Significance
Understanding the role played by disorder in biology is becoming increasingly important. Disordered proteins are central to signaling, development, initiation of transcription, and other vital cellular processes. How and why disordered proteins are used is not entirely clear, but disorder can be important in allostery, facilitate regulatory posttranslational modification, and allow rapid and specific but promiscuous binding. Here, our investigations of biofilm-promoting protein SasG illustrate that disorder can play another role. We show that the intrinsic disorder of one-half of the domains is important for imparting long-range cooperativity in folding of a large multidomain protein—allowing formation of a small local element of structure to precipitate cooperative folding of adjacent disordered domains across a length scale of ∼10 nm.
Keywords: IDP, protein folding, parallel pathways, protein engineering, cooperativity
Abstract
Many human proteins contain intrinsically disordered regions, and disorder in these proteins can be fundamental to their function—for example, facilitating transient but specific binding, promoting allostery, or allowing efficient posttranslational modification. SasG, a multidomain protein implicated in host colonization and biofilm formation in Staphylococcus aureus, provides another example of how disorder can play an important role. Approximately one-half of the domains in the extracellular repetitive region of SasG are intrinsically unfolded in isolation, but these E domains fold in the context of their neighboring folded G5 domains. We have previously shown that the intrinsic disorder of the E domains mediates long-range cooperativity between nonneighboring G5 domains, allowing SasG to form a long, rod-like, mechanically strong structure. Here, we show that the disorder of the E domains coupled with the remarkable stability of the interdomain interface result in cooperative folding kinetics across long distances. Formation of a small structural nucleus at one end of the molecule results in rapid structure formation over a distance of 10 nm, which is likely to be important for the maintenance of the structural integrity of SasG. Moreover, if this normal folding nucleus is disrupted by mutation, the interdomain interface is sufficiently stable to drive the folding of adjacent E and G5 domains along a parallel folding pathway, thus maintaining cooperative folding.
It has been suggested that as much as 20% of the proteome may be intrinsically disordered (1), mainly manifested as intrinsically disordered regions within multidomain proteins, although a few proteins are apparently entirely disordered. Some proteins function as a consequence of disorder: for example, disordered PEVK regions of titin act as an entropic spring (2), whereas in the nuclear pore complex, disordered nucleoporins provide a thick selective barrier controlling nuclear import (3). Disorder can also play other roles: it facilitates posttranslational modification and may promote allostery (4, 5). SasG (Staphylococcus aureus surface protein G) is a cell wall-attached protein from S. aureus that promotes intercellular adhesion during the accumulation phase of biofilm formation via its C-terminal repetitive region (6–8). We previously showed that this part of SasG contains alternating E and G5 domains (Fig. 1A) and that E folds when it is N-terminal of a G5 domain. The disorder of E domains in isolation is essential for formation of a long, stiff, mechanically strong, rod-like structure (9) capable of projecting the N-terminal A domain, which is involved in host colonization (6).
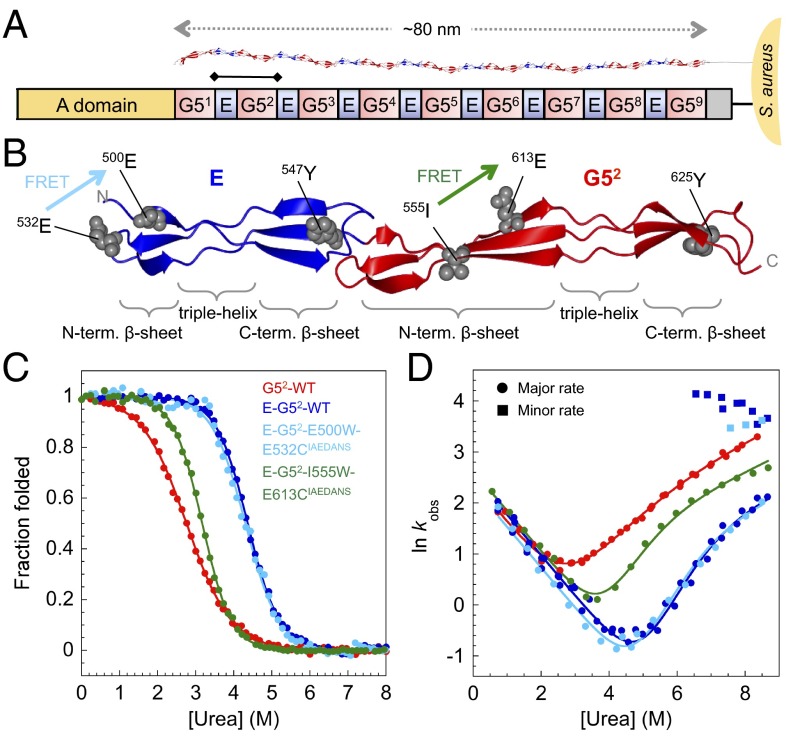
Fig. 1.
