Significance
Leaves, being the prime photosynthetic organ of plants, are critical in many ways to our current biosphere. A defining characteristic, which also optimizes their function, is their flat shape that depends on the correct patterning of their upper and lower tissues during development. Here, we show that the correct patterning of upper and lower leaf tissues depends on two types of transcription factors (class II and class III homeodomain leucine zipper (HD-ZIPs) that act together to repress a set of miRNAs (MIR165/166), which in turn, represses the activity of these transcription factors (class III HD-ZIPs). This three-way interaction maintains the balance of tissue identities during growth, leading to the formation of a flat leaf.
Keywords: organ patterning, leaf morphogenesis, class II HD-ZIP, class III HD-ZIP, MIR165/166
Abstract
A defining feature of plant leaves is their flattened shape. This shape depends on an antagonism between the genes that specify adaxial (top) and abaxial (bottom) tissue identity; however, the molecular nature of this antagonism remains poorly understood. Class III homeodomain leucine zipper (HD-ZIP) transcription factors are key mediators in the regulation of adaxial–abaxial patterning. Their expression is restricted adaxially during early development by the abaxially expressed microRNA (MIR)165/166, yet the mechanism that restricts MIR165/166 expression to abaxial leaf tissues remains unknown. Here, we show that class III and class II HD-ZIP proteins act together to repress MIR165/166 via a conserved cis-element in their promoters. Organ morphology and tissue patterning in plants, therefore, depend on a bidirectional repressive circuit involving a set of miRNAs and its targets.
The morphogenesis of lateral organs in plants and animals is dependent on the specification of distinct cell types early in development. In particular, the correct patterning of adaxial–abaxial tissues in plant organs such as leaves is critical for the generation of a lamina shape and the formation of a polar vascular system (1–4). Adaxial–abaxial cell-type patterning in turn depends on the restricted expression of several genes known to specify these cell types, including the class III homeodomain leucine zipper genes (HD-ZIPIIIs), KANADI genes, HD-ZIPIIs, and microRNA (MIR)165/166 (1, 2, 4–11). In general, genetic analyses have indicated that adaxial and abaxial factors act oppositely in organ patterning (1, 2, 4, 8–11). Hence, loss-of-function mutations in genes promoting adaxial cell identity typically cause an abaxialized phenotype that correlates with the ectopic expression of abaxial genes, whereas loss-of-function mutations in abaxial genes produce an adaxialized phenotype that is accompanied by the expanded expression of adaxial genes. This antagonistic interaction between adaxial and abaxial factors may be mediated by mutually antagonistic regulation (12) or through opposing regulation of common targets (9, 13–16).
A key set of transcription factors involved in plant organ polarity are the HD-ZIPIII proteins, such as REVOLUTA (REV), which specify adaxial cell fate (1, 2, 4, 17). The expression of these genes is restricted specifically to adaxial tissues via the action of two miRNA families, MIR165 and MIR166 (2, 7). In turn, the expression of these miRNAs is restricted to abaxial tissues and this restriction is essential for maintaining proper organ polarity (18).
Here, we address the question of how MIR165/166 are regulated. We show that the HD-ZIPII proteins HAT3 and ATHB4 physically interact with HD-ZIPIII proteins and directly repress MIR165/166 expression via a conserved cis-element located in their promoters. This regulatory interaction largely accounts for HAT3 and ATHB4 function and reveals the molecular nature of a bidirectional repressive circuit essential to maintain balance between adaxial and abaxial tissue specification.
