2 million reads on each culture medium sample using an Illumina HiSeq 2500 platform. The read numbers were counted along the 24 chromosomes with a bin size of 1 Mb, normalized by the mean of the corresponding bin in all samples. As shown in Fig. 2A, even distribution of reads along the chromosomes represents balanced chromosomal contents and thus a normal karyotype of the embryo. A chromosome loss in chromosome 18 results in a 50% decrease of the average read counts mapped to chr18 and was identified by the algorithm, as shown in Fig. 2B. Results for culture medium (Fig. 2, shown in blue) are displayed side by side with the results of the corresponding embryos (Fig. 2, shown in red). Fig. 2 A and B show two examples of matching results obtained from the NICS assay when directly compared with their corresponding embryos, from a normal/transferrable embryo (Fig. 2A) and an embryo with a chromosome loss at chr18 (Fig. 2B), respectively. Fig. 2C shows an example of a false positive, with the embryo showing a normal karyotype and the NICS assay identifying a chromosome gain at chr10 and chr17.
Fig. 2.
Examples of validation of results from the comparison of NICS versus the whole-blastocyst embryos. (A and B) Equivalent karyotype results obtained from NICS and the corresponding blastocyst embryo. Fig. 2A shows consistent results from a normal/transferrable embryo, and Fig. 2B shows consistent results from an embryo with a chromosome loss in chr18. (C) An example of inconsistent results obtained from NICS and the blastocyst embryo, with the embryo showing balanced chromosomal composition and the NICS assay identifying chromosome gains of chr10 and chr17.
Comparison of the Results from NICS and Their Corresponding Blastocyst Embryos.
To validate our results, we performed comparisons between 42 blastocyst culture medium samples and the NGS results of the corresponding blastocyst stage embryos. IRB approvals (Nanjing Jinlin: 2014NZKY-005; Wuxi Maternity: 2014-04-0515-02) were obtained. All of these embryos were voluntarily donated by patients with informed consent obtained before performing the experiments on each embryo. Results are summarized in Table 1.
Table 1.
Summary of the 42 samples profiled by NICS versus their corresponding blastocysts
| 21 Normal embryos (NICS and biopsy consistent) | 15 Abnormal embryos (NICS and biopsy consistent) | ||||
| Sample ID | NICS | Biopsy | Sample ID | NICS | Biopsy |
| EM02 | 46,XX | 46,XX | EM01 | 50,XX,+1,+5,+10,+13 | 47,XX,+4 |
| EM04 | 46,XX | 46,XX | EM10 | 45,XX,−18 | 45,XX,−18 |
| EM06 | 46,XY | 46,XY | EM11 | 46,XY,+5q | 46,XY,−5(p12→qter,∼135M,mos ∼30%) |
| EM07 | 46,XY | 46,XY | EM13 | 46,XX,−1p(pter→p21.1) | 46,XX,−1p(pter→p21.1,∼103M),+18q(q12.3→qter,∼31M, mos) |
| EM09 | 46,XY | 46,XY | EM14 | 45,XY,−18 | 46,XY,+1(p21.1→qter,∼142M),−18(pter→q21.31,∼56M) |
| EM16 | 46,XX | 46,XX | EM15 | 46,XX,+1p(pter→p21.2),−18(q21.32→qter) | 46,XX,+1p(pter→p21.2,∼97M),−18q(q21.32→qter,∼21M) |
| EM21 | 46,XY | 46,XY | EM17 | 55,XY,+5,+6,+8,+11,+17,+19,+20,+21,+22 | 46,XY,+1(p21.1→qter,∼143M),−18(pter→q21.31,∼58M) |
| EM22 | 46,XY | 46,XY | EM18 | 46,XX,+1p(pter→p21.1),−18(q21.32→qter) | 46,XX,+1p(pter→p21.3,∼100M),−18q(q21.31→qter,∼22M) |
| EM23 | 46,XY | 46,XY | EM19 | 46,XX,+14q(q23.3→qter),−15q(q26.1→qte) | 46,XX,+14q(q23.1→qter,∼45M),−15q(q26.1→qter,∼12M) |
| EM24 | 46,XY | 46,XY | EM20 | 46,XY,−1,+15 | 46,XY,−14,+15 |
| EM25 | 46,XX | 46,XX | EM33 | 52,XX,+4,+6,+9,+10,+14,+17 | 44,XX,−4,−14,+15(mos),−18(mos) |
| EM26 | 46,XY | 46,XY | EM35 | 45,XX,−16 | 45,XX,+14(q32.12→qter,∼13M),−16 |
| EM27 | 46,XY | 46,XY | EM37 | 45,XY,−22 | 45,XY,−22 |
| EM28 | 46,XY | 46,XY | EM41 | 50,XX,+15,+17,+18,+20 | 46,XX,−X(mos),+5(p12→q13.1,∼23M,mos),−5q(q13.1→qter,∼112M,mos) |
| EM29 | 46,XX | 46,XX | EM42 | 46,XX,−1p,+18q | 46,XX,−1p,+18q |
| EM30 | 46,XY | 46,XY | 4 False-positive embryos (NICS abnormal, biopsy normal) | ||
| EM31 | 46,XX | 46,XX | |||
| EM32 | 46,XX | 46,XX | EM03 | 48,XX,+10,+17 | 46,XX |
| EM34 | 46,XY | 46,XY | EM08 | 48,XY,+6,+18 | 46,XY |
| EM36 | 46,XY | 46,XY | EM12 | 45,XY,−18 | 46,XY |
| EM38 | 46,XX | 46,XX | EM39 | 47,XY,+22 | 46,XY |
| 2 False-negative embryos (NICS normal, biopsy abnormal) | |||||
| EM05 | 46,XX | 45,XO | |||
| EM40 | 46,XX | 49,XY,+6,+8,+14 | |||
By performing MALBAC-NGS, we successfully obtained information of 24-chromosome ploidy from all 42 samples (100%) using the D3–D5 culture medium as well as their corresponding blastocyst-stage embryos. By profiling whole embryos, 25 samples (59.5%) were identified as normal, in which 21 of them showed concordance with the NICS assay, and the remaining four samples showed chromosomal abnormalities (false positives), converting to a specificity of 84.0%. Out of the 17 embryos that were identified with chromosomal abnormalities under the blastocyst embryo assay, 15 of them showed chromosomal abnormalities with the NICS assay as well, but the remaining two were identified with a normal karyotype with the NICS assay (false negatives), resulting in a sensitivity of 88.2% (Table 1). The positive predictive value (PPV) and negative predictive value (NPV) of NICS in identification of chromosomal abnormalities are 78.9% and 91.3%, respectively. From the four embryos that were identified with chromosomal mosaicism (EM13, EM33, EM11, and EM41), all four were positive for chromosomal imbalance with NICS.
