Major histocompatibility complex (MHC)-related phenotype/genotype correlations are important in study of autoimmune disorders. We previously identified a conserved 33-kb haplotype comprising five genes, lymphotoxin-α (Lta), Tnf, lymphotoxin-β (Ltb), leukocyte-specific transcript 1 (Lst1), and natural cytotoxicity-triggering receptor 3 (Ncr3), in the MHC class-III region regulating arthritis in rats (1). In our study, higher Ltb and Ncr3 expression and lower Lst1 expression correlate with reduced arthritis severity in rats (1). Following these findings, we compared expression of these genes in a cohort of patients with rheumatoid arthritis (RA) and healthy controls and found that the expression of LTB, LST1, and NCR3 was higher in RA cases, but patients with mild RA showed higher NCR3 expression and lower LST1 expression than patients with severe RA (1).
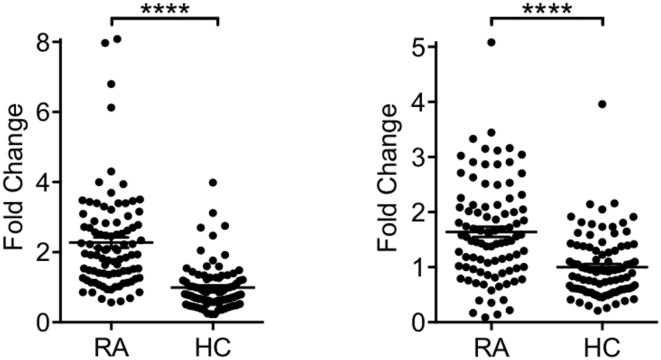
To address genetic variability in the locus, Liu et al. (2) investigate this genetic variability further by analyzing the expression quantitative trait loci (eQTL) dataset from patients with RA in combination with RA genome-wide association studies (GWAS) data. They concluded that increased LST1 and NCR3 were associated with reduced RA susceptibility (2), which could differ from our findings (1) and the findings of another group (3) that LST1 and NCR3 expression was increased in RA cases versus controls (Fig. 1). We believe that this conclusion is not supported by experimental data for several reasons.
Fig. 1.
Expression of LST1 (Left) and NCR3 (Right) in patients with RA (n = 91) and healthy controls (HC; n = 92). ****P < 0.0001.
First, healthy controls were not investigated in this study, and data from patients with RA could not be interpreted in terms of susceptibility.
Second, a cohort of patients with RA in this study consists of only nonresponders to methotrexate (MTX) therapy (2, 4), whereas we included patients with a broader spectrum of treatments and disease activity (1). Because gene expression profiles differ between patients responding and not responding to MTX (5), it is not surprising that findings from two different cohorts could not be cross-replicated. It is also not optimal to make generalized conclusions on RA risk by integration of the eQTL dataset from nonresponding to MTX patients with severe RA with the GWAS dataset, which gives information on disease risk.
Third, Liu et al. (2) do not state if they have controlled for the effect of the DRB1 gene, which is in strong linkage disequilibrium with LST1 and NCR3 (6–8), when selecting eQTLs. We performed association analysis using Immunochip data for SNPs from the study by Liu et al. (2) in our EIRA (Epidemiological Investigation of Rheumatoid Arthritis) cohort (9), which was, in fact, part of the dataset used by Liu et al. (2). In this cohort of 2,762 RA cases and 1,940 controls, we found that after stratification by RA-associated alleles (total for DRB1*01, DRB1*04, and DRB1*10), there is no longer any significant association with RA in LST1-NCR3 after Bonferroni correction (Table 1).
Table 1.