Results and Discussion
HAT3 and ATHB4 Regulate Leaf Polarity by Repressing MIR165/166 Expression.
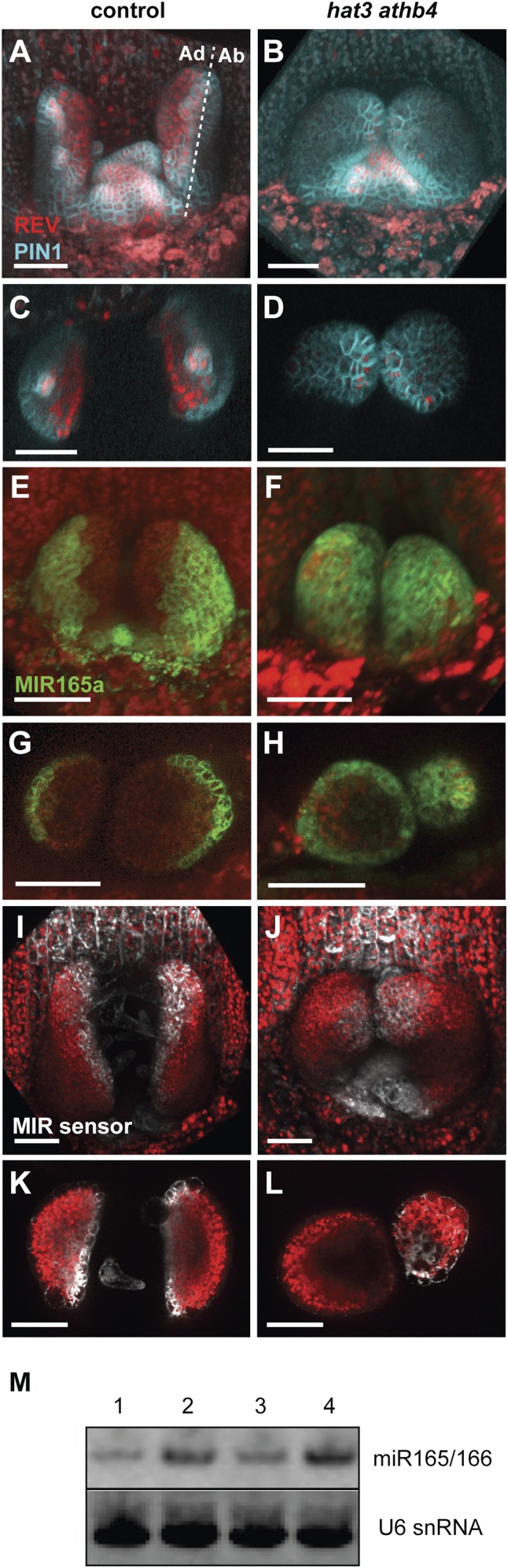
Previous studies have shown that the HD-ZIPII genes HAT3 and ATHB4 play an essential role in establishing leaf polarity by promoting adaxial cell fate (5, 6). Although HAT3 and ATHB4 are known to be downstream targets of the HD-ZIPIII transcription factor REVOLUTA (14), we further investigated the relationship between these genes by monitoring the expression of REV in the hat3 athb4 double mutant (6) using the functional fluorescent reporter pREV::REV-2xYPET. Confocal imaging of 4-d-old hat3 athb4 mutant leaves revealed the REV expression domain to be reduced compared with control seedlings (Fig. 1 A–D), indicating that HAT3 and ATHB4 may be involved in a positive feedback loop in which REV targets reinforce REV expression. Because REV and other HD-ZIPIIIs are regulated by miR165/166 (2, 7), we next examined whether the expression of these miRNAs also depended on HAT3 and ATHB4 function by looking at reporters for their expression in the hat3 athb4 double mutant. We found that transcriptional reporters for both MIR165a and MIR166a (pMIR165a::mTagBFP-ER and pMIR166a::GFP-ER) in the leaves of 4- and 15-d-old hat3 athb4 plants were expressed ectopically throughout the epidermis, instead of being restricted to the abaxial epidermis as in control leaves (Fig. 1 E–H and SI Appendix, Fig. S1 A–H). To test whether the ectopic MIR promoter activity corresponded to ectopic miR activity in the hat3 athb4 mutant, we used a miR165/166 fluorescent biosensor with the miR target sequence from the REV gene (2, 7, 19) (SI Appendix, SI Materials and Methods). This biosensor acts as a negative marker for miR165/166 activity, because it is inactivated in cells where miR165/166 are active. The expression patterns of the miR165/166 biosensor and REV-2xYPET in control leaves were highly similar with expression encompassing the adaxial side (Fig. 1 A, C, I, and K and SI Appendix, Fig. S1 I and J). However, in hat3 athb4 mutant leaves, we found the expression of the miR165/166 biosensor to be similarly reduced compared with REV-2xYPET in the same genetic background (Fig. 1 B, D, J, and L and SI Appendix, Fig. S1 K and L), consistent with the MIR165/166 promoter reporter data. Lastly, we confirmed that HAT3 and ATHB4 are required for repressing MIR expression by small RNA Northern blot analysis, which indicated very high levels of miR165/166 in plants mutant for HAT3 and ATHB4 (Fig. 1M).
Fig. 1.
HAT3 and ATHB4 are required for repressing MIR165/166 expression. (A–D) Expression of pREV::REV-2xYPET (red) in combination with the auxin efflux carrier PIN-FORMED1 (30, 31) fused to GFP (pPIN::PIN1-GFP) (blue) in the shoot apex of 4-d-old control (A and C) and hat3 athb4 plants (B and D). pPIN::PIN1-GFP is used here to outline the tissue. (C and D) Cross-sections of the same leaf primordia shown in A and B, respectively. (E–H) Expression of pMIR165a::mTagBFP-ER (green) in the shoot apex of 4-d-old control (E and G) and hat3 athb4 plants (F and H). (G and H) Cross-sections of the same leaf primordia shown in E and F, respectively. (I–L) Expression of a miR165/166 sensitive biosensor (white) containing the miRNA target sequence from REV fused to the UV-photoconvertible fluorescent protein mEos2FP (pUBQ10::REV-mEos2FP-ER) in the shoot apex of 4-d-old control (I and K) and hat3 athb4 plants (J and L). The sensitive biosensor is inactivated in cells where miR165/166 are active. (K and L) Cross-sections of the same leaf primordia shown in I and J, respectively. Chlorophyll autofluorescence: red (E–L). (Scale bars, 50 μm.) Ad, adaxial side; Ab, abaxial side. (M) Small RNA Northern blot showing expression levels of miR165/166 and U6 snRNA in Col-0 WT (lane 1), p35S::miR165a (lane 2), hat1 hat2 (lane 3), and hat3 athb4 plants (lane 4).