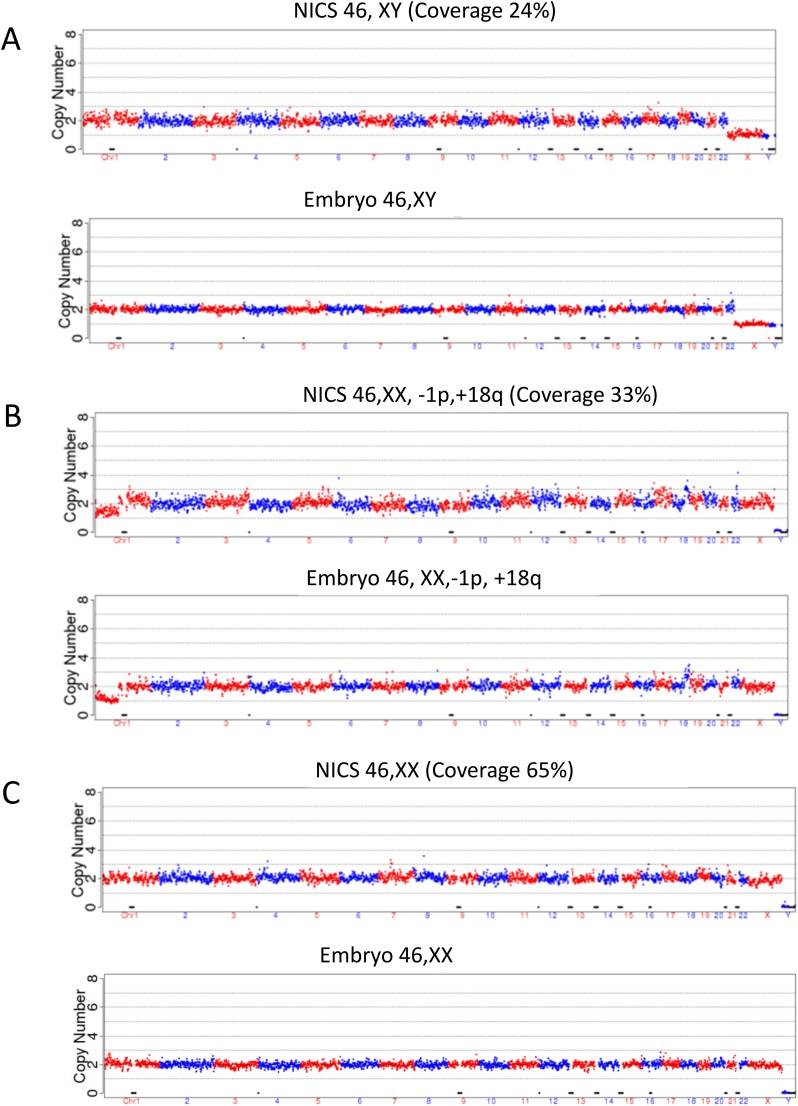
Coverages of 24%, 33%, and 65% were observed with NICS data of three spent cultures. The copy number variation (CNV) results for embryos EM23, EM42, and EM38, by high-depth sequencing (30×) are shown in Fig. S1.
Fig. S1.
CNVs of embryos EM23, EM42, and EM38 determined by high-sequencing depth (30×). Top graph corresponds to spent culture media, and bottom graph is for the corresponding biopsy of embryo at blastocyst stage. (A) NICS and trophectoderm (TE) biopsy results for EM23 with a NICS coverage rate of 24.1%. (B) NICS and TE biopsy results for EM42 with a NICS coverage rate of 32.9%. (C) NICS and TE biopsy results of EM38 with a NICS coverage of 65.0%. Note the consistency of the results in all NICS cases and the corresponding embryo biopsy. As shown in B, the copy number for chromosome 1 was one and not two, and therefore embryo EM42 could not be used for transfer.
Embryo Selection by NICS on the First Patient with Balanced Translocation.
