Significance
Protein aggregation and amyloid formation seem to be at the heart of the pathology of multiple neurodegenerative diseases, including Alzheimer’s disease. protein has long been considered one of the protein components that contributes to the pathogenesis and the progression of the disease. The concepts of energy landscape analysis established in the theory of protein folding are applied here to create a quantitative image of the aggregation energy landscape of . The resulting “amyloid funnel” not only helps visualize the complexity of the early stages of aggregation of WT but also, predicts the effects of mutations at specific sites on aggregation behavior.
Keywords: misfolding, amyloid funnel, nucleation
Abstract
A predictive coarse-grained protein force field [associative memory, water-mediated, structure, and energy model for molecular dynamics (AWSEM)-MD] is used to study the energy landscapes and relative stabilities of amyloid-β protein (1–40) in the monomer and all of its oligomeric forms up to an octamer. We find that an isolated monomer is mainly disordered with a short α-helix formed at the central hydrophobic core region (L17-D23). A less stable hairpin structure, however, becomes increasingly more stable in oligomers, where hydrogen bonds can form between neighboring monomers. We explore the structure and stability of both prefibrillar oligomers that consist of mainly antiparallel β-sheets and fibrillar oligomers with only parallel β-sheets. Prefibrillar oligomers are polymorphic but typically take on a cylindrin-like shape composed of mostly antiparallel β-strands. At the concentration of the simulation, the aggregation free energy landscape is nearly downhill. We use umbrella sampling along a structural progress coordinate for interconversion between prefibrillar and fibrillar forms to identify a conversion pathway between these forms. The fibrillar oligomer only becomes favored over its prefibrillar counterpart in the pentamer where an interconversion bottleneck appears. The structural characterization of the pathway along with statistical mechanical perturbation theory allow us to evaluate the effects of concentration on the free energy landscape of aggregation as well as the effects of the Dutch and Arctic mutations associated with early onset of Alzheimer’s disease.
Alzheimer’s disease is associated with the deposition of amyloid-β (Aβ) protein aggregates in the brain (1). Soluble oligomers, intermediates formed early in the aggregation process, can cause synaptic dysfunction, whereas the later-formed insoluble fibrils may function as reservoirs of the toxic oligomers (2). Owing to their stoichiometric complexity and transience, the early oligomeric forms are difficult to study in the laboratory. Nevertheless, distinct forms of oligomers, described as prefibrillar and fibrillar, have been found to bind differently to conformation-dependent antibodies (3): the fibrillar oligomers and mature fibrils both display a common epitope that is absent from the prefibrillar oligomers. The study of the secondary structure of species using Fourier transform infrared spectroscopy suggests that fibrillar forms of are organized in a parallel β-sheet conformation, much like in the complete fibril structure constructed from solid-state NMR data by Petkova et al. (4), whereas the prefibrillar oligomers contain mainly antiparallel β-sheets (5). Numerous computer simulation studies of both the monomer and higher aggregates using models ranging in complexity from fully atomistic simulations in solvent to lattice models have been undertaken to fill the knowledge gap (6–8). It remains, however, unclear what the exact tertiary arrangements of the β-sheets in the prefibrillar oligomers are as well as how the structures and stabilities of the oligomers change as they grow. To further our understanding, in this paper, we use the AWSEM force field comprehensively to explore the structures and stabilities of oligomers up to the octamer of the full-length molecule. AWSEM has already proved successful in structurally characterizing monomeric protein folding, dimer binding, and misfolding of multidomain proteins (9–12). The coarse-grained nature of this force field allows us to explore in some detail the free energy landscape for oligomer assembly and the interconversion between the prefibrillar and fibrillar forms of full-length oligomers. In addition to enabling the examination of the mechanism of aggregation, the coarse-grained energy landscape simulations allow us to analyze the effects of point mutations, such as those in the Dutch variant (E22Q) and the Arctic variant (E22G), which are associated with early-onset familial forms of Alzheimer’s disease.
Structure of Oligomers Sampled in Simulation
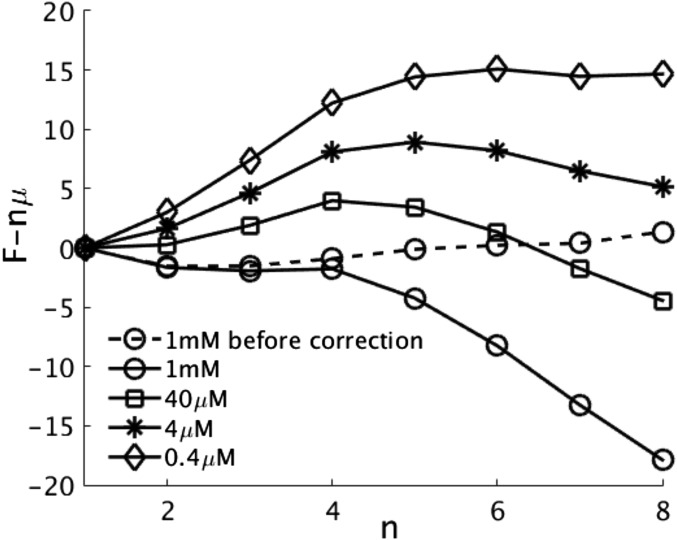