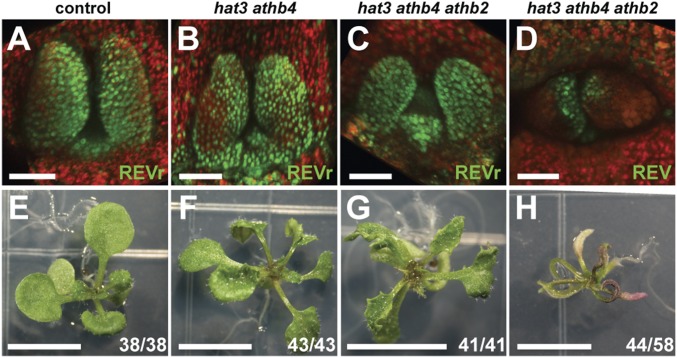
To gauge the relative importance of MIR165/166 regulation to overall HD-ZIPII protein function, we transformed a miR165/166-resistant REV reporter (pREV::REVr-2xVENUS), as well as a miR-sensitive REV reporter (pREV::REV-2xVENUS), into hat3 athb4 and hat3 athb4 athb2 plants (6) and assessed the degree of phenotypic rescue by REVr compared with the control. Compared with the miR-sensitive REV reporter (Fig. 1 A–D, and Fig. 2D), pREV::REVr-2xVENUS expression pattern extended further into the abaxial side of the leaves of 4-d-old hat3 athb4, hat3 athb4 athb2, and control seedlings (Fig. 2 A–C), overlapping with where MIR165a and MIR166a are expressed. Importantly, 15-d-old hat3 athb4 and hat3 athb4 athb2 plants expressing the pREV::REVr-2xVENUS reporter gene developed significantly flatter leaves (Fig. 2 F and G), more similar to control plants (Fig. 2E) compared with the radialized leaves of the miR-sensitive REV control (Fig. 2H). Similarly, when we combined the rev-10d gain-of-function mutation, which disrupts the complementarity between miR165/166 and REV mRNA (2), with the double mutant hat3 athb4, a similar attenuation of the leaf phenotype was apparent (SI Appendix, Fig. S2D). These results indicate that REVr can bypass high levels of miR165/166, as observed in hat3 athb4 mutants and by regulating its other targets, can promote leaf development independent of HAT3 and ATHB4. Thus, HAT3 and ATHB4 primarily function to repress MIR165/166 during leaf development.
Fig. 2.
pREV::REVr-2xVENUS rescues the hat3 athb4 and hat3 athb4 athb2 leaf phenotype. (A–C) Expression pattern of pREV::REVr-2xVENUS (green) in the shoot apex of 4-d-old control (A), hat3 athb4 (B), and hat3 athb4 athb2 plants (C). (D) Expression of a control REV translational reporter (pREV::REV-2xVENUS) (green) in the shoot apex of 4-d-old hat3 athb4 athb2 plants (control). (E–H) Phenotype of 15-d-old control (E), hat3 athb4 (F), and hat3 athb4 athb2 plants (G) transformed with pREV::REVr-2xVENUS. (H) Phenotype of 15-d-old hat3 athb4 athb2 plants transformed with pREV::REV-2xVENUS. Chlorophyll autofluorescence: red (A–D). [Scale bars, 50 μm (A–D) and 5 mm (E–H).]
HAT3 and ATHB4 Physically Interact with REV to Repress MIR165/166 Expression.
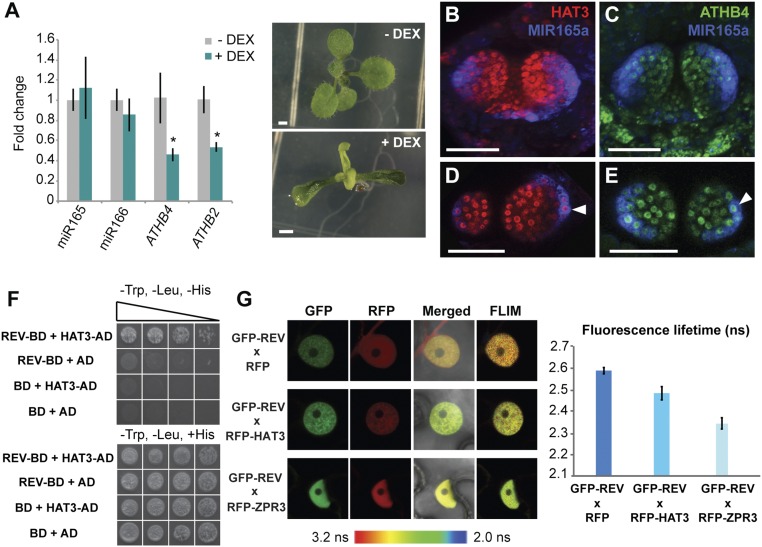
To test whether HAT3 is sufficient to repress MIR165/166 expression, we measured mature miR165/166 levels by RT-qPCR after inducing HAT3 expression ectopically using the two-component GR-LhG4 system (20) driven by the MERISTEM LAYER 1 (ML1) promoter (pML1>>VENUS-HAT3) (21). The chimeric GR-LhG4 construct consists of the ligand-binding site domain of a rat glucorticoid receptor (GR) fused to the synthetic transcription activator LhG4, which comprises the transcription-activation domain-II from Gal4 of Saccharomyces cerevisiae (20). However, whereas we could detect down-regulation of ATHB4 and ATHB2, in agreement with previous studies suggesting negative feedback between HD-ZIPII family members (22), no significant change in mature miR165/166 levels was detected (Fig. 3A). Transgenic pML1>>VENUS-HAT3 plants showed narrow and upward curling leaves (Fig. 3A), which may be a consequence of ATHB4 and ATHB2 down-regulation or regulation of additional adaxial–abaxial factors. Imaging of functional reporters for both HAT3 and ATHB4 proteins also revealed that the expression of HAT3 and ATHB4 extends throughout the abaxial side of the leaf, including abaxial cells in which MIR165a and MIR166a reporter expression was also detected (Fig. 3 B–E and SI Appendix, Fig. S3). We conclude that although either HAT3 or ATHB4 are necessary to restrict MIR165/166 expression to abaxial tissues, neither is sufficient to repress MIR165/166 expression, suggesting that repression of MIR165/166 by HAT3 and ATHB4 involves additional adaxially localized factors.
Fig. 3.