Structure and biophysical data for WT SasG G52 and E–G52. (A) Schematic representation of SasG from S. aureus NCTC 8325. The A domain promotes adhesion to host cells. The core region comprises tandemly arrayed G5 (red) and E (blue) domains (10). The E–G52 fragment of SasG is indicated with a bar. (B) Structure of E–G52 (PDB ID code 3TIP) illustrating the topology of E and G52 domains: two single-layer, triple-stranded β-sheets connected by a central collagen-like triple-helical region. The tyrosines and positions of engineered FRET pairs are shown. FRET pair E500W-E532CIAEDANS (cyan) results in FRET only when E is folded; I555W-E613CIAEDANS (green) results in FRET when G52 is folded. (C) Equilibrium denaturation curves. Data for WT G52, E–G52, and E–G52–E500W–E532CIAEDANS were taken from ref. 9. (D) Urea dependence of the natural logarithm of the observed rate constants (in seconds−1) for proteins shown in C. Circles and squares represent major and minor unfolding rate constants, respectively.
Here, we combine biophysical measurements, protein engineering, and simulation to show that the disorder in the E domains of SasG also promotes cooperative folding and unfolding pathways. We find that SasG domains have a highly polarized transition-state (TS) structure, where formation of a small portion of a three-stranded sheet in the far C-terminal region of a SasG G5 domain is sufficient to drive the folding of structure over a distance of 10 nm. Our studies reveal the importance of the E–G5 interface in driving this cooperativity. Furthermore, when the usual folding nucleus is disrupted by mutation in the multidomain protein, then this interface is sufficiently stable to drive folding of the two adjacent domains. Thus, we propose that disorder can play a key role in ensuring cooperative folding over long distances in multidomain proteins.
Results
SasG Domains Fold Cooperatively at Equilibrium.
SasG domains are highly homologous: the sequence identity between G5 domains (except for the first and last) and between E domains is >97%. Here, we investigated the first E domain and the second G5 domain (G52) either alone or in tandem (E–G52) (Fig. 1). We have previously shown that the E domain is fully unfolded in isolation (10). Because SasG domains have no tryptophans, (un)folding was followed by monitoring intrinsic tyrosine fluorescence. We have shown that urea-induced equilibrium denaturation curves of E–G52 monitored by fluorescence coincide with those recorded by ellipticity at 235 nm (7) and domain-specific FRET probes (9), showing that equilibrium unfolding of the two-domain construct is fully cooperative: two-state with concerted disruption of both domains and secondary and tertiary structure with no accumulation of intermediates (Fig. 1C). The stability of E–G52 is around 3.5 kcal mol−1 greater than that of an isolated G52 domain (6.3 vs. 2.8 kcal mol−1, respectively).
Kinetic Experiments Reveal That SasG Domains Fold and Unfold Cooperatively.
The refolding kinetics of G52 and E–G52 can both be described by a sum of two exponential phases, with a fast folding phase (accounting for at least 30% of the amplitude) and a slower phase that represents proline cis-trans isomerization-limited folding events (E–G52 and G52 have 17 and 8 prolines, respectively). Only the faster phase is discussed here. The rate constant for folding of E–G52 is the same as that of G52 at all denaturant concentrations (Fig. 1D). Under unfolding conditions, at urea concentrations ≤6.5 M, only a single kinetic phase is detected for both G52 and E–G52, but E–G52 unfolds significantly more slowly, and the dependence of the logarithm of the rate constant for unfolding on denaturant concentration (mku) is significantly higher.* The unfolding limbs of the chevron plots are curved (Fig. 1D). To account for nonlinearity in the observed unfolding rate constant, the chevron plot data were fitted to a sequential TSs model (12), in which denaturant induces a switch between two barriers separated by a high-energy intermediate.
At denaturant concentrations below ∼6.5 M urea, all of the evidence suggests that both G52 and the two-domain construct E–G52 fold via a two-state pathway, where the two domains fold and unfold cooperatively: we observe for both constructs that the values of mD–N obtained by combining kinetic m values are the same within error as the equilibrium values (SI Appendix, Table S1). Similarly, the values of free energy of unfolding () calculated from the kinetic data match the equilibrium values (SI Appendix, Table S1). Furthermore, double-jump stopped flow experiments showed no evidence of additional phases that might reveal populated intermediates for either construct.
Cooperative Unfolding Breaks Down at High Denaturant Concentrations.
The unfolding of E–G52 and G52 results in a decrease in tyrosine fluorescence. However, in the unfolding kinetics of E–G52 only, at urea concentrations >7.0 M, we observed a second, faster rate associated with an increase in fluorescence that shows very weak denaturant dependence (Fig. 1D and SI Appendix, Fig. S1). A similar extra phase was also observed for the E–G52 construct labeled with E500W-E532CIAEDANS FRET pair (Fig. 1D), which reports specifically on the (un)folding of E. In contrast, the unfolding kinetics of E–G52 probed by I555W-E613CIAEDANS (resulting in FRET only when G52 is folded) is monophasic (Fig. 1D). We infer that the minor rate detected at high urea concentration is related to unfolding of the E domain, perhaps when the stabilizing interface fails at high denaturant concentrations. Note that two other mutations that strongly destabilized the E domain (G524A and G527A) also decoupled the unfolding of E and G52 (SI Appendix, Fig. S2).