Although aggregation usually occurs in a macroscopic system, simulations are limited to studying thermodynamically small systems. By sampling the configurations of a fixed number of molecules in a periodic box, monitoring the largest cluster, and then, analyzing the resulting distribution of its size, we are able to compute the aggregation free energy as a function of oligomer size. This procedure allows us to obtain an overview of the landscape of stabilities, as shown in Fig. 1. It is computationally challenging to simulate a large number of molecules of the full-length protein on the long timescales relevant to aggregation. Because of the efficiency of the AWSEM simulation code, however, we have managed to extend our simulations with 12 monomers in a box up to the millisecond timescale at an initial concentration of 1 mM. Although this concentration is larger than the physiological range, the simulation can be extrapolated into the range accessed by experiments using statistical mechanical theory. At this simulation size, as aggregation progresses, the effective concentration of monomers in the simulation box decreases when a large cluster forms. We have used Reiss’s nucleation theory (14) to correct for this effect to find stabilities at fixed chemical potential as described in Methods. In addition, we found it necessary to use an enhanced sampling strategy to account for the poor sampling of fibrillar structures in the unbiased simulations as we will discuss in SI Text. Fig. 1 shows the free energy profile reflecting the relative stabilities of oligomers of defined size both with and without the Reiss correction as well as the structural sampling correction. Fig. 1 plots the appropriate thermodynamic potential to describe an aggregate that is the grand ensemble free energy . is the free energy of an oligomer of size n, and μ is the chemical potential of the free monomers that is set by the concentration of the solution (Methods). The grand ensemble free energy profile exhibits downhill characteristics at the simulation concentration 1 mM. Examining the structural ensembles of different oligomers, we see that there is considerable structural complexity along the aggregation pathway as well as malleability under different thermodynamic conditions. For the monomers, two classes of structures can be distinguished (Fig. 2, structure I): in one class, a short helix forms in the central hydrophobic core (CHC) region with the rest of the chain remaining disordered, and in the other class, a β-hairpin structure forms between the CHC region and A30-V36. At 300 K, the helical structure class is more stable than the ensemble of hairpin structures. A similar helical structure was found in an NMR study by Vivekanandan et al. (15), which showed that monomer adopts a compact partially folded structure with the CHC region forming a helix in aqueous solution. In this implementation of AWSEM force field, α-helices are favored over the form. The β-hairpin structures sampled in the monomer simulations are essentially the same as the β-hairpin structure that has been found in the complex of with an affibody protein , which stabilizes the hairpin structure through intermolecular contacts (16). The hairpin structure has also previously been observed in all-atom simulations of (17) and (17, 18). In vitro, these two structural classes are of similar stability as witnessed by there being a reversible α to β conformational transition in monomeric , which occurs on changing the polarity of the solvent (19). In our simulations of the higher oligomers, as shown in the dimer (II) and trimer (III) cases in Fig. 2, the stability of the hairpin structure is greatly increased owing to intermolecular hydrogen bonds. For the trimer and tetramer, cylindrin-shaped structures (20) made up of three or four hairpin-shaped monomers having their CHC regions packed inside the complex dominate the distribution. These cylindrins involve the pairing of strands 17–23 and 30–36 in our simulations. As oligomer size increases, the relative fraction of the fibrillar form increases.
Fig. 1.
We plot the appropriate thermodynamic potential in units of kilocalories per mole for an open grand canonical system. The aggregation free energy for is obtained from simulating 12 monomers in a box with 1 mM concentration at 300 K. The solid line with ○ represents the corrected free energy at the simulated concentration of 1 mM after removing the finite size effect and accounting for the poor sampling of fibril structures in the raw simulation data, which is shown with the dashed line; n is the size of the oligomer. The extrapolated free energy profiles are also shown at (indicated by □) and (indicated by *), the limits of the experimental study (13). We also show the profile at the predicted solubility (indicated by ♢).
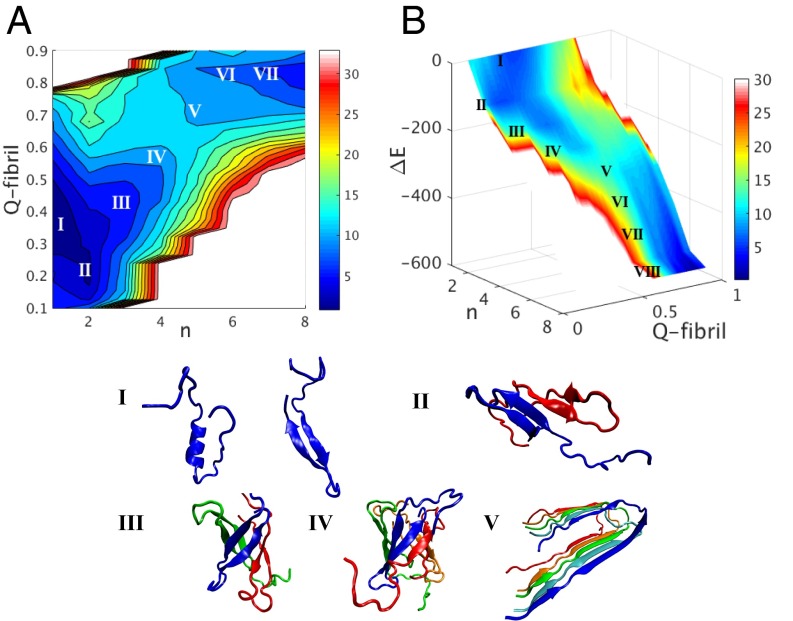
Fig. 2.