HAT3 and ATHB4 physically interact with REV. (A) Levels of mature miR165/166 in pML1>>VENUS-HAT3 plants. Fold changes relative to ACTIN2 (ACT2; AT3G18780) in response to DEX treatment (+DEX; green bars) and in control conditions (0.1% ethanol; −DEX; gray bars) are shown. Data are represented as mean ± SD of three biological replicates. *P ≤ 0.05. Phenotype of 10-d-old pML1>>VENUS-HAT3 plants grown on DEX-GM and control medium is shown. [Scale bars, 1 mm (A).] (B–E) Expression of pHAT3::VENUS-HAT3 (red) (B and D) and pATHB4::VENUS-ATHB4 (green) (C and E) combined with pMIR165a::GFP-ER (blue) in the shoot apex of 3-d-old Col-0 plants. (D and E) Cross-sections of the same leaf primordia shown in B and C, respectively. Colocalization of pHAT3::VENUS-HAT3/pATHB4::VENUS-ATHB4 and pMIR165a::GFP-ER is indicated by arrowheads in D and E, respectively. [Scale bars, 50 μm (B–E).] (F) REV and HAT3 interaction in a yeast two-hybrid assay. Growth of yeast on selective medium (−Trp, −Leu, −His, and +3-AT) for REV–HAT3 combination indicates protein–protein interaction. Five transformed colonies per prey/bait combination were analyzed for their growth on −Trp, −Leu, −His, and +3-AT plates as well as on −Trp, −Leu, +His, and +3-AT plates using dilution series (1:1, 1:5, 1:10, and 1:50). AD, activation domain; BD, binding domain. (G) REV–HAT3 interaction in nuclei of tobacco leaf epidermal cells detected through a FRET–FLIM assay. GFP–REV (green) works as a donor and RFP–HAT3 (red) as an acceptor. GFP–REV (green) x RFP (red) and GFP–REV (green) x RFP–ZPR3 (red) combinations were used as negative and positive controls, respectively. GFP fluorescence lifetime (in nanoseconds, ns) quantification is shown. Error bars show mean ± SE of 10 nuclei.
In a yeast two-hybrid (Y2H) screen using REV as bait, we identified a truncated version of the HAT3 protein, missing the first 88 aa, as a potential binding partner. After confirming this result using a full-length HAT3 cDNA (Fig. 3F), we tested whether interaction between HAT3 and REV could be detected in vivo, using a combination of fluorescence resonance energy transfer and fluorescence lifetime imaging (FRET-FLIM) in tobacco (Nicotiana benthamiana) (Fig. 3G). For this purpose, REV and HAT3 cDNAs were fused to an N-terminal donor (GFP) and to an N-terminal acceptor (RFP), respectively, and cloned under the 35S promoter. Significant reduction in the fluorescence lifetime of GFP was detected when GFP–REV was coexpressed with RFP–HAT3 in nuclei in comparison with the negative control GFP–REV × RFP (Fig. 3G) or with those nuclei with no detectable RFP signal (SI Appendix, Fig. S4), indicating that REV and HAT3 interact in vivo. To validate the technique, we used as a positive control interaction GFP–REV × RFP–ZPR3, where ZPR3 encodes a small leucine zipper-containing protein [LITTLE ZIPPER (ZPR) protein] previously shown to interact with REV (23).
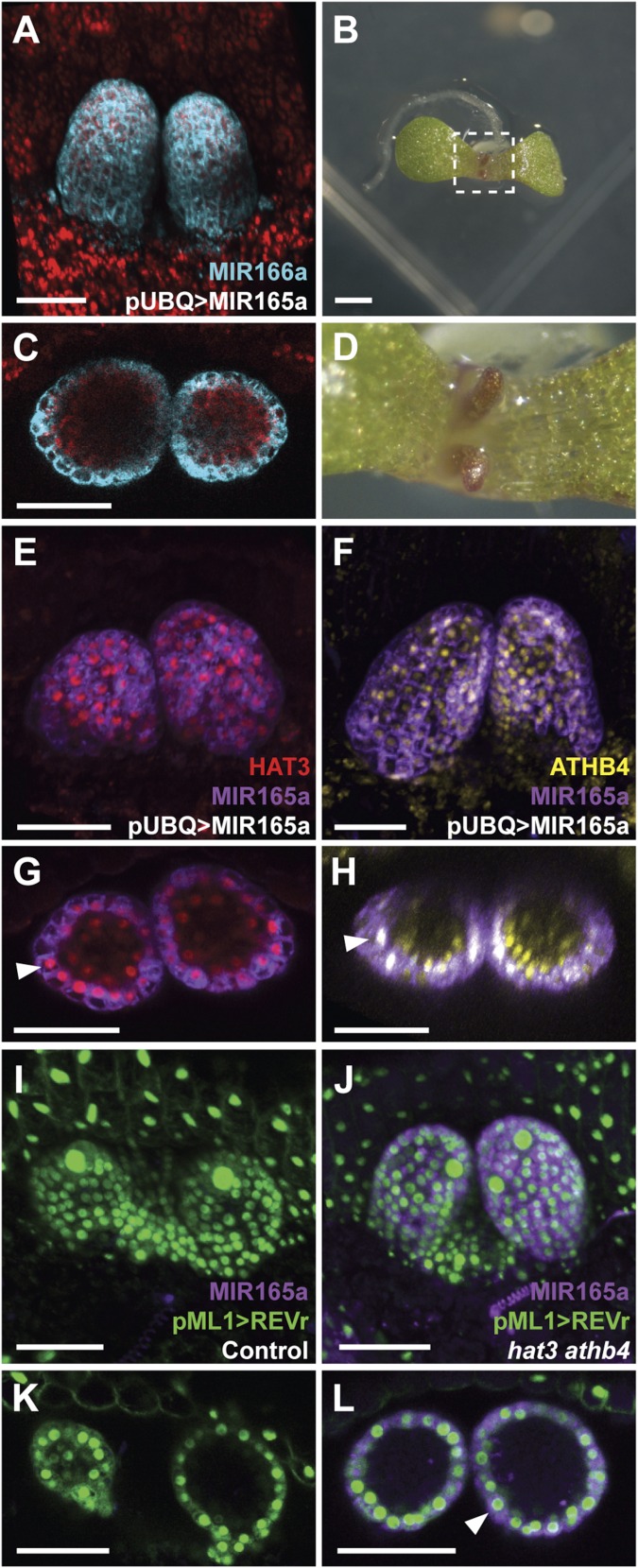