G52 and E–G52 Fold Via the Same Highly Polarized Transition State.
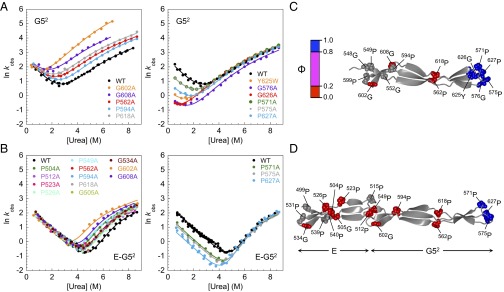
Because G52 and E–G52 fold at the same rate and the dependence of the refolding rate constant on denaturant concentration is the same (Fig. 1D), we infer that they fold via the same rate-limiting TS. To map out which regions are structured early in the folding of G52 and E–G52, a mutational Φ-value analysis was carried out. SasG domains do not have a compact hydrophobic core, and all side chains are exposed to solvent. Mutation of surface residues rarely results in sufficient loss of stability to undertake Φ-value analysis (13). Hence, a series of nonconservative mutations (mainly Pro to Ala and Gly to Ala) was introduced in both G52 and E–G52, and their influence on the thermodynamic stability and kinetics was investigated (SI Appendix, Tables S2–S5). Φ-Values were calculated (SI Appendix, Tables S4 and S5) for mutants where the destabilization energy () was ≥0.7 kcal·mol−1 (14). In general, nonconservative mutations, such as those that we are using here, have to be interpreted with care. However, the resultant chevron plots show that, here, we can be unequivocal (Fig. 2 A and B). Unusually, mutations alter either only the folding kinetics, meaning Φ is close to 1 and the region is fully structured in the TS, or only the unfolding kinetics, meaning Φ ∼ 0, suggesting that the region is as unstructured in the TS as in the D. There are no intermediate Φ-values. When mapped onto the structures, the Φ-value pattern is clear (Fig. 2 C and D). It is only in the extreme C-terminal loop/β-sheet region that any structure is formed at all in the TS (Φ ≥ 0.8) in both G52 and E–G52, suggesting that the rate-limiting TS for folding is common for the two constructs and strongly polarized to the C-terminal region of the G52 domain. The rest of the protein folds only after formation of this initial embryonic structure, formation of which establishes the correct register for the β-strands of the G52 domain.
Fig. 2.
Mapping the structure of TSs for folding of G52 and E–G52. (A) Chevron plots for G52: WT (black) and mutants. (B) Chevron plots for E–G52: WT (black) and mutants. (A, Left and B, Left) Mutants that unfold faster than the WT, but the folding rate is largely unaffected. (A, Right and B, Right) Mutants that fold slower than the WT, but the unfolding rate is unaffected. (C and D) Φ-values of (C) G52, and (D) E–G52 mapped onto the crystal structures. Blue, high Φ-values (>0.8); gray, where ∆∆G was not high enough to obtain reliable Φ-values; red, low Φ-values (<0.2).
Simulations Reveal More Details About the Folding Pathway.
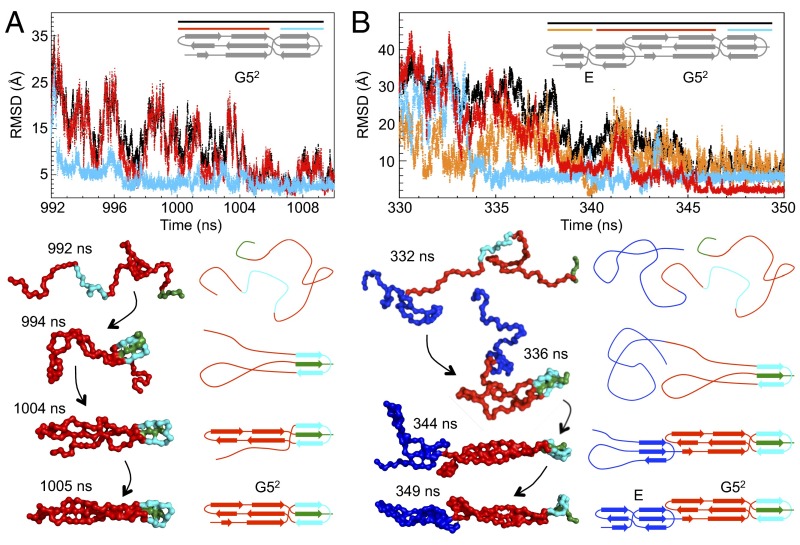
After the main rate-limiting TS, our kinetic experiments are relatively “blind” to the subsequent steps. With simulations, it is possible to probe the entire pathway. Long equilibrium simulations for G52 and E–G52 were carried out using a coarse-grained native-centric model, which allowed us to follow a number of unfolding and folding reactions. In all of these simulations, the first step in the folding of both G52 and E–G52 is formation of the C-terminal β-sheet/loop motif of G52 (Fig. 3). In the case of E–G52, the C-terminal region of E folds concurrently with the N-terminal part of G52, resulting in formation of the E–G5 interface, followed by folding of the N-terminal β-sheet of E, which completes the E–G52 structure (Fig. 3B). Thus, folding of the interface is key to the folding of E (SI Appendix, Fig. S3). At the midpoint temperature, where the proteins are folded 50% of the time (∼320 K for both G52 and E–G52), we observed only a few complete folding events, because the domains are rarely fully unfolded. Hence, we performed a large number of shorter simulations starting from completely unfolded structures (from simulations at high temperature), setting the temperature well below the folding temperature. Folding occurs in most of these short simulations, and in all cases, the sequence of events is that described above. In a few cases, where the E domain folds first, its unfolding is required before the entire E–G52 folds.