With the NICS assay validated by comparison with the voluntarily donated and IRB-approved embryos, we first applied our NICS method on a patient with a balanced translocation. IRB approval (Wuxi Maternity: 2014-04-0515-02) and informed consent were obtained before applying the NICS assay on the embryos. Karyotype analysis of the patient showed a balanced translocation t(14;15)(q22;q24). We obtained a total of three blastocysts from this patient, and we performed NICS on D3–D5 culture medium of all three embryos. Chromosomal abnormalities were detected with the NICS assay in two of them, and therefore those could not be used for transfer (Fig. 3 A and B). To confirm these results, the two embryos were collected and lysed for chromosome screening using the whole embryo, and the same results were obtained, confirming the NICS analyses (Fig. S2). Only one out of the three blastocysts showed a normal karyotype with NICS, and therefore that one was selected for transfer (Fig. 3C). A successful pregnancy resulted from this selected embryo, and a karyotype of the developing embryo was obtained by performing amniocentesis at 19 wk of gestation, confirming the karyotype results previously obtained with the NICS assay. The patient’s pregnancy resulted in the live birth of a chromosomally normal and healthy baby boy on March 5, 2016.
Fig. 3.
Embryo screening and selection using NICS from a patient carrying a balanced translocation of chr14/15. A total of three embryos successfully developed to the blastocyst stage, and D3–D5 culture medium from each embryo was collected for the NICS assay to screen for chromosomal abnormalities. (A and B) Embryos showed chromosomal abnormalities with chr14/15 and therefore could not be transferred. (C) An embryo showed balanced chromosomal composition and was therefore transferred into the uterus of the patient, resulting in a successful pregnancy and a healthy live birth.
Fig. S2.
Chromosomal screen results from whole embryos (A and C), and their corresponding culture media (B and D). E corresponds to a whole live embryo with normal chromosome results using NICS. (B, D, and E) The NICS results for the three blastocysts. E shows a balanced chromosomal composition, and the embryo was therefore transferred into the uterus of the patient. B and D show chromosome gain/loss from chr14/chr15. Therefore, the corresponding embryos were not transferred but were lysed for chromosome screening to confirm the NICS results (A and C). NICS results for both embryos were consistent with chromosomal ploidy information from the blastocyst embryo.
NICS Clinical Application Results in Successful Pregnancies.
After careful and systematic validation of the NICS assay, we have performed NICS on six more patients, in addition to the translocation patient described above. Single-blastocyst transfer was performed on all six patients, and five of these patients achieved successful pregnancies; five of these have already delivered chromosomally normal, healthy newborns, and we continue to follow up on the last currently ongoing pregnancy. Only one patient failed to implant. The clinical indications and outcomes are summarized in Table 2.
Table 2.
Clinical outcome of the first seven patients subjected to NICS
| Patient no. | Maternal age | Clinical indications | Transfer cycles | Clinical outcome |
| P01 | 30 | Reciprocal translocation 46,XY,t(14;15) | 1 | Singleton pregnancy—live birth |
| P02 | 28 | Azoospermia | 1 | Singleton pregnancy—live birth |
| P03 | 34 | Inversion 46,XY,inv(9) | 1 | Singleton pregnancy—live birth |
| P04 | 32 | Reciprocal translocation 46,XX,t(1;18) | 2 | Implantation failure |
| P05 | 26 | Recurrent pregnancy loss | 1 | Singleton pregnancy—live birth |
| P06 | 32 | 47,XYY | 2 | Singleton pregnancy—live birth |
| P07 | 29 | Recurrent implantation failure | 1 | Singleton pregnancy—following up |
Discussion
Blastocyst Biopsy and the Controversy of Extensive PGS for Patients Under IVF/ICSI Treatment.
Blastocyst trophectoderm biopsy has been increasingly used and widely accepted in the PGS field due to its relatively low invasiveness, compared with performing blastomere biopsy at the cleavage stage. Chromosome screening on all 24 chromosomes has been mostly used on patients with advanced maternal age, recurrent pregnancy loss, repeated implantation failure, as well as on patients with abnormal karyotype such as balanced translocation and Robertsonian translocation (32–37). Extensive use of PGS on all IVF/ICSI cycles has been hotly debated in the past few years (38–41) due to the invasiveness of the biopsy procedure itself, particularly regarding the potential harm on the trophectoderm and possible compromise of implantation potential, as well as potential concerns on long-term effects on the offspring, which are very difficult to assess. In addition, the procedure of performing blastocyst-stage biopsy requires considerable training and expertise to perform the sophisticated embryo manipulation, increasing the costs of performing PGS. Of note, the procedure we report here, which simply involves collecting embryo culture medium, requires no special expertise in embryo manipulation and therefore can be potentially used on all IVF/ICSI cycles, thereby holding promise to improve overall clinical success rates.
False Positives and False Negatives in NICS.
We observed two false negatives (2 of 17) and four false positives (4 of 25) with the NICS assay, converting to a false-negative rate and a false-positive rate of chromosomal abnormalities identification of 11.8% and 16.0%, respectively. The two false negatives might have resulted from contamination from the cumulus cells, which are maternal in origin and normally have a balanced chromosomal content (Table 1). In the future, the false-negative rate could be further minimized by carefully and thoroughly removing all cumulus cells before embryo culture.
The false positives most likely arose from mosaicism. It was previously hypothesized that, during embryo development, embryos tend to exclude those cells with mitotic errors to the exterior of the embryo (42). It is therefore possible that the false-positive rate of 16.0% arose from the debris in the culture medium, originating primarily from cells eliminated from the embryo.
Although the false-positives and false-negatives could come from measurements, we note that MALBAC provides better WGA evenness and hence higher precision for CNV determination compared with degenerate oligonucleotide-primed PCR and multiple displacement amplification methods (43).