As REV and HAT3 interact, we next investigated whether HD-ZIPIII activity also contributes to the repression of MIR165/166. For this purpose, we examined pMIR166a::GFP-ER expression after knockdown of the HD-ZIPIIIs using an inducible MIR165a construct based on the two-component GR-LhG4 system (20) driven by the UBQ10 promoter. We found expression of the MIR promoter construct to expand ectopically to adaxial tissues after MIR165a induction (Fig. 4 A and C), correlating with the leaf abaxialization observed in 4-d-old seedlings (Fig. 4 B and D and control conditions in SI Appendix, Fig. S5 A–C). Surprisingly, we also observed that after repressing REV expression via MIR165a induction, both the HAT3 or ATHB4 reporters were still expressed and colocalized with the MIR165a transcriptional reporter (Fig. 4 E–H and control conditions in SI Appendix, Fig. S5 D–G). These results demonstrate that HAT3 and ATHB4 proteins cannot repress MIR165/166 expression in the absence of the transcription factor REV and that REV and other HD-ZIPIIIs are not necessarily required for HD-ZIPII expression. Next, we tested whether ectopic expression of the REVr reporter (pML1>>REVr-2xVENUS) was sufficient to repress pMIR165a::mTagBFP-ER expression in control plants. We found that after REVr induction, expression of the MIR165a promoter reporter was undetectable (Fig. 4 I and K and SI Appendix, Fig. S6 A and B). We then repeated this experiment in the hat3 athb4 mutant background and found that, in contrast to the control, ectopic REVr was not capable of repressing MIR165a reporter expression (Fig. 4 J and L and SI Appendix, Fig. S6 C and D). Also under its own promoter, REVr could not repress pMIR165a::mTagBFP-ER expression in the adaxial domain of hat3 athb4 plants as it does in control plants (SI Appendix, Fig. S7 A–D).
Fig. 4.
REV requires HAT3 and ATHB4 to repress MIR165/166 expression. (A and C) Expression of pMIR166a::GFP-ER (blue) after MIR165a expression driven by the UBQ10 promoter in the shoot apex of Col-0 plants 4 d after germination on DEX-GM medium. (C) Cross-section of the same leaf primordia shown in A. (B and D) Phenotype of 7-d-old plants expressing MIR165a under the UBQ10 promoter. (E–H) Expression of pHAT3::VENUS-HAT3 (red) (E and G) or pATHB4::VENUS-ATHB4 (yellow) (F and H) combined with pMIR165a::GFP-ER (purple) after MIR165a expression driven by the UBQ10 promoter in the shoot apex of Col-0 plants 4 d after germination on DEX-GM medium. (G and H) Cross-sections of the same leaf primordia shown in E and F, respectively. Colocalization of pHAT3::VENUS-HAT3/pATHB4::VENUS-ATHB4 and pMIR165a::GFP-ER is indicated by arrowheads in G and H, respectively. (I–L) Expression of REVr-2xVENUS, which is miR165/166 resistant, driven by the ML1 promoter (green) and pMIR165a::BFP-ER (purple) in the shoot apex of control (I and K) and hat3 athb4 plants (J and L) 4 d after germination on DEX-GM medium. (K and L) Cross-sections of the same leaf primordia shown in I and J, respectively. Colocalization of pML1>>REVr-2xVENUS and pMIR165a::BFP-ER is indicated by an arrowhead in L. Chlorophyll autofluorescence: red (A and C). [Scale bars, 50 μm (A, C, and E–L) and 1 mm (B).]
All together these results demonstrate that both HD-ZIPII and HD-ZIPIII proteins interact in vitro and in vivo and that their combined activities are necessary and sufficient to repress MIR165/166 expression.
HD-ZIPIIs and HD-ZIPIIIs Repress MIR165/166 Expression via a Conserved cis-Element.