Fig. 3.
Probing the folding pathways of SasG using simulations. Simulations of (A) G52 and (B) E–G52 by coarse-grained native-centric model simulations at 320 K. A, Upper and B, Upper show the rmsd as a function of simulation time for a typical refolding event. (A) For G52, rmsd values were calculated for all atoms (black), the C-terminal β-sheet/loop region (cyan), and the N-terminal β-sheet/loop region (red). (B) For E–G52, rmsd values were calculated for all atoms (black), the C-terminal β-sheet/loop region of G52 (cyan), the N-terminal β-sheet/loop region of G52 together with the C-terminal β-sheet/loop region of E (red), and the N-terminal β-sheet/loop region of E (orange). A, Lower and B, Lower illustrate corresponding sequential snapshots from the refolding trajectory and the related schematic topology representation. The G52 domain is shown in red, except for the C-terminal β-sheet/loop region (cyan) and its central C-terminal docking strand (green). The E domain is shown in blue. Additional details from the same trajectory are illustrated in SI Appendix, Fig. S3.
The Stability of the Interface Is Essential to Ensure Cooperative Unfolding of E–G52.
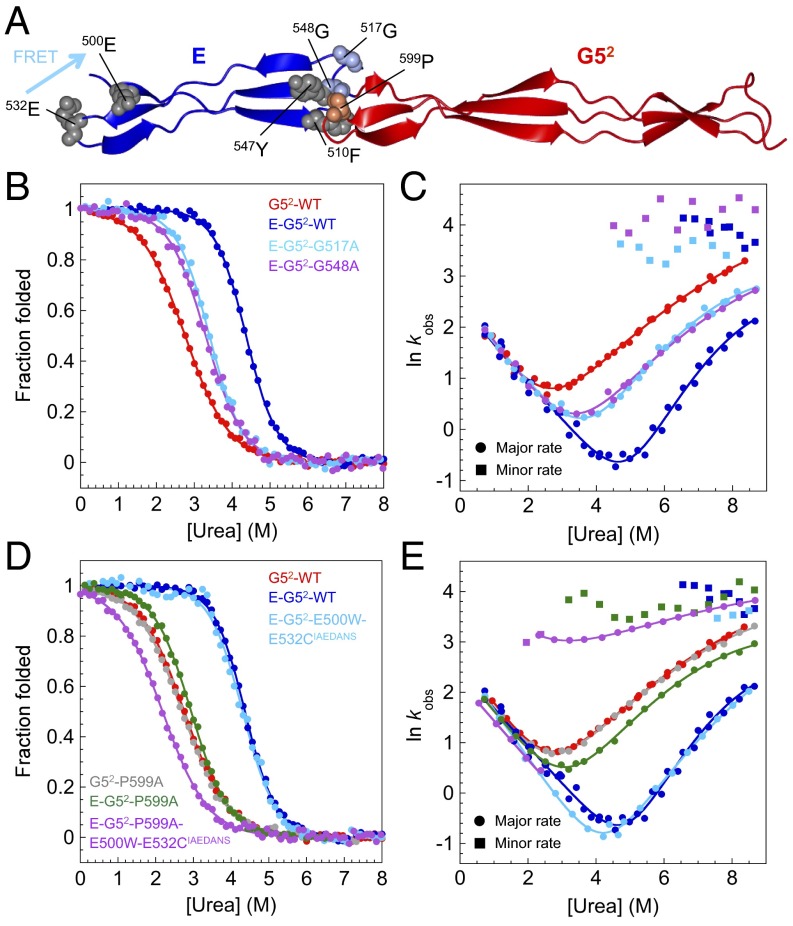
We identified two mutations in the E domain of E–G52 (G517A and G548A) at the interface between the two domains that, although the interface was sufficiently stable to promote the folding of the E domain, resulted in unfolding kinetics that were completely uncoupled; two unfolding phases are observed in all unfolding traces (Fig. 4 A–C). As was seen in WT E–G52, the fast unfolding phase, ascribed to the unfolding of the E domain (which has a low amplitude and is associated with an increase in fluorescence), has a weak dependence on denaturant concentration. Importantly, the slower unfolding phase, associated with the larger fluorescence change, now has the unfolding m value of the G52 domain alone, additional evidence that, for these mutations at the interface, the E and G52 domains now unfold independently.