The NPV of chromosomal abnormalities with the NICS assay is 91.3%, which is substantially higher than the PPV (78.9%) of the assay. The high NPV suggests that the assay is more effective in selecting normal and transferrable embryos than identifying embryos with chromosomal abnormalities. Single-embryo transfer has been increasingly used due to its effectiveness in decreasing multiple pregnancy and miscarriage rates (15, 44). As such, we propose NICS as an additional, and risk-free, procedure for single-embryo transfer.
Regarding the clinical implications of the false positives and false negatives, we note that, in the case of all embryos detected to exhibit aneuploidy by NICS, false positives can in principle be verified by further sequencing a blastocyst biopsy on the sixth day. We are currently developing a system in which NICS can be done rapidly and results can be obtained before embryos are frozen so that a cell or two can be extracted for verification. False negatives, on the other hand, usually do not result in successful pregnancies, and hence are less problematic. Even in the case of leading to pregnancy, they can be detected and avoided by noninvasive prenatal test.
Genome Coverage and Estimated DNA Amount in Spent Culture.
The genome coverages of the spent culture media for the three samples were determined to be 24%, 33%, and 65% by high-sequencing depth of 30× reads. It has been shown that the normal genome coverage for single diploid human cells is about ∼72% with high-sequencing depth (30×) (43). Despite of our low coverage, the CNV results matched exactly their corresponding blastocyst biopsies (a few cells) on the fifth day, in either the normal or aneuploid samples (Fig. S1). Larger cell numbers usually result in saturation of the genome coverage, approaching unity. Our coverage results suggest that the loss or degradation of DNA fragments in the spent culture must occur randomly along the genome and does not affect the inference of the copy number pattern in the embryo. The fact that we observed identical copy number patterns in the spent culture samples and their corresponding embryo biopsies suggests that, under our experimental conditions, the total amount of DNA in each spent culture is equivalent to that of a fraction of a single cell.
Limitations of the NICS Assay for Extensive Chromosome Screening in IVF/ICSI Patients.
We think the limitations of NICS could primarily derive from two aspects: (i) the requirement to be scrupulous in the removal of all cumulus-corona radiata cells (which are of maternal origin and usually with normal chromosomal composition) before performing ICSI. If such removal is not complete, residual cells may release DNA during embryo development, thereby potentially being the cause of false-negative detection. (ii) Similar to PGS, the NICS procedure would be highly recommended to be performed in conjunction with ICSI due to the difficulty of ensuring removal of any supernumerary sperm attached to the zona pellucida. Although culture medium is replaced on D3, which may decrease the likelihood of contamination due to residual cumulus cells and supernumerary sperm, all precautions should be made to reduce such contamination to a minimum if NICS is used routinely in clinical IVF. Of note, in the current stage of technological development, we consider NICS to be a screening assay for chromosomal-level abnormalities instead of being a diagnostic assay for segmental aneuploidies. More validation research is needed to identify segmental aneuploidies with the NICS assay.
In summary, our validation data and initial clinical applications strongly suggest that the NICS assay could help to improve the clinical outcome of IVF embryo selection with ploidy information, in a noninvasive manner. We envision that randomized clinical trials will be designed and performed in the near future, using the NICS assay with single-embryo transfer to evaluate the clinical effectiveness of the assay in different patient groups.
Materials and Methods
Embryo Preparation for the NICS Assay.
We recruited 17 patients from the Reproductive Medicine Centre of Wuxi Maternity and Child Health Hospital and the Reproductive Medical Center of Nanjing Jinling Hospital. IRB approvals (Nanjing Jinlin: 2014NZKY-005; Wuxi Maternity: 2014-04-0515-02) and informed consent were obtained before applying the NICS assay on the embryos. All donated embryos were in excess of clinical needs, and consents on donated D3 embryos were obtained for use in the comparison study. All embryos were fertilized by (ICSI. Donated D3 embryos were previously frozen by vitrification (Cryotop Safety Kit; KITAZATO BioPharma) according to the manufacturer’s instructions and stored in liquid nitrogen. Vitrified embryos were warmed using warming media (Vitrification Thaw Solution; KITAZATO BioPharma). Briefly, the Cryotop strip was quickly immersed into Thawing Solution (containing 1 M sucrose) for 1 min at 37 °C. Embryos were then transferred into dilution solution (containing 0.5 M sucrose) and incubated for 3 min, followed by incubation in two 80-µL droplets of Washing Solution for 3 min each at room temperature (25 °C).
Blastocyst Culture and Transfer.
Warmed D3 embryos were placed in 30-µL droplets of Quinn’s Advantage Protein Plus blastocyst medium (SAGE) containing washed and pregassed mineral oil (SAGE), and they were then further cultured to the blastocyst stage under 5.5% CO2, 5% O2, and balance N2 at 37 °C in Labotect C16 incubators (Labotect). After 2 d in culture, the development and quality of the blastocysts were evaluated according to the blastocyst scoring system. A single blastocyst was selected for transfer to each patient based on the NICS results.
Sample Collection.
To prevent media cross-contamination, different Pasteur pipettes were used for each embryo. Five to 20 μL of blastocyst medium from each embryo was transferred into RNase–DNase-free PCR tubes containing 5 μL of cell lysis buffer (Yikon Genomics). As a negative control, we collected the same amount of blastocyst culture medium but without its being used for embryo culture. All collected samples were frozen immediately in liquid nitrogen and stored at −80 °C until subjected to the NICS assay.
Embryo Selection by NICS on a Patient with Balanced Translocation.