(A) The grand canonical free energy surface in units of kilocalories per mole at the concentration of at 300 K is plotted using an added dimension of Q-fibril that reveals more structural details of the oligomer. Q-fibril is the fraction of contacts formed in a particular structure that matches with those formed in the fibrillar counterpart, which contains only parallel hydrogen bonds. The ideal fibril corresponds with the structure V in Fig. 4. The closer the value of Q-fibril of a structure is to one, the more similar the structure is to a fibril form. Representative structures for the individual basins indicated on the free energy surface are shown (Lower), with the Roman numerals indicating oligomer size. For the monomer, both a disordered structure with fragment L17-D23 being helical and a hairpin-like structure involving the pairing of strands 17–23 and 30–36 are observed and comparably populated in the simulation. The hairpin structure is greatly stabilized after a dimer is formed because of the intermonomer interactions. Trimers and tetramers are largely found with a cylindrin-like structure (20) shown in III and IV. (B) The binding energy for the aggregation of is plotted as a function of the size of the oligomer (n) and its structure similarity compared with a fibril of the same size (Q-fibril) at the concentration of 1 mM and 300 K. The binding energy shown on the z axis is the energy of the entire simulation system subtracted from the total energy of 12 free monomers. This energy provides an amyloid funnel that decreases monotonically as the oligomer size increases. The colors on the surface indicate the grand canonical free energy . The local basins in the landscape indicated in shades of blue are labeled by the number of subunits in each basin. A free energy bottleneck for interconversion between fibrillar and prefibrillar forms occurs in between the tetramer and the pentamer. Although the prefibrillar configurations are more populated when , the fibrillar fractions of the oligomer start to dominate the aggregation pathway on the other side of this small barrier.
Aggregation Energy Landscape of and the Amyloid Funnel
For proteins folding to their native state, the familiar funnel picture of the energy landscape has deepened our understanding with a visually intuitive image and also functioned as the basis of quantitatively accurate modeling. The funneled aspect of the folding landscape is a result of natural selection (21). Similar images of an “amyloid funnel” have been used in discussing aggregation but usually without the benefit of quantification. Presumably, in the case of aggregation, a funneled landscape would not be a favorably evolved feature. The AWSEM simulations in this paper allow us to construct a quantitative image of the aggregation energy landscape for , which shows that the landscape has both funnel-like and rugged characteristics. As always, these images of the landscape are of intrinsically low dimensionality compared with the reality of the full conformational space of aggregates. For clarity, three plots each of higher dimensionality will be introduced in this section sequentially in our discussion. As shown in Fig. 1, at the simulation concentration, the 1D aggregation free energy at the simulation concentration is essentially downhill with a small plateau at after the finite size effects are taken into account. The downhill characteristic of the 1D plot implies that aggregation at the simulated concentration requires only a “monomeric” nucleus in the terminology of Ferrone (22). The full kinetic analysis using simulated free energy profiles, like the laboratory situation, has several tricky points because of the “missing” dimensions that are not included when we describe nucleation using size as the only progress variable. Another complication in the laboratory is that several distinct processes contribute simultaneously to the observed time development of the concentration of aggregated species (13, 23), including not only primary nucleation but also, fragmentation and secondary nucleation. The experimentally fitted parameters would suggest that the nucleus for primary nucleation of can be considered to be monomeric throughout the concentration range of . also seems to be described by a monomeric nucleus (23), although its primary nucleation rate is about two orders of magnitude larger than that of . When we extrapolate the predicted aggregation grand free energy to the experimental concentration range, we reach the apparent limit of a strictly monomeric nucleus after the concentration becomes lower than . Nevertheless, we see that, at 40 , the top of the experimental range of study, the 1D free energy curve has a peak of only about 4 kcal/mol near the tetramer. This barrier is so modest, however, that it is not clear whether it, in fact, would influence the apparent nucleus size as fitted to the kinetics. The barrier does increase further at , which is closer to the predicted solubility limit (). As witnessed by our curve at , which is nearly flat at large n, the predicted solubility limit is in quite good agreement with direct experimental measurement (24). Because the chemical potential depends logarithmically on concentration, discrepancies of a factor of 10 in concentration translate into per monomer errors in binding energy; thus, the modest difference between theory and experiment clearly could well result from a small underestimation of the strength of intermolecular interactions in the AWSEM force field. We also should note, however, that, according to our simulation, the early aggregation steps are kinetically more complex than was considered in the 1D aggregation model that was used in quantitatively fitting the experimental data. The complexity of the early stages of aggregation is evident after we introduce structural variables in the second plot (Fig. 2A) for the aggregation free energy. Up to the pentamer, the unguided simulation samples containing largely “prefibrillar” structures that are not congruent with the experimental structures determined by solid-state NMR on the ultimate fibril form. When we used the fibril structure obtained by Petkova et al. (4) as a template for a hexamer and simulated it with the AWSEM force field, after equilibration, the energy of this fibrillar hexamer turned out to be considerably lower than the energies of the prefibrillar hexamer structures sampled in our unguided simulations of aggregation. For the lower oligomers, when the fibril core is small, the exposed hydrophobic residues at the two ends of the fibril make the fibrillar structure less stable than a cylindrin-shaped prefibrillar structure in which most hydrophobic residues are buried inside. However, after the fibril grows beyond a threshold length ( in our simulations), the portion of exposed residues at the fibril ends becomes small enough so that their unfavorable contribution to the overall stability is outweighed by the favorable contribution from the increasing number of parallel β-hydrogen bonds. This difficulty of direct simulation motivated us to get a better idea of the landscape in the region of stability cross-over by using a biased sampling scheme to explore the route of forming fibrils. This scheme used the quantified structural difference between a selected prefibrillar hexamer and the fibrillar hexamer () as an order parameter as discussed in Methods.