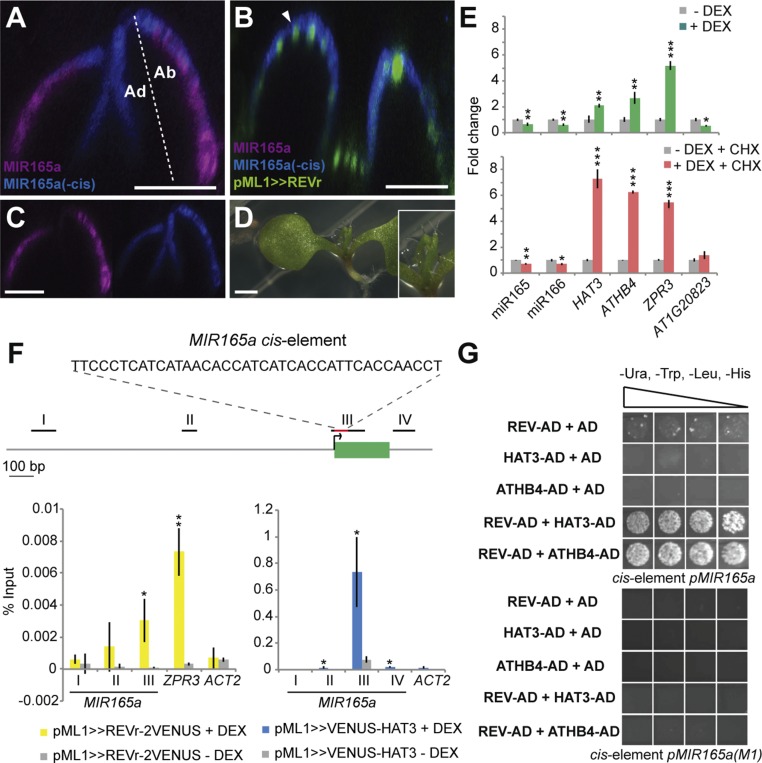
Previous analysis of the MIR165a and MIR166a promoters revealed that repression of promoter activity in adaxial leaf tissues is mediated by a conserved 39-bp-long cis-element located at 1 bp downstream and 22 bp upstream of the MIR165a and MIR166a transcription initiation sites, respectively (24, 25). To test whether repression of MIR165/166 by REV depends on this element, we induced ectopic REVr expression (pML1>>REVr-2xVENUS) in plants expressing pMIR165a::GFP-ER, which contains the cis-element, as well as a MIR165a transcriptional reporter in which this element has been deleted (pMIR165a(−cis)::BFP-ER). We observed that REVr could repress pMIR165a::GFP-ER expression in the leaves of 7-d-old seedlings 2 d after dexamethasone (DEX) treatment; however, it could not repress pMIR165a(−cis)::BFP-ER expression (Fig. 5 A–D), indicating that the previously reported cis-element (25) is essential for the repression of MIR165/166 expression via REV.
Fig. 5.
REV and HAT3 bind a conserved cis-element required to restrict MIR165a expression. (A–C) Expression of pML1>>REVr-2xVENUS (green), pMIR165a::GFP-ER (purple), and pMIR165a(−cis)::BFP-ER (blue) in the shoot apex of 7-d-old Col-0 plants 2 d after transferring to 0.1% ethanol (mock) (A and C) or DEX-GM medium (B). Longitudinal sections of the second pair of leaves are shown (A–C). Colocalization of pML1>>REVr-2xVENUS and pMIR165a(−cis)::BFP-ER is indicated by an arrowhead in B. (D) Phenotype of 10-d-old transgenic pML1>>REVr-2xVENUS_ pMIR165a::GFP-ER_pMIR165a(−cis)::BFP-ER plants grown on DEX-GM medium. [Scale bars, 50 μm (A–C) and 1 mm (D).] Ad, adaxial side; Ab, abaxial side (A). (E) Levels of mature miR165/166 in p35S::GR-REVr plants (14). Fold changes relative to ACT2 in response to DEX treatment (+DEX; green and red bars) and in control conditions (0.1% ethanol; −DEX; gray bars) are plotted. Green bars show expression changes in the absence of the protein biosynthesis inhibitor CHX; red bars show expression changes in the presence of CHX (+CHX). HAT3, ATHB4, ZPR3, and AT1G20823 were tested as known direct or indirect REV targets. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (F) ChIP-qPCR on genomic regions surrounding MIR165a using anti-GFP antibody and the DEX-inducible transgenic lines pML1>>VENUS-HAT3 and pML1>>REVr-2xVENUS. A diagram of the MIR165a genomic region is shown. The black lines, red line, and green box represent the regions amplified by ChIP-qPCR, the cis-element (25), and the MIR165a locus, respectively. ChIP-qPCR data were normalized to the percent of preimmunoprecipitation (pre-IP) input for each sample. Error bars show means ± SD of three biological replicates treated with 10 μM DEX (+DEX; yellow and blue bars) or mock (0.1% ethanol; −DEX; gray bars). ACT2 and ZPR3 were also tested as negative and positive controls, respectively (26, 27). *P ≤ 0.05; **P ≤ 0.01. (G) Interaction of REV and HAT3/ATHB4 with the MIR165a cis-element in a yeast one-hybrid assay. Growth of yeast on selective medium (−Ura, −Trp, −Leu, −His, and +3-AT) for REV–HAT3 and REV–ATHB4 combinations indicates protein–DNA interaction. A mutated version of the cis-element, previously referred to as M1 (25), was used as a negative control. Three transformed colonies per prey/bait combination were analyzed for their growth on −Ura, −Trp, −Leu, −His, and +3-AT selection plates using dilution series (1:5, 1:10, 1:20, 1:50). AD, activation domain.
Next, we investigated whether REV represses MIR165/166 directly by inducing GR-REV in the presence of the protein biosynthesis inhibitor cycloheximide (CHX) and measuring mature miR165/166 levels by RT-qPCR. For this purpose, we treated p35S::GR-REVr plants (23) for 3 h with DEX in the presence of CHX. Significant reduction of miR165/166 levels was detected in the presence or absence of CHX, indicating that no new protein synthesis is required to repress the transcription of these genes (Fig. 5E). Activation of known direct targets of REV such as HAT3, ATHB4, and ZPR3 (14, 23) also occurred in the presence or absence of CHX, whereas repression of the previously reported indirect target AT1G20823 (16) occurred only in the absence of CHX (Fig. 5E), validating our results. To assess the binding of REV to the cis-element in vivo, we performed a ChIP-qPCR assay using primers to amplify several regions surrounding the MIR165a locus. We detected enriched binding near the MIR165a transcriptional start site, corresponding to the location of the cis-element (Fig. 5F, region III), relative to surrounding locations. ACT2 and ZPR3 were also tested as negative and positive controls, respectively (26, 27), and validated our results. Together these data demonstrate that REV represses MIR165/166 directly via a previously characterized conserved cis-element (25).