The patient couple for NICS treatment in Wuxi Maternity reproduction center had 3 y of infertility history. Their chromosome examination showed that the male partner carries a t(14;15)(q22;q24) translocation, and the female has a normal karyotype. We collected five eggs in the IVF cycle (SI Materials and Methods); four MII oocytes were fertilized by ICSI, and three of them developed to the blastocyst stage. All blastocysts were frozen with vitrification freezing protocol after performing NICS (SI Materials and Methods). We obtained normal karyotype from one embryo with the NICS assay (Fig. 3C). We therefore transferred this normal embryo and confirmed clinical pregnancy with enhanced β-HCG followed up by ultrasound. The result of fetal chromosome examination in the amniotic fluid at 19 wk of gestation confirmed the balanced chromosome results previously obtained by NICS. After obtaining informed consent from the patients, the two embryos with chromosome imbalance, according to our NICS results, were lysed and sequenced for validation purposes (Fig. 1).
WGA and DNA Sequencing from Culture Medium of the Embryos.
The MALBAC single-cell WGA method was used to amplify DNA from the culture medium, as well as from the embryos, following the manufacturer’s protocol (catalog no. YK001B; Yikon Genomics), and used to construct the sequencing libraries (NEBNext Ultra DNA Kit; New England Biolabs) for sequencing on an Illumina HiSeq 2500 platform, yielding ∼2 million sequencing reads on each sample. The read numbers were counted along the whole genome with a bin size of 1 Mb. A copy number gain from two to three copies results in a 50% increase in read counts, whereas a copy number loss from two copies to one copy results in a 50% decrease in read counts.
SI Materials and Methods
Embryo Preparation for the NICS Assay.
We recruited 17 patients from the Reproductive Medicine Center of Wuxi Maternity and Child Health Hospital and the Reproductive Medical Center of Nanjing Jinling Hospital. We obtained consents for donated day 3 (D3) embryos used for the comparison study. All embryos were fertilized by intracytoplasmic sperm injection (ICSI), an assisted reproductive technology (ART) used for injection of a single sperm directly into an egg. Donated D3 embryos were previously frozen by vitrification according to manufacturer’s instructions (Cryotop Safety Kit; KITAZATO BioPharma) and stored in liquid nitrogen. Frozen embryos were warmed up using warming media (Vitrification Thaw Solution; KITAZATO BioPharm). Briefly, the Cryotop strip was quickly immersed into the thawing solution (containing 1 M sucrose), for 1 min at 37 °C. Embryos were then transferred into dilution solution (containing 0.5 M sucrose), incubated for 3 min, and rinsed in two 80-µL droplets of washing solution for 3 min each at room temperature (25 °C).
Blastocyst Culture and Transfer.
Warmed D3 embryos were placed in 30-µL droplets of Quinn’s Advantage Protein Plus blastocyst medium (SAGE) containing washed and pregassed mineral oil (SAGE); they were then further cultured to the blastocyst stage under 5.5% (vol/vol) CO2, 5% (vol/vol) O2, and balance N2 at 37 °C in Labotect C16 incubators (Labotect). After 2 or 3 d in culture, the development and quality of the blastocysts were evaluated according to the blastocyst scoring system. A single blastocyst was selected for transfer to each patient based on the noninvasive chromosome screening (NICS) results.
Sample Collection.
To prevent media cross-contamination, different Pasteur pipettes were used for each embryo. For final confirmation, blastocysts were transferred into RNase–DNase-free PCR tubes that contained 5 µL of cell lysis buffer (Yikon Genomics). As a negative control, we collected the same amount of blastocyst culture medium but without the embryo in culture. All collected samples were frozen immediately in liquid nitrogen and stored at −80 °C until the NICS assay was performed.
Embryo Selection by NICS on a Patient with Balanced Translocation.
Embryo 3 showed normal karyotype with the NICS assay, therefore this embryo was selected for transfer during a hormone replacement cycle. Seven days after the embryo was transferred, a blood test revealed normal levels of β-human chorionic gonadotrophin (β-HCG) of 89.69 mIU/mL. After 7 wk of gestation, the presence of a single heartbeat was confirmed by ultrasound. During midstage pregnancy (19 wk), fetal chromosome examination in the amniotic fluid showed translocation carrying as 46, XY, t(14;15). This patient pregnancy is expected to develop to term without predictable medical problems. After informed consent from the patients, the two embryos with chromosome imbalance, according to our NICS results, were lysed and sequenced to validate the results obtained by NICS.
Whole-Genome Amplification and Sequencing of DNA from Culture Medium and Embryos.
To amplify the DNA from the culture medium as well as from the embryos, we used MALBAC for single-cell whole-genome amplification (WGA) following the manufacturer’s protocol (catalog no. YK001B; Yikon Genomics). Briefly, 10 µL of culture medium were lysed followed by MALBAC reaction with a pool of random primers, each having a common 27-nt sequence and 8 variable nt. After eight cycles of quasilinear preamplification, 18 cycles of PCR amplification were performed, and ∼2 µg of DNA were obtained per reaction.
We started the amplification with 10 µL of culture medium for NICS and embryo for testing; 5 µL of lysis buffer were added, and the mix was put in a thermocycler for 90 min at 50 °C and 10 min at 80 °C. After that, 30 µL of preamplification buffer were added to the lysed DNA and incubated on the thermocycler for 3 min at 94 °C and eight cycles of 40 s at 20 °C, 40 s at 30 °C, 30 s at 40 °C, 30 s at 50 °C, 30 s at 60 °C, 4 min at 70 °C, 20 s at 95 °C, 10 s at 58 °C, and then stored at 4 °C indefinitely. The DNA was then further amplified by adding another 30 µL of amplification buffer and incubated for 30 s at 94 °C using 18 cycles of 20 s at 94 °C, 30 s at 58 °C, 3 min at 72 °C, and an additional 5 min at 72 °C.