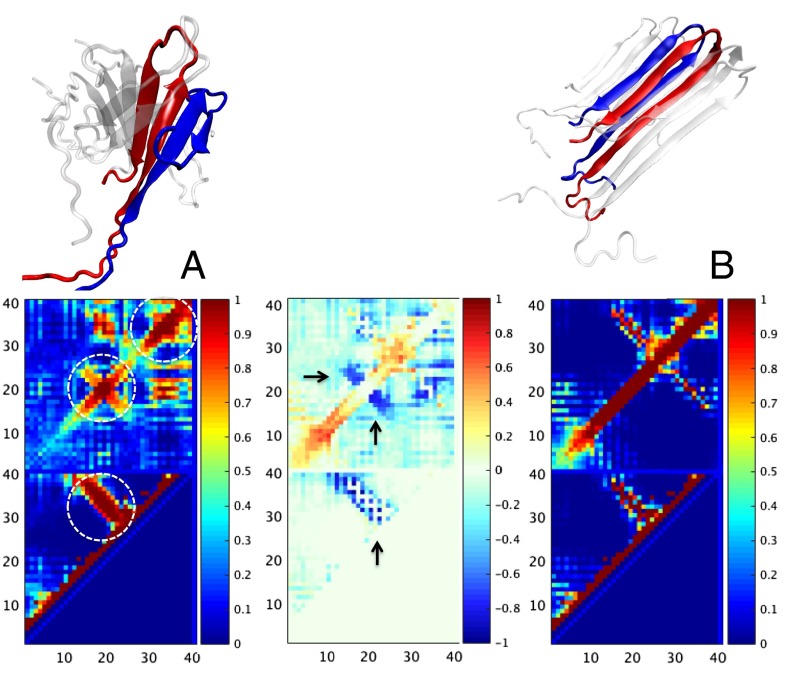
In the hexamer, the topologies of the prefibrillar and fibrillar structural ensembles are, in fact, so different that it is not immediately apparent what kinetic pathways can transform one ensemble into the other. There must be a considerable amount of “backtracking,” in which local stabilizing contacts in the prefibrillar form must be broken followed by significant conformational change occurring so as to form the new set of stabilizing contacts in the fibril form. This sequence of events resembles the mechanism envisioned by Hoyer et al. (16) in their study of the monomeric β-hairpin structure being stabilized by the affibody protein . To elucidate the dynamics of the conversion, we, therefore, carried out a set of umbrella sampling simulations along the progress coordinate , which measures the structural difference by quantifying the similarity of configurations to a selected prefibrillar hexamer that is rich in antiparallel β-sheets along with the similarity to the fibrillar hexamer that contains only parallel β-sheet proposed by Petkova et al. (4). Fig. 3 shows the contact probability maps of the prefibrillar and fibrillar ensembles for the hexamer computed using this order parameter. The difference contact probability map indicates which contacts are broken (blue in Fig. 3) and which ones are formed (red in Fig. 3) in the backtracking step that leads to fibril formation. The simulation results allow us to identify an intermediate state between the prefibrillar and fibrillar basins, in which structures contain strong self-recognizing parallel β-interactions between V12-D23 in one chain with the same string of residues in a neighboring chain rather than this segment interacting with the A30-V36 strand in the same chain as in the prefibrillar form. These self-recognizing interactions resemble those seen in the misfolded structures of a tethered multidomain protein I27-I27 studied in our previous AWSEM simulations of aggregation (11). Indeed, they are shown to be amyloidogenic for the AWSEM force field using the criteria of the AWSEM-based “amylometer” that identifies fragments capable of forming low-energy amyloids (11). These self-recognizing interactions ultimately form the core of the propagating amyloid structure. The energetic and entropic tradeoffs in first aggregating into the prefibrillar forms and then, transforming into the fibril are evident in Fig. 2. We see that the binding free energy of structures monotonically decreases with n in an amyloid funnel but that the decrease is, at first, rather slow in the prefibrillar form; however, the decrease becomes more dramatic after the fibril with parallel β-sheets wins in stability. At the stage of interconversion, a small entropic bottleneck emerges. The precise consequences of this bottleneck for the apparent nucleus size determined in kinetic experiments are hard to assess, because the intrinsic rates of transitions in the Q-fibril direction depend on the dynamic friction in the condensed oligomers, whereas simply adding individual monomers from the solvent may well be diffusion-limited before the rearrangements take place. As shown in Fig. 4, the rearrangements needed to carry out the backtracking are intricate, and the landscape of prefibrillar oligomers is rugged. The self-recognizing interactions are so strong that the N termini of two interacting monomers remain together, whereas the C termini have a chance to break intramolecular β-hydrogen bonds, which then rearrange ultimately to form the complete self-recognizing parallel β-strands. This change leads to the formation of a fibrillar core, which can be stabilized by intermolecular interactions from neighboring complexes, exiting along the energetic amyloid funnel shown in Fig. 2B. This core then grows further when another monomer with a hairpin structure joins the core and goes again through the same dock and lock conformational change (25) as the previous monomer did.
Fig. 3.
Contact probability maps of (A) the prefibrillar hexamer ensemble and (B) the fibrillar hexamer ensemble are created based on the sampled structures. The color indicates the probability that a particular pair of contacts forms in configurations within the structural basin. Lower in each map describes intracontacts, whereas Upper describes intercontacts. The labeling of the two axes is the residue index. Representative structures for A and B are illustrated in Upper, and the two neighboring chains in the structures are colored in red and blue. The circled area in A, Lower shows the predominant hairpin motif within each chain; the circled area in A, Upper shows the self-recognizing interactions for segments V12-D23 and A30-V40. The difference map in the middle is created by subtracting the map in A from the map in B, in which the color indicates formation of new contacts (red) or breakage of existing contacts (blue). The arrows in the difference map point at the regions in which self-recognizing contacts (Upper) or the contacts in the hairpin motif (Lower) will be broken in the process of converting to a fibrillar structure. These locations involved in backtracking are near the residues mutated in the Dutch and Arctic variants (residue 22) associated with early-onset familial Alzheimer’s disease.