Association results for SNPs in the LST1-NCR3 region in the EIRA study
| 2,762 cases/1,940 controls | 708 SE negative cases/878 SE negative controls | |||||||||
| SNP | Chr | Position (hg19) | A1 | A2 | P value* | P valueBONF | OR (95% CI) | P value* | P valueBONF | OR (95% CI) |
| rs2071596 | 6 | 31506691 | A | G | 0.01608 | 0.1447 | 1.13 (1.02–1.25) | 0.6206 | 1 | 0.95 (0.79–1.15) |
| rs2523500 | 6 | 31518354 | G | A | 0.008165 | 0.07348 | 0.89 (0.81–0.97) | 0.5025 | 1 | 0.95 (0.82–1.10) |
| rs6929796 | 6 | 31522669 | A | G | 0.01526 | 0.1373 | 1.13 (1.02–1.25) | 0.5065 | 1 | 0.94 (0.78–1.13) |
| rs2256974 | 6 | 31555392 | A | C | 0.05241 | 0.4717 | 1.11 (1.00–1.22) | 0.6696 | 1 | 0.96 (0.80–1.16) |
| rs2844479 | 6 | 31572956 | C | A | 8.576E-07 | 7.72E-06 | 1.24 (1.14–1.35) | 0.594 | 1 | 1.05 (0.89–1.23) |
| rs2736176 | 6 | 31587561 | C | G | 5.002E-10 | 4.502E-09 | 1.32 (1.21–1.44) | 0.2806 | 1 | 1.10 (0.93–1.30) |
| rs755714 | 6 | 31609813 | A | G | 9.458E-10 | 8.512E-09 | 1.32 (1.20–1.44) | 0.3052 | 1 | 1.09 (0.92–1.30) |
| rs805297 | 6 | 31622606 | A | C | 6.164E-10 | 5.548E-09 | 1.32 (1.21–1.44) | 0.2967 | 1 | 1.10 (0.92–1.30) |
| rs707919 | 6 | 31641139 | G | A | 7.716E-10 | 6.944E-09 | 1.32 (1.21–1.44) | 0.2967 | 1 | 1.10 (0.92–1.30) |
All P values have been calculated for the allelic model. Chr, chromosome; CI, confidence interval; OR, odds ratio; P valueBONF, P value after Bonferroni correction; SE, shared epitope.
Therefore, we believe that the findings by Liu et al. (2) on LST1 and NCR3 should be reevaluated. This discussion again highlights the advantage of using congenic animals to study complex diseases, in which qualitative conclusions can be made and the isolated loci studied experimentally (1, 10).
Footnotes
The authors declare no conflict of interest.
References
- 1.Yau ACY, et al. Conserved 33-kb haplotype in the MHC class III region regulates chronic arthritis. Proc Natl Acad Sci USA. 2016;113(26):E3716–E3724. doi: 10.1073/pnas.1600567113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu G, et al. Cis-eQTLs regulate reduced LST1 and NCR3 gene expression and contribute to increased autoimmune disease risk. Proc Natl Acad Sci USA. 2016;113:E6321–E6322. doi: 10.1073/pnas.1614369113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulcahy H, O’Rourke KP, Adams C, Molloy MG, O’Gara F. LST1 and NCR3 expression in autoimmune inflammation and in response to IFN-γ, LPS and microbial infection. Immunogenetics. 2006;57(12):893–903. doi: 10.1007/s00251-005-0057-2. [DOI] [PubMed] [Google Scholar]
- 4.Weinblatt ME, et al. Intravenous golimumab is effective in patients with active rheumatoid arthritis despite methotrexate therapy with responses as early as week 2: Results of the phase 3, randomised, multicentre, double-blind, placebo-controlled GO-FURTHER trial. Ann Rheum Dis. 2013;72(3):381–389. doi: 10.1136/annrheumdis-2012-201411. [DOI] [PubMed] [Google Scholar]
- 5.Oliveira RDR, et al. Differential gene expression profiles may differentiate responder and nonresponder patients with rheumatoid arthritis for methotrexate (MTX) monotherapy and MTX plus tumor necrosis factor inhibitor combined therapy. J Rheumatol. 2012;39(8):1524–1532. doi: 10.3899/jrheum.120092. [DOI] [PubMed] [Google Scholar]
- 6.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44(3):291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bakker PIW, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38(10):1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vignal C, et al. Genetic association of the major histocompatibility complex with rheumatoid arthritis implicates two non-DRB1 loci. Arthritis Rheum. 2009;60(1):53–62. doi: 10.1002/art.24138. [DOI] [PubMed] [Google Scholar]
- 9.Eyre S, et al. Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate Wellcome Trust Case Control Consortium High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haag S, et al. Positional identification of RT1-B (HLA-DQ) as susceptibility locus for autoimmune arthritis. J Immunol. 2015;194(6):2539–2550. doi: 10.4049/jimmunol.1402238. [DOI] [PubMed] [Google Scholar]