Fig. 4.
Mutations at the interface break the cooperative unfolding of E–G52. (A) Structure of E–G52 showing the location of mutated residues within the E domain (Gly517, Gly548; light blue) and G52 domain (Pro599; orange). Phe510 and Tyr547 (gray) contact Pro599. (B and C) Mutations in the E domain: (B) Equilibrium denaturation curves and (C) urea dependence of the natural logarithm of the observed rate constants for WTs and mutants. (D and E) Mutations in the G52 domain: (D) equilibrium denaturation curves and (E) urea dependence of the natural logarithm of the observed rate constants for WTs and mutants. Circles and squares in C and E represent major and minor rate constants, respectively. Mutations at the interface result in the breakdown of the cooperative unfolding of the E and G52 domains manifested in the presence of a second unfolding rate constant at all denaturant concentrations and a decrease in the dependence of lnku on [urea].
We investigated this further using the interface mutant P599A found in the G52 domain, which has no effect on the thermodynamic stability and kinetics of G52 in isolation but perturbs E–G52 (Fig. 4 D and E). Pro599 is located in the N-terminal loop of G52. In the isolated domain, Pro599 is exposed to solvent, whereas in the context of E–G52, it contributes to the hydrophobic cluster at the E–G5 interdomain interface, where it makes contacts with Phe510 and Tyr547 from the E domain (Fig. 4A). We introduced the E500W-E532CIAEDANS FRET pair (Fig. 4A) in E–G52–P599A, which results in FRET only when E is folded. The unfolding kinetics was monitored by the decrease in 1,5-IAEDANS fluorescence (Fig. 4E), and at high denaturant concentrations that promote unfolding, a single phase was detected, corresponding to the faster unfolding phase found for E–G52–P599A (similar rate constants and the same urea dependence) and clearly representing unfolding of E uncoupled from G52. Note that we still observe the same single refolding phase for this mutant (except around the midpoint) (Fig. 4E) when followed by FRET, because the folding of G52 is the rate-limiting step for folding of the E domain. Thus again, we found that the interface is key to cooperative folding.
Mutations Reveal an Alternative Folding Pathway for E–G52.
We found five destabilizing mutations within the G52 domain that alter the folding pathway in E–G52. Three of these (G576A, Y625W, and G626A) are located in the C-terminal β-sheet/loop region of G52 (Fig. 5A) where, as shown in Fig. 2, folding is nucleated in both G52 and E–G52. These mutations destabilize the proteins by >1 kcal mol−1 relative to WT G52 and E–G52 (Fig. 5 B and C and SI Appendix, Tables S2 and S3). In G52 alone, these three variants all have a Φ-value of ∼1 (that is, they unfold exactly as the WT), and all of the change in stability is reflected in a change in the rate of folding (Fig. 2A, Right). Importantly, the dependence of the rate constant for folding on denaturant concentration (mkf) is exactly the same as for WT G52. In E–G52, however, although these mutants again unfold exactly as the WT, now the folding kinetics are clearly different (Fig. 5D). All still fold more slowly than the WT, but now, the mkf values are significantly increased compared with the WT, suggesting that these variants are folding via a different, significantly more compact, TS with a βT = 0.53 (compared with 0.33 for WT E–G52).†
Fig. 5.
E–G52 can fold by an alternative folding pathway. Mutations in the G5 domain that destabilize the folding nucleus cause a switch in pathway in E–G52 manifested by a change in the dependence of lnkf on [urea]. (A) Structure of E–G52 showing the location of residues mutated or used to engineer the FRET pair. (B and C) Equilibrium denaturation curves for G52 and E–G52, respectively. (D) Chevron plots for WT E–G52 and mutants. Note the change in slope of the folding limb of the chevron plot for all of these mutants. (E) Mutations using Y625W as a pseudo-WT. Chevron plots for WT E–G52 (black), E–G52–Y625W (green), and Pro-to-Ala mutants of E–G52 in the background of Y625W. Note that the interface mutant (P599A) causes the slope to revert to the WT. The other mutants have Φ-values that differ from those in the WT background.
Two other Gly to Ala mutations within the triple-helical region of G52 (G584A and G587A) (Fig. 5A) destabilized the domain so significantly that the mutants are largely disordered at 0 M urea (Fig. 5B and SI Appendix, Table S2). In E–G52, these mutations are also destabilizing, but now, both E and G52 are folded (Fig. 5C and SI Appendix, Table S3). Interestingly, the chevron plots of both E–G52–G584A and E–G52–G587A show the same values as the mutants that destabilize the extreme C-terminal region of E–G52 (Fig. 5D), suggesting that these variants also fold via a new, more compact TS (with a βT of 0.53). Note that folding is still cooperative; in a control experiment, the kinetics of E–G52–G584A recorded using the E500W-E532CIAEDANS FRET pair (reporting specifically on folding of E) was characterized by an identical to the one measured by intrinsic tyrosine fluorescence (Fig. 5D).