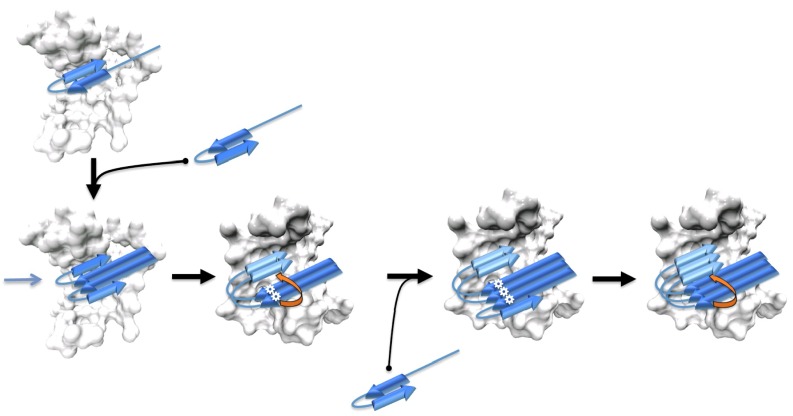
Fig. 4.
A cartoon illustration of the formation and growth of a small fibrillar core. Two monomeric hairpin structures form a dimeric construct with parallel intermolecular β-hydrogen bonds in the N termini. The C-terminal residues will rearrange and form new parallel β-hydrogen bonds through breaking intramolecular antiparallel β-hydrogen bonds, indicated by the orange arrow. White asterisks are used to indicate the location of residue 22, which is at the crucial site where the intramolecular interactions break in backtracking. The resulted dimeric core is so unstable that it has to be stabilized by other intermolecular interactions provided by a neighboring complex, illustrated in the white surface representation. The core then grows farther when another monomer is added to the exposed side of the core via a dock and lock mechanism (25).
Mutational Effects on the Aggregation Free Energy
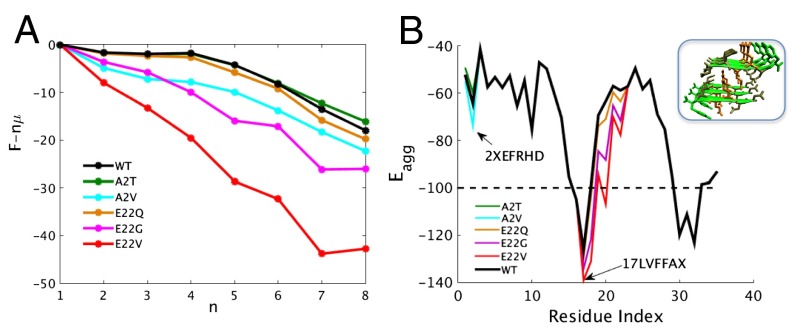
It is likely that, in the absence of solvent, amyloid formation is universal and not highly sequence-dependent. We, therefore, see a variety of peptides and proteins that are not related in sequence, structure, and function that all assemble into highly regular amyloid fibrils. Nevertheless, in solution, sequences containing amyloidogenic regions are needed to initiate the process of aggregation (26–28), and these local sequence patterns are selected against. Most evolved proteins contain only a few such segments. Amyloid formation, especially in the early stages of the process, therefore shows considerable sequence and structural specificity. The hydrophobicity of a single residue has been shown to be crucial for the formation of oligomers and fibrils in . Päiviö et al. (29) showed that the lag phase of aggregation for strongly correlates with the hydrophobicity of residue 22, which is the mutation site that leads to early-onset of Alzheimer’s disease in the so-called Dutch and Arctic variants. Because the coarse-grained AWSEM force field has already preaveraged over solvent degrees of freedom, we can address these mutational effects using the configurations that we have already sampled with AWSEM for the WT . To do this study, we used free energy perturbation calculations on the sampled configurations of the WT sequence to recalculate the grand canonical aggregation free energy first for three mutants at site 22: E22Q, E22A, and E22V. Consistent with the experiment, as shown in Fig. 5, the amyloid funnel becomes increasingly downhill as the degree of hydrophobicity increases at site 22. We note also that site 22 is involved in the crucial backtracking step of rearrangement from prefibrillar to fibrillar forms. Site 22 is only five residues away from the cleavage site for α-secretase, and therefore, genetic mutations on this site also influence the specific peptides produced in vivo (30). Interfering mutations have also been identified near the beginning of the chain at site 2, which we also studied. The mutant A2T has been found to be protective, whereas the mutation A2V is deleterious (31). Simulations show that this site is involved, but much more weakly, in backtracking. The mutations here have less impact on the overall slope of the amyloid funnel for . Nevertheless, A2V is indeed found to favor oligomerization. The effects at this level of the A2T mutation are very small, although they do disfavor slightly the fibrils. Additional kinetic analyses of the landscape would, therefore, be needed to explain the reported modest but puzzling effects of the A2T mutation on aggregation kinetics.
Fig. 5.
(A) Increasing downhill characteristics for the aggregation free energy with increasing hydrophobicity at site 22 in . We also show the weaker effects of mutation at site 2. (B) The amyloidogenicity of the WT sequence and all of the mutants studied. Inset shows the structure of a six-strand amyloid zipper (32) used as the energetic template. We replace the hexameric sequence of the template structure with that from to evaluate the aggregation energy of a new zipper involving self-recognition centered at the given residue index. The amyloidogenic threshold for is (dashed line), below which is considered amyloidogenic. Increased hydrophobicity at site 22 elevates the amyloidogenicity of the hexameric sequence segments that contain the site. This correlation in amyloidogenicity is consistent with the mutational change in the aggregation free energy on the left. The mutations at site 2 have very small effects compared with those at site 22.