The amplified DNA was used for sequencing on an Illumina HiSeq 2500 platform following the protocol previously described in Yan et al. (30), yielding ∼2 million sequencing reads on each sample. The read numbers were counted along the whole genome with a bin size of 1 Mb. A copy number gain from two to three copies results in a 50% increase in read counts, whereas a copy number loss from two copies to one copy results in a 50% decrease in read counts.
We randomly selected three spent culture media samples for NICS to measure the coverage rate. We performed high-depth sequencing (30×) and obtained coverage rates of 24%, 33%, and 65% for embryos EM23, EM43, and EM38, respectively.
Data Analysis.
Raw data from the sequencing library were trimmed with Trimmomatic-0.30 (45); adapters and low-quality bases (quality score less than 20) were removed. High-quality reads were aligned to the University of California, Santa Cruz, human reference genome (hg19) (University of California, Santa Cruz, Genome Browser; genome.ucsc.edu/), using the Burrows–Wheeler Aligner 0.7.4 (46) with default parameters. The aligned reads were sorted with Picard 1.92 (available for download at https://broadinstitute.github.io/picard/). Chromosomal copy number variations (CNVs) were determined with local Perl (available at www.perl.org/) scripts; unique mapped reads were normalized to relative reads number after GC correction in 1,000-kb bins GC correction: unique mapped reads, sequencing reads GC content, genome GC content were calculated in each bin; bins with no reads and zero/100% GC content were removed. Average of sequencing reads GC content and genome GC content was defined as GC_contenti in each bin. Reads of bin i with GC contenti were assigned with a weight wi = M/MGCi, where M is the average number of sequencing reads in each bin on autosomes, MGCi is the average number of sequencing reads in each bin, which is calculated for every 1% GC content. The GC corrected reads number is RNGCi = RNri*wi, where RCri is raw reads number of bin i. The CNVs visualization was performed by R programming language (available at www.r-project.org/). Detailed scripts have been submitted to GitHub: https://github.com/bspatYK/bioinformatics.
History of Patient Treatments.
Patient 1 came to our clinic seeking NICS treatment after 3 y of infertility. Chromosome examination showed that the female has normal karyotype, but the male partner carries a t(14;15)(q22;q24) translocation. The couple had a history of pregnancy loss in 2005 and had naturally conceived a healthy girl in 2007. The female blood hormone test results at day 2 were as follows: follicle-stimulating hormone (FSH), 15.77 mIU/mL; luteinizing hormone (LH), 10.08 mIU/mL; estradiol (E2), 26.26 pg/mL. Ultrasound test showed that antral follicle count was six to seven for the left ovary and five to six for the right ovary. Based on her ovary reservation, we performed the mild stimulation protocol. We collected three eggs in the first cycle of IVF treatment; two MII oocytes among them were fertilized by ICSI, but none of them developed to the cleavage stage. We used the same stimulation protocol in the second cycle and collected four eggs; three MII oocytes among them were fertilized by ICSI, and only one developed to cleavage embryo stage (eight cells). Similarly, we collected five eggs in the third cycle of IVF treatment; four MII oocytes among them were fertilized by ICSI, and three of them developed to the blastocyst stage. All blastocyst embryos were frozen with vitrification freezing protocol after NICS.
Patient 2 had 5 y of infertility history on coming to our clinic for NICS treatment. The female was 28 y of age, and the male, who was 29 y of age, presented with male azoospermia. The karyotype of the male partner was 46, XY, 15p+, and the karyotype of the female was 46, XX. The patient was recommended to undergo ICSI treatment. The female blood hormone test results on cycle day 2 were as follows: FSH, 5.93 mIU/mL; LH, 5.67 mIU/mL; E2, 27.38 pg/mL. The B ultrasound test, a B-Mode 2D ultrasound image display composed of bright dots representing the ultrasound echoes, showed that antral follicle count for both ovaries was about 10. The standard long stimulation protocol was performed according to the ovarian reserve. During the IVF treatment, 10 eggs were collected; of those, 7 MII oocytes were fertilized by ICSI, with 5 of them developing to the blastocyst stage. All blastocysts were frozen with the vitrification freezing protocol after noninvasive screening. One blastocyst was thawed (no. 1) and transferred into the female during hormone replacement cycle. Seven days after the embryo transfer, blood tests were performed, and results showed a β-HCG level of 83.56 mIU/mL, and at 7 wk of gestation we confirmed a fetus with normal heartbeats by B ultrasound. The result of fetal chromosome examination in the amniotic fluid during midstage pregnancy (20 wk + 1 d) showed a normal karyotype of 46,XN. The female patient gave birth to a healthy infant after 38 + 6 wk of pregnancy.