Thus, if we make mutations that significantly destabilize the folding nucleus at the extreme C-terminal end of the G52 domain or mutations that are essential for formation of the triple helix connecting the nucleus to the rest of the protein, we apparently alter the folding pathway—but only when the E domain is present.
Formation of the Interface Is Key to Driving Folding Along the Alternative Pathway.
Crucially, for some of these mutations in the G52 domain (e.g., Y625W and G576A), the folding pathway of isolated G52 does not change; the new pathway is only accessible when the E domain is present, and yet we know that E does not fold in isolation. Given the importance of the interface between the two domains in imparting stability and cooperativity, we hypothesized that the alternative TS (characterized by βT of 0.53) involves formation of a structured E–G52 interface as an early step in this alternative pathway.
If this hypothesis is correct, then residues close to the E–G52 interface, in the E and G52 domains, which all originally have a Φ-value ∼ 0, should have increased Φ-values in this new pathway, and residues in the region with high Φ-values in the WT would have low Φ-values in this alternative pathway. We would also predict that a mutation that destabilized the interface could switch the new pathway back to the original polarized TS in E–G52. Thus, we performed a mutational analysis based on Φ-values, in which E–G52–Y625W was treated as a pseudo-WT (Fig. 5 A and E and SI Appendix, Table S6). [A crystal structure of the protein at 1.6-Å resolution reveals that this substitution does not disrupt the structure of G52 (SI Appendix, Fig. S4 and Table S7).] In that background, we introduced a number of Pro-to-Ala mutations, most of which originally had Φ-values = 0 in the background of WT E–G52. P531A and P540A in E and P618A in G52 (all Φ ∼ 0) were designed to probe the folding of the individual domains, and P512A and P599A (also Φ ∼ 0) were designed to weaken the interface. P571A, which originally had Φ ∼ 1, is found in the C-terminal loop at the center of the nucleation site for the WT pathway. Although one-half of the mutants (P512A, P531A, and P618A) were insufficiently destabilizing to determine Φ-values in the background of E–G52–Y625W, three of the mutants gave us information.
-
i)
The E domain is partly structured in the TS of the alternative pathway; the P540A mutation resulted in a fractional Φ (0.7) in the context of E–G52–Y625W (compared with Φ-values = 0 for Gly to Ala mutations in the same region of the WT E domain). Folding is more affected than unfolding, implying that the triple helix of the E domain is now significantly structured in the TS (Fig. 5E).
-
ii)
The C-terminal loop of G52 is not formed in the TS of the alternative pathway; the P571A mutation now has no effect on the folding rate. The Φ-value is low in the background of E–G52–Y625W (Fig. 5E) (Φ = 0.1 compared with Φ = 1 in the WT).
-
iii)
If the interface is destabilized, then E–G52 reverts to the original folding pathway; the chevron plot of E–G52–Y625W–P599A shows the same as E–G52–P599A and WT E–G52, indicative of the WT-like folding pathway (Fig. 5E). We infer that the mutation P599A at the E–G52 interface destabilizes the new TS and causes folding to revert to the original WT pathway. These results confirm that the new TS involves formation of structure at the interface between the two domains in the alternative folding pathway.
Discussion
SasG is a protein that challenges some of our preconceptions of protein structure and folding. It has an unusual sequence composition typical of an intrinsically disordered protein (∼60% of the residues are charged, Pro or Gly), but it demonstrably folds cooperatively—albeit to an unusual single-sheet extended structure. Despite this unusual structure, the biophysical parameters for folding (m value, stability) are quite unremarkable for a protein of this size (E–G5 and G5 have 132 and 82 residues, respectively). What is remarkable is that G5 domains fold far more rapidly than might be predicted from their relative contact order (15) (SI Appendix, Fig. S5). The interface between the E and G52 domains provides most of the stability for the protein. The importance of the interface is exemplified when we consider the mutation of two highly conserved Gly residues in the triple-helical region of the G52 domain (G584A and G587A), which both cause G52 to be unfolded; when we mutate these same residues in E–G52, the protein folds (Fig. 5 B and C). Thus, we can take an unfolded G52 domain, add an intrinsically unfolded E domain, and produce a folded protein. We have estimated that the interface imparts at least 6 kcal mol−1 to the stability of E–G52 (compared with ∆GD–N for WT G52 and E of 2.8 and ≤−2.5 kcal mol−1, respectively) (9). This interface is also key to maintaining cooperative folding and the long-range cooperativity that imparts stiffness to the SasG structure. Here, we have shown that the interface is essential to ensure that the entire E–G5 motif folds and unfolds in a single cooperative step—mutations at the interface disrupt cooperative folding. However, to our surprise, our data suggest that the interface between E and G52 is completely unformed at the TS for folding (the E domain and the N-terminal region of the G52 domain are both unstructured).