Discussion
The coarse-grained predictive AWSEM force field has allowed us to study the initial oligomerization and later fibrillization of in quantitative detail. We explored the conformational ensembles of specific oligomers using intuitive progress coordinates that monitor the transition between fibrillar and prefibrillar forms as well as how the stabilities of these structural ensembles vary as the number of units in the oligomer changes. The monomer generally adopts a disordered structure with a short helix fragment at the CHC region. When stabilized by intermolecular interactions, an alternative monomeric hairpin structure oligomerizes to form a cylindrin-shaped complex, the dominant prefibrillar form. In the presence of a membrane, a cylindrin-like oligomer may be able to penetrate the membrane like the structurally similar outer membrane pore-forming protein (5). In this way, the prefibrillar forms might act as cytotoxic agents to permeabilize cells. The prefibrillar forms also have unsatisfied hydrophobic actions that may allow additional docking to specific proteins in the cell or on its surface. Our structural library for these oligomers will allow us to assess this possibility in the future for a range of candidate partners. In the initial stages of aggregation, a fibrillar oligomer is not stable on its own but either dissociates or transforms back into a prefibrillar oligomer. A prefibrillar oligomer must locally rearrange by breaking antiparallel β-hydrogen bonds and then, form parallel hydrogen bonds to create a fibrillar core. After the fibrillar core forms, it can be joined by other monomers one by one through a dock and lock mechanism, finally becoming more stable than its prefibrillar counterpart at the pentamer stage. The early-onset Dutch and Arctic variants contain mutations localized in the region involved in this arrangement and indeed, are predicted as perturbation to favor aggregation. The techniques used here can be straightforwardly extended to other fragments, including the more pathologically significant .
Methods
Simulation Model.
AWSEM is a predictive coarse-grained protein folding force field that has been applied to folding, binding, and misfolding problems successfully (9–12). The tertiary interactions are transferable and use parameters optimized by the energy landscape theory learning algorithm. The local in sequence interactions (; are residue indices) are bioinformatic in origin and governed by the associative fragment memory term, . These fragments can be chosen in a variety of ways, ranging from knowledge of homologs to fully atomistic simulations of constituent fragments (33). In this study, we used the de novo structure prediction protocol, where the associative memory term is determined by 20 so-called fragment memories for each segment of sequence with length 9. These fragment memories are sequence homologs found using the sequence alignment tool PSI-BLAST searching through the Protein Data Bank database. General aspects of the AWSEM code are described in greater detail in the supporting information in the work by Davtyan et al. (9).
Simulation Protocol and .
All simulations were performed in the canonical ensemble using the Nose–Hoover thermostat as implemented in the LAMMPS Molecular Dynamics Package. Umbrella sampling simulations were carried out at 320 K and then, extrapolated to 300 K. Structural similarity to a hexameric prefibrillar oligomer formed by monomeric hairpins was selected for primary study of the pathway of the transition from the prefibrillar oligomeric form to the fibrillar form. The progress coordinate for the umbrella sampling simulations is defined as , where with , , and .
Correction for Finite Size Effects and Extrapolation of the Aggregation Free Energy to Different Concentrations.
We simulate N protein monomers in a box of volume V with periodic boundary condition at temperature T. Initially, the N free monomers are distributed evenly in the box. After an equilibration time, snapshots from the rest of the simulation are selected for free energy calculation only when they contain a single cluster with size n and free monomers. We use Reiss’s theory (14) to convert these cluster populations into free energies for n-mers. Let be the partition function for the system when it contains one single n cluster and free monomers; then, the grand canonical partition function of the system can be written as . For a macroscopic system with short-range interactions, the partition sum for the selected configurations can be written as the product of two decoupled terms—one being the partition function for the remaining free monomers and the other being the partition function for the n cluster : . The probability that one finds a single n cluster and free monomers is given by
where is the chemical potential of the free monomers and taken as an invariant when . The grand canonical free energy that governs cluster growth, therefore, is given by
| [1] |
where is the free energy of the cluster (14).
Owing to the finite number of monomers in the simulation box, the free energy directly obtained from our simulation differs from what would be found in an infinite system, because the effective monomer concentration changes. , where , which is the chemical potential of free monomers in the box with volume V and temperature T. The corrected grand canonical free energy for a large system can then be obtained by compensating for the artefactual change of the chemical potential of the free monomers:
We note that, as a function of n, the correction term becomes more dramatic as n increases.
After we have the grand ensemble free energy with an initial free monomer concentration , we can derive the free energy at a different concentration using Eq. 1: , where and are the chemical potentials of the free monomers at concentrations and , respectively. The sampling correction along is described in SI Text. The full free energy profiles, including the finite size correction and the sampling correction, are shown in Fig. S1. More technical details on the evaluation of the mutational effects on the aggregation landscapes are illustrated in Fig. S2.
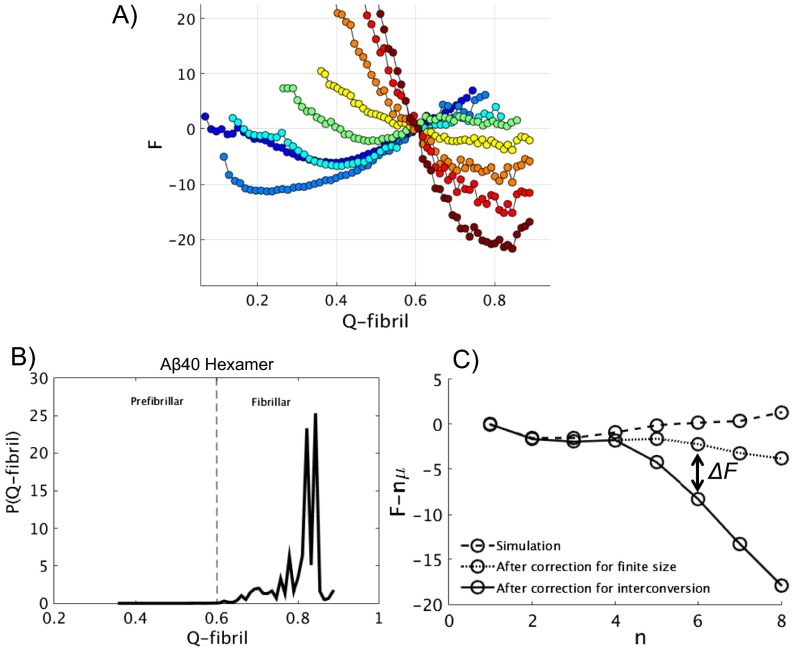
Fig. S1.