Patient 3 was a 34-y-old woman who sought NICS treatment in our clinic after failing conception due to tubal factor. Her husband was 31 y of age. The karyotype of male partner was 46, XY, inv(9)(p12q13), and karyotype of the female was 46, XX. The female blood hormone test results on cycle day 2 were as follows: FSH, 6.74 mIU/mL; LH, 4.55 mIU/mL; E2, 37.42 pg/mL. The B-ultrasound test showed an antral follicle count from both ovaries of about 10. The standard long stimulation protocol was performed based on her ovary reservation. We collected nine eggs in the IVF treatment; eight MII oocytes among them were fertilized by ICSI, and five developed to the blastocyst stage. All blastocysts were vitrified after applying the NICS test. The woman received one thawed blastocyst (no. 4) in hormone replacement cycle, and 8 d after embryo transfer her blood test revealed a β-HCG level of 308.6 mIU/mL. B-ultrasound confirmed a normal heartbeat at 7-wk gestation. The result of fetal chromosome examination in the amniotic fluid during midstage pregnancy (19 wk + 5 d) showed 46, XN, inv(9)(p12q13).
Acknowledgments
We thank Drs. Patty Purcell and Catherine Racowsky for their critical reading and editing of the manuscript and Dr. Luoxing Xiong for her assistance with data analyses. This work was supported by National Basic Research Program of China Grant 2015AA020407; Natural Science Foundation of Jiangsu Province, China, Grant BK20131094; funding from the Beijing Advanced Innovation Center for Genomics at Peking University, Joint Research Program of Medical Science and Technology Development Fund of the Medical Control Center in Wuxi City (Grant YGZX1204); National Natural Science Foundation of China Grant 81503655; and State Key Development Program for Basic Research of China Grant 2013CB945200.
Footnotes
Conflict of interest statement: X.S.X. and S.L. are cofounders of Yikon Genomics Company, Ltd.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. SRP089980).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613294113/-/DCSupplemental.
References
- 1.Battaglia DE, Goodwin P, Klein NA, Soules MR. Influence of maternal age on meiotic spindle assembly in oocytes from naturally cycling women. Hum Reprod. 1996;11(10):2217–2222. doi: 10.1093/oxfordjournals.humrep.a019080. [DOI] [PubMed] [Google Scholar]
- 2.Munné S. Chromosome abnormalities and their relationship to morphology and development of human embryos. Reprod Biomed Online. 2006;12(2):234–253. doi: 10.1016/s1472-6483(10)60866-8. [DOI] [PubMed] [Google Scholar]
- 3.Korenberg JR, et al. Down syndrome phenotypes: The consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994;91(11):4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellani A, Abu-Amero K, Azouri J, El-Akoum S. Successful pregnancies after application of array-comparative genomic hybridization in PGS-aneuploidy screening. Reprod Biomed Online. 2008;17(6):841–847. doi: 10.1016/s1472-6483(10)60413-0. [DOI] [PubMed] [Google Scholar]
- 5.Gutiérrez-Mateo C, et al. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2011;95(3):953–958. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Thornhill AR, et al. Karyomapping—a comprehensive means of simultaneous monogenic and cytogenetic PGD: Comparison with standard approaches in real time for Marfan syndrome. J Assist Reprod Genet. 2015;32(3):347–356. doi: 10.1007/s10815-014-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natesan SA, et al. Live birth after PGD with confirmation by a comprehensive approach (karyomapping) for simultaneous detection of monogenic and chromosomal disorders. Reprod Biomed Online. 2014;29(5):600–605. doi: 10.1016/j.rbmo.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Treff NR, et al. Single nucleotide polymorphism microarray-based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil Steril. 2011;95(5):1606–1612.e1,2. doi: 10.1016/j.fertnstert.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Natesan SA, et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med. 2014;16(11):838–845. doi: 10.1038/gim.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treff NR, et al. Development and validation of an accurate quantitative real-time polymerase chain reaction-based assay for human blastocyst comprehensive chromosomal aneuploidy screening. Fertil Steril. 2012;97(4):819–824. doi: 10.1016/j.fertnstert.2012.01.115. [DOI] [PubMed] [Google Scholar]
- 11.Martín J, et al. The impact of next-generation sequencing technology on preimplantation genetic diagnosis and screening. Fertil Steril. 2013;99(4):1054–1061.e3. doi: 10.1016/j.fertnstert.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155(7):1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 13.Scott RT, Jr, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: A randomized controlled trial. Fertil Steril. 2013;100(3):697–703. doi: 10.1016/j.fertnstert.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 14.Forman EJ, et al. In vitro fertilization with single euploid blastocyst transfer: A randomized controlled trial. Fertil Steril. 2013;100(1):100–107.e1. doi: 10.1016/j.fertnstert.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, et al. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: Results from a randomized pilot study. Mol Cytogenet. 2012;5(1):24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keltz MD, et al. Preimplantation genetic screening (PGS) with comparative genomic hybridization (CGH) following day 3 single cell blastomere biopsy markedly improves IVF outcomes while lowering multiple pregnancies and miscarriages. J Assist Reprod Genet. 2013;30(10):1333–1339. doi: 10.1007/s10815-013-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cimadomo D, et al. The impact of biopsy on human embryo developmental potential during preimplantation genetic diagnosis. BioMed Res Int. 2016;2016:7193075. doi: 10.1155/2016/7193075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, et al. Blastomere biopsy influences epigenetic reprogramming during early embryo development, which impacts neural development and function in resulting mice. Cell Mol Life Sci. 2014;71(9):1761–1774. doi: 10.1007/s00018-013-1466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H-C, et al. Aberrant epigenetic modification in murine brain tissues of offspring from preimplantation genetic diagnosis blastomere biopsies. Biol Reprod. 2013;89(5):117. doi: 10.1095/biolreprod.113.109926. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Y, et al. Preimplantation genetic diagnosis (PGD) influences adrenal development and response to cold stress in resulting mice. Cell Tissue Res. 2013;354(3):729–741. doi: 10.1007/s00441-013-1728-1. [DOI] [PubMed] [Google Scholar]