Although we could not determine whether repression by HAT3 is also direct using inducible expression because HAT3 is already expressed broadly (Fig. 3 A–E), we used ChIP-qPCR to assess whether HAT3 also binds the region containing the MIR165a cis-element in vivo, and found that the binding of HAT3 is enriched in the same region as for REV (Fig. 5F, region III), supporting the proposal that HAT3 also regulates MIR165/166 directly and, therefore, that REV and HAT3 bind the cis-element as a complex.
Next, we tested the ability of HAT3 and REV to bind the cis-element in a yeast one-hybrid (Y1H) assay and found no evidence of protein–DNA interaction for both proteins when tested individually (Fig. 5G). However, when HAT3 or ATHB4 were included in combination with REV, we detected interaction with the MIR cis-element and not with a mutated version of this cis-element that had previously been shown to be defective in directing abaxial MIR expression (25) (Fig. 5G). These results further indicate that direct interaction between these proteins promotes their binding to the cis-element. Because additional HD-ZIPIII genes such as PHABULOSA (PHB) and PHAVOLUTA (PHV) work redundantly with REV to promote adaxial cell fate (2, 7), we also tested whether HAT3 and ATHB4 interact with these two HD-ZIPIIIs to bind the cis-element by yeast one-hybrid assays. PHV–HAT3 and PHV–ATHB4 combinations also tested positive for DNA binding, although with weaker specificity in comparison with REV–HAT3 and REV-ATHB4 combinations (SI Appendix, Fig. S8A). In addition, yeast one-hybrid assays showed that PHB and PHV interact with the HD-ZIPII ATHB2 to bind the cis-element (SI Appendix, Fig. S8A), suggesting that interactions between additional members of these two gene families may also regulate MIR165/166 expression, which is consistent with the stronger phenotype associated with the hat3 athb4 athb2 triple mutant (6).
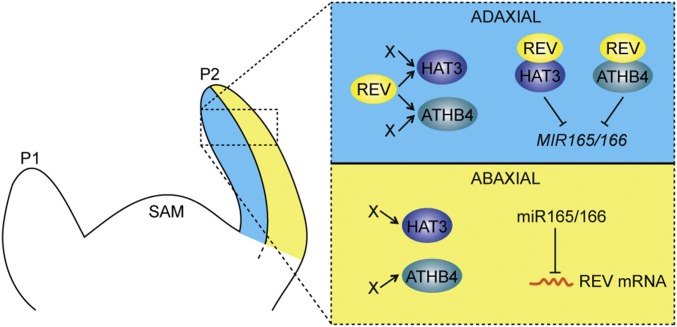
Overall, our data demonstrate that maintenance of leaf polarity involves physical interaction between class III HD-ZIPs and their target genes, class II HD-ZIPs, to establish direct repression of MIR165/166 (Fig. 6). In addition to REV, HAT3, and ATHB4, another factor recently shown to repress MIR166a is the adaxial transcription factor ASSYMETRIC LEAVES 2 (AS2). However, this regulation occurs at later stages of leaf development via a binding site located further upstream of the cis-element reported here (28). In turn, AS2 is directly repressed by the abaxial factor KANADI1 (12), and KAN1 together with KAN2 help repress HD-ZIPIII expression (1). Hence, besides the antagonistic HD-ZIPII/III-MIR165/166 relationship described here, several other antagonistic interactions help maintain distinct adaxial and abaxial patterns of gene expression across the leaf. An important challenge for the future will be to determine how adaxial and abaxial gene expression patterns are initially specified in young primordia and how equal partitioning is maintained during rapid cell proliferation and growth.
Fig. 6.
A regulatory network involving MIR165/166, class II and class III HD-ZIP genes controls adaxial–abaxial patterning of the leaf. P1 and P2, leaf primordia; SAM, shoot apical meristem.
Materials and Methods
Plant Material and Treatments.
Arabidopsis thaliana (L.) Heyhn plants were in Columbia-0 (Col-0) background. Additional details regarding the mutant and reporter lines generated in this background as well as the plant treatments are provided in SI Appendix.
Constructions for the Transgenic Plants.
All plasmids used in this study were constructed following standard molecular biology techniques. Additional experimental details are provided in SI Appendix.
Confocal Microscopy and Image Analysis.
Live imaging analyses were performed on a Leica SP5 confocal microscope using a water-dipping 25× objective. Additional details regarding the live imaging settings are described in SI Appendix.
Small RNA Northern Blot.
Total RNAs were isolated from 2-wk-old Col-0, p35S::miR165a, hat1 hat2, and hat3 athb4 plants. Additional analysis details are described in SI Appendix.
RT-qPCR.
For quantification of mature miR165/166 levels after inducing HAT3 or REVr ectopically, transgenic pML1::GR-LhG4_p6xOp::VENUS-HAT3 and p35S::GR-REVr (14, 23) plants were used. Additional analysis details are provided in SI Appendix.
Yeast Two-Hybrid Assay.
The yeast two-hybrid screening was performed by Hybrigenics Services (www.hybrigenics-services.com) using a mating approach with Y187 (Clontech library) and L40ΔGal4 (MATa) yeast strains as previously described (30). Additional experimental details are described in SI Appendix.
FRET–FLIM.