Our data show that folding of SasG is initiated at the far C-terminal end of the G52 domain. At this point, there is a turn between the two outer β-strands, and the terminal “docking” strand is inserted between these into the loop (Fig. 3). Assembly of this small structural element in one domain is sufficient to drive folding of the entire E–G5 molecule over a distance of more than 10 nm. However, folding at the interface is clearly an option, because destabilization of the C-terminal nucleation site allows folding via a higher-energy TS, where formation of the interface is key. E–G52 can thus fold via parallel pathways, but the lowest-energy pathway involves formation of the C-terminal nucleus. It is unclear why this WT pathway should be lower in energy than a pathway involving formation of the interface, the most stable region of the structure and essentially, the only region where there is any significant burial of hydrophobic residues. It may be because the entropic cost of forming the interface is larger; it involves bringing together loops from the E and the G5 domains that are distant in sequence (∼85 residues apart), although the interactions in the C-terminal nucleus are by no means short range (∼50 residues between the C-terminal residues of the final strand and the turn). Alternatively, the intrinsic disorder of the E domain may again be key. The formation of the interface involves the folding, at least in part, of the E domain, a process that is inherently costly in terms of free energy. Importantly, however, cooperative folding is a feature of both pathways, because the E domain cannot fold in the absence of G5.
In WT protein (except under very destabilizing conditions as described), the protein folds and unfolds as a single unit; no intermediates are populated in folding, in unfolding, or at equilibrium, which is, by definition, cooperative folding. Such tight and robust cooperativity in folding has not been seen previously in multidomain proteins. Even where there are significant interfaces between domains, kinetics reveals that the domains fold in a non–two-state manner, with each domain behaving as an independent folding unit (16, 17). The “obligate” cooperativity of SasG arises, because E can only fold in the presence of folded G5, but once folded, the entire domain is very significantly more stable than the sum of the stability of the two domains individually.
The kind of cooperativity that we are observing in the SasG protein (obligate folding cooperativity) is reminiscent of the folding of repeat proteins. These proteins comprise tandem arrays of small repeats (20–40 residues) that are unstable on their own and fold, apparently cooperatively, through formation of interfaces between the repeats (18–25). However, tandem repeats are very different to SasG, where contacts within the domains themselves and between domains are very long range, whereas contacts in repeat proteins are very local (SI Appendix, Fig. S5). Although there is a dominant folding pathway in SasG, parallel pathways are a key feature of repeat proteins, in particular as the number of repeats increases. Despite each subunit being intrinsically unstable alone, kinetic cooperativity is not generally maintained beyond three to four subunits in repeat proteins, but SasG is able to maintain cooperative folding across a distance of ∼12 nm.
Conclusion
The importance of intrinsic disorder in biology is becoming increasingly apparent; however, why would nature choose disordered domains to form a multidomain protein? We had previously shown that disorder-mediated thermodynamic cooperativity allows SasG to adopt long, mechanically strong, rod-like structures (9). Now, we have shown how this disorder coupled with the remarkable stability of the interdomain interface can result in cooperative folding kinetics, with no populated intermediates, across long distances. The folding of classic multidomain proteins is highly cooperative but only within the relatively local confines of a single domain. In repeat proteins, short-range cooperativity is apparent between three and four individually unstable repeats. SasG provides a paradigm for much longer-range cooperative folding—by the obligatory folding of alternate intrinsically disordered domains with their folded neighbors.
Materials and Methods
All experimental procedures are described in detail in SI Appendix.
Analysis of Kinetic Data.
For some mutants, kinetic data were fitted to a model allowing for parallel pathways (details are in SI Appendix, Fig. S6).
Simulations.
Simulations were performed using a coarse-grained model where only Cα atoms are represented and interactions depend on the native reference structure and the residue type. Details are given in SI Appendix.
Determination of the Structure of E–G52–Y625W.
Details of the crystallization and structure determination of E–G52–Y625W can be found in SI Appendix. The coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with ID code 5DBL.
Supplementary Material
Acknowledgments
This research was supported by Biotechnology and Biological Sciences Research Council Grants BB/J006459/1 (to D.T.G. and J.C.) and BB/J005029/1 (to F.W. and J.R.P). Also, C.A.T.F.M is supported by the Cambridge Trust and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) Science Without Borders Cambridge Scholarship. J.R.P holds British Heart Foundation Senior Basic Science Fellowship FS/12/36/29588. J.C. is a Wellcome Trust Senior Research Fellow WT/095195.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5DBL).
*The dependence of folding/unfolding rate constants on [urea] (kinetic m values mkf and mku) is determined by the change in solvent-accessible surface area (SASA) between the denatured state (D) and the TS (in folding) and between the TS and the native state (N; for unfolding) (11). Thus, because E–G52 and G52 have the same folding m values, we can assume that they fold via the same TS. The unfolding m value (mku) is higher for E–G52 than for G52, because the entire E domain plus a significant proportion of the G52 domain unfold between N and TS.