- 21.Assou S, Aït-Ahmed O, El Messaoudi S, Thierry AR, Hamamah S. Non-invasive pre-implantation genetic diagnosis of X-linked disorders. Med Hypotheses. 2014;83(4):506–508. doi: 10.1016/j.mehy.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Galluzzi L, et al. Extracellular embryo genomic DNA and its potential for genotyping applications. Future Science OA. 2015;1(4):FSO62. doi: 10.4155/fso.15.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palini S, et al. Genomic DNA in human blastocoele fluid. Reprod Biomed Online. 2013;26(6):603–610. doi: 10.1016/j.rbmo.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Gianaroli L, et al. Blastocentesis: A source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil Steril. 2014;102(6):1692–1699.e6. doi: 10.1016/j.fertnstert.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 25.Stigliani S, Anserini P, Venturini PL, Scaruffi P. Mitochondrial DNA content in embryo culture medium is significantly associated with human embryo fragmentation. Hum Reprod. 2013;28(10):2652–2660. doi: 10.1093/humrep/det314. [DOI] [PubMed] [Google Scholar]
- 26.Stigliani S, et al. Mitochondrial DNA in day 3 embryo culture medium is a novel, non-invasive biomarker of blastocyst potential and implantation outcome. Mol Hum Reprod. 2014;20(12):1238–1246. doi: 10.1093/molehr/gau086. [DOI] [PubMed] [Google Scholar]
- 27.Wu H, et al. Medium-based noninvasive preimplantation genetic diagnosis for human α-thalassemias-SEA. Medicine (Baltimore) 2015;94(12):e669. doi: 10.1097/MD.0000000000000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J, et al. Validation of multiple annealing and looping-based amplification cycle sequencing for 24-chromosome aneuploidy screening of cleavage-stage embryos. Fertil Steril. 2014;102(6):1685–1691. doi: 10.1016/j.fertnstert.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Yan L, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci USA. 2015;112(52):15964–15969. doi: 10.1073/pnas.1523297113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, et al. Validation of a next-generation sequencing–based protocol for 24-chromosome aneuploidy screening of blastocysts. Fertil Steril. 2016;105(6):1532–1536. doi: 10.1016/j.fertnstert.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 32.Gui B, et al. Chromosomal analysis of blastocysts from balanced chromosomal rearrangement carriers. Reproduction. 2016;151(4):455–464. doi: 10.1530/REP-16-0007. [DOI] [PubMed] [Google Scholar]
- 33.Schoolcraft W, et al. Comprehensive chromosome screening (CCS) with vitrification results in improved clinical outcome in women >35 years: A randomized control trial. Fertil Steril. 2012;98(3):S1. [Google Scholar]
- 34.Hodes-Wertz B, et al. Idiopathic recurrent miscarriage is caused mostly by aneuploid embryos. Fertil Steril. 2012;98(3):675–680. doi: 10.1016/j.fertnstert.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Margalioth EJ, Ben-Chetrit A, Gal M, Eldar-Geva T. Investigation and treatment of repeated implantation failure following IVF-ET. Hum Reprod. 2006;21(12):3036–3043. doi: 10.1093/humrep/del305. [DOI] [PubMed] [Google Scholar]
- 36.Frydman N, et al. Assisting reproduction of infertile men carrying a Robertsonian translocation. Hum Reprod. 2001;16(11):2274–2277. doi: 10.1093/humrep/16.11.2274. [DOI] [PubMed] [Google Scholar]
- 37.Fragouli E, et al. Comprehensive chromosome screening of polar bodies and blastocysts from couples experiencing repeated implantation failure. Fertil Steril. 2010;94(3):875–887. doi: 10.1016/j.fertnstert.2009.04.053. [DOI] [PubMed] [Google Scholar]
- 38.Chen M, Wei S, Hu J, Quan S. Can comprehensive chromosome screening technology improve IVF/ICSI outcomes? A meta-analysis. PLoS One. 2015;10(10):e0140779. doi: 10.1371/journal.pone.0140779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Twisk M, et al. No beneficial effect of preimplantation genetic screening in women of advanced maternal age with a high risk for embryonic aneuploidy. Hum Reprod. 2008;23(12):2813–2817. doi: 10.1093/humrep/den231. [DOI] [PubMed] [Google Scholar]
- 40.Hardarson T, et al. Preimplantation genetic screening in women of advanced maternal age caused a decrease in clinical pregnancy rate: A randomized controlled trial. Hum Reprod. 2008;23(12):2806–2812. doi: 10.1093/humrep/den217. [DOI] [PubMed] [Google Scholar]
- 41.Schoolcraft WB, Katz-Jaffe MG, Stevens J, Rawlins M, Munne S. Preimplantation aneuploidy testing for infertile patients of advanced maternal age: A randomized prospective trial. Fertil Steril. 2009;92(1):157–162. doi: 10.1016/j.fertnstert.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 42.Taylor TH, et al. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update. 2014;20(4):571–581. doi: 10.1093/humupd/dmu016. [DOI] [PubMed] [Google Scholar]
- 43.Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-cell whole-genome amplification and sequencing: Methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 44.Ryan GL, et al. A mandatory single blastocyst transfer policy with educational campaign in a United States IVF program reduces multiple gestation rates without sacrificing pregnancy rates. Fertil Steril. 2007;88(2):354–360. doi: 10.1016/j.fertnstert.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]