(A) The free energies as a function of Q- for oligomers with size n = 1, …, 8. The value of n coincides with the color spectrum, the monomer being in dark blue and the octamer being in dark red. (B) The probability distribution of various structural ensembles of hexameric is plotted along coordinate Q- . We choose Q- =0.6 as the cutoff for distinguishing prefibrillar and fibrillar components to calculate the cumulative probability and , respectively. (C) The aggregation free energy of at 1 mM concentration and 300 K before and after corrections.
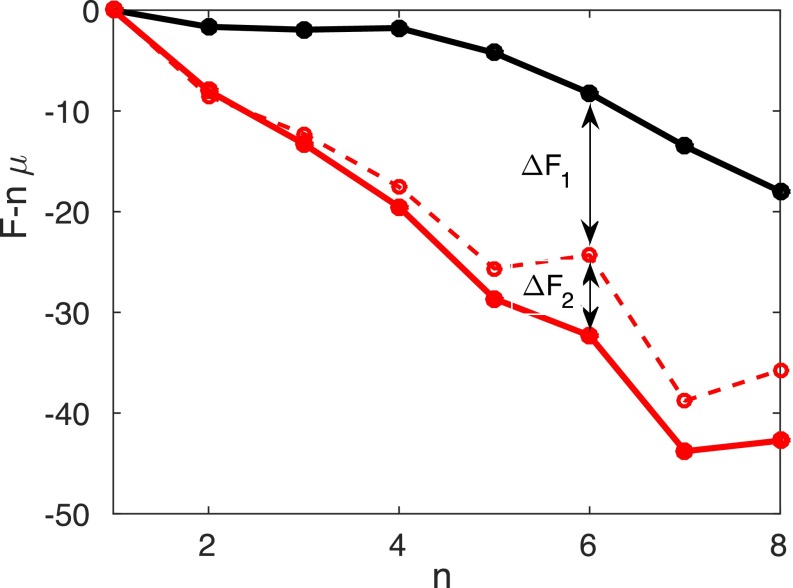
Fig. S2.
The mutational effects on the free energy are calculated from two sets of simulations. The free energy for the WT is in black. The dashed curve represents the free energy after incorporating the changes calculated from the simulations probing the growth of the oligomer; the solid red curve represents the final free energy after further including the changes calculated from the interconversion simulations.
SI Text
Correction to the Aggregation Free Energy Because of Insufficient Sampling of the Fibrillar Structures in Unbiased Runs.
In our unbiased simulations, which we use initially to calculate the aggregation free energy profile, the process of the interconversion from a prefibrillar oligomer to its fibrillar counterpart is kinetically slow because of the backtracking and does not occur in the unbiased sampling. As a result of this slowness, the fibrillar oligomers are not properly sampled in an unbiased simulation, although they are the more stable species for , with n being the oligomer size. To correct for this sampling problem, we carried out an additional eight sets of umbrella sampling simulations, in which we applied biasing potentials to enforce a thorough probing of the conformational space for each species of all eight different sizes (n = 1, …, 8), exploring the fibrillar region along with the kinetically accessible prefibrillar region. The obtained free energies are shown in Fig. S1A. This information is then used to make additional correction to the aggregation free energy after the effects of finite size are corrected for. As shown in Fig. S1B, in the example of the hexamer, we use Q-fibril = 0.6 as the cutoff to calculate the cumulative probability for the prefibrillar ensemble to compare with that for the fibrillar ensemble . We note that for n = 1, …, 8. The correction to the free energy can then be written as as shown in Fig. S1C. This correction is negligible when n < 5, because is very close to one, but it becomes more evident when n increases.
Calculation of Mutational Effects on the Aggregation Landscapes Using Free Energy Perturbation.
The free energy perturbation calculation is applied to the sets of both biased and unbiased simulations mentioned above to evaluate the mutational effects on the aggregation landscapes. We recalculate the energy of the already sampled structures with the mutated sequence to evaluate the change of energy: . The change of free energy can then be written as . As shown in Fig. S2, the change of the free energy on mutation (E22V) includes both the calculated from the unbiased simulations that probe the growth of the oligomer and the calculated from the biased simulations that explore the interconversion process.