†The Tanford β-value, , is a measure of the position of the TS (in terms of SASA or compactness) between D and N (11). An alternative explanation for a switch in mkf is that a mutation results in destabilization of a TS that falls later on the same single pathway. Several lines of evidence suggest that this is a less reasonable explanation than parallel pathways. Only mutations that destabilize the WT pathway (with Φ ∼1) are affected; the same mutations in G52 alone do not result in a change in mkf; a residue with Φ ∼ 1 in the WT has Φ ∼ 0 in Y625W (see Formation of the Interface Is Key to Driving Folding Along the Alternative Pathway, point ii).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608762113/-/DCSupplemental.
References
- 1.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Tskhovrebova L, Trinick J. Titin: Properties and family relationships. Nat Rev Mol Cell Biol. 2003;4(9):679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 3.Milles S, et al. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell. 2015;163(3):734–745. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shammas SL, Travis AJ, Clarke J. Allostery within a transcription coactivator is predominantly mediated through dissociation rate constants. Proc Natl Acad Sci USA. 2014;111(33):12055–12060. doi: 10.1073/pnas.1405815111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Law SM, Gagnon JK, Mapp AK, Brooks CL., 3rd Prepaying the entropic cost for allosteric regulation in KIX. Proc Natl Acad Sci USA. 2014;111(33):12067–12072. doi: 10.1073/pnas.1405831111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153(Pt 8):2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 7.Geoghegan JA, et al. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol. 2010;192(21):5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Formosa-Dague C, Speziale P, Foster TJ, Geoghegan JA, Dufrêne YF. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc Natl Acad Sci USA. 2016;113(2):410–415. doi: 10.1073/pnas.1519265113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruszka DT, et al. Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein. Nat Commun. 2015;6:7271. doi: 10.1038/ncomms8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruszka DT, et al. Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc Natl Acad Sci USA. 2012;109(17):E1011–E1018. doi: 10.1073/pnas.1119456109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fersht AR. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. Freeman; New York: 1999. [Google Scholar]
- 12.Bachmann A, Kiefhaber T. Apparent two-state tendamistat folding is a sequential process along a defined route. J Mol Biol. 2001;306(2):375–386. doi: 10.1006/jmbi.2000.4399. [DOI] [PubMed] [Google Scholar]
- 13.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224(3):771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 14.Fersht AR, Sato S. Phi-value analysis and the nature of protein-folding transition states. Proc Natl Acad Sci USA. 2004;101(21):7976–7981. doi: 10.1073/pnas.0402684101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277(4):985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 16.Batey S, Nickson AA, Clarke J. Studying the folding of multidomain proteins. HFSP J. 2008;2(6):365–377. doi: 10.2976/1.2991513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batey S, Clarke J. Apparent cooperativity in the folding of multidomain proteins depends on the relative rates of folding of the constituent domains. Proc Natl Acad Sci USA. 2006;103(48):18113–18118. doi: 10.1073/pnas.0604580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werbeck ND, Rowling PJ, Chellamuthu VR, Itzhaki LS. Shifting transition states in the unfolding of a large ankyrin repeat protein. Proc Natl Acad Sci USA. 2008;105(29):9982–9987. doi: 10.1073/pnas.0705300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werbeck ND, Itzhaki LS. Probing a moving target with a plastic unfolding intermediate of an ankyrin-repeat protein. Proc Natl Acad Sci USA. 2007;104(19):7863–7868. doi: 10.1073/pnas.0610315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe AR, Itzhaki LS. Biophysical characterisation of the small ankyrin repeat protein myotrophin. J Mol Biol. 2007;365(4):1245–1255. doi: 10.1016/j.jmb.2006.10.060. [DOI] [PubMed] [Google Scholar]
- 21.Tang KS, Fersht AR, Itzhaki LS. Sequential unfolding of ankyrin repeats in tumor suppressor p16. Structure. 2003;11(1):67–73. doi: 10.1016/s0969-2126(02)00929-2. [DOI] [PubMed] [Google Scholar]
- 22.Tripp KW, Barrick D. Rerouting the folding pathway of the Notch ankyrin domain by reshaping the energy landscape. J Am Chem Soc. 2008;130(17):5681–5688. doi: 10.1021/ja0763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrick D, Ferreiro DU, Komives EA. Folding landscapes of ankyrin repeat proteins: Experiments meet theory. Curr Opin Struct Biol. 2008;18(1):27–34. doi: 10.1016/j.sbi.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradley CM, Barrick D. The notch ankyrin domain folds via a discrete, centralized pathway. Structure. 2006;14(8):1303–1312. doi: 10.1016/j.str.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Mello CC, Bradley CM, Tripp KW, Barrick D. Experimental characterization of the folding kinetics of the notch ankyrin domain. J Mol Biol. 2005;352(2):266–281. doi: 10.1016/j.jmb.2005.07.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.