For the FRET–FLIM studies, GFP–REVOLUTA, RFP–HAT3, and RFP–ZPR3 (positive control) (23) were expressed under the control of the 35S promoter in tobacco plants (N. benthamiana). Additional experimental details are provided in SI Appendix.
ChIP-qPCR.
For ChIP-qPCR, transgenic pML1::GR-LhG4_p6xOp::VENUS-HAT3 and pML1::GR-LhG4_p6xOp::REVr-2xVENUS plants were treated with DEX or 0.1% ethanol (vol/vol) for 4 h as previously described (15). Additional analysis details are described in SI Appendix.
Yeast One-Hybrid Assay.
Full-length cDNAs for HAT3, ATHB4, ATHB2, REV, PHB, and PHV were cloned, to be used as the prey proteins. A DNA stretch containing four repeats of a 80-nt-long DNA sequence located 149 bp upstream of the MIR165a exon was synthesized and used as the bait DNA. Additional experimental details are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank P. N. Benfey, J.-Y. Lee, K. Nakajima, and J. L. Bowman for sharing seeds and plasmids as well as A. Obrdlik, M. Ghosh Dastidar, and A. Vilches-Barro for scientific discussion. M.G.H. acknowledges the European Research Council (GA 261081) and Australian Research Council for current funding. The laboratory of S.W. is supported by grants from the Deutsche Forschungsgemeinschaft (WE4281/7-1), the European Research Council (GA 336295), and start-up funding from the Copenhagen Plant Science Centre. F.O. was supported by Max Planck Society funds to D. Weigel.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.A.M. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516110113/-/DCSupplemental.
References
- 1.Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;11(16):1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- 2.Emery JF, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13(20):1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 3.Waites R, Hudson A. phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development. 1995;121(7):2143–2154. [Google Scholar]
- 4.McConnell JR, et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411(6838):709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 5.Bou-Torrent J, et al. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal Behav. 2012;7(11):1382–1387. doi: 10.4161/psb.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turchi L, et al. Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development. 2013;140(10):2118–2129. doi: 10.1242/dev.092833. [DOI] [PubMed] [Google Scholar]
- 7.Mallory AC, et al. MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 2004;23(16):3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development. 2004;131(12):2997–3006. doi: 10.1242/dev.01186. [DOI] [PubMed] [Google Scholar]
- 9.Izhaki A, Bowman JL. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell. 2007;19(2):495–508. doi: 10.1105/tpc.106.047472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411(6838):706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- 11.McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125(15):2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- 12.Wu G, et al. KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc Natl Acad Sci USA. 2008;105(42):16392–16397. doi: 10.1073/pnas.0803997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilegems M, et al. Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development. 2010;137(6):975–984. doi: 10.1242/dev.047662. [DOI] [PubMed] [Google Scholar]
- 14.Brandt R, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012;72(1):31–42. doi: 10.1111/j.1365-313X.2012.05049.x. [DOI] [PubMed] [Google Scholar]
- 15.Merelo P, et al. Genome-wide identification of KANADI1 target genes. PLoS One. 2013;8(10):e77341. doi: 10.1371/journal.pone.0077341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhart BJ, et al. Establishing a framework for the Ad/abaxial regulatory network of Arabidopsis: Ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell. 2013;25(9):3228–3249. doi: 10.1105/tpc.113.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman JL, Eshed Y, Baum SF. Establishment of polarity in angiosperm lateral organs. Trends Genet. 2002;18(3):134–141. doi: 10.1016/s0168-9525(01)02601-4. [DOI] [PubMed] [Google Scholar]
- 18.Tatematsu K, Toyokura K, Miyashima S, Nakajima K, Okada K. A molecular mechanism that confines the activity pattern of miR165 in Arabidopsis leaf primordia. Plant J. 2015;82(4):596–608. doi: 10.1111/tpj.12834. [DOI] [PubMed] [Google Scholar]
- 19.Floyd SK, Bowman JL. Gene regulation: Ancient microRNA target sequences in plants. Nature. 2004;428(6982):485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 20.Craft J, et al. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 2005;41(6):899–918. doi: 10.1111/j.1365-313X.2005.02342.x. [DOI] [PubMed] [Google Scholar]
- 21.Sessions A, Weigel D, Yanofsky MF. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 1999;20(2):259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- 22.Ciarbelli AR, et al. The Arabidopsis homeodomain-leucine zipper II gene family: Diversity and redundancy. Plant Mol Biol. 2008;68(4-5):465–478. doi: 10.1007/s11103-008-9383-8. [DOI] [PubMed] [Google Scholar]
- 23.Wenkel S, Emery J, Hou BH, Evans MM, Barton MK. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell. 2007;19(11):3379–3390. doi: 10.1105/tpc.107.055772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Z, et al. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138(4):2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao X, et al. Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes. FEBS Lett. 2009;583(22):3711–3717. doi: 10.1016/j.febslet.2009.10.076. [DOI] [PubMed] [Google Scholar]
- 26.Brandt R, et al. Control of stem cell homeostasis via interlocking microRNA and microProtein feedback loops. Mech Dev. 2013;130(1):25–33. doi: 10.1016/j.mod.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi N, et al. PROTOCOLS: Chromatin immunoprecipitation from Arabidopsis tissues. Arabidopsis Book. 2014;12:e0170. doi: 10.1199/tab.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Husbands AY, Benkovics AH, Nogueira FTS, Lodha M, Timmermans MCP. The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell. 2015;27(12):3321–3335. doi: 10.1105/tpc.15.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Gen. 1997;16(3):277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 30.Gälweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282(5397):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 31.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12(4):507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.