Acknowledgments
We thank the Data Analysis and Visualization Cyberinfrastructure funded by National Science Foundation Grant OCI-0959097. This work was supported by National Institute of General Medical Sciences Grant R01 GM44557. Additional support was provided by D. R. Bullard-Welch Chair at Rice University Grant C-0016.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612362113/-/DCSupplemental.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat Rev Mol Cell Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 3.Kayed R, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2(1):18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45(2):498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerf E, et al. Antiparallel β-sheet: A signature structure of the oligomeric amyloid beta-peptide. Biochem J. 2009;421(3):415–423. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 6.Nasica-Labouze J, et al. Amyloid β protein and Alzheimer’s disease: When computer simulations complement experimental studies. Chem Rev. 2015;115(9):3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Straub JE, Thirumalai D. Toward a molecular theory of early and late events in monomer to amyloid fibril formation. Annu Rev Phys Chem. 2011;62(1):437–463. doi: 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morriss-Andrews A, Shea JE. Computational studies of protein aggregation: Methods and applications. Annu Rev Phys Chem. 2015;66(1):643–666. doi: 10.1146/annurev-physchem-040513-103738. [DOI] [PubMed] [Google Scholar]
- 9.Davtyan A, et al. AWSEM-MD: Protein structure prediction using coarse-grained physical potentials and bioinformatically based local structure biasing. J Phys Chem B. 2012;116(29):8494–8503. doi: 10.1021/jp212541y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng W, Schafer NP, Davtyan A, Papoian GA, Wolynes PG. Predictive energy landscapes for protein-protein association. Proc Natl Acad Sci USA. 2012;109(47):19244–19249. doi: 10.1073/pnas.1216215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng W, Schafer NP, Wolynes PG. Frustration in the energy landscapes of multidomain protein misfolding. Proc Natl Acad Sci USA. 2013;110(5):1680–1685. doi: 10.1073/pnas.1222130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng W, Schafer NP, Wolynes PG. Free energy landscapes for initiation and branching of protein aggregation. Proc Natl Acad Sci USA. 2013;110(51):20515–20520. doi: 10.1073/pnas.1320483110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meisl G, et al. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc Natl Acad Sci USA. 2014;111(26):9384–9389. doi: 10.1073/pnas.1401564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiss H, Bowles RK. Some fundamental statistical mechanical relations concerning physical clusters of interest to nucleation theory. J Chem Phys. 1999;111(16):7501. [Google Scholar]
- 15.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. A partially folded structure of amyloid-β (1-40) in an aqueous environment. Biochem Biophys Res Commun. 2011;411(2):312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyer W, Gronwall C, Jonsson A, Stahl S, Hard T. Stabilization of a beta-hairpin in monomeric Alzheimer’s amyloid-beta peptide inhibits amyloid formation. Proc Natl Acad Sci USA. 2008;105(13):5099–5104. doi: 10.1073/pnas.0711731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenman DJ, Connors CR, Chen W, Wang C, García AE. Aβ monomers transiently sample oligomer and fibril-like configurations: Ensemble characterization using a combined MD/NMR approach. J Mol Biol. 2013;425(18):3338–3359. doi: 10.1016/j.jmb.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ball KA, Phillips AH, Wemmer DE, Head-Gordon T. Differences in beta-strand populations of monomeric Aβ40 and Aβ42. Biophys J. 2013;104(12):2714–2724. doi: 10.1016/j.bpj.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomaselli S, et al. The α-to-β conformational transition of Alzheimer’s Aβ (1-42) peptide in aqueous media is reversible: A step by step conformational analysis suggests the location of beta conformation seeding. ChemBioChem. 2006;7(2):257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 20.Laganowsky A, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolynes PG. Evolution, energy landscapes and the paradoxes of protein folding. Biochimie. 2015;119:218–230. doi: 10.1016/j.biochi.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrone FA. Assembly of Aβ proceeds via monomeric nuclei. J Mol Biol. 2015;427(2):287–290. doi: 10.1016/j.jmb.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 23.Cohen SI, et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci USA. 2013;110(24):9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiang W, Kelley K, Tycko R. Polymorph-specific kinetics and thermodynamics of β-amyloid fibril growth. J Am Chem Soc. 2013;135(18):6860–6871. doi: 10.1021/ja311963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han W, Schulten K. Fibril elongation by aβ (17-42): Kinetic network analysis of hybrid-resolution molecular dynamics simulations. J Am Chem Soc. 2014;136(35):12450–12460. doi: 10.1021/ja507002p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanova MI, Sawaya MR, Gingery M, Attinger A, Eisenberg D. An amyloid-forming segment of beta2-microglobulin suggests a molecular model for the fibril. Proc Natl Acad Sci USA. 2004;101(29):10584–10589. doi: 10.1073/pnas.0403756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DuBay KF, et al. Prediction of the absolute aggregation rates of amyloidogenic polypeptide chains. J Mol Biol. 2004;341(5):1317–1326. doi: 10.1016/j.jmb.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 28.Esteras-Chopo A, Serrano L, Lopez de la Paz M. The amyloid stretch hypothesis: Recruiting proteins toward the dark side. Proc Natl Acad Sci USA. 2005;102(46):16672–16677. doi: 10.1073/pnas.0505905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Päiviö A, Jarvet J, Gräslund A, Lannfelt L, Westlind-Danielsson A. Unique physicochemical profile of β-amyloid peptide variant Aβ1–40e22g protofibrils: Conceivable neuropathogen in arctic mutant carriers. J Mol Biol. 2004;339(1):145–159. doi: 10.1016/j.jmb.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 30.Selkoe DJ. Alzheimer’s disease: G, proteins, and therapy. Physiol Rev. 2001;81(2):741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 31.Das P, Murray B, Belfort G. Alzheimer’s protective A2t mutation changes the conformational landscape of the Aβ1–42 monomer differently than does the A2v mutation. Biophys J. 2015;108(3):738–747. doi: 10.1016/j.bpj.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson R, Eisenberg D. Recent atomic models of amyloid fibril structure. Curr Opin Struct Biol. 2006;16(2):260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Chen M, Lin X, Zheng W, Onuchic JN, Wolynes PG. Protein folding and structure prediction from the ground up: The atomistic associative memory, water mediated, structure and energy model. J Phys Chem B. 2016;120(33):8557–8565. doi: 10.1021/acs.jpcb.6b02451. [DOI] [PMC free article] [PubMed] [Google Scholar]